Abstract
Activation induced cytidine deaminase (AID) is required for somatic hypermutation and immunoglobulin class switching in activated B cells. Because AID possesses no known target site specificity, there have been efforts to identify non-immunoglobulin AID targets. We show that AID acts promiscuously, generating widespread DNA double strand breaks (DSB), genomic instability and cytotoxicity in B cells with diminished homologous recombination (HR) capability. We demonstrate that the HR factor XRCC2 suppresses AID-induced off-target DSBs, promoting B cell survival. Finally, we suggest that aberrations affecting human chromosome 7q36, including XRCC2, correlate with genomic instability in B cell cancers. Our findings demonstrate that AID has promiscuous genomic DSB-inducing activity, identify HR as a safeguard against off-target AID action, and have implications for genomic instability in B cell cancers.
Development of functional adaptive immunity is driven by a series of genomic DNA rearrangements in lymphocytes that are required for the generation of antigen recognition diversity, receptor editing, and lymphoid effector function 1. In mature, naïve B lymphocytes, immunoglobulin class switch recombination is initiated by Activation induced cytidine deaminase (AID) (2. Mutations in the gene encoding AID (Aicda) lead to immunoglobulin class switching and somatic hypermutation defects in mice and humans, and underlie human hyper IgM syndrome 3. Within the immunoglobulin heavy chain (Igh) locus, AID-dependent DNA double strand breaks (DSBs) primarily occur in transcribed, repetitive sequences, termed switch (S) regions, upstream of isotype-specific coding exons. Unlike site-specific recombination activating gene (RAG) activity, AID is target sequence independent 4. In this context, two recent studies found evidence that AID can induce rare point mutations or DSBs, respectively, in select non-Ig genes 5, 6. These findings have spurred efforts to identify off-target AID activity and other putative non-Ig AID targets.
It has recently been reported that AID can exhibit off-target point mutation activity in stimulated B cells 5, 7. Moreover, the identification of chromosomal translocation breakpoints in or near Igh S regions in some B lymphoid cancers has suggested that AID could potentially stimulate ectopic chromosomal rearrangements, leading to oncogene activation 8. Consistent with this notion, it was recently found that AID can generate potentially oncogenic DSBs at the Myc proto-oncogene locus, albeit very rarely and at a substantially lower rate than class switch recombination (CSR)-initiating DSBs in Igh S-regions 6. These off-target AID-induced point mutations and rare translocations have been collectively coined “collateral damage” 9.
Homologous recombination (HR) is a high-fidelity DNA double strand break repair pathway important for maintaining general genome stability in many eukaryotic cells. XRCC2 is a key member of the RAD51 family of mammalian HR factors 10–12. XRCC2 deficient cells exhibit proliferation defects, hypersensitivity to ionizing radiation and other DNA damaging agents, and spontaneous chromosomal instability 12–14. Mice harboring a homozygous knockout (KO) of the Xrcc2 gene exhibit mid-gestational embryonic lethality associated with widespread cellular apoptosis 12, 15, 16. We previously showed that XRCC2 is required for successful proliferation and genomic integrity in early developing B cells 17. Here we have investigated the interplay between AID and XRCC2 in mature, activated B cells. We now show that AID can promiscuously attack the genome, leading to widespread, highly cytotoxic DSBs in activated, XRCC2-defective mature B cells. Our findings highlight HR as a critical genome-protective pathway in activated B cells, important for resisting the potentially deleterious effects of AID off-target activity.
RESULTS
Activation is toxic to XRCC2 defective B cells
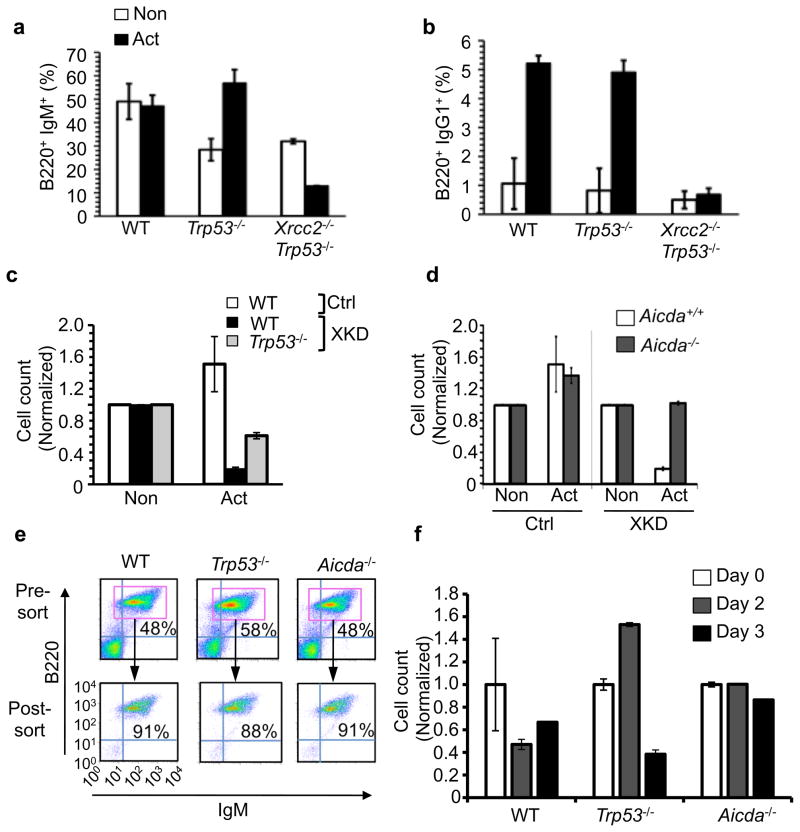
We initially found that XRCC2, like its paralogue RAD51, was transcriptionally upregulated following B cell activation (Supplementary Fig. 1a–b) 18. To investigate the interrelationship between B cell activation, immunoglobulin class switching, and XRCC2 we next tested whether Xrcc2-deficient B cells were impaired for immunoglobulin class switching. Because, Xrcc2 nullizygosity in mice produces embryonic lethality 12, 14, 16, we used a fetal liver cell culture system to obtain Xrcc2-null primary B cells (Supplementary Fig. 2) 17, 19. The resulting mature, naïve IgM-expressing B cells were transferred to media containing either anti-CD40 antibody alone (non-activated), to induce proliferation without class switching; or anti-CD40 plus interleukin (IL)-4 (activated), to induce proliferation and class switch recombination (Fig. 1a–b, Supplementary Figs. 3–4). After 3 days of activation, control (Xrcc2+/+) cultures showed efficient isotype switching from IgM to IgG1 (Fig. 1a–b, Supplementary Fig. 4). By contrast, activated Xrcc2-deficient cultures were severely compromised for class switching, showing no increase in IgG1+ cells (Fig. 1b). Importantly, we also observed a decrease in the fraction of IgM+ B cells in activated Xrcc2-null cultures, but not Xrcc2+/+ cultures (Fig. 1a; Supplementary Fig. 4). Even partial reduction of XRCC2 function conferred activation-dependent cytotoxicity. Using a lentivirus-based system to deliver Xrcc2 shRNA (XKD) or scrambled control shRNA (Ctrl) constructs to primary splenic B cells or B cell lines (Fig. 1c, Supplementary Fig. 5–9), we found that XRCC2 knockdown, like nullizygosity, sensitized B cells to activation (Fig. 1c). Previously, we showed that deficiency for p53 (encoded by Trp53) could rescue early B cell development in an Xrcc2-mutant fetal liver culture assay 17. Here, using the lentivirus-based shRNA system, we were able to demonstrate a partial B-cell rescue in the absence of p53, indicating that there may be p53 dependent and independent responses to activation in mature B cells.
Figure 1.
Activation is cytotoxic to HR-defective B cells. B cells from primary fetal liver cultures (FLCs) were stimulated for 3 days with anti-CD40 (Non) or anti-CD40 plus IL-4 (Act) and analyzed. (a) FLC-derived B cells analyzed by flow cytometry for expression of B220 and IgM. (b) B cells from (a) analyzed for B220 and IgG1 to assess class switching. (c) WT or Trp53−/− splenocytes transduced with either XKD or Ctrl-expressing shRNA constructs and stimulated after one day. Relative number of transduced cells in each culture was determined by flow cytometric measurement of EGFP positive cells 3 days post-stimulation. (d) WT or Aicda−/− cells were transduced with either XKD or Ctrl constructs and relative cell numbers with or without activation was determined as in (c). (e) Splenic B cells from WT, Trp53−/−, or Aicda−/− mice were purified by magnetic bead sorting. The purified cells (post-sort) were used as starting cultures for the experiments shown in (f). (f) Purified WT, Trp53−/− or Aicda−/− B cells were transduced with either XKD or Ctrl-expressing shRNA constructs, stimulated, and analyzed for relative cell number after activation. All data are normalized to anti-CD40 (Non) controls. For FLC assays (a–b) data represent five replicates each from WT and Trp53−/− samples and two replicates from each of two Xrcc2−/− Trp53−/− samples. For knockdown experiments data represent three (c) or four (d–f) replicates for all samples. Error bars in all plots indicate standard error.
AID-dependent activation-induced B cell cytotoxicity
Because XRCC2 defective (KO and XKD cells) B cells were sensitive to activation we hypothesized that cytotoxicity might be mediated by AID. To test this, we carried out stimulation and survival assays in XKD cells derived from either normal (Aicda+/+) or AID-deficient (Aicda−/−) mice (Fig. 1d–f, Supplementary Fig. 8). Consistent with the results above, XRCC2 knockdown in Aicda+/+ B cells compromised survival following activation (Fig. 1d–f). By contrast, activation of XKD B cells from Aicda−/− mice produced no detectable change in viability or survival relative to non-activation. To further verify that activation directly induced AID-dependent B cell cytotoxicity, we repeated the survival assays using starting cell populations that were enriched specifically for IgM+ B cells and assayed at various timepoints after activation (Fig. 1e–f). Whereas purified normal or Trp53−/− B cells with the XKD construct showed activation induced cytotoxicity, purified Aidca−/− XKD B cells were resistant to the deleterious effects of activation (Fig. 1e–f). Taken together with the data above, these results demonstrate that AID induces significant cellular toxicity in activated, HR-defective primary B cells.
AID induces widespread genomic breaks in B cells
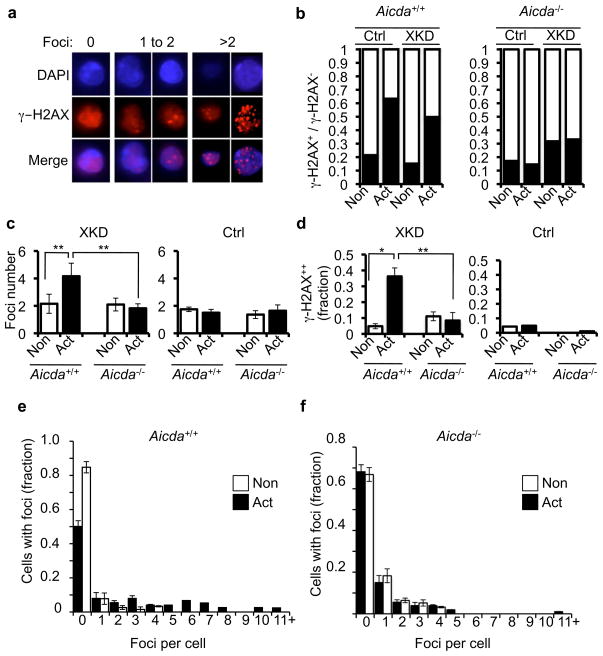
Because XRCC2 has well known DSB repair functions 20, we next tested whether AID-induced toxicity was associated with unrepaired DSBs in Xrcc2-defective B cells. To test this prediction, Aicda+/+ or Aicda−/− B cell cultures harboring the XKD construct were supplemented with either anti-CD40 alone (non-activated) or with anti-CD40 plus IL-4 (activated), and then stained for foci of γ–H2AX, a marker of unrepaired DSBs (Fig. 2a) 21. Both control and XKD B cell cultures showed an AID-dependent increase in the fraction of γ–H2AX- positive cells (cells containing 1 or more γ–H2AX foci) after activation (Fig. 2b). As expected Aicda−/− cultures, with either control or the XKD shRNAs, exhibited no change in γ–H2AX positivity upon activation (Fig. 2b). However, when we quantified the number of foci per cell in the γ–H2AX-positive fraction, XKD cells showed a specific and significant increase in the number of γ–H2AX foci per cell (from approximately 2 foci per cell to more than 4 foci per cell) upon activation (Fig 2c). Importantly, this effect was abrogated in Aicda−/− cells, and thus we conclude was dependent on AID (Fig. 2c). We next measured the fraction of γ–H2AX-positive cells that specifically contained supernumerary (more than 2) γ–H2AX foci, reasoning that such foci might represent potential off-target, non-Igh DSBs. Consistent with this prediction, we observed a significant increase in the fraction of XKD cells that contained AID-dependent, supernumerary γ–H2AX foci, supporting the possibility of AID-related DSBs at genomic locations other than the two copies of the Igh locus (Fig. 2d). Finally, further analyzing the distribution of foci revealed a significant shift toward a higher number of foci per cell in activated Aicda+/+, but not Aicda−/−, XKD cultures (Fig. 2e–f). Critically, this effect was not due to differential cell cycle responses to anti-CD40 versus anti-CD40 plus IL-4, as both culture conditions elicited similar overall cell cycle profiles (Supplementary Fig. 3). We also observed a similar γ–H2AX response in the class-switch competent mouse B cell line, CH12-F322, containing the same Xrcc2 knockdown construct used in primary cells (we term these cells CH12/XKD) (Supplementary Figs. 7, 9–10). Finally, in independent assays we also observed an accumulation of activation-induced foci of the DNA damage response factor 53BP1, another marker of DNA double strand breaks, specifically in XKD cells, but not in control cells (Supplementary Fig. 11).
Figure 2.
XRCC2-defective B cells exhibit numerous AID-dependent DSBs (a) Activated primary cells expressing the XKD construct were stained for γ–H2AX. Images show cells stained with anti γ–H2AX (red), nuclei counterstained with DAPI (blue), and the merge. Examples show cells harboring 0, 1 to 2, or >2 γ–H2AX foci. (b) Aicda+/+ or Aicda−/− XKD splenic B cells were grown in non-activated (Non) or activated (Act) conditions and analyzed for γ–H2AX foci. The γ–H2AX negative fraction (no foci; open bar) versus γ–H2AX positive (one or more foci; filled bar) is shown for each sample (c) The average number of foci in γ–H2AX positive fraction of each sample from (b) was determined. (d) Fraction of cells with supernumerary γ–H2AX foci (greater than 2 foci per cell) was determined for the γ–H2AX-positive cells in (b). (e–f) Data from (b) were separated into unitary bins (0, 1, 2, 3, etc.) up to 10 foci/cell. Cells with greater numbers of foci were grouped in the >10 foci/cell bin. Plotted are the data for Non versus Act samples of either (e) WT or (f) Aicda−/− cultures. Error bars indicate standard error of the mean. Significance was determined by two sample t-test. **p<0.01 *p<0.05.
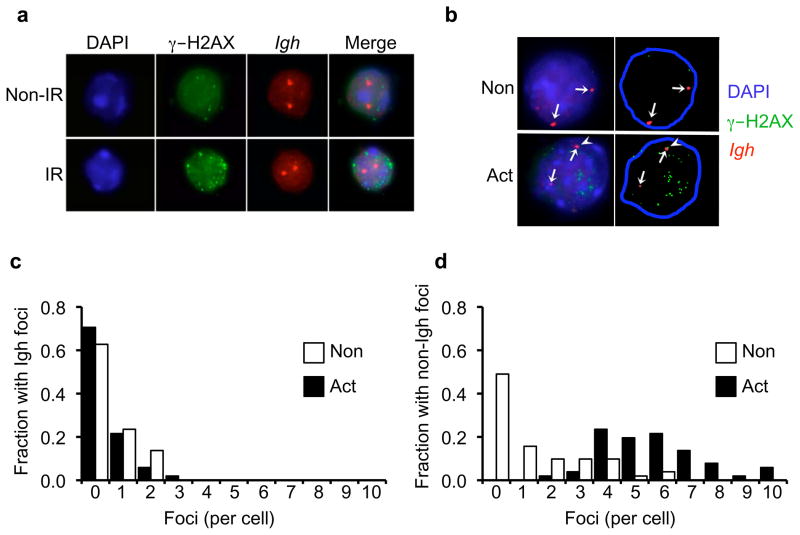
The significant increases in DNA damage foci in activated XRCC2-defective cells strongly suggested AID off-target activity outside the Igh locus. To directly evaluate whether activation induced DSBs occurred at genomic locations outside Igh, a fluorescence in situ hybridization (FISH)-immunofluorescence (IF) analysis was performed to simultaneously localize Igh loci and γ–H2AX foci. Activated (and non-activated control) CH12/XKD cells were stained with a FISH probe overlapping the 3′ end of the Igh locus (containing Igh constant region exons with AID-targeted switch regions), and with the anti-γ–H2AX antibody (Fig. 3; Supplementary Figs. 12–13). This analysis confirmed that supernumerary foci in activated XKD cells occurred at locations outside of, and often distant from, the Igh gene. Quantification of supernumerary foci revealed a strong increase in the number of non-Igh foci per cell, and the fraction of cells with supernumerary foci, in activated XKD cultures (Fig. 3c–d). Altogether, these data show that AID can induce a high level of off-target DNA double strand breaks at non-Igh locations, in an XRCC2-mutant context.
Figure 3.
AID generates widespread off-target DSBs. (a) Fluorescence in situ hybridization (FISH) detection of Igh (red) and immunofluorescent detection of γ–H2AX (green) in B cells either non-irradiated (Non-IR) or exposed to 5 Gy (IR) ionizing radiation. Nuclei counterstained with DAPI. (b) FISH analysis of CH12/XKD cells under non-activated (Non) or activated (Act) culture conditions showing the localization of γ–H2AX foci (green) and the Igh loci (red). Arrows indicate Igh signals. Flare in the activated culture indicates a γH2AX focus co-localized with one Igh FISH signal. (c–d) γH2AX foci were counted in cells from (b) and visually scored as colocalized with the Igh locus (Igh foci; c) or not localized to the Igh locus (non-Igh foci;d). Data were separated into unitary bins up to 10 foci, and the fraction of non-activated (blue bars; N=51 nuclei) or activated (red bars; N=51 nuclei) cells showing Igh foci (c) or non-Igh foci (d) is plotted. Magnified images are shown in Supplementary Fig. 12. Detailed segmentation is shown in Supplementary Fig. 13.
AID induces S-phase accumulation
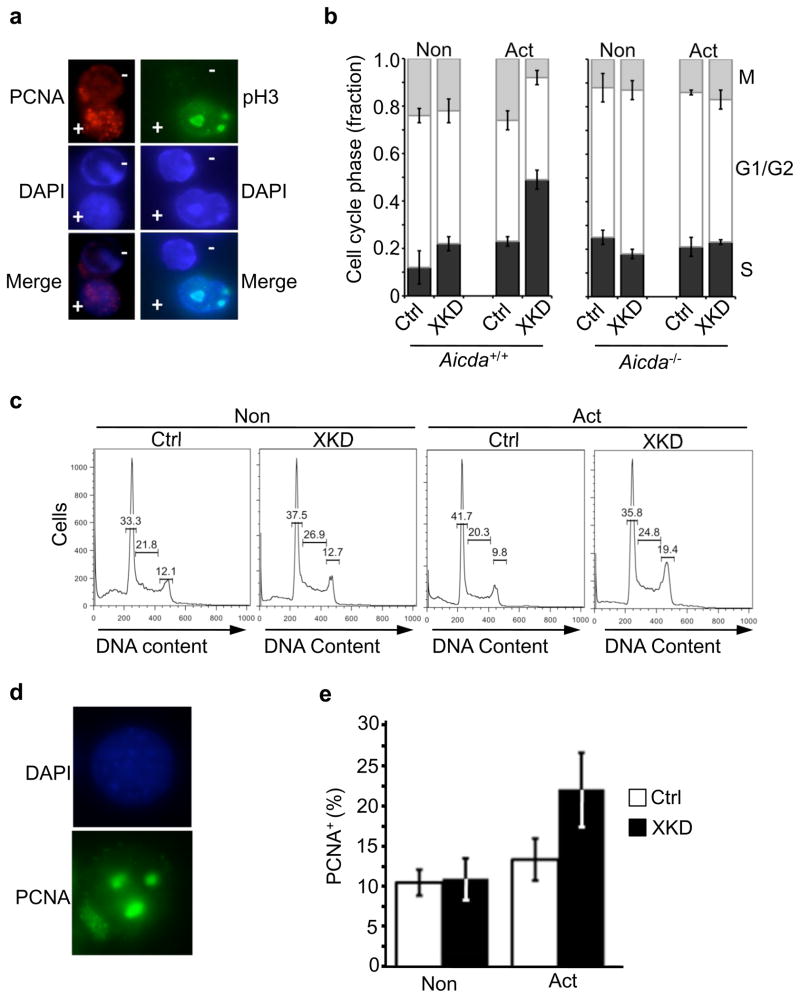
Because we previously showed a role for XRCC2 in S-phase transit in early B cells, we also measured cell-cycle responses in XRCC2-defective mature B cells17. Aicda+/+ or Aicda−/− B cells, transduced with either a control (EGFP) or XKD construct, were cultured under activating (anti-CD40 plus IL-4) or non-activating (anti-CD40 alone) conditions, and analyzed for cell cycle distribution using a quantitative immunofluorescence microscopy assay. Cells from each culture were fixed, stained, and quantified for proliferating cell nuxclear antigen (PCNA) a marker of S-phase or phosphorylated histone H3 (pH3) a marker of mitosis (Fig. 4a–b). This analysis revealed a significant increase in the fraction of cells in the S-phase of the cell cycle, and a corresponding decrease in the M-phase fraction, specifically in activated XKD cells (Fig. 4b). Moreover, this effect was observed only in Aicda+/+ B cells, but not in identically treated Aicda−/− B cells, indicating that the cell cycle response was directly due to AID activity (Fig. 4b). An activation induced S-phase delay was also observed in CH12/XKD, but not in CH12/Ctrl, cells (Fig. 4c–e, Supplementary Fig. 14)
Figure 4.
Activation induces AID-dependent S-phase accumulation in XRCC2-defective B cells. (a) Immunofluorescence detection of either the S-phase marker PCNA (red) or the M-phase marker phosphorylated histone H3 (pH3; green). Shown are images with positively (+) and negatively (−) staining cells in the same field of view (top panels). DAPI DNA counterstained (blue; middle panels) and merged images (bottom panels) are also shown. (b) Quantification of PCNA and pH3 immunostaining data for Aicda+/+ or Aicda−/− B cell cultures with either the XKD or Ctrl (EGFP) constructs that were cultured with anti-CD40 plus IL-4 (Act) or anti-CD40 alone (Non). The fractions of PCNA positive cells (black bars) indicative of S-phase, pH3 positive cells (gray bars) indicative of M-phase, or the double negative cells, indicating G1 or G2 (open bars) are shown. (c) Representative flow cytometry showing cell cycle distribution. Histograms of propidium iodide (PI) staining in Ctrl or XKD cells under non-activating or activating conditions, with percentage of G1, S, and G2/M cells shown. (d–e) Fractions of cells from (c) showing punctate PCNA staining, indicating S-phase, was determined. Error bars indicate S.E.M. from three independent experiments.
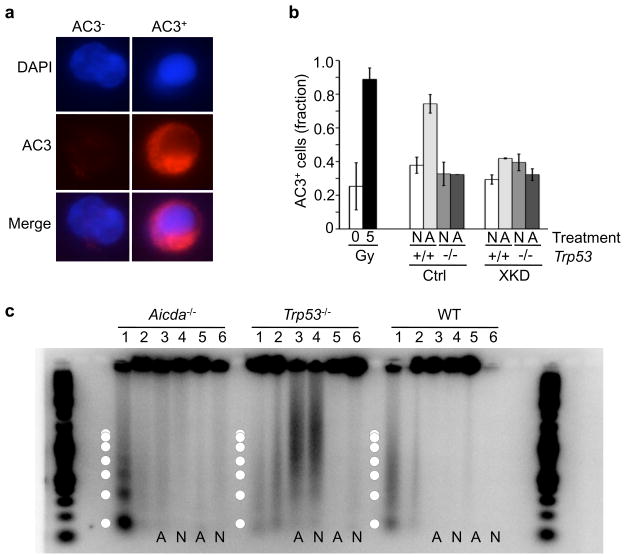
We next evaluated whether cytotoxicity or AID-dependent off-target DSBs were associated with apoptotic induction, using several different measures of apoptosis. First, activated and non-activated primary XKD cells from either wild-type or Trp53−/− mice were analyzed for the presence of activated caspase 3 (AC3), an early apoptotic marker, by immunofluorescence imaging and quantitative flow cytometry (Fig. 5a–b). As controls for baseline and DNA damage-induced AC3 signals were measured in primary splenocytes before and after exposure to 5 Gy of ionizing radiation (IR), respectively (Fig. 5b). Consistent with previously published accounts of activation-mediated B cell apoptosis, an increase in AC3-positivity was observed in activated cells with the control (Scrambled) shRNA construct (Fig. 5b). By contrast, the AC3+ fraction was indistinguishable in activated and non-activated XKD cells from either wild-type or Trp53−/− mice (Fig. 5b). To independently analyze apoptosis genomic DNA laddering, an indicator of late-stage apoptosis, was measured in primary XKD or control cells on either a wild-type, Trp53−/−, or Aicda−/− genetic background. As observed with AC3 staining, exposure to 5Gy of IR led to detectable DNA laddering and low molecular weight DNA production in wild-type and Aicda−/− cells, but not in apoptosis-resistant Trp53−/− cells (Fig. 5c, lanes 1,2). By contrast, activation of XKD cells did not produce DNA laddering or a significant increase in low molecular weight DNA in either WT or Aicda−/− cells (Fig. 5c, lanes 3,4). Xrcc2 knockdown did result in the generation of electrophoretically mobile DNA fragments in Trp53−/− mice, but this was not specific to activation (Fig. 5c, lanes 3,4 versus 5,6). Altogether, these results indicate that activation-induced DSBs in HR-defective B cells are directly mediated by AID off-target activity, leading to S-phase delay and/or arrest but not apoptosis.
Figure 5.
AID-induced cytotoxicity in HR-defective B cells is apoptosis-independent. (a) Immunofluorescence detection of activated caspase 3 (AC3) a marker of apoptotic cells. Non-apoptotic (AC3−) and an apoptotic (AC3+) splenocyte, DAPI counterstained nuclei (blue), activated caspase 3 (AC3; red). (b) Primary splenocytes from Trp53+/+ or Trp53−/− mice transduced with either XKD or Ctrl shRNA construct analyzed for AC3 by flow cytometry after culture in non-activating (N; anti-CD40-alone) or activating (A; anti-CD40 plus IL-4) conditions. As controls, freshly isolated wild-type splenocytes were analyzed after exposure to 0 or 5Gy ionizing irradiation. (c) Genomic DNA from Aicda−/−, Trp53−/−, or wild-type (WT) primary splenocytes. Each sample was either non-transduced (lanes 1,2 each), transduced with the XKD construct (lanes 3,4) or transduced with the Ctrl construct (lanes 5,6). As controls for DNA damage-induced apoptotic laddering, untransduced samples (Aicda−/−, Trp53−/−, and WT) were analyzed either after (lanes 1) or before (lanes 2) exposure to 5Gy of IR. Transduced (XKD or scrambled) samples were run after activating (A: lanes 3 and 5) or non-activating (N: lanes 4 and 6) culture. Superimposed circles designate approximate band sizes expected for apoptotic DNA laddering. AC3 data (b) represent mean of four replicates for each sample. DNA laddering assays (c) were performed on two independent mice for each genotype.
AID off-target activity produces chromosomal instability
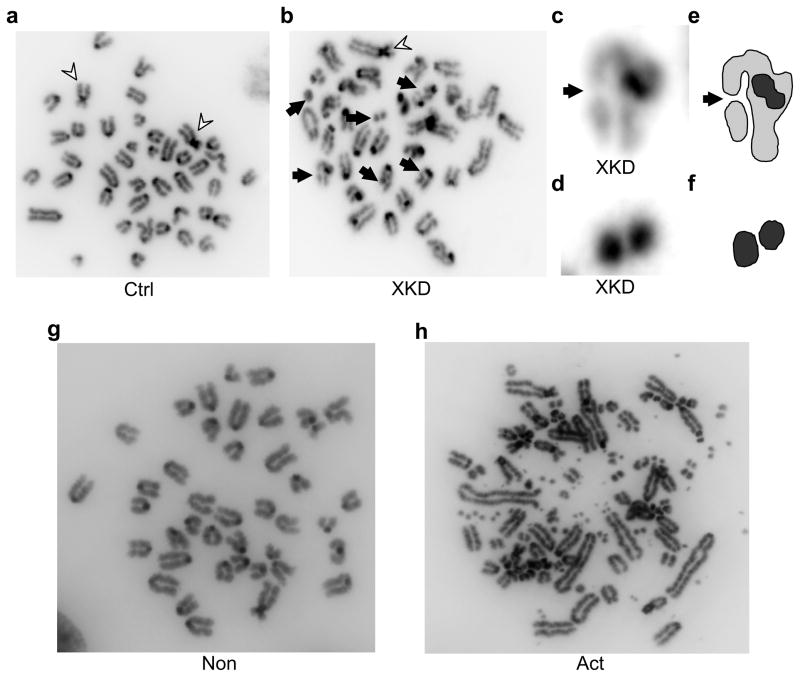
One implication of these findings is that AID off-target assault on the genome might confer a risk of chromosomal instability, with potentially oncogenic consequences. Indeed, one recent study has suggested that AID can stimulate translocations between Igh and the Myc proto-oncogene 6. Thus, we tested whether XRCC2 functions to suppress activation-induced chromosome aberrations in B cells. CH12/XKD cultures showed a higher level of chromosome damage than CH12/Ctrl cultures, even under non-activating conditions: 29% of untreated and 29% of non-activated XKD cells contained chromosomal damage, versus 13% for untreated and 9% of non-activated Ctrl cells (Fig. 6, Table 1). However, after activation, 79% of CH12/XKD cells contained chromosome and chromatid damage with some cells showing evidence of extreme genome damage. By contrast, 21% of activated CH12/Ctrl cells contained chromosome breaks or fragments (Fig. 6, Table 1; Supplementary Fig. 15). These data demonstrate that B cell activation can induce (sometimes extreme) chromosomal instability, and point to a key role for XRCC2 in suppressing spontaneous and especially activation-induced chromosomal damage, in B lymphocytes. Taken together, these findings support a model in which AID promiscuously and broadly attacks the B cell genome, leading to widespread DSBs, termed collateral damage (Supplementary Fig. 16). The homologous recombination pathway of DSB repair is necessary to prevent collateral damage-induced genomic instability or B cell cytotoxicity.
Figure 6.
B cell activation induces chromosomal instability in HR-defective cells. CH12-F3 cells were stably transduced with either the XKD or Ctrl shRNA construct (CH12/XKD and CH12/Ctrl, respectively), and were karyotyped after exposure to 1 μg/ml anti-CD40 alone (Non) or to 1 μg/ml anti-CD40 plus 25 ng/ml IL-4 and 1ng/ml TGFβ (Act). Shown are images of (a) activated CH12/Ctrl cells or (b) activated CH12/XKD cells. Flares indicate two Robertsonian-type chromosome translocations present in the parental cell line; arrows indicate induced de novo chromosome and chromatid breaks. (c) Magnified image of a chromatid break in (b). Arrow indicates a chromatid break. (d) Magnified image of chromosome fragment from (b). (e,f) Segmentation of images from c,d respectively. (g,h) Karyotypes from non-activated (g) or activated (h) CH12/XKD cells. Example of activation-induced extreme instability is shown in (h).
Table 1.
Summary of chromosome damage in XKD cells
| Ctrl | XKD | |||||
|---|---|---|---|---|---|---|
| Untreated | Non | Act | Untreated | Non | Act | |
| Cells with breaks (%) | 13 | 9 | 21 | 29 | 29 | 76 |
| Average chromosomes | 40 | 41 | 40 | 40 | 41 | 39 |
| Metaphases | 23 | 22 | 24 | 21 | 21 | 21 |
Karyotype data quantified for the CH12/Ctrl versus CH12/XKD cells under untreated (no culture supplement), non-activated (anti-CD40) or activated conditions (anti-CD40 plus IL-4 plus TGFβ).
7q36 is linked to genome instability in B cell cancers
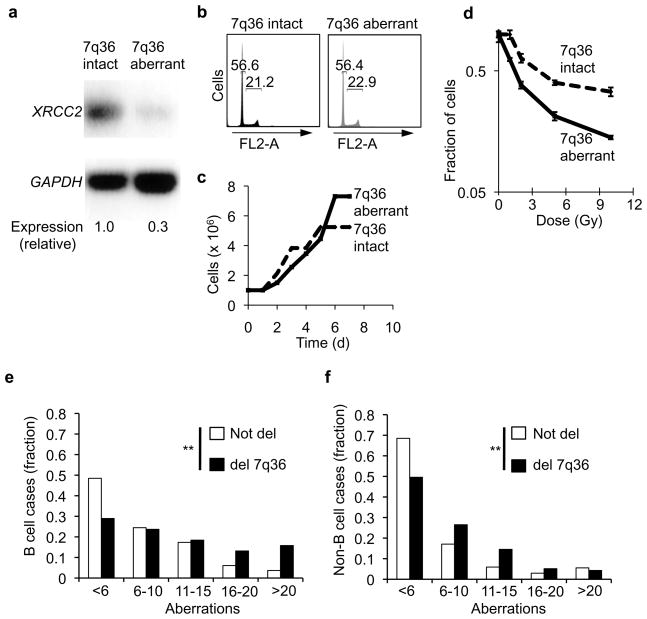
In humans, XRCC2 is located on chromosome 7 cytoband q36 23, a region that is frequently rearranged in various cancers (Supplementary Fig. 17) 24,25,26. Our data suggest that XRCC2 might be particularly important for suppression of activation-induced genomic instability in B cells. To test this, we obtained human B cell-derived lines (Coriell Institute), that contained either intact or cytogenetically aberrant 7q36 (GM07323 and GM13689, respectively). The 7q36-aberrant line exhibited approximately 3-fold lower expression of XRCC2 than its 7q36-intact counterpart, but, similar overall cell cycle and proliferation phenotypes (Fig. 7a–c). Consistent with reduced XRCC2 expression, the 7q36-aberrant line also showed increased sensitivity to ionizing radiation at all doses tested over a range from 0 to 10 Gy (Fig. 7d). This is consistent with the idea that 7q36 aberrations can functionally impair XRCC2. Next, to evaluate whether 7q36 aberrations might associate with high chromosome instability in B cell cancers, we analyzed karyotype data from 46,075 case reports in the Mitelman Database of Chromosome Aberrations in Cancer (CGAP; http://cgap.nci.nih.gov/Chromosomes/Mitelman). Individual cases were sub-categorized, comparing case reports for all B cell cancers (N=11,994) versus all non-B cell cancers (N=34,081), either with or without deletions affecting 7q36 (Fig. 7e–f). We found that 7q36 deletion was significantly associated with overall higher levels of karyotypic instability in both B cell (p<0.001) and non-B cell tumors (p<0.001), but that the effect was especially strong in the B-lineage neoplasms (Fig 7e–f). This is consistent with the notion that XRCC2 plays a general role in DNA repair and genome stability, and that spontaneous chromosomal instability associated with 7q36 aberrations might be exacerbated in B cell neoplasms. We propose that AID off-target activity might potentially be one source of genome destabilization in B cell cancers with 7q36 aberrations. In this context, AID expression has been shown to correlate with blast crisis B lineage leukemia and therapy resistance in myeloid leukemia, and to associate with generally poor prognosis in chronic B lymphocytic leukemia26, 27. Our data now suggest that synergy between AID action and 7q36 defects might be especially deleterious in patients with such tumors.
Figure 7.
Chromosome 7q36 aberrations correspond with decreased XRCC2 expression and karyotype instability in human B-lymphoid cancer cells. (a) Reverse transcription (RT)-PCR analysis of XRCC2 and GAPDH (loading control) transcripts in matched human B cell cells lines (Coriell Institute) either with intact 7q36 (GM07323) or cytogenetically aberrant 7q36 (GM13689 (b) Cell cycle profile analysis of 7q36-intact and 7q36-aberrant cell lines as determined by flow cytometry. (c) Proliferation analysis of 7q36 intact and 7q36 aberrant cells. (d) 7q36-aberrant cell line shows hypersensitivity to ionizing irradiations, an expected phenotype associated with diminished XRCC2 expression. (e,f) Karyotype data were analyzed from 46,075 case reports in the Mitelman Database of Chromosome Aberrations in Cancer (CGAP). (e) All B-cell cancer case reports (N=11,994) were analyzed for the total number of chromosome aberrations per cell. Plots show the fraction of cases within the indicated bin for tumors with 7q36 deletions or without 7q36 deletions. (f) All non-B cell cancer case reports in the Mitelman database (N=34,081) were scored for chromosome aberrations as in panel (f). Case reports with 7q36 deletions or without 7q36 deletions are plotted. Statistical significance was determined by chi-square test. ** p<0.001
DISCUSSION
Here we have investigated the interplay between AID activity and DNA double strand break repair in activated B cells. We demonstrate that, in addition to class switching and somatic hypermutation activities, AID also generates a high level of off-target DSBs in activated B cells, which endanger the B cell genome when DNA break repair is compromised. It will be of interest to identify non-Ig genomic locations targeted for AID attack. One recent study showed that AID could induce rare off-target point mutations at specific non-Ig genes 5. Thus, it is possible that these same non-Ig targets are also subject to AID-induced DSBs. This model would predict that off-target breakage at specific non-Ig genes should occur during the G1 cell cycle phase, when AID is thought to also act on the Igh locus. However, this model might also predict p53-dependent G1-to-S phase checkpoint activation in response to off-target DSBs. However, we observe a small overall effect of p53 on AID-induced collateral damage. Therefore, we propose an alternative model in which that DNA replication forks represent attractive off-target substrates for inappropriate AID activity. Replication forks contain abundant single stranded DNA (ssDNA), a known AID substrate, and replication protein A (RPA), a known AID co-factor 27. In this context, AID-dependent cytidine deamination of ssDNA at replication forks could create single stranded nicks that induce fork collapse and ultimately DSBs, which would be natural substrates for HR-mediated replication restart, as has been shown in other contexts 28, 29.
At multiple stages of their development, B cells must undergo programmed, DSB-stimulated gene rearrangements–V(D)J recombination and immunoglobulin class switch recombination. To successfully complete their developmental program, B cells must properly repair these DSBs and must proliferate 30–32. These developmental dynamics in the B cell genome pose the risk of genomic instability. While Non-Homologous End-Joining and Alternative End-Joining are known to function in the resolution of programmed DSBs, the pathways that safeguard the genome at-large are less understood 32. We previously showed a role for HR in proliferation and genome stability in early B cell development 17. Here we demonstrate a specific role for the HR factor XRCC2 in genome protection in mature B cells in which the AID has been activated. In this context, we propose that HR is essential to resist the genome destabilizing effects of AID-induced collateral damage, and is thus essential to promote B cell development. Importantly, we show that even a partial reduction of XRCC2 expression is sufficient to confer extreme sensitivity to AID off-target damage and cytotoxicity. Polymorphic variants of human XRCC2 have been identified, which might be associated with moderately reduced XRCC2 function 33. Our data now suggest that B cells in such individuals might be more prone to AID off-target damage, potentially leading to B cell developmental deficiencies or higher lymphoid cancer risk.
The functions of XRCC2 in early B cell development appear to differ from its roles in mature, activated B cells. We previously proposed that XRCC2 collaborates with p53 to ensure the integrity of DNA replication forks, or to restart stalled forks, in rapidly proliferating pro- or pre-B cells. In the absence of XRCC2, p53 enforces a prolonged early S-phase arrest in developmentally immature B cells. However, ablation of p53 permits XRCC2-null B-precursor cells to transit S-phase and complete development, albeit at the cost of high genomic instability. By contrast, the sensitivity of XRCC2-null mature B cells to activation is largely p53-independent. Survival following activation of XRCC2 and p53 double null B cells was severely compromised, indicating that the genotoxicity was not exclusively mediated by pathways downstream of p53. Interestingly, in the knockdown experiments, where XRCC2 levels were reduced but not eliminated, inactivation of p53 led to a partial restoration of viability after stimulation. Therefore, we suggest that XRCC2 plays a dual role in activated B cells: p53-independent suppression of collateral damage and p53-dependent support of DNA replication. In this context, we also observed an S-phase cell cycle delay in stimulated CH12/XKD cells. This could reflect a replication defect in the absence of XRCC2, or might indicate the involvement of p53-independent checkpoints, such as Ataxia Telangiectasia and Rad3-related (ATR) in the response to collateral damage.
In summary, we show that AID can promiscuously attack the B cell genome, producing widespread DSBs, that have been termed collateral damage9. We further demonstrate that the HR factor XRCC2 is critical to resist AID-induced collateral damage ensuring B cell viability and genome stability. Our findings suggest that HR has co-evolved with the adaptive immune system, to provide a genomic safeguard against DNA damaging mechanisms that have been co-opted for programmed genome rearrangements in lymphocytes. In this context, it is interesting to note that chicken B cells undergo AID-induced immunoglobulin diversification by an XRCC2-dependent gene conversion mechanism. In the absence of XRCC2, chicken B cells resort to a mammalian-like somatic hypermutation mechanism34. One intriguing possibility is that off-target AID-induced DNA breakage represents a vestigial antigen receptor diversification process that is revealed when HR-mediated repair is defective. Alternatively, it is possible that AID-initiated gene conversion in avian lymphocytes represents a mode of immunoglobulin rearrangement that is more mechanistically distinct from mammalian SHM and CSR than previously thought. Our data may also provide a molecular explanation for the recent finding that AID can control germinal center size by inducing B cell apoptosis 35. AID induces off-target damage, the extent of which is controlled by XRCC2-mediated repair. However, a fraction of cells with off-target damage might evade XRCC2-mediated repair, and be eliminated by apoptosis, thus controlling GC size. This study also has important implications for understanding immune system development, and the sources (and repair) of potentially genome-damaging DNA double strand breaks. Specifically, our findings indicate that the same mechanisms that introduce developmentally programmed DSBs can carry an inherent and simultaneous risk to the genome at large. This now suggests that HR is important, not only for normal lymphocyte development 17, but also for preventing lymphoid pathologies such as immunodeficiency or cancer.
Methods
Mice
Xrcc2 and Trp53 knockout mice, and colony maintenance were as previously described Supp. Refs. 1–2. Xrcc2+/−, Trp53+/− and Aicda−/− mouse colonies were separately maintained Supp. Ref. 3. Xrcc2 and Trp53 heterozygous mice were intercrossed to generate compound mutant, and control strains. All mice were maintained in pressurized, individually ventilated caging and fed standard lab diet. All mouse work was carried out according to IACUC approved protocols.
Cell and tissue culture
For fetal liver cell cultures, embryos were isolated at E13 to E15. Single-cell suspensions of fetal liver cells (FLC) were prepared and maintained as previously described Supp. Ref. 1. Adult splenocytes were obtained from 4–6 month old C57BL6/J mice, and maintained in RPMI medium with L-glutamine (Gibco), supplemented with 10% heat inactivated FBS (Gibco). NIH3T3 cells were maintained in complete DMEM with glucose, and supplemented with 10% heat inactivated FBS (Gibco). CH12-F3 cells were maintained in RPMI media with L-glutamine (Gibco), supplemented with 10% heat inactivated FBS (Omega Scientific) and 5% NCTC 109 media (Gibco) Supp. Ref. 4.
Stimulation of B cells
For immunoglobulin class switching assays, B cells were obtained either directly from adult spleen, or from ex vivo development of FLC, as described above. 106 FLC were cultured for 5 days, then removed from the adherent T220 feeder layer, and 105 cells were transferred to RPMI-1640 media containing 10% FBS and supplemented with either 1μg/ml ant̃i CD40 (Pharmingen or ebioscience) alone or with 1μg/ml ant̃i CD40 plus 25 ng/ml IL-4 (Peprotech) for 2 days Supp. Fig. 1. The media and cytokines were replenished after 2 days. Total splenocytes were for transduction or further purified for B220+/IgM+ B cells by depletion of CD43-expressing cells via magnetic bead-based cell sorting (MACS; Miltenyi, Inc. Cat. No. 120-000-302). 106 total splenocytes or sorted B220+/IgM+ B cells were then stimulated and re-stimulated with as described above. For stimulation of the CH12-F3 line, or its derivatives, 105 cells per mL were treated as splenic B cells, but also 1ng/ml TGFβ (R&D Systems).
Lentiviral transduction and shRNA
Lentiviral shRNA vectors targeting the Xrcc2 (TRCN0000071023, 71024, and 71027) were obtained from Sigma, Inc. CH12-F3 derivative lines stably expressing the either anti-Xrcc2 shRNA (CH12/XKD) or a control (Ctrl) shRNA with scrambled sequence (CH12/Ctrl), were generated by direct transduction with individual shRNA vectors, followed selection for puromycin resistance. For transduction of primary splenocytes, the original vectors were modified by replacing the puromycin resistance cassette with EGFP (Clontech). For controls, either an empty pLKO.1-EGFP or a scrambled (Ctrl) shRNA-expressing vector was used. Vectors were packaged into pseutotyped viruses and, were used to transduce primary cells as previously described Supp. Ref. 5.
Ionizing radiation sensitivity
To measure IR sensitivity 0.5×105 transduced splenocytes,104 transduced CH12 cells or 104 human B cells were exposed to ionizing radiation from Cs-137 irradiator (Shepard), at doses ranging from 0.5 to 5 Gy. Cells were incubated for 4 days, counted for viability by Trypan Blue exclusion, and survival normalized to unirradiated controls.
RT-PCR
Reverse transcription (RT)-PCR was performed using the Qiagen One-Step™ RT-PCR kit according to manufacturers protocols. Oligonucleotide primer and PCR conditions used to detect Xrcc2 and Gapdh were as described Supp. Ref. 6. As a specificity control for the knockdown studies, RT-PCR detection of the related gene Rad51 was performed. Oligonucleotides to detect Rad51 transcript were 5′-GCTTATGCACCGAAGAAGGA-3′ (forward) and 5′-AGCTTGTCCAGCTCTTTGGA-3′ (reverse) generating a 188 bp product. The RT-PCR products were separated by electrophoresis on a 1.0% agarose gel (with ethidium bromide), and visualized using a BioRad Gel Doc system.
Immunoblotting
Whole cell extracts lysed with RIPA buffer for 30 minutes on ice. Whole cell extracts were separated by SDS-PAGE electrophoresis (Pierce), transferred to a PVDF membrane (Invitolon PVDF, Cat# LC2005, Invitrogen), and probed for XRCC2 (Clone N-20; 1:100 Santa Cruz Biotechnology) or ACTIN (Cat#. Ab8227-50; 1:5000 Abcam). HRP-labeled secondary antibody was used to detect the primary antibody, via respectively (Cat#. SC2020 1:5000 Santa Cruz Biotechnology). HRP detection was performed using either Pierce Supersignal Dura West (Cat# 34076) or GE chemiluminescence (Cat#RPN2132) systems.
Flow cytometry and immunofluorescence
For flow cytometry, 105–106 cells were stained with an antibody-fluorophore cocktail containing B220-phycoerythrin (PE)-Cy7 (eBioscience, 1:100), IgG1-PE (Pharmingen, 1:100) and IgA-PE (Pharmingen,1:100) to analyze cells for CSR. Cell cycle distribution was detected as previously described Supp. Ref. 1. All flow cytometry procedures were performed using a FACSCalibur flow cytometer running Cell Quest Pro (BD Biosciences) acquisition software and analyzed using FlowJo software (V.8.4.6; Treestar, Inc.).
For immunofluorescence microscopy, cells were fixed, permeabilized, blocked and incubated with the following primary antibodies: phosphorylated (γ)-H2AX (Bethyl; 1:400), 53BP1 (Bethyl 1:100), Active Caspase 3 (Abcam 1:100), phosphorylated histone H3 serine 10 (pH3; Millipore 1:1000), or PCNA (Abcam; 1:100) as previously described Supp. Ref. 1. Slides were then treated and imaged as previously described Supp. Ref. 1. Cells stained for Active Caspase 3 (not fixed on coverslips) were also measured for positivity via flow cytometry.
Genomic DNA electrophoresis analysis
Cells were digested with Proteinase-K Solution [10mg/mL Proteinase-K, 10mM Tris-Cl (pH8.0) 5mM EDTA (pH8.0) 50mM NaCl] for 2 hours at 56°C, briefly centrifuged to remove cell debris. Genomic DNA was precipitated by isopropanol, and resulting pellets were resuspended in Tris-EDTA (pH8.0). DNA was quantified, and 2.5 μg was loaded per lane in a 1.5% agarose gel containing the DNA binding dye ethidium bromide. DNA was electrophoresed and imaged by Bio-Rad Quantity One gel documentation system. For controls, cells from each genotype were irradiated at 0 or 4Gy (CS137 irradiator; Shepard).
Fluorescence in-situ Hybridization (FISH)
Cells were fixed, permeabilized, freeze/thawed/soaked, denatured, and stored in 70% ethanol for FISH as previously described Supp. Refs. 7–8. Prior to hybridization, cells were neutralized, washed with 2X SSC (pH7.0), denatured, blocked with salmon sperm DNA/tRNA/formamide, and probed with Cy3-labeled BAC probe (RP23-109B20, Igh locus) as previously described Supp. Ref. 7–8. Coverslips were washed with 50% formamide/2XSSC, and probed for γ–H2AX by immunofluorescence.
Cytogenetic analyses
Cells were incubated for 4 hours in the presence of 40 ng/ml colcemid (Karyomax; Invitrogen), to enforce metaphase arrest. After colcemid treatment, cells treated as previously described Supp. Ref. 1 and imaged as described herein.
Supplementary Material
Acknowledgments
We thank T. Honjo, R. Maser, S. La Salle and A. Y. Tsygankov for sharing reagents and mice. We are grateful to C. Boboila, B. Schwer, F. Alt and, J. Chaudhuri for reagents and technical assistance. We acknowledge the Jackson Laboratory Gene Expression, Molecular Biology, and Flow Cytometry Services for technical support. This work was supported by grants from the National Institutes of Health R01CA138646, P20RR018789-06, T32DK07449-26, P30CA034196, and the Maine Cancer Foundation.
Footnotes
Author contributions
M.G.H. designed and performed experiments, and wrote manuscript; J.A., E.C., N.M.D., J.M., and K.J.S. performed experiments and contributed data; Y.H., B.L.K., and R.Y.W. are members of Jackson Laboratory Scientific Services and contributed data; K.D.M. designed experiments and wrote manuscript.
References
- 1.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 3.Durandy A, Taubenheim N, Peron S, Fischer A. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv Immunol. 2007;94:275–306. doi: 10.1016/S0065-2776(06)94009-7. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 5.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 6.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Tsai AG, et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahowald GK, Baron JM, Sleckman BP. Collateral damage from antigen receptor gene diversification. Cell. 2008;135:1009–1012. doi: 10.1016/j.cell.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Thacker J. The role of homologous recombination processes in the repair of severe forms of DNA damage in mammalian cells. Biochimie. 1999;81:77–85. doi: 10.1016/s0300-9084(99)80041-8. [DOI] [PubMed] [Google Scholar]
- 11.Braybrooke JP, Spink KG, Thacker J, Hickson ID. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. The Journal of biological chemistry. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- 12.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. Embo J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu N. XRCC2 is Required for the Formation of Rad51 Foci Induced by Ionizing Radiation and DNA Cross-Linking Agent Mitomycin C. J Biomed Biotechnol. 2002;2:106–113. doi: 10.1155/S1110724302204040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deans B, Griffin CS, O’Regan P, Jasin M, Thacker J. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 2003;63:8181–8187. [PubMed] [Google Scholar]
- 15.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci U S A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adam J, Deans B, Thacker J. A role for Xrcc2 in the early stages of mouse development. DNA Repair (Amst) 2007;6:224–234. doi: 10.1016/j.dnarep.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Caddle LB, Hasham MG, Schott WH, Shirley BJ, Mills KD. Homologous recombination is necessary for normal lymphocyte development. Mol Cell Biol. 2008;28:2295–2303. doi: 10.1128/MCB.02139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li MJ, et al. Rad51 expression and localization in B cells carrying out class switch recombination. Proc Natl Acad Sci U S A. 1996;93:10222–10227. doi: 10.1073/pnas.93.19.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borzillo GV, Endo K, Tsujimoto Y. Bcl-2 confers growth and survival advantage to interleukin 7-dependent early pre-B cells which become factor independent by a multistep process in culture. Oncogene. 1992;7:869–876. [PubMed] [Google Scholar]
- 20.Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 21.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 23.Thacker J, Tambini CE, Simpson PJ, Tsui LC, Scherer SW. Localization to chromosome 7q36.1 of the human XRCC2 gene, determining sensitivity to DNA-damaging agents. Hum Mol Genet. 1995;4:113–120. doi: 10.1093/hmg/4.1.113. [DOI] [PubMed] [Google Scholar]
- 24.Dohner K, et al. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92:4031–4035. [PubMed] [Google Scholar]
- 25.Simmons HM, et al. Cytogenetic and molecular heterogeneity of 7q36/12p13 rearrangements in childhood AML. Leukemia. 2002;16:2408–2416. doi: 10.1038/sj.leu.2402773. [DOI] [PubMed] [Google Scholar]
- 26.Mao X, et al. A case of adult T-cell leukaemia/lymphoma characterized by multiplex-fluorescence in situ hybridization, comparative genomic hybridization, fluorescence in situ hybridization and cytogenetics. The British journal of dermatology. 2001;145:117–122. doi: 10.1046/j.1365-2133.2001.04294.x. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 28.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 29.Nakano T, et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance to DNA-protein crosslinks in mammalian cells. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couedel C, et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alt F, Honjo T. AID for Immunoglobin Diversity. Vol. 94. Academic Press; London: 2007. [Google Scholar]
- 32.Soulas-Sprauel P, et al. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene. 2007;26:7780–7791. doi: 10.1038/sj.onc.1210875. [DOI] [PubMed] [Google Scholar]
- 33.Danoy P, Sonoda E, Lathrop M, Takeda S, Matsuda F. A naturally occurring genetic variant of human XRCC2 (R188H) confers increased resistance to cisplatin-induced DNA damage. Biochem Biophys Res Commun. 2007;352:763–768. doi: 10.1016/j.bbrc.2006.11.083. [DOI] [PubMed] [Google Scholar]
- 34.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 35.Zaheen A, et al. AID constrains germinal center size by rendering B cells susceptible to apoptosis. Blood. 2009;114:547–554. doi: 10.1182/blood-2009-03-211763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.