Abstract
We consider the problem of clustering observations using a potentially large set of features. One might expect that the true underlying clusters present in the data differ only with respect to a small fraction of the features, and will be missed if one clusters the observations using the full set of features. We propose a novel framework for sparse clustering, in which one clusters the observations using an adaptively chosen subset of the features. The method uses a lasso-type penalty to select the features. We use this framework to develop simple methods for sparse K-means and sparse hierarchical clustering. A single criterion governs both the selection of the features and the resulting clusters. These approaches are demonstrated on simulated data and on genomic data sets.
1 Introduction
Let X denote an n×p data matrix, with n observations and p features. Suppose that we wish to cluster the observations, and we suspect that the true underlying clusters differ only with respect to some of the features. We propose a method for sparse clustering, which allows us to group the observations using only an adaptively-chosen subset of the features. This method is most useful for the high-dimensional setting where p ≫ n, but can also be used when p < n. Sparse clustering has a number of advantages. If the underlying groups differ only in terms of some of the features, then it might result in more accurate identification of these groups than standard clustering. It also yields interpretable results, since one can determine precisely which features are responsible for the observed differences between the groups or clusters. In addition, fewer features are required to assign a new observation to a pre-existing cluster.
As a motivating example, we generated 500 independent observations from a bivariate normal distribution. A mean shift on the first feature defines the two classes. The resulting data, as well as the clusters obtained using standard 2-means clustering and our sparse 2-means clustering proposal, can be seen in Figure 1. Unlike standard 2-means clustering, our proposal for sparse 2-means clustering automatically identifies a subset of the features to use in clustering the observations. Here it uses only the first feature, and consequently agrees quite well with the true class labels. 1 In this example, one could use an elliptical metric in order to identify the two classes without using feature selection. However, this will not work in general.
Figure 1.
In a two-dimensional example, two classes differ only with respect to the first feature. Sparse 2-means clustering selects only the first feature, and therefore yields a superior result.
Clustering methods require some concept of the dissimilarity between pairs of observations. Let d(xi, xi′) denote some measure of dissimilarity between observations xi and xi′, which are rows i and i′ of the data matrix X. Throughout this paper, we will assume that d is additive in the features. That is, , where di,i′,j indicates the dissimilarity between observations i and i′ along feature j. All of the data examples in this paper take d to be squared Euclidean distance, di;i′;j = (Xij − Xi′j)2. However, other dissimilarity measures are possible, such as the absolute difference di,i′,j = |Xij − Xi′j |.
The rest of this paper is organized as follows. In Section 2, we briey review existing methods for sparse clustering and we present the general framework of our proposal. We present sparse K-means clustering in Section 3 and sparse hierarchical clustering in Section 4. Section 5 contains an application of our method to a gene expression data set, and Section 6 contains an application to a single nucleotide polymorphism data set. The discussion is in Section 7.
2 An overview of sparse clustering
2.1 Past work on sparse clustering
A number of authors have noted the necessity of specialized clustering techniques for the high-dimensional setting. Here, we briefly review previous proposals for feature selection and dimensionality reduction in clustering.
One way to reduce the dimensionality of the data before clustering is by performing a matrix decomposition. One can approximate the n × p data matrix X as X ≈ AB where A is a n × q matrix and B is a q × p matrix, q ≪ p. Then, one can cluster the observations using A as the data matrix, rather than X. For instance, Ghosh & Chinnaiyan (2002) and Liu et al. (2003) propose performing PCA in order to obtain a matrix A of reduced dimensionality; then, the n rows of A can be clustered. Similarly, Tamayo et al. (2007) suggest decomposing X using the non-negative matrix factorization (Lee & Seung 1999, Lee & Seung 2001), followed by clustering the rows of A. However, these approaches have a number of drawbacks. First of all, the resulting clustering is not sparse in the features, since each of the columns of A is a function of the full set of p features. Moreover, there is no guarantee that A contains the signal that one is interested in detecting via clustering. In fact, Chang (1983) studies the effect of performing PCA to reduce the data dimension before clustering, and finds that this procedure is not justified since the principal components with largest eigenvalues do not necessarily provide the best separation between subgroups.
The model-based clustering framework has been studied extensively in recent years, and many of the proposals for feature selection and dimensionality reduction for clustering fall in this setting. An overview of model-based clustering can be found in McLachlan & Peel (2000) and Fraley & Raftery (2002). The basic idea is as follows. One can model the rows of X as independent multivariate observations drawn from a mixture model with K components; usually a mixture of Gaussians is used. That is, given the data, the log likelihood is
| (1) |
where fk is a Gaussian density parametrized by its mean μk and covariance matrix Σk. The EM algorithm (Dempster et al. 1977) can be used to fit this model.
However, when p ≈ n or p ≫ n a problem arises because the p × p covariance matrix Σk cannot be estimated from only n observations. Proposals for overcoming this problem include the factor analyzer approach of McLachlan et al. (2002) and McLachlan et al. (2003), which assume that the observations lie in a low-dimensional latent factor space and that Σk is low rank. This leads to dimensionality reduction but not sparsity, since the latent factors are linear combinations of all of the features.
It turns out that model-based clustering lends itself easily to feature selection. Rather than seeking μk and Σk that maximize the log likelihood (1), one can instead maximize the log likelihood subject to a penalty that is chosen to yield sparsity in the features. This approach is taken in a number of papers, including Pan & Shen (2007), Wang & Zhu (2008), and Xie et al. (2008). For instance, if we assume that the features of X are centered to have mean zero, then Pan & Shen (2007) propose maximizing the penalized log likelihood
| (2) |
where Σ1 = …. = ΣK is taken to be a diagonal matrix. That is, an L1 (or lasso) penalty is applied to the elements of μk. When the non-negative tuning parameter λ is large, then some of the elements of μk will be exactly equal to zero. If, for some variable j, μkj = 0 for all k = 1, …, K, then the resulting clustering will not involve feature j. Hence, this yields a clustering that is sparse in the features.
Raftery & Dean (2006) also present a method for feature selection in the model-based clustering setting, using an entirely different approach. They recast the variable selection problem as a model selection problem: models containing nested subsets of variables are compared. The nested models are sparse in the features, and so this yields a method for sparse clustering. A related proposal is made in Maugis et al. (2009).
Friedman & Meulman (2004) propose clustering objects on subsets of attributes (COSA). Let Ck denote the indices of the observations in the kth of K clusters. Then, the COSA criterion is
| (3) |
(Actually, this is a simplified version of the COSA proposal, which allows for different feature weights within each cluster.) Here, ak is some function of the number of elements in cluster k, w ∈ ℝp is a vector of feature weights, and λ ≥ 0 is a tuning parameter. It can be seen that this criterion is related to a weighted version of K-means clustering. Unfortunately, this proposal does not truly result in a sparse clustering, since all variables have non-zero weights. An extension of (3) is proposed in order to generalize the method to other types of clustering, such as hierarchical clustering. The proposed optimization algorithm is quite complex, and involves multiple tuning parameters.
Our proposal can be thought of as a much simpler version of (3). It is a general framework that can be applied in order to obtain sparse versions of a number of clustering methods. The resulting algorithms are efficient even when p is quite large.
2.2 The proposed sparse clustering framework
Suppose that we wish to cluster n observations on p dimensions; recall that X is of dimension n×p. In this paper, we take a general approach to the problem of sparse clustering. Let Xj ∈ ℝn denote feature j. Many clustering methods can be expressed as optimizing criteria of the form
| (4) |
where fj(Xj, Θ) is some function that involves only the jth feature of the data, and Θ is a parameter restricted to lie in a set D. K-means and hierarchical clustering are two such examples, as we show in the next few sections. (With K-means, for example, fj turns out to be the between cluster sum of squares for feature j, and Θ is a partition of the observations into K disjoint sets.) We define sparse clustering as the solution to the problem
| (5) |
where wj is a weight corresponding to feature j. We make a few observations about (5):
The L1, or lasso, penalty on w results in sparsity for small values of the tuning parameter s: that is, some of the wj’s will equal zero. The L2 penalty also serves an important role, since without it, at most one element of w would be non-zero in general. A criterion involving a linear objective subject to both an L1 and an L2 constraint was also used in Witten et al. (2009).
The value of wj can be interpreted as the contribution of feature j to the resulting sparse clustering: a large value of wj indicates a feature that contributes greatly, and wj = 0 means that feature j is not involved in the clustering.
In general, for the formulation (5) to result in a non-trivial sparse clustering, it is necessary that fj(Xj, Θ) > 0 for some or all j. That is, if fj(Xj, Θ) ≤ 0, then wj = 0. If fj(Xj, Θ) > 0, then the non-negativity constraint on wj has no effect.
We optimize (5) using an iterative algorithm: holding w fixed, we optimize (5) with respect to Θ, and holding Θ fixed, we optimize (5) with respect to w. In general, we do not achieve a global optimum of (5) using this iterative approach; however, we are guaranteed that each iteration increases the objective function. The first optimization typically involves application of a standard clustering procedure to a weighted version of the data. To optimize (5) with respect to w with Θ held fixed, we note that the problem can be re-written as
| (6) |
where aj = fj(Xj, Θ). This is easily solved by soft-thresholding, as detailed next.
Proposition
The solution to the convex problem (6) is , where x+ denotes the positive part of x and where Δ = 0 if that results in ||w||1 ≤ s; otherwise, Δ > 0 is chosen to yield ||w||1 = s. Here, S is the soft-thresholding operator, defined as S(x, c) = sign(x)(|x| − c)+.
The proposition follows from the Karush-Kuhn-Tucker conditions (see e.g. Boyd & Vandenberghe 2004).
In the next two sections we show that K-means clustering and hierarchical clustering optimize criteria of the form (4). We then propose sparse versions of K-means clustering and hierarchical clustering using (5). The resulting criteria for sparse clustering take on simple forms, are easily optimized, and involve a single tuning parameter s that controls the number of features involved in the clustering. Moreover, our proposal is a general framework that can be applied to any clustering procedure for which a criterion of the form (4) is available.
In this paper we consider the sparse clustering criterion (5), where sparsity in the features results from the L1 penalty on w. Other penalties on w could be used that would also yield sparsity (see e.g. Fan & Li 2001, Lv & Fan 2009). Such alternatives are not considered in this paper.
3 Sparse K-means clustering
3.1 The sparse K-means method
K-means clustering minimizes the within-cluster sum of squares (WCSS). That is, it seeks to partition the n observations into K sets, or clusters, such that the WCSS
| (7) |
is minimal, where nk is the number of observations in cluster k and Ck contains the indices of the observations in cluster k. In general, di,i′,j can denote any dissimilarity measure between observations i and i′ along feature j. However, in this paper we will take di,i′,j = (Xij − Xi′j)2; for this reason, we refer to (7) as the within-cluster sum of squares. Note that if we define the between-cluster sum of squares (BCSS) as
| (8) |
then minimizing the WCSS is equivalent to maximizing the BCSS.
One could try to develop a method for sparse K-means clustering by optimizing a weighted WCSS, subject to constraints on the weights: that is,
| (9) |
Here, s is a tuning parameter. Since each element of the weighted sum is negative, the maximum occurs when all weights are zero, regardless of the value of s. This is not an interesting solution. We instead maximize a weighted BCSS, subject to constraints on the weights. Our sparse K-means clustering criterion is as follows:
| (10) |
The weights will be sparse for an appropriate choice of the tuning parameter s. Note that if w1 = … = wp, then (10) simply reduces to the standard K-means clustering criterion. We observe that (8) and (10) are special cases of (4) and (5) where Θ = (C1, …, CK), , and D denotes the set of all possible partitions of the observations into K clusters.
The criterion (10) assigns a weight to each feature, based on the increase in BCSS that the feature can contribute. First, consider the criterion with the weights w1, … wp fixed. It reduces to a clustering problem, using a weighted dissimilarity measure. Second, consider the criterion with the clusters C1, … CK fixed. Then a weight will be assigned to each feature based on the BCSS of that feature; features with larger BCSS will be given larger weights. We present an iterative algorithm for maximizing (10).
Algorithm for sparse K-means clustering
Initialize w as .
-
Iterate until convergence:
-
Holding w fixed, optimize (10) with respect to C1, … CK. That is,
(11) by applying the standard K-means algorithm to the n × n dissimilarity matrix with (i, i′) element Σj wjdi,i′,j.
-
Holding C1, … CK fixed, optimize (10) with respect to w by applying the Proposition: where
(12) and Δ = 0 if that results in ||w||1 < s; otherwise, Δ > 0 is chosen so that ||w||1 = s.
-
The clusters are given by C1, … CK, and the feature weights corresponding to this clustering are given by w1, … wp.
When d is squared Euclidean distance, Step 2(a) can be optimized by performing K-means on the data after scaling each feature j by . In our implementation of sparse K-means, we iterate Step 2 until the stopping criterion
| (13) |
is satisfied. Here, wr indicates the set of weights obtained at iteration r. In the examples that we have examined, this criterion tends to be satisfied within no more than 5 to 10 iterations. However, we note that the algorithm generally will not converge to the global optimum of the criterion (10), since the criterion is non-convex and uses in Step 2(a) the algorithm for K-means clustering, which is not guaranteed to find a global optimum (see e.g. MacQueen 1967).
Note the similarity between the COSA criterion (3) and (10): when in (3), then both criteria involve minimizing a weighted function of the WCSS, where the feature weights reflect the importance of each feature in the clustering. However, (3) does not result in weights that are exactly equal to zero unless λ = 0, in which case only one weight is non-zero. The combination of L1 and L2 constraints in (10) yields the desired effect.
The Appendix contains an additional remark about our criterion for sparse K-means clustering.
3.2 Selection of tuning parameter for sparse K-means
The sparse K-means clustering algorithm has one tuning parameter, s, which is the L1 bound on w in (10). We assume that K, the number of clusters, is fixed. The problem of selecting K is outside of the scope of this paper, and has been discussed extensively in the literature for standard K-means clustering; we refer the interested reader to Milligan & Cooper (1985), Kaufman & Rousseeuw (1990), Tibshirani et al. (2001), Sugar & James (2003), and Tibshirani & Walther (2005).
A method for choosing the value of s is required. Note that one cannot simply select s to maximize the objective function in (10), since as s is increased, the objective will increase as well. Instead, we apply a permutation approach that is closely related to the gap statistic of Tibshirani et al. (2001) for selecting the number of clusters K in standard K-means clustering.
Algorithm to select tuning parameter s for sparse K-means
Obtain permuted data sets X1, … XB by independently permuting the observations within each feature.
-
For each candidate tuning parameter value s:
Compute , the objective obtained by performing sparse K-means with tuning parameter value s on the data X.
For b = 1, 2, … B, compute Ob(s), the objective obtained by performing sparse K-means with tuning parameter value s on the data Xb.
Calculate .
Choose s* corresponding to the largest value of Gap(s). Alternatively, one can choose s* to equal the smallest value for which Gap(s*) is within a standard deviation of log(Ob(s*)) of the largest value of Gap(s).
Note that while there may be strong correlations between the features in the original data X, the features in the permuted data sets X1, … Xb are uncorrelated with each other. The gap statistic measures the strength of the clustering obtained on the real data relative to the clustering obtained on null data that does not contain subgroups. The optimal tuning parameter value occurs when this quantity is greatest.
In Figure 2, we apply this method to a simple example with 6 equally-sized classes, where n = 120, p = 2000, and 200 features differ between classes. In the figure we have used the classification error rate (CER) for two partitions of a set of n observations. This is defined as follows. Let P and Q denote the two partitions; P might be the true class labels, and Q might be a partition obtained by clustering. Let 1P(i,i′) be an indicator for whether partition P places observations i and i′ in the same group, and define 1Q(i,i′) analogously. Then, the CER (used for example in Chipman & Tibshirani 2005) is defined as . The CER equals 0 if the partitions P and Q agree perfectly; a high value indicates disagreement. Note that CER is one minus the Rand index (Rand 1971).
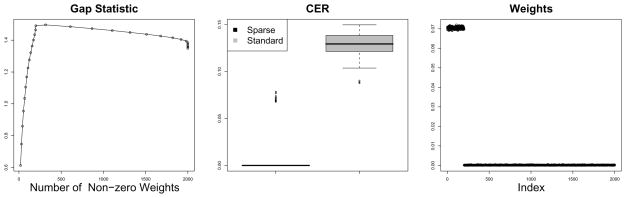
Figure 2.
Sparse and standard 6-means clustering are applied to a simulated 6-class example. Left: The gap statistics obtained using the sparse 6-means tuning parameter selection method, as a function of the number of features with non-zero weights, averaged over 10 simulated data sets. Center: Boxplots of the CERs obtained using sparse and standard 6-means clustering on 100 simulated data sets. Right: The weights obtained using sparse 6-means clustering, averaged over 100 simulated data sets.
3.3 A simulation study of sparse K-means
3.3.1 Simulation 1: A comparison of sparse and standard K-means
We compare the performances of standard and sparse K-means in a simulation study where q = 50 features differ between K = 3 classes. Xij ~ N(μij, 1) independent; μij = μ(1i∈C1,j≤q − 1i∈C2,j≤q). Data sets were generated with various values of μ and p, with 20 observations per class. The results can be seen in Tables 1, 2, and 3. In this example, when p > q, sparse 3-means tends to outperform standard 3-means, since it exploits the sparsity of the signal. On the other hand, when p = q, then standard 3-means is at an advantage, since it gives equal weights to all features. The value of the tuning parameter s for sparse 3-means was selected to maximize the gap statistic. As seen in Table 3, this generally resulted in more than 50 features with non-zero weights. This reflects the fact that the tuning parameter selection method tends not to be very accurate. Fewer features with non-zero weights would result from selecting the tuning parameter at the smallest value that is within one standard deviation of the maximal gap statistic, as described in Section 3.2.
Table 1.
Standard 3-means results for Simulation 1. The reported values are the mean (and standard error) of the CER over 20 simulations. The μ/p combinations for which the CER of standard 3-means is significantly less than that of sparse 3-means (at level α = 0:05) are shown in bold.
| p = 50 | p = 200 | p = 500 | p = 1000 | |
|---|---|---|---|---|
| μ = 0.6 | 0.07(0.01) | 0.184(0.015) | 0.22(0.009) | 0.272(0.006) |
| μ = 0.7 | 0.023(0.005) | 0.077(0.009) | 0.16(0.012) | 0.232(0.01) |
| μ = 0.8 | 0.013(0.004) | 0.038(0.007) | 0.08(0.005) | 0.198(0.01) |
| μ= 0.9 | 0.001(0.001) | 0.013(0.005) | 0.048(0.008) | 0.102(0.013) |
| μ = 1 | 0.002(0.002) | 0.004(0.002) | 0.013(0.004) | 0.05(0.006) |
Table 2.
Sparse 3-means results for Simulation 1. The reported values are the mean (and standard error) of the CER over 20 simulations. The μ/p combinations for which the CER of sparse 3-means is significantly less than that of standard 3-means (at level α = 0:05) are shown in bold.
| p = 50 | p = 200 | p = 500 | p = 1000 | |
|---|---|---|---|---|
| μ = 0.6 | 0.146(0.014) | 0.157(0.016) | 0.183(0.015) | 0.241(0.017) |
| μ = 0.7 | 0.081(0.011) | 0.049(0.008) | 0.078(0.013) | 0.098(0.013) |
| μ = 0.8 | 0.043(0.008) | 0.031(0.007) | 0.031(0.005) | 0.037(0.006) |
| μ = 0.9 | 0.015(0.006) | 0.005(0.003) | 0.014(0.004) | 0.014(0.004) |
| μ = 1 | 0.009(0.004) | 0.004(0.002) | 0.001(0.001) | 0.002(0.002) |
Table 3.
Sparse 3-means results for Simulation 1. The mean number of non-zero feature weights resulting from the method for tuning parameter selection of Section 3.2 is shown; standard errors are given in parentheses. Note that 50 features differ between the three classes.
| p = 50 | p = 200 | p = 500 | p = 1000 | |
|---|---|---|---|---|
| μ = 0.6 | 41.35(0.895) | 167.4(7.147) | 243.1(31.726) | 119.45(41.259) |
| μ = 0.7 | 40.85(0.642) | 195.65(2.514) | 208.85(19.995) | 130.15(17.007) |
| μ = 0.8 | 38.2(0.651) | 198.85(0.654) | 156.35(13.491) | 106.7(10.988) |
| μ = 0.9 | 38.7(0.719) | 200(0) | 204.75(19.96) | 83.7(9.271) |
| μ = 1 | 36.95(0.478) | 200(0) | 222.85(20.247) | 91.65(14.573) |
3.3.2 Simulation 2: A comparison with other approaches
We compare the performance of sparse K-means to a number of competitors:
-
The COSA proposal of Friedman & Meulman (2004). COSA was run using the R code available from the website http://www-stat.stanford.edu/~jhf/COSA.html, in order to obtain a reweighted dissimilarity matrix. Then, two methods were used to obtain a clustering:
3-medoids clustering (using the partitioning around medoids algorithm described in Kaufman & Rousseeuw 1990) was performed on the reweighted dissimilarity matrix.
Hierarchical clustering with average linkage was performed on the reweighted dissimilarity matrix, and the dendrogram was cut so that 3 groups were obtained.
The model-based clustering approach of Raftery & Dean (2006). It was run using the R package clustvarsel, available from http://cran.r-project.org/.
The penalized log-likelihood approach of Pan & Shen (2007). R code implementing this method was provided by the authors.
PCA followed by 3-means clustering. Only the first principal component was used, since in the simulations considered the first principal component contained the signal. This is similar to several proposals in the literature (see e.g. Ghosh & Chinnaiyan 2002, Liu et al. 2003, Tamayo et al. 2007).
The set-up is similar to that of Section 3.3.1, in that there are K = 3 classes and Xij ~ N(μij, 1) independent; μij = μ(1i∈C1,j≤q − 1i∈C2,j≤q). Two simulations were run: a small simulation with p = 25, q = 5, and 10 observations per class, and a larger simulation with p = 500, q = 50, and 20 observations per class. The results are shown in Table 4. The quantitities reported are the mean and standard error (given in parentheses) of the CER and the number of non-zero coefficients, over 25 simulated data sets. Note that the method of Raftery & Dean (2006) was run only on the smaller simulation for computational reasons.
Table 4.
Results for Simulation 2. The quantities reported are the mean and standard error (given in parentheses) of the CER, and of the number of non-zero coefficients, over 25 simulated data sets.
| Simulation | Method | CER | Num. Non-zero Coef. |
|---|---|---|---|
| Small Simulation: p = 25, q = 5, 10 obs. per class | Sparse K-means | 0.112(0.019) | 8.2(0.733) |
| K-means | 0.263(0.011) | 25(0) | |
| Pan and Shen | 0.126(0.017) | 6.72(0.334) | |
| COSA w/Hier. Clust. | 0.381(0.016) | 25(0) | |
| COSA w/K-medoids | 0.369(0.012) | 25(0) | |
| Raftery and Dean | 0.514(0.031) | 22(0.86) | |
| PCA w/K-means | 0.16(0.012) | 25(0) | |
| Large Simulation: p = 500, q = 50, 20 obs. per class | Sparse K-means | 0.106(0.019) | 141.92(9.561) |
| K-means | 0.214(0.011) | 500(0) | |
| Pan and Shen | 0.134(0.013) | 76(3.821) | |
| COSA w/Hier. Clust. | 0.458(0.011) | 500(0) | |
| COSA w/K-medoids | 0.427(0.004) | 500(0) | |
| PCA w/K-means | 0.058(0.006) | 500(0) | |
We make a few comments about Table 4. First of all, neither variant of COSA performed well in this example, in terms of CER. This is somewhat surprising. However, COSA allows the features to take on a different set of weights with respect to each cluster. In the simulation, each cluster is defined on the same set of features, and COSA may have lost power by allowing different weights for each cluster. The method of Raftery & Dean (2006) also does quite poorly in this example, although its performance seems to improve somewhat as the signal to noise ratio in the simulation is increased (results not shown). The penalized model-based clustering method of Pan & Shen (2007) resulted in low CER as well as sparsity in both simulations. In addition, the simple method of PCA followed by 3-means clustering yielded quite low CER. However, since the principal components are linear combinations of all of the features, the resulting clustering is not sparse in the features and thus does not achieve the stated goal in this paper of performing feature selection.
In both simulations, sparse K-means performed quite well, in that it resulted in a low CER and sparsity. The tuning parameter was chosen to maximize the gap statistic; however, greater sparsity could have been achieved by choosing the smallest tuning parameter value within one standard deviation of the maximal gap statistic, as described in Section 3.2. Our proposal also has the advantage of generalizing to other types of clustering, as described in the next section.
4 Sparse hierarchical clustering
4.1 The sparse hierarchical clustering method
Hierarchical clustering produces a dendrogram that represents a nested set of clusters: depending on where the dendrogram is cut, between 1 and n clusters can result. One could develop a method for sparse hierarchical clustering by cutting the dendrogram at some height and maximizing a weighted version of the resulting BCSS, as in Section 3. However, it is not clear where the dendrogram should be cut, nor whether multiple cuts should be made and somehow combined. Instead, we pursue a simpler and more natural approach to sparse hierarchical clustering in this section.
Note that hierarchical clustering takes as input a n × n dissimilarity matrix U. The clustering can use any type of linkage - complete, average, or single. If U is the overall dissimilarity matrix {Σj di,i′,j}i,i′, then standard hierarchical clustering results. In this section, we cast the overall dissimilarity matrix {Σj di,i′,j}i,i′ in the form (4), and then propose a criterion of the form (5) that leads to a reweighted 12 dissimilarity matrix that is sparse in the features. When hierarchical clustering is performed on this reweighted dissimilarity matrix, then sparse hierarchical clustering results.
Since scaling the dissimilarity matrix by a factor does not affect the shape of the resulting dendrogram, we ignore proportionality constants in the following discussion. Consider the criterion
| (14) |
Let U* optimize (14). It is not hard to show that , and so performing hierarchical clustering on U* results in standard hierarchical clustering. So we can think of standard hierarchical clustering as resulting from the criterion (14). To obtain sparsity in the features, we modify (14) by multiplying each element of the summation over j by a weight wj, subject to constraints on the weights:
| (15) |
The U** that optimizes (15) is proportional to {Σj di,i′,jwj}i,i′. Since w is sparse for small values of the tuning parameter s, U** involves only a subset of the features, and so performing hierarchical clustering on U** results in sparse hierarchical clustering. We refer to (15) as the sparse hierarchical clustering criterion. Observe that (14) takes the form (4) with Θ = U, fj(Xj, Θ) = Σi,i′di,i′,jUi,i′ and Θ ∈ D corresponding to . It follows directly that (15) takes the form (5), and so sparse hierarchical clustering fits into the framework of Section 2.2.
By inspection, (15) is bi-convex in U and w: with w fixed, it is convex in U, and with U fixed, it is convex in w. This leads to an extremely simple algorithm for optimizing it. However, before we present this algorithm, we introduce some additional notation that will prove useful. Let D ∈ ℝn2 × p be the matrix in which column j consists of the elements {di,i′,j}i,i′, strung out into a vector. Then, Σj wjΣi,i′di,i′.jUi,i′ = uTDw where u ∈ ℝn2 is obtained by stringing out U into a vector. It follows that the criterion (15) is equivalent to
| (16) |
We now present our algorithm for sparse hierarchical clustering.
Algorithm for sparse hierarchical clustering
Initialize w as .
-
Iterate until convergence:
Update .
Update where a = DTu and Δ = 0 if this results in ||w||1 ≤ s; otherwise, Δ > 0 is chosen such that ||w||1 = s.
Re-write u as a n × n matrix, U.
Perform hierarchical clustering on the n × n dissimilarity matrix U.
Observe that (16) is the sparse principal components (SPC) criterion of Witten et al. (2009), with an additional non-negativity constraint on w. If di,i′,j ≥ 0, as is usually the case, then the non-negativity constraint can be dropped. In fact, Steps 1 and 2 of the algorithm for sparse hierarchical clustering are essentially the SPC algorithm of Witten et al. (2009).
When viewed in this way, our method for sparse hierarchical clustering is quite simple. The first SPC of the n2 × p matrix D is denoted w. Then u ∝ Dw can be re-written as a n × n matrix U, which is a weighted linear combination of the feature-wise dissimilarity matrices. When s is small, then some of the wj will equal zero, and so U will depend on only a subset of the features. We then perform hierarchical clustering on U in order to obtain a dendrogram that is based only on an adaptively-chosen subset of the features.
In our implementation of this algorithm, we used (13) as a stopping criterion in Step 2. In the examples that we considered, the stopping criterion generally was satisfied within 10 iterations. As mentioned earlier, the criterion (16) is bi-convex in u and w, and we are not guaranteed convergence to a global optimum using this iterative algorithm.
4.2 A simple underlying model for sparse hierarchical clustering
We study the behaviors of sparse and standard hierarchical clustering under a simple model. Suppose that the n observations fall into two classes, C1 and C2, which differ only with respect to the first q features. The elements Xij are independent and normally distributed with a mean shift between the two classes in the first q features:
| (17) |
Note that for i ≠ i′,
| (18) |
Let di,i′,j = (Xij − Xi′j)2; that is, the dissimilarity measure is squared Euclidean distance. Then, for i ≠ i′,
| (19) |
where denotes the noncentral distribution with noncentrality parameter λ. This means that the overall dissimilarity matrix used by standard hierarchical clustering has off-diagonal elements
| (20) |
and so for i ≠ i′,
| (21) |
and
| (22) |
We now consider the behavior of sparse hierarchical clustering. Suppose that wj ∝ 1j≤q; this corresponds to the ideal situation in which the important features have equal non-zero weights, and the unimportant features have weights that equal zero. Then the dissimilarity matrix used for sparse hierarchical clustering has elements
| (23) |
So in this ideal setting, the dissimilarity matrix used for sparse hierarchical clustering (23) is a denoised version of the dissimilarity matrix used for standard hierarchical clustering (20). Of course, in practice, wj is not proportional to 1j≤q.
We now allow w to take a more general form. Recall that w is the first SPC of D, obtained by writing {di,i′,j}i,i′,j as a n2 × p matrix. To simplify the discussion, suppose instead that w is the first SPC of E(D). Then, w is not random, and
| (24) |
from (21). To see this latter point, note that by the sparse hierarchical clustering algorithm, w is obtained by repeating the operation w = S(E(D)TE(D)w, Δ)/||S(E(D)TE(D)w, Δ)||2 until convergence, for Δ > 0 chosen to yield ||w||1 = s in each iteration. Initially, w1 = … = wp. By inspection, in each subsequent iteration, (24) holds true.
From (21), the expectations of the off-diagonal elements of the dissimilarity matrix used for sparse hierarchical clustering are therefore
| (25) |
By comparing (22) to (25), and using (24), we see that the expected dissimilarity between observations in different classes relative to observations in the same class is greater for sparse hierarchical clustering than for standard hierarchical clustering. Note that we have taken the weight vector w to be the first SPC of E(D), rather than the first SPC of D.
4.3 Selection of tuning parameter for sparse hierarchical clustering
We now consider the problem of selecting a value for s, the L1 bound for w in the sparse hierarchical clustering criterion. We take an approach similar to that of Section 3.2 for sparse K-means, in this case letting O(s) = Σj wjΣi,i′di,i′.jUi,i′. Since the details of this method are the same as in Section 3.2, we do not write out the algorithm here.
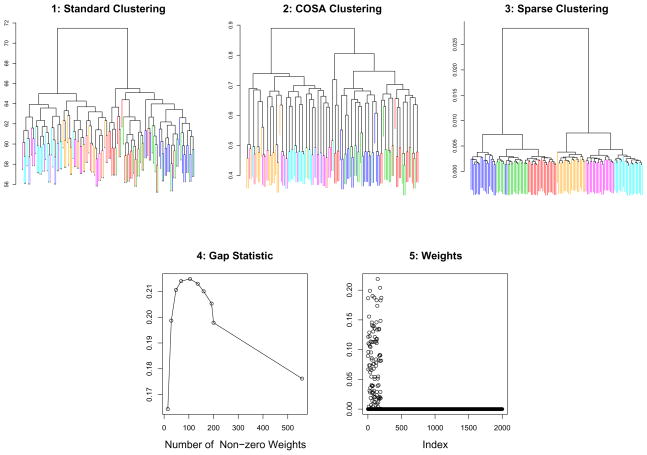
We demonstrate the performance of this tuning parameter selection method on the simulated 6-class data set used for Figure 2. We performed standard, COSA, and sparse hierarchical clustering with complete linkage. The results can be seen in Figure 3. Sparse hierarchical clustering results in better separation between the subgroups. Moreover, the correct features are given non-zero weights.
Figure 3.
Standard hierarchical clustering, COSA, and sparse hierarchical clustering with complete linkage were performed on simulated 6-class data. 1, 2, 3: The color of each leaf indicates its class identity. CERs were computed by cutting each dendrogram at the height that results in 6 clusters: standard, COSA, and sparse clustering yielded CERs of 0.169, 0.160, and 0.0254. 4: The gap statistics obtained for sparse hierarchical clustering, as a function of the number of features included for each value of the tuning parameter. 5: The w obtained using sparse hierarchical clustering; note that the six classes differ with respect to the first 200 features.
In this example and throughout this paper, we used di,i′,j = (Xij −Xi′j)2; that is, the dissimilarity measure used was squared Euclidean distance. However, in many simulated examples, we found that better performance results from using absolute value dissimilarity, di,i′,j = |Xij − Xi′j |.
4.4 Complementary sparse clustering
Standard hierarchical clustering is often dominated by a single group of features that have high variance and are highly correlated with each other. The same is true of sparse hierarchical clustering. Nowak & Tibshirani (2008) propose complementary clustering, a method that allows for the discovery of a secondary clustering after removing the signal found in the standard hierarchical clustering. Here we provide a method for complementary sparse clustering, an analogous approach for the sparse clustering framework. This simple method follows directly from our sparse hierarchical clustering proposal.
As in Section 4.1, we let D denote the n2×p matrix of which column j consists of {di,i′,j}i,i′ in vector form. Let u1, w1 optimize (16); that is, U1 (obtained by writing u1 in matrix form) is a weighted linear combination of the feature-wise dissimilarity matrices, and w1 denotes the corresponding feature weights. Then, the criterion
| (26) |
results in a dissimilarity matrix U2, obtained by writing u2 as a n × n matrix, that yields a complementary sparse clustering. The feature weights for this secondary clustering are given by w2. Note that (26) is simply the proposal of Witten et al. (2009) for finding the second SPC of D subject to orthogonality constraints, with an additional non-negativity constraint on w.
Observe that U2 is symmetric with zeroes on the diagonal, and that due to the constraint that , some elements of U2 will be negative. However, since only the off-diagonal elements of a dissimilarity matrix are used in hierarchical clustering, and since the shape of the dendrogram is not affected by adding a constant to the off-diagonal elements, in practice this is not a problem. The algorithm for complementary sparse clustering is as follows:
Algorithm for complementary sparse hierarchical clustering
Apply the sparse hierarchical clustering algorithm, and let u1 denote the resulting linear combination of the p feature-wise dissimilarity matrices, written in vector form.
Initialize w2 as .
-
Iterate until convergence:
Update .
Update where a = DTu2 and Δ = 0 if this results in ||w2||1 ≤ s; otherwise, Δ > 0 is chosen such that ||w2||1 = s.
Re-write u2 as a n × n matrix, U2.
Perform hierarchical clustering on U2.
Of course, one could easily extend this procedure in order to obtain further complementary clusterings.
5 Re-analysis of a breast cancer data set
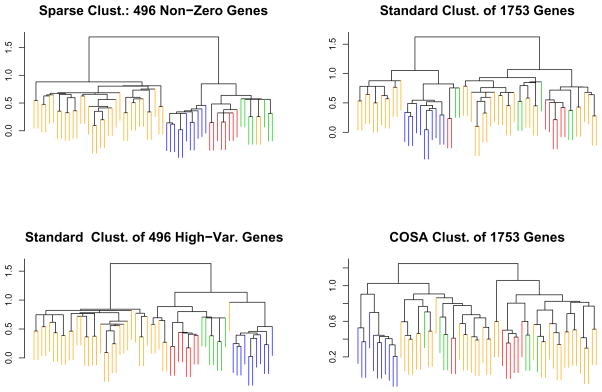
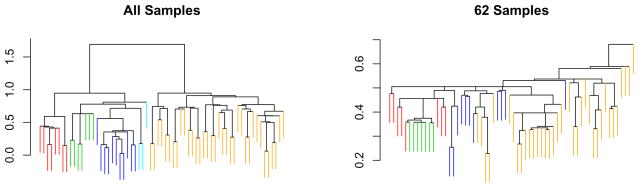
In a well-known paper, Perou et al. (2000) used gene expression microarrays to profile 65 surgical specimens of human breast tumors. Some of the samples were taken from the same tumor before and after chemotherapy. The data are available at http://genome-www.stanford.edu/breast_cancer/molecularportraits/download.shtml. The 65 samples were hierarchically clustered using what we will refer to as “Eisen” linkage; this is a centroid-based linkage that is implemented in Michael Eisen’s Cluster program (Eisen et al. 1998). Two sets of genes were used for the clustering: the full set of 1753 genes, and an intrinsic gene set consisting of 496 genes. The intrinsic genes were defined as having the greatest level of variation in expression between different tumors relative to variation in expression between paired samples taken from the same tumor before and after chemotherapy. The dendrogram obtained using the intrinsic gene set was used to identify four classes – basal-like, Erb-B2, normal breast-like, and ER+ – to which 62 of the 65 samples belong. It was determined that the remaining three observations did not belong to any of the four classes. These four classes are not visible in the dendrogram obtained using the full set of genes, and the authors concluded that the intrinsic gene set is necessary to observe the classes. In Figure 5, two dendrograms obtained by clustering on the intrinsic gene set are shown. The first was obtained by clustering all 65 observations, and the second was obtained by clustering the 62 observations that were assigned to one of the four classes. The former figure is in the original paper, and the latter is not. In particular, note that the four classes are not clearly visible in the dendrogram obtained using only 62 observations.
Figure 5.
Four hierarchical clustering methods were used to cluster the 62 observations that were assigned to one of four classes in Perou et al. (2000). Sparse clustering results in the best separation between the four classes. The color coding is as in Figure 4.
We wondered whether our proposal for sparse hierarchical clustering could yield a dendrogram that reflects the four classes, without any knowledge of the paired samples or of the intrinsic genes. We performed four versions of hierarchical clustering with Eisen linkage on the 62 observations that were assigned to the four classes:
Sparse hierarchical clustering of all 1753 genes, with the tuning parameter chosen to yield 496 non-zero genes.
Standard hierarchical clustering using all 1753 genes.
Standard hierarchical clustering using the 496 genes with highest marginal variance.
COSA hierarchical clustering using all 1753 genes.
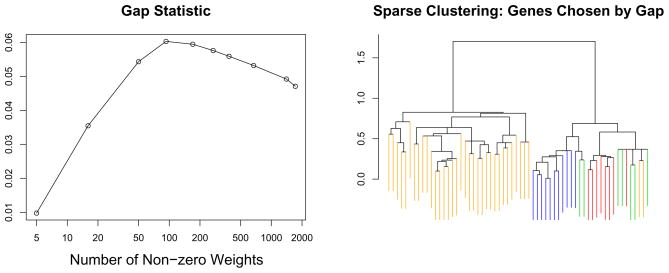
The resulting dendrograms are shown in Figure 5. Sparse clustering of all 1753 genes with the tuning parameter chosen to yield 496 non-zero genes does best at capturing the four classes; in fact, a comparison with Figure 4 reveals that it does quite a bit better than clustering based on the intrinsic genes only! Figure 6 displays the result of performing the automated tuning parameter selection method. This resulted in 93 genes having non-zero weights.
Figure 4.
Using the intrinsic gene set, hierarchical clustering was performed on all 65 observations (left panel) and on only the 62 observations that were assigned to one of the four classes (right panel). Note that the classes identified using all 65 observations are largely lost in the dendrogram obtained using just 62 observations. The four classes are basal-like (red), Erb-B2 (green), normal breast-like (blue), and ER+ (orange). In the left-hand panel, observations that do not belong to any class are shown in light blue.
Figure 6.
The gap statistic was used to determine the optimal value of the tuning parameter for sparse hierarchical clustering. Left: The largest value of the gap statistic corresponds to 93 genes with non-zero weights. Right: The dendrogram corresponding to 93 non-zero weights. The color coding is as in Figure 4.
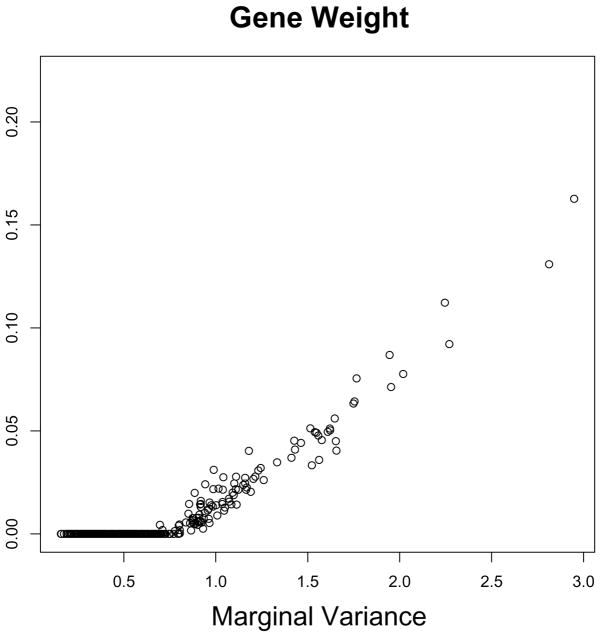
Figure 7 shows that the gene weights obtained using sparse clustering are highly correlated with the marginal variances of the genes. However, the results obtained from sparse clustering are different from the results obtained by simply clustering on the high-variance genes (Figure 5). The reason for this lies in the form of the criterion (15). Though the non-zero wj’s tend to correspond to genes with high marginal variances, sparse clustering does not simply cluster the genes with highest marginal variances. Rather, it weights each gene-wise dissimilarity matrix by a different amount.
Figure 7.
For each gene, the sparse clustering weight is plotted against the marginal variance.
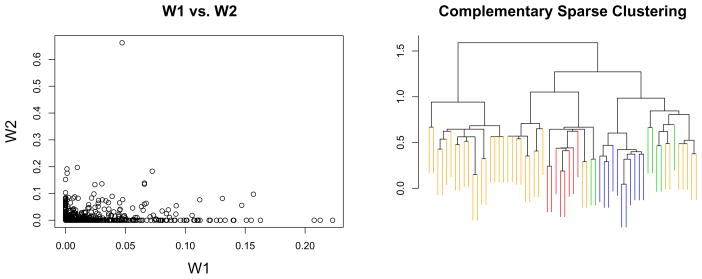
We also performed complementary sparse clustering on the full set of 1753 genes, using the method of Section 4.4. Tuning parameters for the initial and complementary sparse clusterings were selected to yield 496 genes with non-zero weights. The complementary sparse clustering dendrogram is shown in Figure 8, along with a plot of w1 and w2 (the feature weights for the initial and complementary clusterings). The dendrogram obtained using complementary sparse clustering suggests a previously unknown pattern in the data. (Recall that the dendrogram for the initial sparse clustering can be found in Figure 5.)
Figure 8.
Complementary sparse clustering was performed. Tuning parameters for the initial and complementary clusterings were selected to yield 496 genes with non-zero weights. Left: A plot of w1 against w2. Right: The dendrogram for complementary sparse clustering. The color coding is as in Figure 4.
In response to a reviewer’s inquiry about the robustness of sparse hierarchical clustering, we repeatedly resampled the 62 observations that belong to one of the four classes and performed sparse hierarchical clustering on the resampled data sets. (A small jitter was added to the resampled observations in order to avoid samples having correlation one with each other.) We found that for the most part, the resulting clusters accurately reflected the true class labels.
6 SNP data example
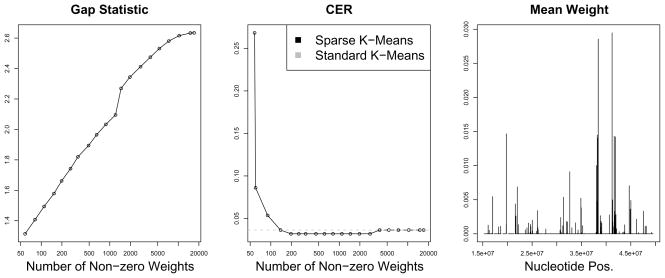
We wondered whether one could use sparse clustering in order to identify distinct populations in single nucleotide polymorphism (SNP) data, and also to identify the SNPs that differ between the populations. A SNP is a nucleotide position in a DNA sequence at which genetic variability exists in the population. We used the publicly-available Haplotype Map (“HapMap”) data of the International HapMap Consortium (International HapMap Consortium 2005, International HapMap Consortium 2007). The Phase III SNP data for eleven populations are available from http://ftp.hapmap.org/genotypes/2008-07_phaseIII/hapmap_format/forward/ we used only the data for chromosome 22. We restricted the analysis to three of the populations: African ancestry in southwest USA, Utah residents with European ancestry, and Han Chinese from Beijing. We used the SNPs for which measurements are available in all three populations. The resulting data have dimension 315×17026. We coded AA as 2, Aa as 1, and aa as 0. Missing values were imputed using 5-nearest neighbors (Troyanskaya et al. 2001). Sparse and standard 3-means clustering were performed on the data. The CERs obtained using standard 3-means and sparse 3- means are shown in Figure 9; CER was computed by comparing the clustering class labels to the true population identity for each sample. When the tuning parameter in sparse clustering was chosen to yield between 198 and 2809 SNPs with non-zero weights, sparse clustering resulted in slightly lower CER than standard 3-means clustering. The main improvement of sparse clustering over standard clustering is in interpretability, since the non-zero elements of w determine the SNPs involved in the sparse clustering. We can use the weights obtained from sparse clustering to identify SNPs on chromosome 22 that distinguish between the populations (Figure 9). SNPs in a few genomic regions appear to be responsible for the clustering obtained.
Figure 9.
Left: The gap statistics obtained as a function of the number of SNPs with non-zero weights. Center: The CERs obtained using sparse and standard 3-means clustering, for a range of values of the tuning parameter. Right: Sparse clustering was performed using the tuning parameter that yields 198 non-zero SNPs (this is the smallest number of SNPs that resulted in minimal CER in the center panel). Chromosome 22 was split into 500 segments of equal length. The average weights of the SNPs in each segment are shown, as a function of the nucleotide position of the segments.
In this example, the tuning parameter selection method of Section 3.2 does not perform well. Rather than selecting a tuning parameter that yields between 198 and 2809 SNPs with non-zero weights (resulting in the lowest CER), the highest gap statistic is obtained when all SNPs are used. The one standard deviation rule described in Section 3.2 results in a tuning parameter that yields 7160 genes with non-zero weights. The fact that the gap statistic seemingly overestimates the number of features with non-zero weights may reflect the need for a more accurate method for tuning parameter selection, or it may suggest the presence of further population substructure beyond the three population labels.
In this example, we applied sparse clustering to SNP data for which the populations were already known. However, the presence of unknown subpopulations in SNP data is often a concern, as population substructure can confound attempts to identify SNPs that are associated with diseases and other outcomes (see e.g. Price et al. 2006). In general, one could use sparse clustering to identify subpopulations in SNP data in an unsupervised way before further analyses are performed.
7 Extensions and discussion
We have proposed a general framework for sparse clustering, and have introduced methods for sparse K-means clustering and sparse hierarchical clustering that are special cases in this framework. Other clustering methods can also be made sparse in this way; for instance, a method for sparse K-medoids is presented in the Appendix.
The approach of Section 4.1 involves reweighting a dissimilarity matrix. Hence it can be applied to any method that takes a dissimilarity matrix as its input, such as multidimensional scaling. Since this method involves computations on a n2 × p matrix, it has the potential to become quite slow if n, in addition to p, is very large.
Though our sparse clustering methods seem to perform well on real and simulated data, the performance of the gap statistic of Section 3.2 for selecting the tuning parameter is mixed. This is not surprising, since tuning parameter selection in the unsupervised setting is known to be a very difficult problem. This is an area in which more work is needed.
The R package sparcl implementing the methods proposed in this paper will be made available at http://cran.r-project.org/.
Acknowledgments
This manuscript was improved by helpful comments from the editor, an associate editor, and two anonymous referees. We thank Jerome Friedman, Nema Dean, Wei Pan, and Hui Zhou for making R code available and for useful comments. Trevor Hastie provided helpful suggestions. Daniela Witten was partially supported by a National Defense Science and Engineering Graduate Fellowship. Robert Tibshirani was partially supported by National Science Foundation Grant DMS-9971405 and National Institutes of Health Contract N01-HV-28183.
Appendix
An additional remark on sparse K-means clustering
In the case where d is squared Euclidean distance, the K-means criterion (7) is equivalent to
| (27) |
where μk is the centroid for cluster k. However, if d is not squared Euclidean distance - for instance, if d is the sum of the absolute differences - then (7) and (27) are not equivalent. We used the criterion (7) to define K-means clustering, and consequently to derive a method for sparse K-means clustering, for simplicity and consistency with the COSA method of Friedman & Meulman (2004). But if (27) is used to define K-means clustering and the dissimilarity measure is not squared Euclidean distance (but is still additive in the features), then an analogous criterion and algorithm for sparse K-means clustering can be derived instead. In practice, this is not an important distinction, since K-means clustering is generally performed using squared distance as the dissimilarity measure.
Sparse K-medoids clustering
In Section 2.2, we mentioned that any clustering method of the form (4) could be modified to obtain a sparse clustering method of the form (5). (However, for the resulting sparse method to have a non-zero weight for feature j, it is necessary that fj(Xj, Θ) > 0.) In addition to K-means and sparse hierarchical clustering, another method that takes the form (4) is K-medoids. Let ik ∈ {1, …, n} denote the index of the observation that serves as the medoid for cluster k, and let Ck denote the indices of the observations in cluster k. The K-medoids criterion is
| (28) |
or equivalently
| (29) |
where i0 ∈ {1, …, n} is the index of the medoid for the full set of n observations. Since (29) is of the form (4), the criterion
| (30) |
results in a method for sparse K-medoids clustering, which can be optimized by an iterative approach.
References
- Boyd S, Vandenberghe L. Convex Optimization. Cambridge University Press; 2004. [Google Scholar]
- Chang W-C. On using principal components before separating a mixture of two multivariate normal distributions. Journal of the Royal Statistical Society, Series C (Applied Statistics) 1983;32:267–275. [Google Scholar]
- Chipman H, Tibshirani R. Hybrid hierarchical clustering with applications to microarray data. Biostatistics. 2005;7:286–301. doi: 10.1093/biostatistics/kxj007. [DOI] [PubMed] [Google Scholar]
- Dempster A, Laird N, Rubin D. Maximum likelihood from incomplete data via the EM algorithm (with discussion) J R Statist Soc B. 1977;39:1–38. [Google Scholar]
- Eisen M, Spellman P, Brown P, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci, USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J Amer Statist Assoc. 2001;96:1348–1360. [Google Scholar]
- Fraley C, Raftery A. Model-based clustering, discriminant analysis, and density estimation. J Amer Statist Assoc. 2002;97:611–631. [Google Scholar]
- Friedman J, Meulman J. Clustering objects on subsets of attributes. J Roy Stat Soc, Ser B. 2004;66:815–849. [Google Scholar]
- Ghosh D, Chinnaiyan AM. Mixture modelling of gene expression data from microarray experiments. Bioinformatics. 2002;18:275–286. doi: 10.1093/bioinformatics/18.2.275. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw P. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley; New York: 1990. [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Algorithms for non-negative matrix factorization. Advances in Neural Information Processing Systems, (NIPS 2001) 2001 [Google Scholar]
- Liu JS, Zhang JL, Palumbo MJ, Lawrence CE. Bayesian clustering with variable and transformation selections. Bayesian statistics. 2003;7:249–275. [Google Scholar]
- Lv J, Fan Y. A unified approach to model selection and sparse recovery using regularized least squares. Annals of Statistics 2009 [Google Scholar]
- MacQueen J. Some methods for classification and analysis of multivariate observations. In: LeCam LM, Neyman J, editors. Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability. Univ. of California Press; 1967. pp. 281–297. [Google Scholar]
- Maugis C, Celeux G, Martin-Magniette M-L. Variable selection for clustering with Gaussian mixture models. Biometrics. 2009;65:701–709. doi: 10.1111/j.1541-0420.2008.01160.x. [DOI] [PubMed] [Google Scholar]
- McLachlan GJ, Bean RW, Peel D. A mixture model-based approach to the clustering of microarray expression data. Bioinformatics. 2002;18:413–422. doi: 10.1093/bioinformatics/18.3.413. [DOI] [PubMed] [Google Scholar]
- McLachlan GJ, Peel D. Finite Mixture Models. John Wiley & Sons; New York, NY: 2000. [Google Scholar]
- McLachlan GJ, Peel D, Bean RW. Modelling high-dimensional data by mixtures of factor analyzers. Computational Statisitics and Data Analysis. 2003;41:379–388. [Google Scholar]
- Milligan GW, Cooper MC. An examination of procedures for determining the number of clusters in a data set. Psychometrika. 1985;50:159–179. [Google Scholar]
- Nowak G, Tibshirani R. Complementary hierarchical clustering. Biostatistics. 2008;9(3):467–483. doi: 10.1093/biostatistics/kxm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Shen X. Penalized model-based clustering with application to variable selection. Journal of Machine Learning Research. 2007;8:1145–1164. [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, Rijn MVD, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-dale A, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Raftery A, Dean N. Variable selection for model-based clustering. J Amer Stat Assoc. 2006;101:168–178. [Google Scholar]
- Rand WM. Objective criteria for the evaluation of clustering methods. Journal of the American Statistical Association. 1971;66:846–850. [Google Scholar]
- Sugar CA, James GM. Finding the number of clusters in a dataset: an information-theoretic approach. Journal of the American Statistical Association. 2003;98:750–763. [Google Scholar]
- Tamayo P, Scanfeld D, Ebert BL, Gillette MA, Roberts CWM, Mesirov JP. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. PNAS. 2007;104:5959–5964. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Walther G. Cluster validation by prediction strength. J Comp Graph Stat. 2005;14(3):511–528. [Google Scholar]
- Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a dataset via the gap statistic. J Royal Statist Soc B. 2001;32(2):411–423. [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman R. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;16:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhu J. Variable selection for model-based high-dimensional clustering and its application to microarray data. Biometrics. 2008;64:440–448. doi: 10.1111/j.1541-0420.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- Witten D, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10(3):515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Pan W, Shen X. Penalized model-based clustering with cluster-specific diagonal covariance matrices and grouped variables. Electronic Journal of Statistics. 2008;2:168–212. doi: 10.1214/08-EJS194. [DOI] [PMC free article] [PubMed] [Google Scholar]