Abstract
Peripheral blood mononuclear cells (PBMCs), saliva, seminal plasma, and dried blood spots were evaluated as specimen types for the APTIMA HIV-1 RNA Qualitative Assay (APTIMA HIV-1 Assay), which employs a target capture step to recover HIV-1-specific sequences from complex specimen types. Analytical sensitivity studies were carried out using samples that were either diluted or eluted with a buffered detergent and spiked with different concentrations of HIV-1 ranging from 1 to 10,000 copies/mL. PBMC samples spiked with HIV-1 had comparable analytical sensitivity to HIV-1 spiked plasma with a 95% limit of detection of 13.1 and 17.2 copies/mL, respectively. Analytical sensitivity in seminal plasma specimens diluted 1:5 and saliva diluted 1:2 was comparable to HIV-1 spiked dilution buffer alone. Whole blood and dried blood spot specimens spiked with HIV-1 had equivalent reactivity at 250 copies/spot (5000 copies/mL). However, the 95% limit of detection values were significantly different (293.7 copies/mL for whole blood and 2384 copies/mL for dried blood spot specimens). No significant effect on analytical sensitivity was observed when one HIV-1 positive dried blood spot punch was pooled with up to 9 HIV-1 negative dried blood spot punches. Together, these studies demonstrate that the APTIMA HIV-1 RNA Qualitative Assay can be used to process a diverse array of specimen types with minimal impact on analytical sensitivity for most specimen types.
Keywords: RNA detection, HIV-1, Dried blood spots
1. Introduction
More than 90% of new HIV-1 infections occur in developing parts of the world where there is limited access to equipment for processing clinical specimens. These settings often have inadequate storage facilities for preserving the integrity of traditional specimen types such as plasma or serum. Acquisition of serum and plasma require a trained phlebotomist, which further limits the ability to obtain specimens to diagnose HIV-1 infection using most current testing methods. In addition, public health initiatives are moving away from the use of plasma or serum as a specimen to determine HIV-1 infection in high-risk populations in favor of alternative specimen types which can be obtained without venipuncture or used for point-of-care screening, such as dried blood spots or saliva. This poses new challenges in the diagnosis of pre-seroconversion primary infection, which currently relies on testing of plasma specimens for HIV-1 RNA (Zetola and Pilcher, 2007). HIV-1 primary infection is increasingly recognized as a particularly infectious stage, and its recognition may be important for prevention efforts and clinical purposes (Hollingsworth et al., 2008; Pilcher et al., 2007). Therefore, the ability to diagnose pre-seroconversion HIV-1 without the use of phlebotomy specimens may have important public health and clinical applications in the future. Development of methods to detect HIV-1 in alternative specimen types would also be of great benefit in HIV-1 transmission studies, which also requires the use of specimen types other than plasma or serum (Hollingsworth et al., 2008).
The APTIMA HIV-1 RNA Qualitative Assay (APTIMA HIV-1 Assay) is licensed by the FDA for use as an aid in the diagnosis of acute HIV-1 infection in blood plasma specimens and as a confirmatory test for HIV-1 infection in specimens repeatedly reactive for HIV-1 antibodies. The experiments described here were performed to determine if the APTIMA HIV-1 Assay could be adapted to detect HIV-1 in clinical specimens other than blood plasma such as peripheral blood mononuclear cells (PBMCs), seminal plasma, saliva and dried blood spots. Detection of HIV-1 RNA in these diverse specimen types could have important research as well as diagnostic applications.
2. Materials and methods
2.1. APTIMA HIV-1 Assay
The APTIMA HIV-1 Assay (Gen-Probe Incorporated, San Diego, CA) utilizes three main technologies: (1) target capture-based sample preparation, (2) Transcription-Mediated Amplification (TMA) and (3) chemiluminescent detection via a Hybridization Protection Assay (HPA) (Giachetti et al., 2002). All experiments described here follow the testing protocol as described in the assay package insert (APTIMA HIV-1 Qualitative Assay, Gen-Probe Incorporated, San Diego, CA). Briefly, 0.5 mL of specimen is incubated with target capture reagent, which solubilizes the HIV-1 RNA genome and separates it from plasma. Amplification is then performed using TMA, which is an isothermal process that utilizes MMLV transcriptase and T7 RNA polymerase. Amplicons are then subjected to HPA using chemiluminescent-labeled probes that are specific for the selected HIV-1 genomic region. Selection reagent differentiates between hybridized and unhybridized probes by inactivation of the label on unhybridized probes. Results are interpreted by a luminometer and reported as Relative Light Units. An internal control RNA transcript is added to each reaction at the target capture step to control for proper capture, amplification and detection. The package insert can be found online at http://www.genprobe.com/inserts/.
2.2. Specimen collection and handling
All protocols were approved by an institutional review board and informed consent was obtained from study participants. All samples were tested after removal of specific subject identification.
2.2.1. PBMCs
PBMCs were isolated from whole blood by Ficoll-Hypaque gradient centrifugation. After rinsing with phosphate-buffered saline (PBS), PBMC aliquots of 5 × 106 cells were cryopreserved at −80 °C for no longer than 1 month. PBMCs were thawed and resuspended in 500 μL of PBS prior to spiking with HIV-1.
2.2.2. Saliva
Whole saliva was obtained in-house at Gen-Probe Incorporated from subjects who were asked to chew a flavorless gum base and expectorate into a tube for up to 4 min or until 2 mL of saliva was obtained. Specimens were aliquotted in 1.5 mL cryo-vials and stored at −20 °C until use. Thawed saliva was diluted 1:2 (1 part to 1 part specimen transfer media) in specimen transfer media prior to spiking with HIV-1. Specimen transfer media is a buffered detergent and is commercially available as a component of the APTIMA Specimen Transfer Kit (Gen-Probe Incorporated, San Diego, CA).
2.2.3. Seminal plasma
Semen collected from donors was incubated at room temperature for at least 30 min but no longer than 4 h prior to centrifugation at 600 × g for 15 min. Seminal plasma was harvested and stored in 500 μL aliquots at −70 °C prior to use. Thawed seminal plasma was diluted 1:5 (1 part to 4 parts specimen transfer media) with specimen transfer media prior to spiking with HIV-1.
2.2.4. Dried blood spots
Whole blood was collected from donors in plastic K2EDTA tubes (Becton Dickinson, Franklin Lakes, NJ) and stored at room temperature for no longer than 2 h prior to use. Whatman 903 ProteinSaver Cards (VWR Scientific, West Chester, PA) were spotted with 5 × 50 μL aliquots of HIV-1 spiked or non-spiked whole blood (50 μL per spot, 5 spots per card). Cards were stored desiccated in plastic bags in the dark at room temperature prior to use in experiments. Whole dried blood spots (13 mm in diameter) or hole punches (6 mm in diameter) of the dried blood spots were excised and incubated at 95 °C for 20 min in specimen transfer media and gently agitated every 5 min. Following this elution step, 500 μL of the eluate was tested with the APTIMA HIV-1 Assay.
2.3. Generation of sensitivity panels
Plasma infected with HIV-1 subtype B was used to spike specimens to prepare analytical sensitivity panels. HIV-1 infected plasma was quantitated using a validated, TMA-based HIV-1 quantitative assay calibrated against the Virology Quality Assurance Laboratory standard of the AIDS Clinical Trials Group (Virology Quality Assurance Laboratory, Rush-Presbyterian St. Luke’s Medical Center, Chicago, IL). Pools of HIV-1 negative PBMCs, specimen transfer media-diluted saliva, specimen transfer media-diluted seminal plasma, whole blood specimens (used for preparation of dried blood spots), and specimen transfer media or EDTA plasma (as controls) were spiked with various concentrations of HIV-1 and stored at −20 °C until use.
2.4. Statistical methods
Confidence intervals and statistical significance for positivity percentages were calculated using Stat2 v9.0 (Prentice Hall, Upper Saddle River, NJ). Probit analysis for the predicted 95% probability of detection of different specimen types and ANOVA of single and pooled dried blood spot punches were calculated using SAS v9.1 (SAS Institute, Cary, NC).
3. Results
The analytical sensitivity of the APTIMA HIV-1 Assay is 98.5% in serum or blood plasma specimens at 30 HIV-1 RNA copies/mL with a 95% confidence interval of 97.3–99.2% (Giachetti et al., 2002). However, it was unknown whether this level of sensitivity would be achieved with other specimen types. The following experiments were performed to adapt the assay to different specimen types and determine the assay analytical sensitivity for each specimen type.
3.1. PBMC specimens
Analytical sensitivity was evaluated by testing spiked PBMCs or blood plasma control panels containing the indicated concentration of HIV-1 RNA in the APTIMA HIV-1 Assay. All replicates were 100% positive at 50 copies/mL in both HIV-1 spiked PBMCs and blood plasma (Fig. 1). Positivity remained at 100% for spiked blood plasma samples down to 10 HIV RNA copies/mL, and spiked PBMC sample positivity was 85% at this concentration. A difference of 15% in sensitivity was observed at 10 and 1 HIV-1 RNA copies/mL. However, these differences were not statistically significant (p = 0.23). In addition, the predicted 95% probability of detection for PBMCs was 17.2 HIV-1 RNA copies/mL, which was not significantly different from the predicted 95% probability of detection of HIV-1 in spiked plasma (Table 1).
Fig. 1.

Comparison of HIV-1 spiked in peripheral blood mononuclear cells (PBMCs) and blood plasma. Error bars represent the 95% confidence interval around each data point. For PBMCs ( ), n = 17 for 50 copies/mL, n = 20 for 20 and 10 copies/mL, n = 19 for 2 copies/mL, n = 20 for 1 copy/mL. For plasma (●), n = 20 for all copy levels.
), n = 17 for 50 copies/mL, n = 20 for 20 and 10 copies/mL, n = 19 for 2 copies/mL, n = 20 for 1 copy/mL. For plasma (●), n = 20 for all copy levels.
Table 1.
Comparison of the predicted 95% probability of detection between specimen types.
| Specimen type | 95% Limit of detection (95% fiducial limits) (copies/mL) |
|---|---|
| Plasma | 13.1 (12.1–14.4) |
| PBMCs | 17.2 (10.6–38.9) |
| 1:5 Seminal plasma | 65.4 (37.0–182.5) |
| 1:2 Saliva | 25.7 (18.8–40.6) |
| 1:10 Whole blood | 293.7 (205.2–812.7) |
| Whole dried blood spot | 2384 (1567–4897) |
| Single punch dried blood spot | 10217 (5874–26645) |
3.2. Seminal plasma specimens
Initial studies using pooled seminal plasma showed that undiluted seminal plasma samples, either neat or spiked with HIV-1, strongly inhibited the APTIMA HIV-1 Assay, in accordance with earlier observations that semen or seminal plasma can inhibit nucleic acid amplification (Dyer et al., 1996). Dilution of seminal plasma with specimen transfer media, a detergent-based solution, was therefore explored to determine if this effect could be overcome. Diluting the seminal plasma 1:5 with specimen transfer media prior to spiking with HIV-1 alleviated the inhibitory effects (data not shown). Analytical sensitivity using serial diluted concentrations of HIV-1 spiked samples of seminal plasma diluted 1:5 in specimen transfer media was compared to HIV-1 spiked specimen transfer media alone. Analytical sensitivity was comparable between serial concentrations of HIV-1 spiked specimen transfer media and seminal plasma specimens diluted 1:5 in specimen transfer media (Fig. 2), and the predicted 95% probability of detection was 65.4 HIV-1 RNA copies/mL (Table 1).
Fig. 2.

Comparison of HIV-1 spiked 1:5 diluted seminal plasma and specimen transfer media. Error bars represent the 95% confidence interval around each data point. For diluted seminal plasma (●), n = 30 for 100 and 50 copies/mL, n = 70 for 30 and 10 copies/mL, and n = 15 for 3, 1 and 0 copies/mL. For specimen transfer media ( ), n = 10 for 100 copies/mL, n = 30 for 50 copies/mL, n = 70 for 30 and 10 copies/mL and n = 15 for 3, 1 and 0 copies/mL.
), n = 10 for 100 copies/mL, n = 30 for 50 copies/mL, n = 70 for 30 and 10 copies/mL and n = 15 for 3, 1 and 0 copies/mL.
3.3. Saliva specimens
Initial experiments using undiluted saliva were also performed with the APTIMA HIV-1 Assay. As with seminal plasma, inhibition of amplification was observed using whole saliva specimens (data not shown). However, diluting saliva with specimen transfer media at a ratio of 1:2 eliminated this inhibition. Analytical sensitivity of the assay was comparable in saliva diluted 1:2 with specimen transfer media and then spiked with HIV-1 and HIV-1 spiked specimen transfer media at the indicated concentrations and there were no significant differences between specimen types (p = 0.31, Fig. 3). The predicted 95% probability of detection for 1:2 diluted saliva was 25.7 HIV-1 RNA copies/mL (Table 1).
Fig. 3.

Comparison of HIV-1 spiked 1:2 diluted saliva and specimen transfer media. Error bars represent the 95% confidence interval around each data point. For diluted saliva ( ), n = 10 for 100 copies/mL, 30 for 50 copies/mL, n = 70 for 30 and 10 copies/mL and n = 20 for 3, 1 and 0 copies/mL. For specimen transfer media (●), n = 20 for 100 copies/mL, 30 for 50 copies/mL, n = 70 for 30 and 10 copies/mL and n = 20 for 3, 1 and 0 copies/mL.
), n = 10 for 100 copies/mL, 30 for 50 copies/mL, n = 70 for 30 and 10 copies/mL and n = 20 for 3, 1 and 0 copies/mL. For specimen transfer media (●), n = 20 for 100 copies/mL, 30 for 50 copies/mL, n = 70 for 30 and 10 copies/mL and n = 20 for 3, 1 and 0 copies/mL.
3.4. Dried blood spot specimens
Eluates from dried blood spot specimens were compared to a second set of samples which were generated by spiking 50 μL of whole blood containing HIV-1 at the indicated copy level into 450 μL of specimen transfer media. These samples were tested with the dried blood spot specimens. Analytical sensitivity for detection of HIV-1 was 100% in both dried blood spots and whole blood diluted 1:10 with specimen transfer media at 250 copies of HIV-1/spot (or 5000 copies/mL) (Fig. 4). Significant differences in sensitivity began to occur at 25 copies of HIV-1/spot (500 copies/mL), where blood samples were detected at a rate of 100% while sensitivity dropped to 65% in dried blood spot samples (p < 0.01), suggesting poor elution of the HIV-1 RNA from the dried blood spot paper. This observation was supported when the 95% limit of detection was compared between these samples types: for 1:10 diluted whole blood 95% detection is predicted at 293.7 HIV-1 RNA copies/mL, significantly lower than the observed value for dried blood spots (2384 HIV-1 RNA copies/mL, Table 1).
Fig. 4.
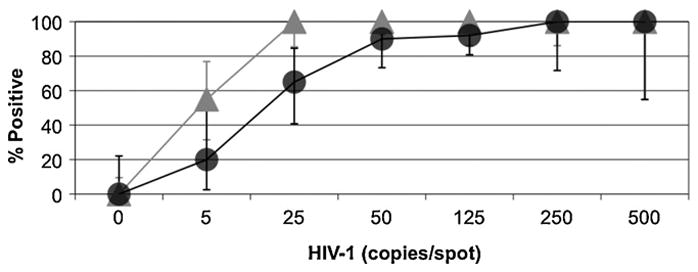
Comparison of HIV-1 spiked 1:10 diluted whole blood and dried blood spots. Error bars represent the 95% confidence interval around each data point. For spiked whole blood ( ), n = 5 for 500 copies/spot, 20 for 250 and 125 copies/spot, 40 for 50 copies/spot, 19 for 25 copies/spot, 20 for 5 copies/spot and 30 for 0 copies/spot. For dried blood spots (●), n = 5 for 500 copies/spot, 9 for 250 copies/spot, 50 for 125 copies/spot, 35 for 50 copies/spot, 20 for 25 copies/spot, 10 for 5 copies/spot and 12 for 0 copies/spot.
), n = 5 for 500 copies/spot, 20 for 250 and 125 copies/spot, 40 for 50 copies/spot, 19 for 25 copies/spot, 20 for 5 copies/spot and 30 for 0 copies/spot. For dried blood spots (●), n = 5 for 500 copies/spot, 9 for 250 copies/spot, 50 for 125 copies/spot, 35 for 50 copies/spot, 20 for 25 copies/spot, 10 for 5 copies/spot and 12 for 0 copies/spot.
Assay sensitivity for conventional single hole punches (approximately 6 mm in diameter) was also evaluated and directly compared to whole dried blood spots (approximately 13 mm in diameter). As expected, lower sensitivity was observed in single punch samples due to the reduction in specimen size and thus HIV-1 RNA input level (Fig. 5). Analytical sensitivity for detection of HIV-1 RNA was 100% in both specimen types at 500 and 250 HIV-1 copies/spot (10,000 and 5000 copies/mL, respectively) Significant differences in sensitivity were observed beginning at 125 copies of HIV-1/spot (2500 copies/mL), where sensitivity was 92% in whole spot samples and 63% in single punch samples (p < 0.01). As expected the 95% predicted probability of detection for a single punch dried blood spot sample was significantly higher than the observed value for whole spots (10,217 HIV-1 RNA copies/mL, Table 1).
Fig. 5.

Comparison of whole dried blood spots to a single punch. Error bars represent the 95% confidence interval around each data point. For the whole dried blood spots (●), n = 5 for 500 copies/spot, 20 for 250 and 125 copies/spot, 40 for 50 copies/spot, 19 for 25 copies/spot, 20 for 12.5 copies/spot and 30 for 0 copies/spot. For the single punch spots ( ), n = 5 for 500 copies/spot, 14 for 250 copies/spot, 40 for 125 copies/spot, 40 for 50 copies/spot, 25 for 25 copies/spot, 20 for 12.5 copies/spot and 12 for 0 copies/spot.
), n = 5 for 500 copies/spot, 14 for 250 copies/spot, 40 for 125 copies/spot, 40 for 50 copies/spot, 25 for 25 copies/spot, 20 for 12.5 copies/spot and 12 for 0 copies/spot.
Single punches were also tested in pools to determine the effect of combining single punches containing or not containing HIV-1 RNA on analytical sensitivity. A single punch spiked with HIV-1 at the indicated copy level was combined with either 4 or 9 HIV-1 negative single punches prior to elution in specimen transfer media. The single punch samples and the pooled samples were all 100% positive at 250 HIV-1 copies/spot (Fig. 6). Analytical sensitivity was similar in single spots and pools with lower copy levels of HIV-1, regardless of pool size, and differences in positivity were not significant (p = 0.4821), indicating that pooling of dried blood spots will not affect the sensitivity of the APTIMA HIV-1 Assay.
Fig. 6.

Comparison of pooled and single dried blood spot punches. Error bars represent the 95% confidence interval around each data point. For the pooled dried blood spots (5 pool  10 pool
10 pool  ), n = 10 for 250 copies/spot (5 pooled and 10 pooled), n = 30 and n = 20 for 125 and 50 copies/spot (5 pooled and 10 pooled, respectively), n = 20 and n = 10 for 25 copies/spot (5 pooled and 10 pooled, respectively) and 10 replicates for 12.5 and 5 copies/spot. For the single punch spots (●), n = 14 for 250 copies/spot, 40 for 125 and 50 copies/spot, 25 for 25 copies/spot, and 20 replicates for 12.5 and 5 copies per spot.
), n = 10 for 250 copies/spot (5 pooled and 10 pooled), n = 30 and n = 20 for 125 and 50 copies/spot (5 pooled and 10 pooled, respectively), n = 20 and n = 10 for 25 copies/spot (5 pooled and 10 pooled, respectively) and 10 replicates for 12.5 and 5 copies/spot. For the single punch spots (●), n = 14 for 250 copies/spot, 40 for 125 and 50 copies/spot, 25 for 25 copies/spot, and 20 replicates for 12.5 and 5 copies per spot.
4. Discussion
Methods used to measure HIV-1 analytical sensitivity in specimen types from biological compartments other than blood have not been standardized. Investigators have used a variety of methods in studies to determine the clinical sensitivity of different nucleic acid tests in different biological compartments or sample types (Ayele et al., 2007; Bourlet et al., 2001; Pilcher et al., 2007; Shepard et al., 2000). Analysis of some sample types like saliva and semen are difficult because these fluids contain inhibitors for some nucleic acid tests (Dyer et al., 1996; Shepard et al., 2000). To eliminate these inhibitors, investigators have used the Boom extraction technique (Boom et al., 1990) in which hydroxyapatite separates nucleic acid from inhibitors and concentrates it before amplification (Fiscus, 2005). In studies described in this report, all of the sample types except PBMCs were tested after an elution or dilution step with specimen transfer media, followed by APTIMA assay target capture. This method significantly reduced the number of reagents and processing time that is required prior to nucleic acid amplification when compared to procedures requiring Boom extraction. Equivalent reactivity was observed whether specimens were spiked with HIV-1 prior to dilution or after dilution with specimen transfer media (data not shown).
No significant loss in HIV-1 detection was observed in diluted samples of PBMCs, seminal plasma, and saliva when directly compared to spiked control samples containing the same amount of virus. The predicted 95% probability of detection (with 95% confidence intervals) for the APTIMA HIV-1 Assay is 13.1 (12.1–14.4) copies of HIV-1/mL for blood plasma (Giachetti et al., 2002). The necessary dilution steps (1:2 or 1:5), therefore, places the 95% probability of detection 2- to 5-fold higher for saliva and seminal plasma, respectively, when compared to blood plasma (Table 1). Analytical sensitivity for dried blood spots was considerably lower, where samples containing less than 250 (5000 copies/mL) showed less than 100% reactivity. This was likely due to the reduced amount of HIV-1 recovered from the dried blood spots. Future studies will address whether elution efficiency can be improved using specimen transfer media or other extraction buffers.
The ability to reliably detect HIV-1 in PBMCs, saliva, seminal plasma and dried blood spots has different implications dependent on the sample type. For example, PBMC testing permits detection of intracellular expression of viral RNA rather than proviral DNA, which is particularly important when assessing viral transcriptional activity (Hatano et al., 2009; Deeks and Walker, 2007). Use of saliva as an alternative to blood or serum for HIV-1 RNA testing, may be useful at specimen collection sites where phlebotomy is not available, as well as in epidemiological studies for early transmission (Klein et al., 2003). Qualitative testing using seminal plasma is useful to determine presence or absence of sexual partner infectivity risk, as well as a screening method to monitor sperm washing techniques in instances where HIV-1 positive or serodiscordant couples are undergoing in vitro fertilization therapy (Coll et al., 1999). Dried blood spot samples can be collected without venipuncture using fingersticks and can be processed, shipped and stored at room temperature prior to testing either separately or in pools. Pooled blood plasma sample testing has been used in blood bank screening for several years as a way to reduce costs (Stramer et al., 2004). Application of the same concept to pooled spots is advantageous, especially considering that no statistically significant difference in analytical sensitivity was observed. In addition, there are no dilution effects, as in the case of pooling liquid-based specimens, since up to 10 spots can be combined in a single volume of specimen transfer media. Even though samples comprised of 6 mm punches had less sensitivity than 13 mm whole punch samples, use of the 6 mm sample would allow for retesting and pool discrimination. Although specimens used in the studies described here tested HIV-1 subtype B, comparable detectability has been observed with non-B subtypes (A, C, D, E, F, G, N and O), making these observations also relevant in geographic areas where B is not the most prevalent subtype (Package Insert, APTIMA HIV-1 Qualitative Assay). Future experiments will compare analytical sensitivity of the different subtypes spiked into the specimen types described in this publication. In conclusion, HIV-1 RNA can accurately be detected by the APTIMA HIV-1 Assay in multiple specimen types besides blood plasma and serum and may be useful as an aid in future transmission and epidemiological studies (Pilcher et al., 2007).
Acknowledgments
The authors would like to thank Caroline Magno and John Major for technical assistance, Judy Jin for statistical assistance and Mike Deras for valuable discussions.
References
- Ayele W, Schuurman R, Messele T, Dorigo-Zetsma W, Mengistu Y, Goudsmit J, Paxton WA, de Baar MP, Pollakis G. Use of dried spots of whole blood, plasma, and mother’s milk collected on filter paper for measurement of human immunodeficiency virus type 1 burden. J Clin Microbiol. 2007;45:891–896. doi: 10.1128/JCM.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlet T, Cazorla C, Berthelot P, Grattard F, Cognasse F, Fresard A, Defontaine C, Lucht FR, Genine C, Pozzetto B. Compartmentalization of HIV-1 according to antiretroviral therapy: viral loads are correlated in blood and semen but poorly in blood and saliva. AIDS. 2001;15:284–285. doi: 10.1097/00002030-200101260-00025. [DOI] [PubMed] [Google Scholar]
- Coll O, Vidal R, Martinez de Tejada B, Ballescá JL, Azulay M, Vanrell JA. Management of HIV serodiscordant couples. The clinician point of view. Contracept Fertil Sex. 1999;27:399–404. [PubMed] [Google Scholar]
- Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Dyer JR, Gilliam BL, Eron JJ, Jr, Grosso L, Cohen MS, Fiscus SA. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA with Amplicor reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- Fiscus SA. Quantitation of HIV-1 viral RNA in blood plasma and genital secretions. Methods Mol Biol. 2005;304:201–213. doi: 10.1385/1-59259-907-9:201. [DOI] [PubMed] [Google Scholar]
- Giachetti C, Linnen JM, Kolk DP, Dockter J, Gillotte-Taylor K, Park M, Ho-Sing-Loy M, McCormick MK, Mimms LT, McDonough SH. Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J Clin Microbiol. 2002;40:2408–2419. doi: 10.1128/JCM.40.7.2408-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Delwart EL, Norris PJ, Lee TH, Dunn-Williams J, Hunt PW, Hoh R, Stramer SL, Linnen JM, McCune JM, Martin JN, Busch MP, Deeks SG. Evidence for persistent low-level viremia in individuals who control HIV in the absence of antiretroviral therapy. J Virol. 2009;83:329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth TD, Anderson RM, Fraser C. HIV-1 transmission, by stage of infection. J Infect Dis. 2008;198:687–693. doi: 10.1086/590501. [DOI] [PubMed] [Google Scholar]
- Klein D, Hurley LB, Merrill D, Quesenberry CP., Jr Consortium for HIV/AIDS Interregional Research. Review of medical encounters in the 5 years before a diagnosis of HIV-1 infection: implications for early detection. J Acquir Immune Defic Syndr. 2003;32:143–152. doi: 10.1097/00126334-200302010-00005. [DOI] [PubMed] [Google Scholar]
- Pilcher CD, Joaki G, Hoffman IF, Martinson FE, Mapanje C, Stewart PW, Powers KA, Galvin S, Chilongozi D, Gama S, Price MA, Fiscus SA, Cohen MS. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS. 2007;21:1723–1730. doi: 10.1097/QAD.0b013e3281532c82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard RN, Schock J, Robertson K, Shugars DC, Dyer J, Vernazza P, Hall C, Cohen MS, Fiscus SA. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol. 2000;38:1414–1418. doi: 10.1128/jcm.38.4.1414-1418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer SL, Glynn SA, Kleinman SH, Strong DM, Caglioti S, Wright DJ, Dodd RY, Busch MP National Heart, Lung, and Blood Institute Nucleic Acid Test Study Group. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- Zetola NM, Pilcher CD. Diagnosis and management of acute HIV infection. Infect Dis Clin N Am. 2007;21:19–48. doi: 10.1016/j.idc.2007.01.008. [DOI] [PubMed] [Google Scholar]


