Our genes code for the building blocks of our body, and variations in our genome predispose us to distinct traits, including disease. To add complexity to the interpretation and regulation of our genetic code, the DNA packed into the chromatin forming our chromosomes is modified by factors that alter its expressivity but without changing its sequence. These factors are referred to as epigenetic factors. A recent definition states: “An epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence.”1.
Many factors are involved in establishing epigenetic traits, including DNA methylation, histone variants, chemical modification of histones, position of nucleosomes, and three-dimensional organization of genes in the nucleus, for example. These modifications often act in concert to alter the expression of genes, or to effect allele-specific inheritance. Epigenetic regulation can be established cell-autonomously, through intercellular signaling, and by environmental influences. All cells in the body are subject to epigenetic regulation, and thus understanding this important layer of regulation is key to elucidating mechanisms of cardiovascular development, physiology, and disease.
This review series in Circulation Research will explore recent advances in epigenetic research, with an outlook toward the cardiovascular system. The reviews will explore the current state of the art and will focus on the exciting future of epigenetic research as it relates to the cardiovascular system.
Epigenetic regulation is commonly considered to include the chemical modification of histone residues that accompany regulated gene expression. Many have noted that the definition of epigenetics quoted above implies heritability of a phenotype through either mitosis or meiosis. At first glance, this might seem to exclude the changes in histone modifications that are established during gene regulation, but are thought to be transient due to histone turnover. However, some of these changes are in fact actively maintained 2, and thus histone modifications can be included in a broader definition of epigenetics.
To enact a concerted phenotypic program during differentiation, or remodeling in disease, several broad programs of gene expression must be co-regulated. Epigenetic regulation through histone modifications is an important aspect of this co-regulation 3. The unstructured tails of histones—the proteins that assemble into the nucleosomes around which chromosomal DNA is wound—are subject to myriad chemical modifications, including acetylation, methylation, phosphorylation, ubiquitinylation, and sumoylation. In combination, these modifications are thought to result in a histone “code” that is read and translated into signals for activation or repression of associated genes 3, 4. For example, certain histone modifications are most often associated with repressed genes, and others with active genes.
The widespread coordinated deployment of particular histone modifications ensures a stable and efficiently enacted regulatory mechanism that can be targeted to specific sets of genes. In embryonic stem cells, to cite a notable example, most developmentally relevant transcription factor genes are silenced by the imposition of a repressive histone code 5, 6; this is thought to ensure that embryonic stem cells remain pluripotent. Another example is the epigenetic repression of the entire HoxD cluster and its gradual activation by the removal of repressive histone modifications 7. The Hox locus is also the site of an exciting regulatory link between long non-coding RNAs (lincRNAs) and the establishment of histone modifications at specific loci 8. The function of lincRNAs in recruiting chromatin modifying complexes 8, 9 provides an attractive mechanistic link between cis- and trans-regulation within the genome at the level of histone modifications.
It will be of considerable interest to determine whether similar broad regulatory mechanisms are involved in cardiovascular biology. Some clues indicate that this might be the case. The histone methyltransferase WHSC1 interacts with an important cardiac transcription factor, Nkx2-5, to regulate the normal development of the heart 10. Jarid2, also known as Jumonji, is an integral component of the Polycomb repressor complex, which deposits repressive histone marks 11–13. Jarid2 has long been known to function in heart development 14, 15, but its mechanism of action was unknown. Thus, epigenetic regulation is likely to be as important a mechanism for the cardiovascular system as it is in other systems, and it is likely to have important and widespread roles in normal physiology as well as in disease. A review in this series by Ching-Pin Chang and colleagues will explore in detail the role of epigenetic and chromatin-based regulation of cardiovascular development and physiology.
The techniques used to study the epigenome are complex. Thanks to the emergence of high-throughput technologies, it is now possible to efficiently and completely evaluate on a genomic scale the epigenetic modifications that accompany the status of a cell 16. Currently, most technologies involve immunoprecipitation of chromatin associated with specific epitopes, such as histone modifications, histone variants, or DNA methylation, followed by the identification of the associated DNA sequences by hybridization to microarrays, or more commonly now by direct sequencing. New sequencing platforms, while not widely available, allow the scaling down of these techniques, so that smaller samples such as those one would obtain from a human cardiac biopsy can provide equivalent rich information. These new sequencing chemistries also allow, for example, direct measures of DNA methylation 17, which will enable the mapping of this important epigenetic mark on an unprecedented scale. Other exciting developments involve the study of three-dimensional organization of genes in the nucleus, forming interchromosomal transcription “factories”. The three-dimensional organization of chromosomal segments is thought to be regulated by epigenetic mechanisms, and is emerging as an important means to coregulate widely separated genes 18. Keji Zhao and colleagues will review current and future approaches for studying epigenetic modifications.
Epigenetic changes are also thought to be at the root of cellular reprogramming, the process by which a differentiated cell type can be induced to adopt an alternate cell fate. The most well-known and spectacular example of this is the generation of induced pluripotent cells (iPS cells) from fully differentiated somatic cells 19, 20. iPS cells are functionally similar, if not identical 21, to embryonic stem cells, and the spectacular change in status from a “terminally” differentiated somatic cell to a fully pluripotent cell involves epigenetic reprogramming of the DNA methylation of pluripotency genes, amongst certainly many others. The recent elucidation of the mechanism of these changes in DNA methylation will open the door to a broader understanding of mechanisms of reprogramming as well as regulation by DNA methylation 22.
Other forms of reprogramming have been described, including the induction of insulin-producing pancreatic beta cells from exocrine cells 23 and the generation of functional neurons from skin fibroblasts 24. In the cardiovascular system, the ability to generate new cardiomyocytes, endothelial cells, or smooth muscle cells from other cell types would be of considerable benefit for strategies aimed at regeneration of diseased cardiovascular tissues. Approaches to reprogramming somatic cells into cardiovascular cell types have not yet been described, but the promise offered by the success in reprogramming other cell types brings hope that this avenue will be broadly successful in the near future. In this series, Deepak Srivastava and Shinya Yamanaka will review this exciting field.
In summary, the field of epigenetics has provided important insights into broadly applicable aspects of differentiation, physiology, and disease in many contexts. Large-scale research efforts, in the form of large consortia and funding initiatives, are being deployed to further understand epigenetic regulation. As epigenetics research expands its horizons toward the cardiovascular system, our understanding of cardiovascular biology will be greatly enhanced. Importantly, since epigenetic regulators are mainly enzymes whose functions can be altered by natural and designed compounds, epigenetic regulation of the cardiovascular system may emerge as an exciting and important arena for drug development.
Figure.
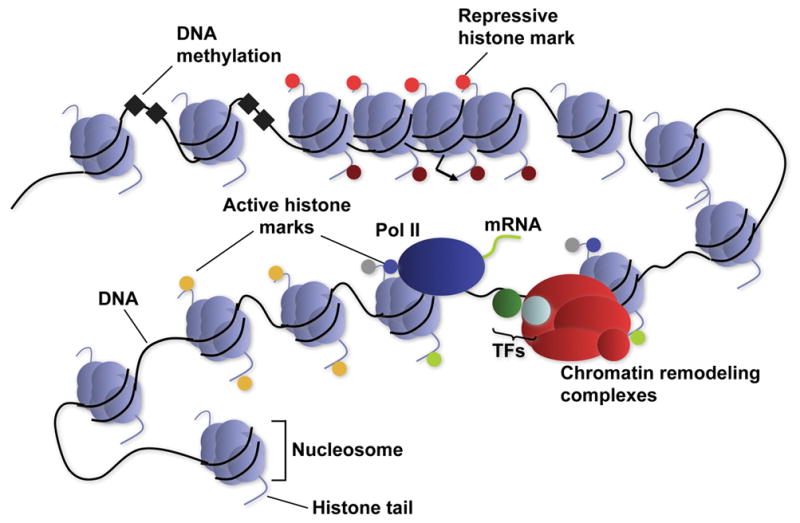
Diagrammatic representation of chromatin, and chromatin-mediated gene regulation. TFs: transcription factors.
Acknowledgments
I thank Jeffrey Alexander for artwork, and Gary Howard and Stephen Ordway for editorial assistance.
Sources of Funding
Work in my laboratory is supported by grants from NIH/NHLBI, the California Institute for Regenerative Medicine, and the Lawrence J. and Florence A. DeGeorge Charitable Trust/American Heart Association Established Investigator Award.
Footnotes
Disclosure
B.G.B. serves on the scientific advisory board of iPierian, Inc.
References
- 1.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 6.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soshnikova N, Duboule D. Epigenetic temporal control of mouse hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 8.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human hox loci by noncoding rnas. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding rnas associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone h3 lysine 36 trimethyltransferase links nkx2-5 to wolf-hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 11.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. Jarid2 regulates binding of the polycomb repressive complex 2 to target genes in es cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 12.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/jumonji coordinates control of prc2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- 15.Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. Jumonji downregulates cardiac cell proliferation by repressing cyclin d1 expression. Dev Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- 16.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sexton T, Bantignies F, Cavalli G. Genomic interactions: Chromatin loops and gene meeting points in transcriptional regulation. Seminars in cell & developmental biology. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qf1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires aid-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008 doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]


