Abstract
Modeling and analysis of MR images of the developing human brain is a challenge due to rapid changes in brain morphology and morphometry. We present an approach to the construction of a spatiotemporal atlas of the fetal brain with temporal models of MR intensity, tissue probability and shape changes. This spatiotemporal model is created from a set of reconstructed MR images of fetal subjects with different gestational ages. Groupwise registration of manual segmentations and voxelwise nonlinear modeling allow us to capture the appearance, disappearance and spatial variation of brain structures over time. Applying this model to atlas-based segmentation, we generate age-specific MR templates and tissue probability maps and use them to initialize automatic tissue delineation in new MR images. The choice of model parameters and the final performance are evaluated using clinical MR scans of young fetuses with gestational ages ranging from 20.57 to 24.71 weeks. Experimental results indicate that quadratic temporal models can correctly capture growth-related changes in the fetal brain anatomy and provide improvement in accuracy of atlas-based tissue segmentation.
Keywords: Structural MRI, Fetal imaging, Atlas building, Tissue segmentation
Introduction
Magnetic resonance (MR) imaging is an essential tool for the study of early human brain development in utero (Girard et al., 1995; Prayer et al., 2006; Rutherford et al., 2008; Glenn, 2010). Recently developed methods of image reconstruction permit the formation of motion-corrected 3D volumes of the fetal brain from 2D MR scans acquired in utero (Rousseau et al., 2006; Jiang et al., 2007; Kim et al., 2010). Accurate and efficient automatic segmentation of individual tissues (Habas et al., in press) is then the next key step in quantification of fetal brain development. This allows us to extract patterns of normal brain tissue growth in utero and correctly identify cases that may be related to abnormal neurodevelopmental outcomes.
Fetal MRI is usually performed after 20 weeks gestational age (GA) when the main steps of organogenesis are completed. At this stage, the fetal brain consists of a mixture of developing gray matter, developing white matter and other transient structures such as the germinal matrix (Prayer et al., 2006). During embryology and early fetal life, the germinal matrix is a site of production of both neurons and glial cells which then migrate out to their final locations. The volume of the germinal matrix reaches its peak at about 23 weeks GA and decreases subsequently (Battin et al., 1998; Kinoshita et al., 2001).
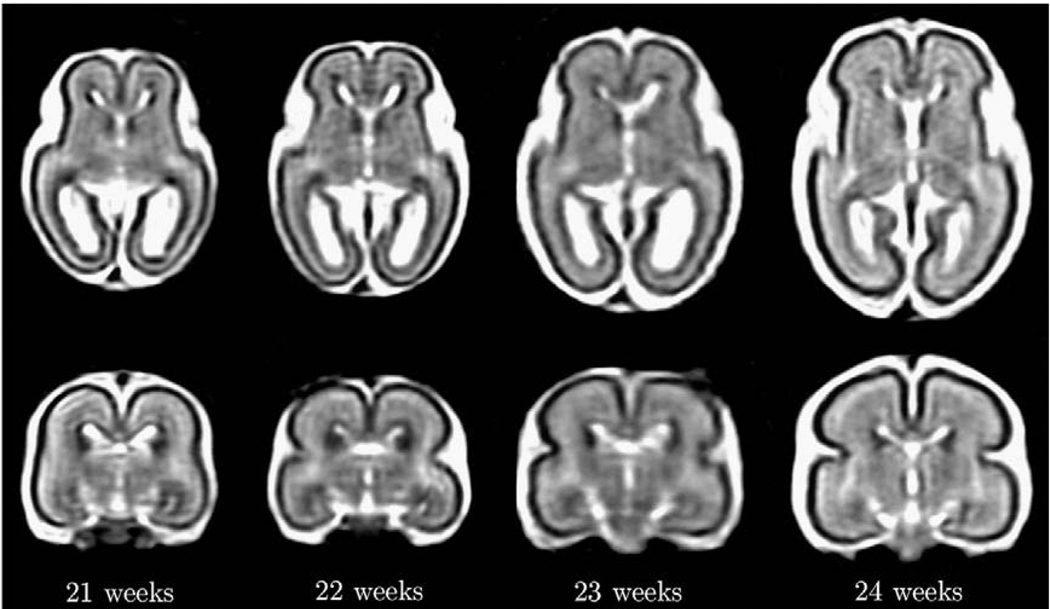
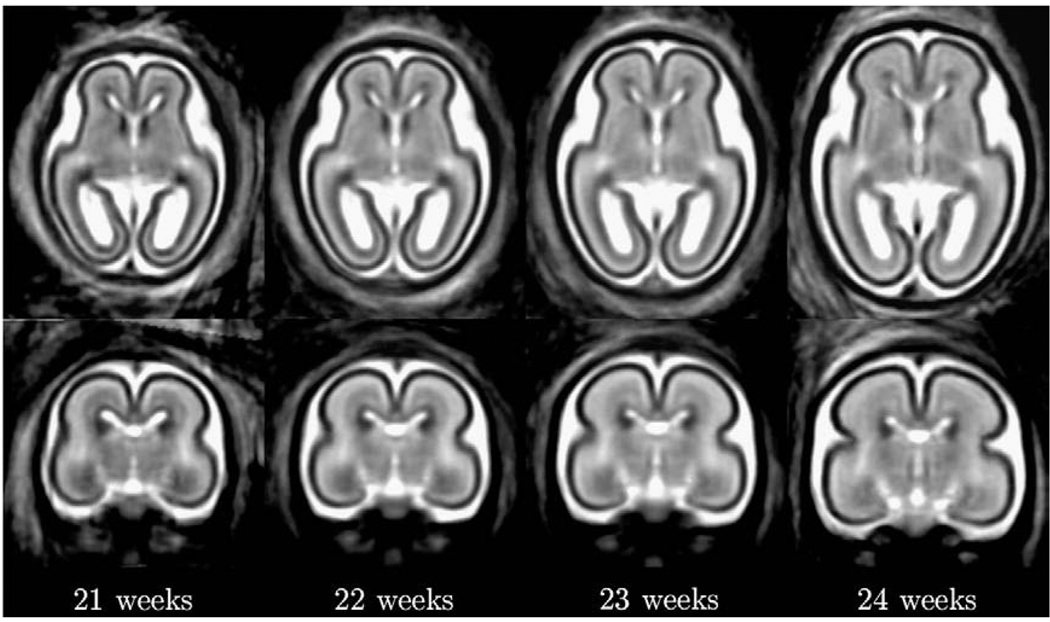
The key challenge in automated analysis of fetal brain MRI data is rapid changes occurring over very short timescales as illustrated in Fig. 1. These include growth-related changes in the size and shape of the brain, maturation-related changes in MR intensities of developing gray and white matter and the appearance and complete disappearance of the germinal matrix from different brain regions. Therefore, to meaningfully label tissues present within a given MR image of the fetal brain, it is necessary to interpret the underlying anatomy in close relation to its developmental stage.
Fig. 1.
Axial and coronal views from reconstructed T2w MR images of typical fetal brain anatomies at around 21, 22, 23 and 24 weeks gestational age.
Numerous studies with older anatomies suggested that the use of age-specific atlases can significantly improve the results of automated analysis of brain MRI data. In multiple neonatal segmentation studies (Warfield et al., 2000; Prastawa et al., 2005; Xue et al., 2007; Weisenfeld and Warfield, 2009), specific templates for newborn brains were used to correctly label developing tissues, including myelinated and non-myelinated white matter. Murgasova et al. (2007) showed that the use of age-specific atlases significantly improves the accuracy of the expectation-maximization (EM) segmentation algorithm for brain structures in 1-year-old and 2-year-old children. Yoon et al. (2009) constructed an age-matched intensity template and tissue probability maps from a set of pediatric MR images and demonstrated their beneficial effect on morphometric analysis of brain MR data of 8-year-old children. Similarly, the results of a study with adult brain anatomies (Studholme et al., 2001) indicated the importance of using an age-appropriate template for the group being analyzed.
In our previous work (Habas et al., in press), we focused on an early developmental stage (20.5–22.5 weeks GA) and demonstrated the feasibility of atlas-based segmentation of developing tissues in the human brain from motion-corrected in utero MRI. As the study was based on a relatively homogeneous group of young fetuses, a single probabilistic atlas of tissue distribution was constructed and applied to drive automatic segmentations. Extending this framework to a wider range of gestational ages, however, given the dynamics of changes occurring in the young fetal brain, separate atlases should be constructed and used for different weeks of early brain development. Such an approach is not practical due to potentially insufficient availability of MR scans of subjects with normal brain development at certain gestational ages as well as time and expertise required to perform accurate manual segmentations.
Instead, in this paper, we present an approach to the construction of a complete spatiotemporal atlas of the human fetal brain with temporal parametric models of MR intensity, tissue probability and shape changes. Spatiotemporal registration and modeling have been previously applied for cardiac MRI (Meyer et al., 1996; Heller et al., 2001; Perperidis et al., 2005) and are becoming popular in the domain of brain image analysis (Davis et al., 2007; Pohl et al., 2007). However, building an atlas of brain tissue growth poses significantly different challenges than temporal modeling of soft tissue deformation. Here, we demonstrate that a spatiotemporal atlas of the fetal brain with an appropriate degree of nonlinearity is able to correctly capture the spatial and temporal variation in presence of developing brain tissues as well as their appearance in MR images. We also show that age-specific MR intensity templates and tissue probability maps generated from the spatiotemporal atlas can be applied for improved tissue segmentation in MR images of new fetal subjects.
Methods
Overview
In order to construct a spatiotemporal atlas of the fetal brain, multiple time point imaging of the same fetal anatomy is not a viable route due to physical and emotional burden for the mother. It is not ethically appropriate to scan pregnant women with short enough time intervals to capture subtle changes in morphology and morphometry of the developing fetal brain. As a result, we have to use a collection of clinical MR images of different fetuses with different gestational ages. This approach, on the other hand, allows our model to capture also between-subject variability which is important for building statistical priors for the analysis of new subject images. To bring a set of fetal anatomies into spatial correspondence, we take advantage of their accurate manual segmentations and perform groupwise registration of tissue probability maps. After registration, we model the effect of the gestational age on probabilities of developing cortical gray matter, developing white matter, the germinal matrix and ventricles at each voxel of the fetal brain. In a similar fashion, we create spatiotemporal models of MR intensity, shape and global scale changes to build a complete four-dimensional atlas of the fetal brain anatomy. From this spatiotemporal atlas, we generate age-specific MR intensity templates and corresponding tissue probability maps and apply them for automatic segmentation of developing tissues in MR images of new fetal subjects.
Fetal MR imaging protocol
Fetal imaging is performed in our institution on a 1.5T scanner (GE Healthcare, Milwaukee, WI) without sedation of the mother or the fetus. For each subject, multiple stacks of single-shot fast spin-echo (SSFSE) T2-weighted (T2w) slice images (pixel size 1mm×1mm, slice thickness ≈3mm) are acquired in the approximately axial, sagittal and coronal planes with respect to the fetal brain. The MR sequence parameters (TR=4500 ms, TE=91 ms) were originally designed for clinical scans and cannot be adjusted for image analysis purposes. After acquisition, image stacks are registered using the slice intersection motion correction (SIMC) technique (Kim et al., 2010) to account for spontaneous fetal movement during scanning and reconstructed into 3D volumes with isotropic voxel size of 0.5 mm. Reconstructed volumes are manually segmented into regions of developing cortical gray matter (GM), developing white matter (WM) also including the deep gray nuclei, the germinal matrix (GMAT), ventricles (VENT) and non-brain structures (NB).
Groupwise registration of tissue label maps
In order to accurately relate data from S training anatomies with different gestational ages, we need to define meaningful and consistent mapping between subjects with different shapes and tissue morphologies. Due to limited contrast of T2w MR images used in clinical practice, there is a substantial intensity overlap between tissues in the growing fetal brain, especially between developing cortical gray matter and the germinal matrix as shown in Fig. 1. As this may cause incorrect alignment of developing tissues during intensity-based registration, we take advantage of existing manual segmentations of all subjects and perform groupwise registration of tissue maps rather than MR intensity images. Unlike fully data-driven approaches, this allows us to resolve registration ambiguities between different tissue types that appear with very similar intensities on T2w MR images used in the study.
We are still, however, faced with the problem of temporally inconsistent structural features such as the border between the germinal matrix and developing white matter gradually disappearing with brain growth. This can induce artifactually large deformations during nonlinear registration and lead to extreme warping between subjects with and without the germinal matrix in different brain regions. In our approach, we exclude such temporally inconsistent tissue boundaries from the registration model and rely only on three basic classes that exist at all gestational ages, namely
developing cortical gray matter (GM),
developing white matter and the germinal matrix combined (WM+GMAT), and
“others” including ventricles (VENT), extraventricular cerebrospinal fluid (CSF) and non-brain structures (NB) (e.g., skull, amniotic fluid, etc.).
To provide continuous band-limited images which can be interpolated and transformed without introducing aliasing artifacts as well as to reduce the influence of any voxel errors potentially present in the manual tracings, we generate a set of S smooth maps
| (1) |
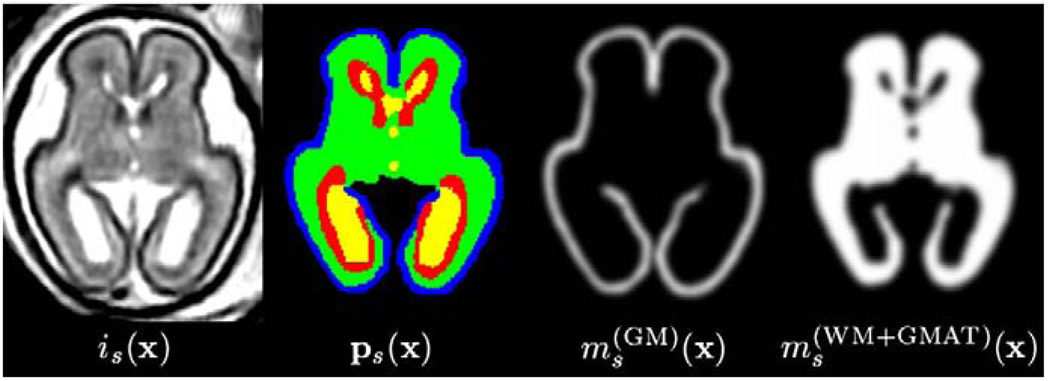
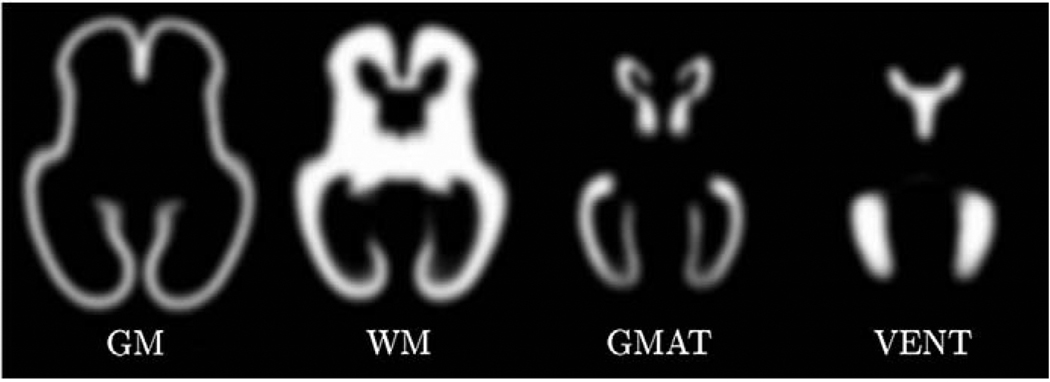
by blurring of binary regions extracted from the full manual segmentation of each subject s. The scalar map components, , range from 0 to 1 and represent the presence of each class at spatial location x as shown in Fig. 2.
Fig. 2.
Axial views from an MR intensity image is(x), its full manual segmentation ps(x) as well as components of a smooth tissue map ms(x) used for spatial normalization.
To initialize groupwise registration, we select a reference anatomy and perform a global linear registration of scalar maps to estimate subjects-specific rotations, translations and scaling parameters as mapping from the reference coordinates to each of the S training anatomies. As a global transformation, this affine registration can only approximately align the main tissue boundaries of interest even when both components of ms(x) are used. Starting from this initial alignment, fine scale differences between brain anatomies including changes in cortical folding are then estimated by a more localized deformable template-free groupwise registration (Studholme and Cardenas, 2004), here using full tissue maps ms(x) rather than MRI intensities. This template-free approach avoids both the use of a single target brain and the construction of an intermediate blurred target image by forming a collective measure of alignment for all subject anatomies.
To define such a collective measure of alignment for multiple tissue classes, we begin by considering the match between a pair of subject anatomies (s,q) from the training set. The squared difference
| (2) |
between the subjects' tissue maps ms(x) and mq(x) transformed to the intermediate average space by subject-specific displacement fields us(x) and uq(x), respectively, is used as the alignment measure of the pair. The derivative of d(s,q)(x) with respect to us(x) is used as the registration force. The collective force ds(x) bringing subject s closer to all other subjects from the training set is then the summation of these force fields.
| (3) |
To regularize the displacement fields us(x), we use an elastic deformation model (Bajcsy et al., 1983; Shen and Davatzikos, 2002) in which the tissue difference force ds(x) is balanced by linear elastic energy
| (4) |
where μ and λ are constants determining the strength of regularization. The value of β determines the influence of a symmetric penalty term derived from the current average displacement
| (5) |
which drives the average deformation of all subjects toward zero. An iterative gradient descent is used to solve Eq. 4 over all subjects. This seeks a set of S displacement fields us(x) that balance the elastic deformation energy with the image dissimilarity while seeking an average anatomy equidistant from each of the S subjects. With the parameters λ and μ experimentally set to suitable values, the resulting set of S subject-specific displacement fields us(x) provides an invertible mapping between the group's average shape and each of the S training anatomies.
Voxelwise temporal modeling from registered anatomies
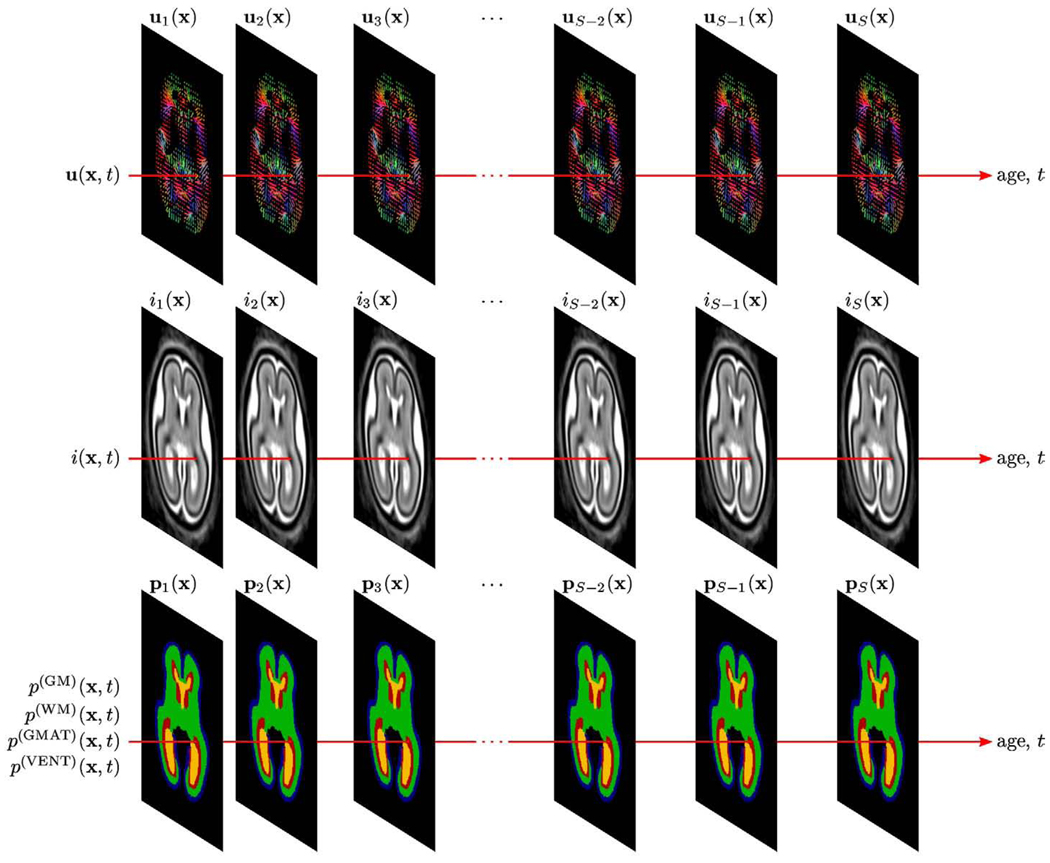
After registration, for each subject s with gestational age ts, we use its global linear scaling as and the diffeomorphic map paremeterized by the displacement field us(x) to warp the full manual segmentation to the average space and obtain an age-specific tissue label map ps(x). Also using as and us(x), we transform the reconstructed MR volume of the subject to the average space to create an age-specific intensity image is(x). From this set of S measurements at specific time points ts, we construct a continuous spatiotemporal atlas of shape, MR intensity and tissue probability capable of representing the fetal brain anatomy at any given developmental age. For each voxel x, we use the gestational age t as an independent variable and voxel values extracted from age-specific maps us(x), is(x) and ps(x) as targets for temporal modeling of shape changes u(x,t), MR intensity i(x,t) as well as probabilities of developing cortical gray matter p(GM)(x,t), developing white matter p(WM)(x,t), the germinal matrix p(GMAT)(x, t) and the ventricles p(VENT)(x,t) as shown in Fig. 3.
Fig. 3.
Voxelwise modeling of temporal changes in shape, MR intensity and tissue probabilities from a set of S registered fetal brain anatomies with different gestational ages t.
Temporal modeling of anatomical deformations
The growth-related changes in the size and shape of the fetal brain are captured firstly by the three directional components of the initial affine scaling and then by the displacement fields us(x) estimated during the elastic groupwise registration.
For each linear scaling component, a parametric temporal model a(j)(t) is obtained by polynomial fitting to values corresponding to the gestational ages ts of the S training subjects. Specifically, a(j)(t) is modeled using a temporal polynomial of degree Da
| (6) |
with coefficients 𝒜(jd) estimated through least squares fitting to the S spatial scaling factors
| (7) |
| (8) |
Temporal modeling of shape deformations is performed separately for each component of the three-dimensional displacement vector u(x). For each deformation direction j, changes in u(j)(x,t) are modeled using a temporal polynomial of degree Du
| (9) |
with coefficients 𝒰(jd)(x) found through least squares fitting to the S values extracted from the age-specific displacement fields us(x).
| (10) |
| (11) |
Temporal modeling of MR intensities
The MRI data of each individual are transformed to the group coordinate system allowing the evaluation of age-related intensity changes across the atlas population. The relative contrasts of different tissues are known to vary spatially across the brain even within a given tissue class and over time as the fetal brain develops. As a result, we have to capture not only the average intensity of each tissue class but also the temporal changes at each voxel location. MR intensity changes at voxel x are modeled using a temporal polynomial of degree Di
| (12) |
with coefficients ℐ(d)(x) found through least squares fitting to the S observed intensity values is(x).
| (13) |
| (14) |
Temporal modeling of tissue probabilities
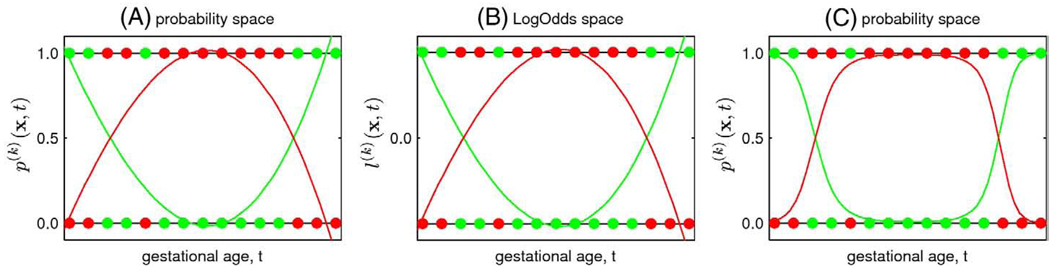
The final component of the spatiotemporal atlas is provided by spatially normalized manual tissue segmentations of each subject s with gestational age ts. From these discrete segmentations , we generate a set of continuous models p(k)(x,t) representing the probability of class k∈{GM, WM, GMAT, VENT} occurring at voxel x at a given time t during brain development. However, if the unconstrained modeling used for global linear scaling, shape deformations and MR intensities is applied directly to the probability values, the resulting model estimates may not be in the range of valid probabilities as illustrated in Fig. 4. Therefore, we need to constrain the fitting of temporal models of tissue probabilities or use an alternative representation of probabilities such as LogOdds previously used for shape representation (Pohl et al., 2006b) and building of probabilistic atlases (Pohl et al., 2007). Unlike other functions of probability maps, LogOdds form a vector space with a probabilistic interpretation and can be composed using linear methods. This allows for flexible and computationally efficient statistical modeling and inference directly in the space of LogOdds.
Fig. 4.
Fitting of temporal polynomials p(1)(x,t) (green line) and p(2)(x,t) (red line) to subject-specific probabilities of two abstract mutually exclusive classes (green and red dots, respectively). (A) Unconstrained modeling directly in the probability space may result in p(k)(x,t) values that are not valid probabilities (p(k)(x,t)<0 or p(k)(x,t)>1). (B) Alternatively, data points ps(k)(x) can be transferred to the space of LogOdds using 𝒪(·) where unconstrained temporal modeling is performed. (C) Age-specific p(k)(x,t) values transferred back from the space of LogOdds using 𝒪−1(·) and normalized across all tissue classes are, unlike in (A), legitimate probabilities. (For interpretation of the references to colour in this figure legend, the reader is referred to the web of this article.)
In the multinomial LogOdds representation suitable for medical imaging problems with multiple labels (Pohl et al., 2007), the LogOdds 𝒪(·) of probability is the logarithm of the ratio between and the last entry of ps(x)
| (15) |
where k∈{GM, WM, GMAT, VENT}. In the context of fetal brain tissue segmentation, is the probability that voxel x in manual segmentation of subject anatomy s is assigned to tissue class k whereas is the probability of assignment of voxel x to non-brain structures. The inverse LogOdds function 𝒪−1(·) is the generalized logistic function
| (16) |
where
| (17) |
is a normalizing factor.
After transformation of the age-dependent tissue probabilities ps(x) to the space of LogOdds, the LogOdds of class k∈{GM, WM, GMAT, VENT} are modeled using a temporal polynomial of degree Dp
| (18) |
with coefficients ℒ(kd)(x) found through least squares fitting to the S observed LogOdds values .
| (19) |
| (20) |
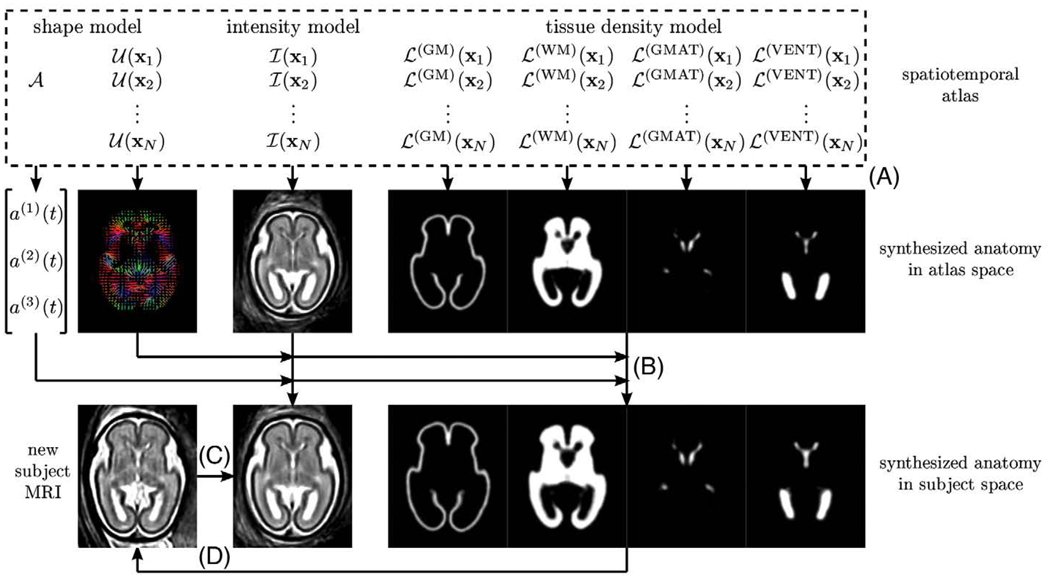
Synthesis of fetal brain anatomies
A complete age-specific fetal brain anatomycan be synthesized from the estimated model parameters 𝒜, 𝒰, ℐ and ℒ by discretizing the continuous temporal models of global linear scaling, local deformation, MR intensity and tissue probability, respectively, for a given gestational age t (Fig. 5). A synthetic age-specific MR image i(x,t) is composed in the space of the atlas from intensity estimates for each voxel x at gestational age t (Fig. 5A). To obtain a corresponding vector of expected tissue probabilities p(x,t), the continuous temporal models l(k)(x,t) are discretized in the space of LogOdds and the resulting values are transferred to the probability space using the inverse LogOdds function O−1(·).
Fig. 5.
Synthesis of a complete fetal brain anatomy with application to segmentation of new fetal MRI. (A) An age-specific MR intensity template and tissue probability maps are synthesized from model parameters in the space of the spatiotemporal atlas. (B) Age-specific shape deformation and linear scaling transform the MR image and tissue maps to the expected size and shape of the new subject. (C) The new subject MRI is registered to the synthetic MR template. (D) Using an inverse transformation, the age-specific tissue probability maps are warped to the space of the new subject and used as priors for atlas-based segmentation of the new subject MRI.
The inverse of the age-specific shape deformation u(x,t) and global linear scaling a(t) calculated from respective temporal models need to be applied to both the synthesized MR image i(x,t) and the tissue probability map p(x,t) to transform them to the expected size and shape of the fetal brain at the gestational age t (Fig. 5B).
Application to atlas-based tissue segmentation
Age-specific MR intensity templates and tissue probability maps generated from the spatiotemporal atlas can be used for tissue segmentation of new subject anatomies as also shown in Fig. 5. First, the MR image of a new subject to be segmented needs to be registered to the age-matched synthetic MR intensity template (Fig. 5C). This is achieved by a sequence of global linear transformations followed by multiple elastic deformations. Then, based on the inverse of the estimated spatial transformation, the age-specific tissue probability map is warped to the space of the new subject (Fig. 5D) and used as a source of spatially varying priors for atlas-based tissue segmentation of the new fetal anatomy.
Results
Population
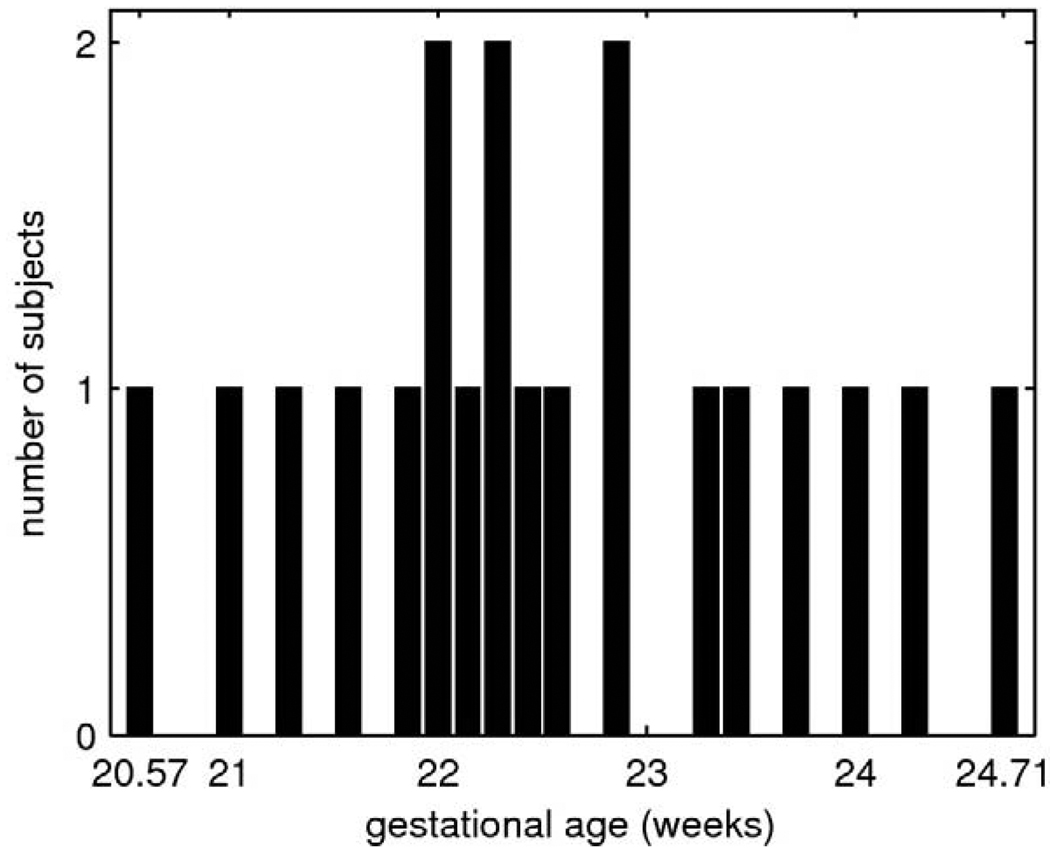
The study was performed using clinical MR scans of 20 fetal subjects with normal brain development selected for reconstruction and manual tracing from a larger pool collected at University of California San Francisco. The mothers were referred for fetal MRI due to questionable abnormalities on prenatal ultrasound, a prior abnormal pregnancy or recruited as volunteers. The gestational ages ts of the fetuses were almost uniformly distributed between 20.57 and 24.71 weeks as shown in Fig. 6.
Fig. 6.
The number of training fetal anatomies as a function of the gestational age.
MR image processing
For each subject, a high-resolution MR volume was formed using the slice intersection motion correction (SIMC) technique (Kim et al., 2010). The reconstructed MR volumes were manually segmented into regions of developing cortical gray matter (GM), developing white matter (WM), the germinal matrix (GMAT), ventricles (VENT) and non-brain structures (NB). The resulting tissue label maps served as reference segmentations for further modeling and validation. The reconstructed 3D images were also corrected for intensity in homogeneity commonly present in fetal MRI data. Low-degree polynomial models of the bias field were estimated using the EM algorithm (Van Leemput et al., 1999) with fixed binary tissue probabilities given by the reference manual segmentations. Finally, the intensities of all images were linearly adjusted based on the average intensity of ventricular CSF.
Spatial normalization
To bring all 20 fetal anatomies into spatial correspondence, tissue maps ms(x) were extracted from full manual segmentation of each subject s and smoothed with a 2-mm full width at half maximum (FWHM) Gaussian kernel. The component of each tissue map was first globally aligned to an average shape anatomy from a previous segmentation study (Habas et al., in press) by maximization of normalized mutual information (Studholme et al., 1999). The obtained 20 age-specific estimates of global linear scaling as in three orthogonal directions (left–right, anterior–posterior and superior–inferior) were used for temporal modeling of changes in the overall size of the fetal brain.
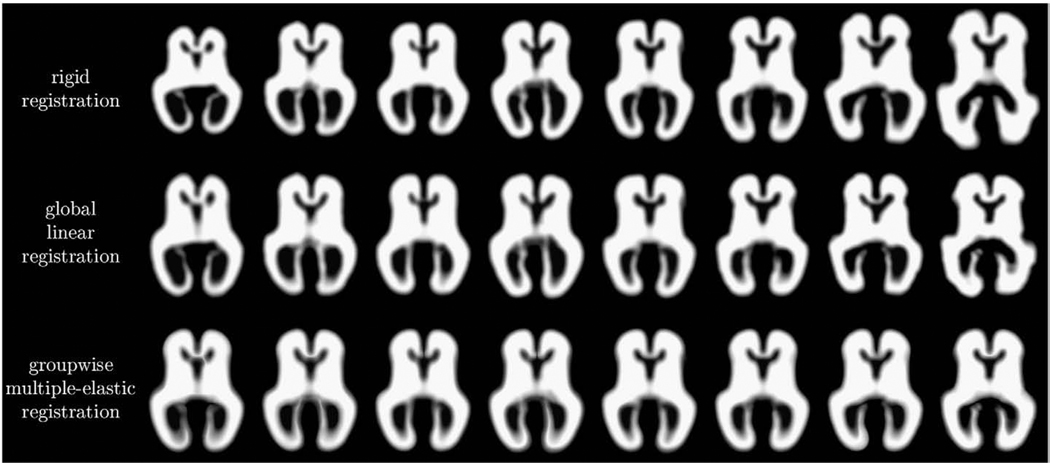
This initial linear registration also provided a starting point for further collective alignment of the full tissue maps ms(x) into a common average space. To resolve large shape differences between fetal brain anatomies with different gestational ages and keep the mapping diffeomorphic, we used a composition of two elastic deformations. For each of them, we set λ=0 as suggested by Modersitzki (2004) and experimentally chose the value of μ=15 to ensure that folding of the deformations did not occur during registration. The value of β=200 was also selected empirically and the final transformations were not found to be very sensitive to this parameter. Typical registration results for a representative subset of 8 subjects with different gestational ages are presented in Fig. 7.
Fig. 7.
Tissue maps of 8 fetal subjects after rigid registration (top row), global linear registration (middle row) and groupwise multiple elastic registration (bottom row).
After groupwise registration, the 20 age-specific displacement fields us(x) were directly used for spatiotemporal modeling of shape changes. Based on as and us(x), the full manual segmentations of each subject s were transformed to the average space using nearest-neighbor interpolation and provided 20 data points ps(x) for spatiotemporal modeling for tissue probabilities. The MR intensity images of all subjects were warped to the average space using linear interpolation and provided 20 data points is(x) for spatiotemporal modeling of MR intensity.
Modeling of anatomical changes
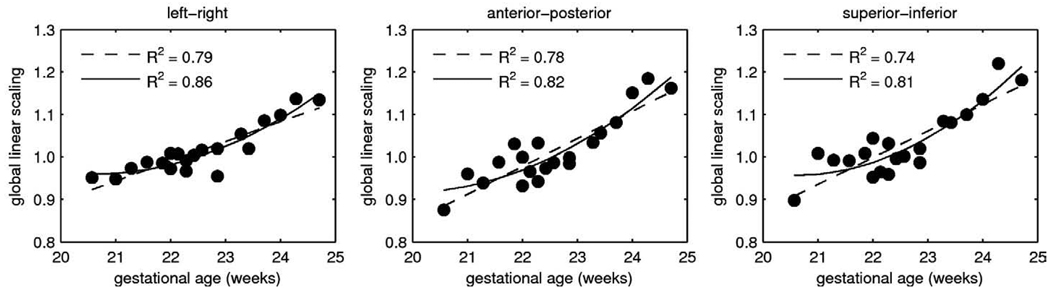
Temporal modeling of global size changes was performed independently for each of the three orthogonal directions as shown in Fig. 8. The ability of polynomial models with different degrees Da to capture temporal patterns present in the training data was evaluated by the coefficient of determination, R2. The value of R2 represents the fraction of total variability in a data set that is accounted for by a statistical model. For each direction, quadratic models (Da=2) captured much more variance (R2∈[0.81, 0.86]) than linear models (Da=1, R2∈[0.74, 0.79]). However, increasing the degree of the temporal models to Da=3 did not bring further improvement of the coefficient of determination (R2∈[0.81, 0.86]). Therefore, for all subsequent experiments, quadratic models of global linear scaling were used for all three directions.
Fig. 8.
Temporal modeling of global linear scaling in three orthogonal directions using linear models (Da=1, dashed lines) and quadratic models (Da=2, solid lines).
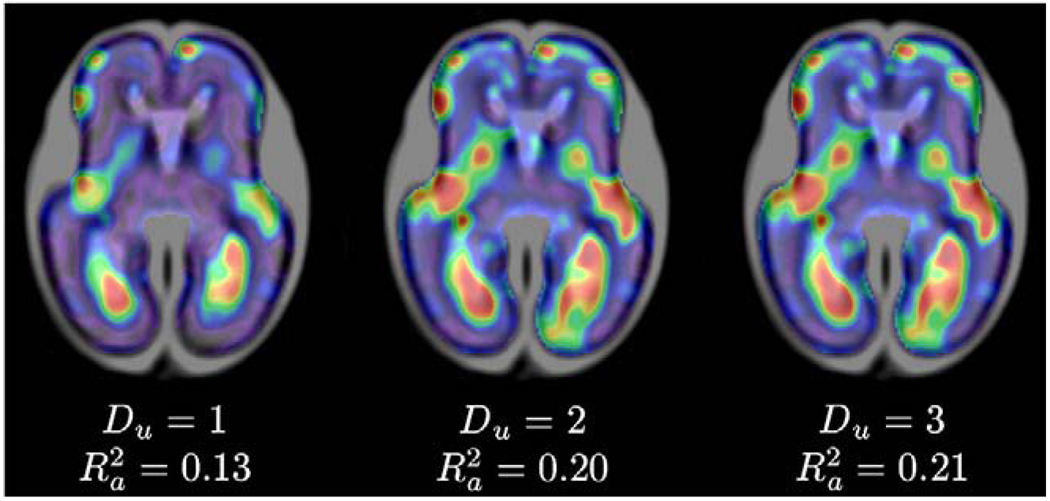
For evaluation of temporal models of shape changes, spatial maps of the R2 coefficient were generated from polynomial shape models u(x,t) with degrees Du=1, Du=2 and Du=3, as shown in Fig. 9. The main patterns captured by the shape models are the formation of the lateral sulcus, changes in shape of the frontal lobe, and shrinking of ventricles with respect to the overall size of the brain, especially in the area of the occipital lobe. Low R2 values in other areas of the fetal brain indicate that these regions are sites of anatomical variability rather than consistent temporal shape changes. The average values of the R2 coefficient calculated in the brain area indicate that age-related changes in the fetal brain anatomy can be better described by quadratic temporal models than by linear models . As cubic temporal models did not offer significantly higher values, quadratic models of temporal shape changes were used for all subsequent modeling.
Fig. 9.
Smoothed maps of R2 coefficient for temporal models of shape changes with Du=1, Du=2, and Du=3. Cooler colors represent lower R2 values while warmer colors represent higher values of the R2 coefficient. Maps were automatically masked to the brain region where values were calculated.
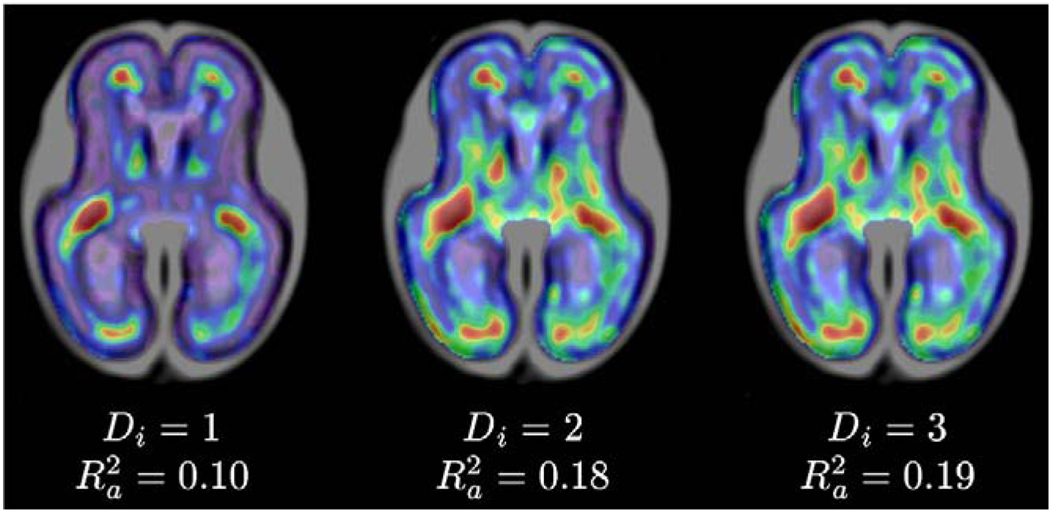
Modeling of MR intensities
Fig. 10 shows spatial maps of the R2 coefficient obtained from temporal models of intensity changes i(x,t) with degrees Di=1, Di=2 and Di=3. Linear intensity changes occur mainly in the areas of the frontal and occipital lobe where the germinal matrix fades out and ultimately disappears with age. In addition, a quadratic model captures intensity changes in the developing white matter. Other areas of the brain exhibit only variations of intensity around the group mean that cannot be captured by low-degree polynomial models. This is reflected by low average values for the whole brain volume–. As the cubic intensity model did not yield substantially higher value than the quadratic model, temporal polynomials with Di=2 were used for all subsequent modeling. Fig. 11 presents synthetic age-specific MR intensity images for gestational ages t between 21 to 24 weeks generated from the quadratic intensity model. For each time point, an MR image i(x,t) was generated first in the space of the spatiotemporal atlas and then transformed using the inverse of estimated age-specific shape deformation u(x,t) and global linear scaling a(t).
Fig. 10.
Smoothed maps of R2 coefficient for temporal models of intensity changes with Di=1, Di=2, and Di=3. Cooler colors represent lower R2 values while warmer colors represent higher values of the R2 coefficient. Maps were automatically masked to the brain region where values were calculated.
Fig. 11.
Axial and coronal views from age-specific T2w MR images i(x,t) of the fetal brain synthesized from a spatiotemporal atlas with Da=2, Du=2, and Di=2 for gestational ages t from 21 to 24 weeks. Images were transformed from the average space using age-specific shape deformation u(x,t) and global linear scaling a(t).
Modeling of tissue probabilities
For evaluation of tissue probability models, we performed qualitative assessment of age-specific tissue probability maps generated from temporal models with different degrees Dp and quantitative analysis of their impact on atlas-based tissue segmentation of new fetal anatomies. Initially, we created a spatiotemporal atlas with the degree of tissue probability model Dp=0. This is equivalent to simple averaging of spatially normalized tissue label maps as is often performed in atlas building. The spatially varying tissue probability maps p(x) generated from such an atlas are not time-dependent and are shown in Fig. 12.
Fig. 12.
Average tissue probability maps p(k)(x) for developing gray matter (GM), developing white matter (WM), the germinal matrix (GMAT) and ventricles (VENT) generated in the average space from a spatiotemporal atlas with Dp=0 (no temporal dependency).
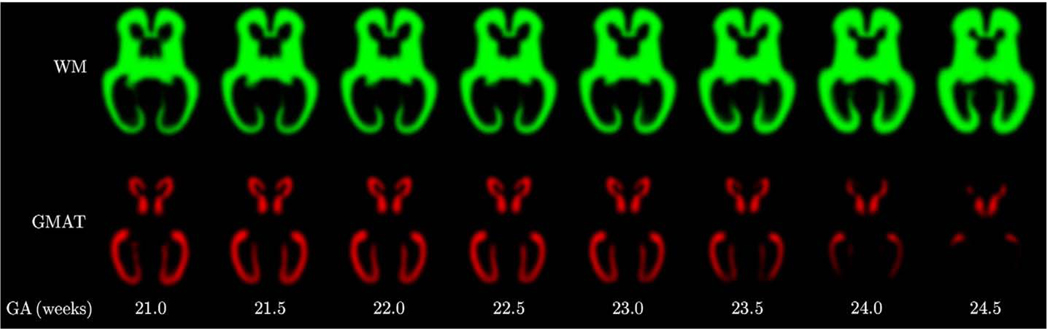
Then, to model temporal changes in tissue presence at different regions of the fetal brain, we also created spatiotemporal atlases with Dp=1 and Dp=2. From these atlases, we generated sets of age-specific tissue probability maps p(x,t) for gestational ages t from 21.0 to 24.5 weeks. Visual inspection of these maps confirms that the quadratic temporal model can correctly capture age-related changes in tissue morphology of the fetal brain. For example, the tissue probability maps presented in Fig. 13 show that before 23 weeks GA there is a substantial amount of the germinal matrix present in various areas of the fetal brain. After the 23rd gestational week, however, the germinal matrix starts to disappear rapidly from the occipital lobe where is almost completely absent after the 24th gestational week. At this stage, this transient structure also starts to disappear from the area of the temporal lobe. The observed pattern is very consistent with previously described changes in tissue morphology based on clinical qualitative evaluation of 2D MR scans of the fetal brain (Battin et al., 1998; Kinoshita et al., 2001; Prayer et al., 2006). Fig. 14 shows 3D visualization of the main tissue boundaries in the fetal brain for gestational ages ranging from 21.0 to 24.5 weeks.
Fig. 13.
Age-specific tissue probability maps p(k) (x,t) for developing white matter (WM) and the germinal matrix (GMAT) generated in the average space from a spatiotemporal atlas with Dp=2.
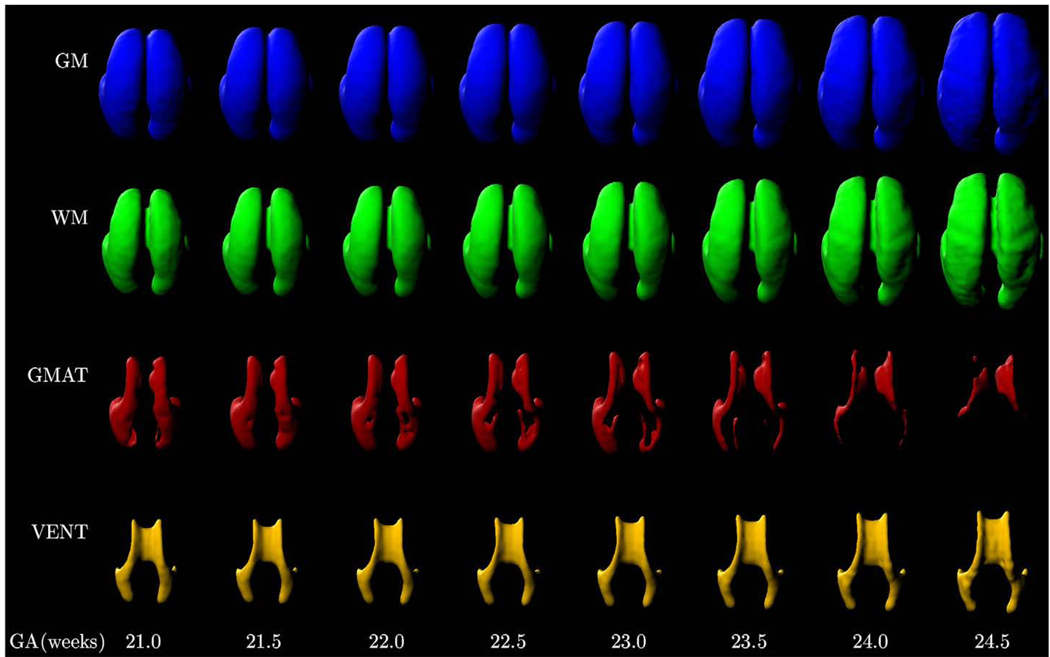
Fig. 14.
3D visualization of main tissue types in the fetal brain for gestational ages from 21.0 to 24.5 weeks. The geometry of each age-specific surface was derived from a spatiotemporal atlas with Da=2, Du=2 and Dp=2.
Atlas-based tissue segmentation
Based on previously reported results for neonates and young children (Murgasova et al., 2007; Yoon et al., 2009), we expected that the use of age-specific atlases may improve the results of atlas-based tissue segmentation of the fetal brain. To verify this hypothesis, we performed the following leave-one-out validation experiment.
After groupwise registration of the entire study population (20 subjects), each subject was held out once for testing purposes. The remaining 19 subjects were used to build a set of spatiotemporal atlases with Da=2, Du=2, Di=2 and different degrees of the tissue probability model—Dp=0, Dp=1 and Dp=2. From each such atlas, we generated a synthetic age-specific MR intensity template and an age-specific tissue probability map for the gestational age of the excluded subject. The MRI of the excluded subject was aligned to the age-matched MR template using a sequence of global linear registrations driven by maximization of normalized mutual information (Studholme et al., 1999) followed by multiple elastic deformations driven by maximization of mutual information (Viola and Wells, 1997) within a fixed mask. Then, based on the inverse of the estimated spatial transformation, all three age-matched tissue probability maps (for Dp=0, Dp=1 and Dp=2) were deformed to the space of the excluded subject. Finally, each tissue map was used as a source of spatially varying tissue priors for automatic atlas-based EM segmentation of the subject MRI (Habas et al., in press). The results of automatic segmentation were evaluated in terms of Dice similarity coefficient (DSC) (Dice, 1945) calculated with respect to the manual segmentation of the excluded subject.
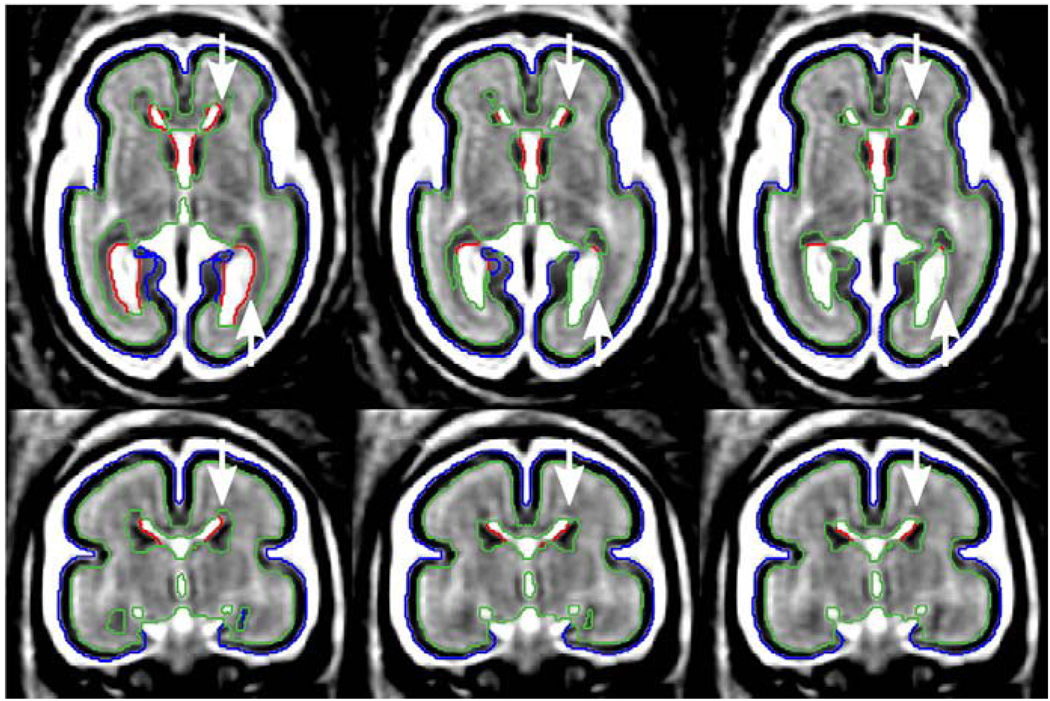
Table 1 presents summary results of the segmentation experiment. For all degrees of the tissue probability model, the accuracy of automatic atlas-based segmentation for developing cortical gray matter, developing white matter and ventricles (DSC>0.8) is similar to DSC values previously reported in segmentation studies with neonates (Prastawa et al., 2005; Xue et al., 2007) and young children (Murgasova et al., 2007). The inferior average accuracy for segmentation of the germinal matrix with a single atlas (Dp=0, DSC=0.675) arises mainly from oversegmenting of this structure in older subjects. Fig. 15 shows results of atlas-based tissue segmentation for a fetal subject at 24 weeks GA. For Dp=0, due to a strong prior from the atlas, a substantial amount of developing white matter is incorrectly labeled as the germinal matrix in regions where this structure is no longer present (frontal and occipital lobe as indicated by arrows). On the other hand, the germinal matrix is undersegmented for subjects younger than 21 weeks.
Table 1.
Average DSC accuracy of atlas-based EM segmentation with probabilistic atlases generated from spatiotemporal models with Dp=0, Dp=1 and Dp=2 (20 subjects).
| Tissue or structure | Dp=0 | Dp=1 | Dp=2 |
|---|---|---|---|
| GM | 0.832±0.034 | 0.833±0.033 | 0.834±0.034 |
| GMAT | 0.675±0.158 | 0.742±0.091 | 0.772±0.082 |
| WM | 0.908±0.027 | 0.918±0.022 | 0.921±0.023 |
| VENT | 0.878±0.046 | 0.879±0.046 | 0.880±0.046 |
Fig. 15.
The impact of the degree of the tissue probability model (Dp) on automatic atlas-based tissue segmentation of a fetal subject at 24 weeks GA. Arrows indicate areas where the use of age-specific atlases generated from models with Dp=1 and Dp=2 improves segmentation of the germinal matrix.
The use of age-specific atlases generated from tissue probability models with Dp=1 and Dp=2 improves segmentation accuracy for all fetal brain tissues. Particular refinement can be observed for the germinal matrix where the DSC index increases from 0.675 for Dp=0 to 0.772 for Dp=2 (p=0.046). Moreover, the standard DSC error for GMAT is reduced almost by half, from 0.158 for Dp=0 to 0.082 for Dp=2, due to more consistent segmentation accuracy across all gestational ages.
Discussion
We presented an approach to modeling of early human brain growth from clinical MRI. Using a set of reconstructed 3D MR images and their manually defined tissue segmentations, we created a spatiotemporal atlas of MR intensity, tissue probability and shape changes in the human fetal brain during early in utero development. From this continuous mathematical model, we can synthesize maps of MR intensity and tissue occurrence for any gestational age covered by the atlas and apply them for improved tissue segmentation in new fetal anatomies.
As multiple time point imaging of the same fetal anatomy is not feasible, the spatiotemporal atlas was built using MR images of different fetuses with different gestational ages. To bring all anatomies into collective alignment in the average space of the group, we applied groupwise registration of tissue maps extracted from fixed manual segmentations. Although this approach did not define full correspondence between images with different gestational ages, it allowed us to consistently align the main tissue boundaries present in the developing fetal brain and resolve matching ambiguities between multiple tissue types that appear with similar intensities on T2w MR images. Extending this framework to a wider range of gestational ages, we plan to replace the single average space with temporarily parameterized average geometry. Another possible direction of future work is to combine the groupwise registration with simultaneous tissue segmentation and atlas building (Ashburner and Friston, 2005; Pohl et al., 2006a).
After registration, we performed parametric modeling of changes in MR intensity, tissue probability and shape of the developing fetal brain. Although additional features such as gender or disease group could be also considered, we used gestational age as the only independent variable for this initial work. For all analyzed properties of the fetal brain anatomy, quadratic temporal models provided a better description of patterns observed for the study population than linear models. For modeling of tissue probabilities, quadratic polynomials allowed us to represent both appearance and disappearance of the germinal matrix from different regions of the brain. As tissue occurrence or MR intensity are not expected to oscillate, the use of higher order temporal models was neither justified nor necessary.
Our morphology model included three main tissue types present in the young fetal brain (developing cortical gray matter, developing white matter, the germinal matrix) and ventricles. Other structures, such as the basal ganglia or the subplate, are difficult to see at this early stage (20–24 weeks GA) but become more visible as the fetuses mature. As a temporary solution, we combined these regions of the fetal brain with developing white matter. In the future, however, they will be fully incorporated into the modeling process as separate and important structures.
Spatiotemporal modeling of all properties was performed in a voxelwise manner, without explicit constraints on spatial smoothness of synthesized maps. Although not an issue for MR intensity and tissue probabilities, this may potentially cause problems in modeling of shape changes. In practice, however, given the spatial smoothness of all subject maps, we found that the shape model provided invertible diffeomorphic mappings for all time points. In the future, we plan to compare our voxelwise approach to alternative techniques such as kernel regression for large deformation models (Davis et al., 2007) and explore extensions to global modeling using ridge regression (Sjostrand et al., 2008).
Finally, we presented a practical application of the spatiotemporal atlas for tissue segmentation in new MR images. As the synthesis of age-matched MR templates reduces the geometric differences that have to be resolved during registration, the anatomy to be segmented can be precisely aligned with the source of tissue priors. The use of age-specific probabilistic atlases improves the overall accuracy of automatic tissue segmentation, as measured by the average DSC values, and allows for correct delineation of transient brain structures that evolve rapidly with gestational age. In clinical applications, accurate quantification of the fetal brain anatomy in relation to its developmental stage may help detect subtle abnormalities that otherwise could be missed.
Acknowledgments
This work was primarily funded by NIH/NINDS Grant R01 NS 055064. Imaging studies were partially funded by NIH/NINDS Grant K23 NS 052506 and NIH/NCRR UCSF-CTSI Grant UL1 RR 024131. The work of F. Rousseau was supported by the European Research Council under the European Community Seventh Framework Programme (FP7/2007-2013 Grant Agreement 207667).
The authors thank Dr. Julia A. Scott for help with editing of the manuscript.
References
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bajcsy R, Lieberson R, Reivich M. A computerized system for the elastic matching of deformed radiographic images to idealized atlas images. J. Comput. Assist. Tomogr. 1983;7(4):618–625. doi: 10.1097/00004728-198308000-00008. [DOI] [PubMed] [Google Scholar]
- Battin MR, Maalouf EF, Counsell SJ, Herlihy AH, Rutherford MA, Azzopardi D, Edwards AD. Magnetic resonance imaging of the brain in very preterm infants: visualization of the germinal matrix, early myelination, and cortical folding. Pediatrics. 1998;101(6):957–962. doi: 10.1542/peds.101.6.957. [DOI] [PubMed] [Google Scholar]
- Davis BC, Fletcher PT, Bullitt E, Joshi SC. Population shape regression from random design data; Proc. International Conference on Computer Vision; 2007. pp. 1–7. [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- Girard N, Raybaud C, Poncet M. In vivo MR study of brain maturation in normal fetuses. Am. J. Neuroradiol. 1995;16(2):407–413. [PMC free article] [PubMed] [Google Scholar]
- Glenn OA. MR imaging of the fetal brain. Pediatr. Radiol. 2010;40(1):68–81. doi: 10.1007/s00247-009-1459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Atlas-based segmentation of developing tissues in the human brain with quantitative validation in young fetuses. Hum. Brain Mapp. doi: 10.1002/hbm.20935. in press. doi:10.1002/hbm.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EN, Staib LH, Dione DP, Constable RT, Shi CQ, Duncan JS, Sinusas AJ. A new method for quantification of spatial and temporal parameters of endocardial motion: evaluation of experimental infarction using magnetic resonance imaging. Can. J. Cardiol. 2001;17(3):309–318. [PubMed] [Google Scholar]
- Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Trans. Med. Imaging. 2007;26(7):967–980. doi: 10.1109/TMI.2007.895456. [DOI] [PubMed] [Google Scholar]
- Kim K, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Intersection based motion correction of multislice MRI for 3-D in utero fetal brain image formation. IEEE Trans. Med. Imaging. 2010;29(1):146–158. doi: 10.1109/TMI.2009.2030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita Y, Okudera T, Tsuru E, Yokota A. Volumetric analysis of the germinal matrix and lateral ventricles performed using MR images of postmortem fetuses. Am. J. Neuroradiol. 2001;22(2):382–388. [PMC free article] [PubMed] [Google Scholar]
- Meyer FG, Constable RT, Sinusas AJ, Duncan JS. Tracking myocardial deformation using phase contrast MR velocity fields: a stochastic approach. IEEE Trans. Med. Imaging. 1996;15(4):453–465. doi: 10.1109/42.511749. [DOI] [PubMed] [Google Scholar]
- Modersitzki J. Numerical methods for image registration. Oxford Science Publications; 2004. [Google Scholar]
- Murgasova M, Dyet L, Edwards AD, Rutherford M, Hajnal JV, Rueckert D. Segmentation of brain MRI in young children. Acad. Radiol. 2007;14(11):1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Perperidis D, Mohiaddin RH, Rueckert D. Spatio-temporal free-form registration of cardiac MR image sequences. Med. Image Anal. 2005;9(5):441–456. doi: 10.1016/j.media.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Pohl KM, Fisher J, Grimson WEL, Kikinis R, Wells WM. A Bayesian model for joint segmentation and registration. Neuroimage. 2006a;31(1):228–239. doi: 10.1016/j.neuroimage.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Pohl KM, Fisher J, Shenton M, McCarley RW, Grimson WEL, Kikinis R, Wells WM. Logarithm odds maps for shape representation. Medical Image Computing and Computer-Assisted Intervention: LNCS. 2006b;4191:955–963. doi: 10.1007/11866763_117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl KM, Fisher J, Bouix S, Shenton M, McCarley RW, Grimson WEL, Kikinis R, Wells WM. Using the logarithm of odds to define a vector space on probabilistic atlases. Med. Image Anal. 2007;11(5):465–477. doi: 10.1016/j.media.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M, Gilmore JH, Lin W, Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med. Image Anal. 2005;9(5):457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Prayer D, Kasprian G, Krampl E, Ulm B, Witzani L, Prayer L, Brugger PC. MRI of normal fetal brain development. Eur. J. Radiol. 2006;57(2):199–216. doi: 10.1016/j.ejrad.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza CE, Vigneron DB, Barkovich AJ, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Acad. Radiol. 2006;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Rutherford M, Jiang S, Allsop J, Perkins L, Srinivasan L, Hayat T, Kumar S, Hajnal J. MR imaging methods for assessing fetal brain development. Dev. Neurobiol. 2008;68(6):700–711. doi: 10.1002/dneu.20614. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Sjostrand K, Cardenas VA, Larsen R, Studholme C. A generalization of voxel-wise procedures for high-dimensional statistical inference using ridge regression. Medical Imaging 2008; Image Processing: Proc. SPIE; 2008. p. 69140A. [Google Scholar]
- Studholme C, Cardenas VA. A template free approach to volumetric spatial normalization of brain anatomy. Pattern Recognit. Lett. 2004;25(10):1191–1202. [Google Scholar]
- Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognit. 1999;32(1):71–86. [Google Scholar]
- Studholme C, Cardenas VA, Weiner MW. Multiscale image and multiscale deformation of brain anatomy for building average brain atlases. Medical Imaging 2001; Image Processing: Proc. SPIE; 2001. pp. 557–568. [Google Scholar]
- Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Trans. Med. Imaging. 1999;18(10):885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- Viola P, Wells WM. Alignment by maximization of mutual information. Int. J. Comput. Vision. 1997;24(2):137–154. [Google Scholar]
- Warfield S, Kaus M, Jolesz FA, Kikinis R. Adaptive, template moderated, spatially varying statistical classification. Med. Image Anal. 2000;4(1):43–55. doi: 10.1016/s1361-8415(00)00003-7. [DOI] [PubMed] [Google Scholar]
- Weisenfeld NI, Warfield SK. Automatic segmentation of newborn brain MRI. Neuroimage. 2009;47(2):564–572. doi: 10.1016/j.neuroimage.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Srinivasan L, Jiang S, Rutherford M, Edwards AD, Rueckert D, Hajnal JV. Automatic segmentation and reconstruction of the cortex from neonatal MRI. Neuroimage. 2007;38(3):461–477. doi: 10.1016/j.neuroimage.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Yoon U, Fonov VS, Perusse D, Evans AC. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45(3):769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]