Abstract
Background
Aberrant activation of tyrosine kinases, caused by either mutation or gene fusion, is of major importance for the development of many hematologic malignancies, particularly myeloproliferative neoplasms. We hypothesized that hitherto unrecognized, cytogenetically cryptic tyrosine kinase fusions may be common in non-classical or atypical myeloproliferative neoplasms and related myelodysplastic/myeloproliferative neoplasms.
Design and Methods
To detect genomic copy number changes associated with such fusions, we performed a systematic search in 68 patients using custom designed, targeted, high-resolution array comparative genomic hybridization. Arrays contained 44,000 oligonucleotide probes that targeted 500 genes including all 90 tyrosine kinases plus downstream tyrosine kinase signaling components, other translocation targets, transcription factors, and other factors known to be important for myelopoiesis.
Results
No abnormalities involving tyrosine kinases were detected; however, nine cytogenetically cryptic copy number imbalances were detected in seven patients, including hemizygous deletions of RUNX1 or CEBPA in two cases with atypical chronic myeloid leukemia. Mutation analysis of the remaining alleles revealed non-mutated RUNX1 and a frameshift insertion within CEBPA. A further mutation screen of 187 patients with myelodysplastic/myeloproliferative neoplasms identified RUNX1 mutations in 27 (14%) and CEBPA mutations in seven (4%) patients. Analysis of other transcription factors known to be frequently mutated in acute myeloid leukemia revealed NPM1 mutations in six (3%) and WT1 mutations in two (1%) patients with myelodysplastic/myeloproliferative neoplasms. Univariate analysis indicated that patients with mutations had a shorter overall survival (28 versus 44 months, P=0.019) compared with patients without mutations, with the prognosis for cases with CEBPA, NPM1 or WT1 mutations being particularly poor.
Conclusions
We conclude that mutations of transcription and other nuclear factors are frequent in myelodysplastic/myeloproliferative neoplasms and are generally mutually exclusive. CEBPA, NPM1 or WT1 mutations may be associated with a poor prognosis, an observation that will need to be confirmed by detailed prospective studies.
Keywords: RUNX1, AML1, CEBPA, MDS, MPN, myeloproliferative
Introduction
Myeloproliferative neoplasms (MPN) are clonal hematopoietic stem cell disorders characterized by abnormal proliferation and survival of one or more myeloid cell types in the bone marrow and increased numbers of mature and immature cells in the peripheral blood.1 In addition to the four classical MPN, polycythemia vera, essential thrombocythemia, primary myelofibrosis and chronic myeloid leukemia (CML), there are rarer subtypes referred to as non-classical or atypical MPN such as chronic eosinophilic leukemia (CEL), hypereosinophilic syndrome (HES), chronic neutrophilic leukemia or MPN unclassifiable (MPN-U).2 These disorders may overlap with myelodysplastic/myeloproliferative neoplasms (MDS/MPN) including BCR-ABL-negative atypical CML, chronic myelomonocytic leukemia (CMML) and MDS/MPN unclassifiable (MDS/MPN-U), in which proliferation is accompanied by dysplastic features or ineffective hematopoiesis in other lineages.3
The molecular pathogenesis of atypical MPN and MDS/MPN is only partially understood. In many patients, aberrant activation of tyrosine kinase signaling has been found as a consequence of four principal mechanisms: (i) activating tyrosine kinase mutations, e.g. FLT3 and JAK2,4 (ii) mutations in downstream signaling components, e.g. RAS,5,6 (iii) mutations in negative regulators, e.g. CBL7–9 and (iv) constitutively active tyrosine kinase fusion genes arising as a consequence of genomic rearrangements.10 Collectively, however, these abnormalities account for well under 50% of cases.
Of the tyrosine kinase fusions, FIP1L1-PDGFRA in CEL is unique in that it results from a cytogenetically invisible 800 kb interstitial deletion at chromosome band 4q12.11 All other fusions in atypical MPN and MDS/MPN, most of which are extremely rare, are associated with visible karyotypic aberrations. However cytogenetically cryptic tyrosine kinase fusions have been described in acute lymphoblastic leukemia, either as a consequence of translocations involving regions that cannot be distinguished visually (EML1-ABL)12 or, most remarkably, episomal amplification (NUP214-ABL).13 The discovery of cryptic fusions relied on a fortuitous case with an incidental visible translocation (FIP1L1-PDGFRA) or the use of fluorescence in situ hybridization to screen for disruption of ABL (EML1-ABL and NUP214-ABL). It is, therefore, possible that many other similar abnormalities remain to be discovered.
In this study, we hypothesized that hitherto unrecognized, cytogenetically cryptic tyrosine kinase fusions might be common in patients with atypical MPN or MDS/MPN. Since cryptic fusions are frequently associated with DNA copy number changes, we performed a targeted screen of all tyrosine kinases as well as tyrosine kinase signaling components, known translocation targets and transcription factors using custom designed, high-resolution targeted array comparative genomic hybridization (CGH). We sought to determine whether copy number changes characterize atypical MPN and MDS/MPN of unknown molecular etiology and whether they could be used as a tool to help identify novel fusion genes or other driver mutations.
Design and Methods
Patients and clinical samples
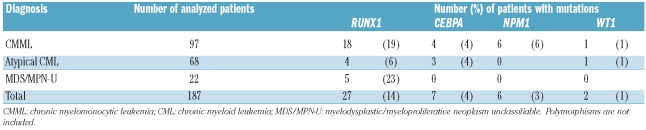
Pretreatment leukocyte genomic DNA from 68 patients was studied by targeted array CGH. Patients had MDS/MPN (CMML, n=9; atypical CML, n=8; MDS/MPN-U, n=14); CEL/HES, n=17; MPN-U, n=16; chronic neutrophilic leukemia, n=3, or acute basophilic leukemia, n=1. All samples tested negative for BCR-ABL, FIP1L1-PDGFRA, JAK2V617F and none had karyotypic abnormalities suggestive of other known tyrosine kinase fusions. Nine patients with CEL/HES showed a significant response to imatinib treatment in the absence of any known imatinib-sensitive abnormality. A further 187 patients with MDS/MPN (CMML, n=97; atypical CML, n=68; MDS/MPN-U, n=22) were analyzed for sequence variants by direct sequencing. The study was approved by the Internal Review Boards from participating institutions and informed consent was provided according to the Declaration of Helsinki.
Targeted array comparative genomic hybridization
We designed Agilent HD-CGH Microarrays (Agilent Technologies, Palo Alto, CA, USA) using the 4x44K format. Slides consisted of four individual arrays, each containing 44,000 60-mer oligonucleotide probes. Individual oligonucleotides were selected from the Agilent eArray online database (https://earray.chem.agilent.com/erray). Each array targeted 500 genes including all 90 tyrosine kinases plus downstream tyrosine kinase signaling components, other translocation targets, transcription factors, and other factors known to be important for myelopoiesis (for a full list of targeted genes see Online Supplementary Table S1). For each target gene, 50–100 probes were selected that spanned the gene plus flanking sequences of up to 200 kb, providing a resolution of up to 5–10 kb. Samples were processed according to the Oligonucleotide Array-based CGH for Genomic DNA Analysis Protocol (version 4.0, Agilent Technologies). Briefly, 1.5 μg of the patients’ and reference DNA were labeled with either Cy3 or Cy5 by random priming after restriction enzyme digestion. Differentially labeled patients’ and control DNA was mixed, denatured and hybridized to array slides for 24 h under stringent conditions. Slides were then washed, dried and scanned using an Agilent G2505B microarray scanner and Agilent Scan Control software (version A.7.0.1). Microarray images were analyzed by Agilent Feature Extraction software (version 9.5.3.1) and the data were subsequently imported into CGH Analytics software (version 3.4.40, Agilent Technologies) for downstream analysis.
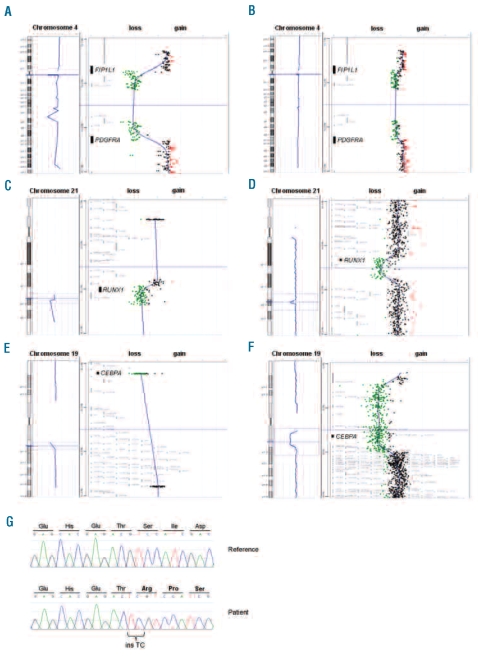
Since the proportion of background normal cells in our samples was unknown and likely to be variable between cases, it was important to determine that our targeted arrays could detect heterozygous deletions in a background of normal cells. We found that FIP1L1-PDGFRA was readily detectable when EOL1 DNA was diluted 1:2.5 with normal DNA (Figure 1A and B). Since EOL1 cells have two copies of the del(4) and one normal 4, this is equivalent to detection of a heterozygous deletion when normal cells constitute 50% of the sample.
Figure 1.
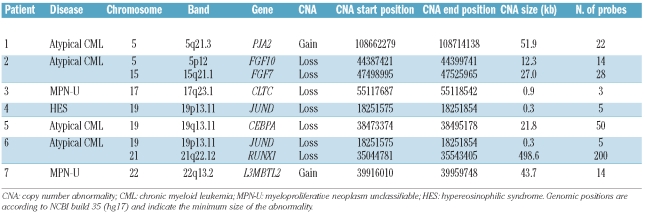
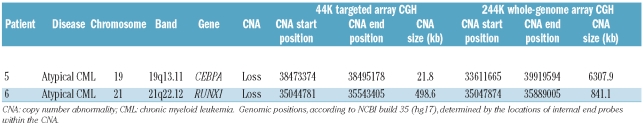
Targeted array CGH and sequencing profiles. (A) Control experiments demonstrating that targeted arrays were readily able to identify the 800kb FIP1L1-PDGFRA deletion in EOL1 cells in a background of normal cells. EOL1 cells harboring two copies of the del(4) and one normal chromosome 4. (B) 1:2.5 mixtures of EOL1 DNA with normal DNA, simulating a heterozygous deletion with 50% background normal cells. (C) RUNX1 deletion in a patient with atypical CML. Targeted array CGH profile of chromosome 21 (left) with zoom of the respective region (right) showing a hemizygous deletion of RUNX1. (D) Whole-genome 244K array CGH results confirm this observation and further characterize the deletion as a 841kb deletion including RUNX1. (E) CEBPA deletion in a patient with atypical CML. Targeted array CGH profile of chromosome 19 (left) with zoom of the respective region (right) showing a hemizygous deletion of CEBPA. (F) Whole-genome 244K array CGH results further characterize the deletion as a 6.3Mb deletion including CEBPA and several other genes. (G) Sequencing result of the atypical CML patient with a CEBPA deletion. A homozygous 2 bp insertion was observed resulting in a frameshift at amino acid threonine 60 (bottom panel). The CEBPA reference sequence is shown in the top panel.
Human genome-wide array comparative genomic hybridization
In some cases in which targeted array CGH revealed copy number changes that extended beyond the boundaries of the probes targeting a gene of interest, commercially available Agilent Human Genome CGH Microarrays in 244K format (containing 244,000 coding and non-coding human sequences with a genome-wide resolution of approximately 7–9 kb) were used to identify the full extent of the copy number abnormalities. The sample processing was performed as described for the targeted arrays except that 2 μg of patients’ and reference DNA were used.
Scoring criteria
Copy number changes less than 10 Mb were regarded as cytogenetically cryptic. Abnormalities in three consecutive oligonucleotide probes were required to call a copy number imbalance by array CGH. Copy number changes were compared to the Database of Genomic Variants14 to exclude known constitutional copy number variants.
Mutation analysis
Mutation analysis was performed on whole-genome amplified DNA material using the IllustraTM GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Buckinghamshire, UK). The entire coding regions of RUNX1 (exons 3–8) (ENST00000344691) and CEBPA (exon 1) (ENST00000328368) and selected exons of NPM1 (exon 12) (ENST00000296930) and WT1 (exons 7 and 9) (ENST00000332351) were analyzed by direct sequencing of polymerase chain reaction products using published primer sequences15–18 and standard techniques on an ABI 3130 Genetic Analyzer (Applied Biosystems, Warrington, UK) with Mutation Surveyor software (SoftGenetics, State College, PA, USA). A dropping factor (relative intensity drop of the wild-type allele peak relative to that seen in a concurrently run normal sample) of 60% or more was considered indicative of a biallelic mutation whereas a dropping factor of less than 60% was considered as monoallelic. This is a conservative definition and since we did not know the relative proportion of malignant and non-malignant cells in our samples the true incidence of homozygosity may be underestimated. All mutations were confirmed on unamplified genomic DNA by sequencing in both directions.
Results
Detection of copy number imbalances in the majority of patients
Sixty-six of 68 patients (97%) harbored detectable genomic imbalances with a median of five (range, 1–19) copy number abnormalities per patient. Of the 424 copy number imbalances detected, 252 (59%) were amplifications and 172 (41%) were deletions involving 41 of the 500 targeted genes. The median size of the copy number changes was 3.0 kb (range, 0.1 – 498.6 kb) involving a median of six oligonucleotide probes (range, 3–200). Forty patients showed either gain or loss of DNA material and 26 patients had both. One patient with CMML and one patient with MPN-U did not show any copy number abnormality within the targeted genes. Of the 41 regions showing copy number imbalance, 20 were within regions of known copy number variants. Thirteen additional regions with a median size of 1.2 kb (range, 0.1 – 18.2 kb) were identified that showed copy number loss in some individuals and copy number gain in others strongly suggesting that they are copy number variants, despite not being listed in the Database of Genomic Variants (Online Supplementary Table S2).
Identifications of novel copy number abnormalities
After eliminating copy number variants, nine cytogenetically cryptic imbalances at eight loci remained in seven patients (Table 1). Their median size was 21.8 kb (range, 0.3 – 498.6kb) involving a median of 14 oligonucleotide probes (range, 3–200). Seven copy number changes represented loss of DNA material including a 499 kb deletion of RUNX1 at 21q22.12 (Figure 1C) and a 22 kb deletion of CEBPA at 19q13.11 (Figure 1E) in two patients with atypical CML. Two copy number imbalances represented gain of DNA material (PJA2 and L3MBT2L). None of the imbalances affected tyrosine kinases.
Table 1.
Copy number abnormalities found by targeted array CGH.
Breakpoint analysis of RUNX1 and CEBPA deletions
For this initial study we focused on the RUNX1 and CEBPA deletions because of their known prognostic significance in AML; the other abnormalities are still under investigation. In patients with RUNX1 or CEBPA deletions the copy number abnormalities extended beyond the limits of the probe set targeting these genes. Whole-genome array CGH was, therefore, performed to characterize the genomic boundaries of these abnormalities (Table 2). An 841 kb-deleted region including only RUNX1 was identified in the patient with RUNX1 deletion (Figure 1D). In the patient with the CEBPA deletion, a region of 6.3 Mb was identified with involvement of adjacent genes which were not represented on the initial targeted array (Figure 1F).
Table 2.
Copy number boundaries obtained using human genome-wide array CGH.
Mutation analysis of candidate genes reveals frequent mutations of transcription factors
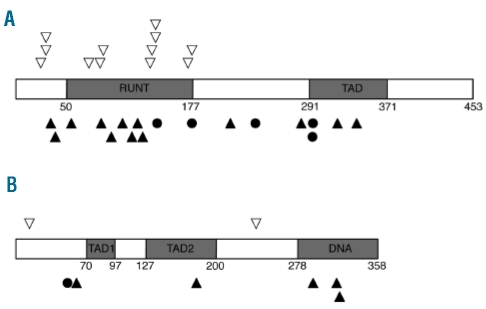
We focused on RUNX1 and CEBPA because of their known involvement in AML. Mutation analysis of the remaining alleles in the two patients with deletions revealed non-mutated RUNX1 and a 2 bp insertion within CEBPA leading to a frameshift at amino acid threonine 60 (Figure 1G). A mutation screen of a further 187 patients with MDS/MPN identified RUNX1 mutations in 27 (14%; CMML, n=18; atypical CML, n=4; MDS/MPN-U, n=5) and CEBPA mutations in 7 (4%; CMML, n=4; atypical CML, n=3) patients. Three patients had two RUNX1 mutations leading to a total of 30 RUNX1 mutations (frameshift ins/del, n=13; missense, n=12; nonsense, n=5). Four mutations (13%) were homozygous. Eighteen RUNX1 mutations (60%) were located within the RUNT domain (amino acids 50–177) and four (13%) were within the transactivation domain (amino acids 291–371) (Figure 2A). One patient had two CEBPA mutations leading to a total of eight CEBPA mutations (frameshift ins/del, n=5; missense, n=2; nonsense, n=1). Three mutations (38%) were homozygous and mutations were spread throughout the coding sequence (Figure 2B). A recurrent in-frame insertion of 6 bp (H196_P197dup) within the TAD2 domain of CEBPA was identified in 13 cases but has previously been identified as a polymorphism.19,20
Figure 2.
Schematic presentation of (A) RUNX1 protein and (B) CEBPA protein with locations of mutations. The majority (60%) of RUNX1 mutations were found within the RUNT domain. CEBPA mutations were spread throughout the coding sequence. ▵ Missense mutation; ▴ frameshift insertion/deletion mutation; ● nonsense mutation; TAD, transactivation domain; DNA, DNA binding domain.
Mutation analysis of other transcription factors known to be frequently mutated in AML revealed NPM1 mutations in 6/187 (3%) and WT1 mutations in 2/187 (1%) MDS/MPN patients. Taken together, mutations in transcription factors were identified in 41/187 (22%) MDS/MPN patients. Of these, only one patient had mutations in two different genes (CEBPA and WT1) indicating that transcription factor mutations are generally mutually exclusive (Table 3 and Online Supplementary Table S3).
Table 3.
Total number of patients with mutations of transcription factors.
Prognostic impact of transcription factor mutations
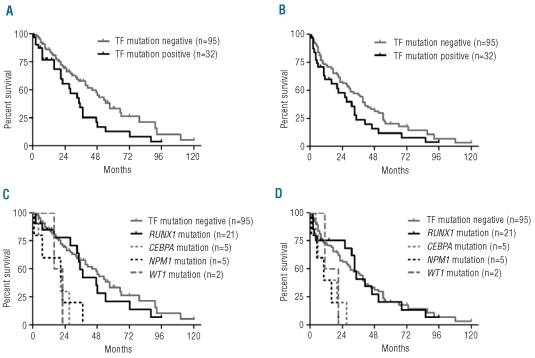
Transcription factor mutations have prognostic significance in AML and we, therefore, assessed the impact of mutational status on survival in the 127 MDS/MPN patients for whom data were available. On univariate analysis patients with mutations of transcription factors had a shorter overall survival and progression-free survival compared with mutation-negative cases (overall survival: 28 versus 44 months, P=0.019, log-rank test; progression-free survival: 21 versus 30 months, P=n.s). (Figure 3A and B). Patients with CEBPA, NPM1, or WT1 mutations had a particularly poor prognosis (P=0.0047, P=0.0024 and P=0.058, respectively, with regard to overall survival). In contrast, there was no significant difference between RUNX1-mutated cases and those without transcription factor mutations (Figure 3C and D). It should be emphasized, however, that our cohort of patients was derived from different centers, their treatment was heterogeneous, the numbers of CEBPA, WT1 and NPM1 mutated cases were small and the analysis was done retrospectively. Clearly, therefore, prospective studies that take into account all relevant prognostic variables will be necessary to confirm the prognostic significance of transcription factor mutations.
Figure 3.
Clinical significance of transcription factor (TF) mutations in MDS/MPN. Kaplan-Meier estimates of (A) overall survival and (B) progression-free survival for 127 MDS/MPN patients with or without mutations of transcription factors. Overall survival was significantly lower in mutation-positive compared to mutation-negative cases (P=0.0190, log-rank test). Kaplan-Meier estimates of (C) overall survival and (D) progression-free survival for each specific mutation. Patients with CEBPA, NPM1 or WT1 mutations collectively showed a highly significant shorter overall survival (P<0.001) and progression-free survival (P<0.001) compared to cases without transcription factor mutations. Considering each gene individually, the P values for CEBPA, NPM1 and WT1 were 0.0047, 0.0024 and 0.0583, respectively, for overall survival and 0.0096, 0.0018 and 0.174, respectively for progression-free survival.
Discussion
In this study, we performed a systematic search for cytogenetically cryptic abnormalities in patients with atypical MPN and MDS/MPN using custom designed, high-resolution targeted array CGH. Array CGH has the potential to pick up small genomic imbalances that are below the resolution of conventional cytogenetics. Targeted arrays were designed to focus on all 90 tyrosine kinases plus a range of other genes known to be involved in leukemia and myelopoiesis, e.g. known translocation genes, transcription factors as well as selected cytokines and receptors. Our aim was to detect small DNA copy number changes that might indicate the presence of novel fusion genes or other driver mutations.
After excluding all known or likely copy number variants, nine cytogenetically cryptic copy number imbalances were detected in seven patients, including hemizygous deletions of RUNX1 or CEBPA genes. A further mutation screen of 187 MDS/MPN patients revealed frequent RUNX1 and CEBPA mutations. RUNX1 (also named AML1 or CBFA2) is located on chromosome band 21q22.12 and encodes the alpha subunit of the core-binding factor (CBF) complex.21 This complex activates and represses transcription of key regulators of growth, survival and differentiation pathways. RUNX1 is one of the most frequent targets of chromosome translocations in leukemia and somatic mutations have also been identified, especially in AML M0 subtype,22,23 de novo high-risk MDS,24 and therapy-related MDS/AML.15,25 Until recently only a small number of patients with CMML had been examined for RUNX1 mutations and this gene has not been examined at all in atypical CML or other related diseases. In CMML, Harada et al. did not find RUNX1 mutations in four patients,24 nor did Preudhomme et al. in 27 patients.23 Here, we report RUNX1 mutations in 27 of 187 MDS/MPN patients (14%) with a high mutation frequency seen in CMML (19%). Three patients had two RUNX1 mutations leading to a total of 30 RUNX1 mutations. Eighteen mutations were likely causative changes predicted to result in premature chain termination and 12 were missense substitutions that have not been reported as single nucleotide polymorphisms. Similar to our results, two recent studies reported RUNX1 mutations in 9 of 30 (33%) and 30 of 81 (37%) CMML patients, respectively.26,27 In accordance with these studies, we also detected the majority (60%) of RUNX1 mutations within the N-terminal RUNT domain, which is the most conserved region of RUNX family members and is directly involved in DNA binding and interactions with CBFβ.28 Kuo et al. described a higher risk of AML progression in CMML patients with RUNX1 mutations at the C-terminal compared to the N-terminal region.27 Although we observed a similar trend in our cohort (data not shown), the number of patients with C-terminal RUNX1 mutations was small in our study (n=6) as well as in that by Kuo et al. (n=9). Overall we saw no difference in outcome between RUNX1-mutated cases and those without transcription factor mutations. In contrast, RUNX1 mutations have been shown to be associated with a poor prognosis in de novo and therapy-related MDS.15,24 A recent study on 470 adult AML patients found that RUNX1 mutations were a marker for a significantly lower complete remission rate and shorter disease-free and overall survival.29
The CEBPA gene, located on chromosome band 19q13.11, encodes the transcription factor CCAAT/enhancer-binding protein-alpha which is essential for normal differentiation of granculocytes.30 The involvement of CEBPA in leukemogenesis has been confirmed in many studies, with inactivating mutations reported predominately in AML M0, M1 and M2.16,31–34 Mutations are usually acquired but can occasionally be inherited.35 Only a small number of CMML patients have been investigated for CEBPA mutations to date. Kaeferstein et al. found no CEBPA mutations in five CMML patients,36 whereas Shih et al. reported CEBPA mutations in three of 15 CMML patients (20%).37 We identified eight different CEBPA mutations in seven of 187 MDS/MPN patients (4%). Six mutations were likely causative changes predicted to result in premature chain termination and two were missense substitutions that have not been reported as single nucleotide polymorphisms.
Following our finding of RUNX1 and CEBPA mutations in MDS/MPN we screened for mutations in two other transcription factors known to be mutated frequently in AML. We detected NPM1 mutations in six (3%) and WT1 mutations in two (1%) MDS/MPN patients. Only one patient had mutations in two different genes (CEBPA and WT1) indicating that transcription factor mutations are generally mutually exclusive in MDS/MPN. This is similar to findings from a study on AML in which only 17/312 patients (5%) had more than one mutation in hypothetical class II genes (NPM1, CEBPA, and MLL) and 12/241 patients (5%) had more than one class I mutation (FLT3-ITD, FLT3-TKD, and NRAS).38 All NPM1 mutations we found were identical and correspond to mutation type A according to Falini et al.17 Type A mutations are the most frequent NPM1 mutations in AML and the resulting shift in the reading frame alters the C-terminal portion of the NPM protein.
In a preliminary analysis we observed a shorter overall survival in patients with mutations of transcription factors compared with mutation-negative cases. This effect was accounted for principally by cases with CEBPA, NPM1, or WT1 mutations. None of these patients had a concomitant FLT3-ITD. In contrast to our results, CEBPA and NPM1 mutations (without FLT3-ITD) are linked to a favorable outcome in AML16,33,38 whereas the prognostic impact of WT1 mutations in AML is controversial.18,39,40 Three recent studies found that only CEBPA double mutations but not single mutations (as predominately observed in our study) are associated with a favorable prognosis in AML.34,41,42
We did not find any cytogenetically cryptic abnormality involving tyrosine kinases in our cohort, which included nine patients with FIP1L1-PDGFRA negative CEL/HES who responded to imatinib.43 Due to our focus on tyrosine kinases and the high resolution of our arrays, it seems unlikely that we missed any tyrosine kinase fusions, unless they were perfectly or nearly perfectly balanced. As reported elsewhere, we did find an unusual constitutional polymorphic TFG-GPR128 fusion using our targeted arrays but this was not associated with any obvious phenotype.44
Since our approach was hypothesis-driven and focused on selected genes we only covered less than 2% of human genes on our targeted arrays and thus other important abnormalities may have been missed. Human genome-wide array CGH or single nucleotide polymorphism array analysis are useful tools for finding alterations that can point to novel candidate genes and pathways and new abnormalities have been detected using these approaches. Gelsi-Boyer et al. identified novel mutations of the polycomb-associated gene ASXL1 in 17 out of 39 patients with CMML (43%).45 Delhommeau et al. found frequent TET2 mutations in about 15% of patients with various myeloid cancers including two of nine patients (22%) with CMML.46 A high incidence of TET2 mutations in CMML patients was confirmed in three other recent studies.47–49 It is likely that mutations of these genes will also be found in atypical CML and related atypical MPN.
We conclude that mutations of transcription and other nuclear factors (particularly RUNX1) are frequent in MDS/MPN patients and thereby give molecular-based support for the grouping of those diseases into the same World Health Organization category (MDS/MPN).2 In contrast, cryptic tyrosine kinase fusion genes are rare. Transcription factors are promising targets for future developments of novel targeted therapies.
Footnotes
Funding: this work was supported by a Leukaemia Research (UK) Specialist Programme Grant. TE was supported by the Dr. Mildred Scheel Stiftung für Krebsforschung (Deutsche Krebshilfe e.V., Germany). AH was supported by the German José Carreras Foundation (H 03/01).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
TE, AC and NCPC designed the study and performed experiments; KW, CH-C, JS, AJ and FG performed or assisted with the laboratory work and data analysis; KZ, AR and AH provided samples and clinical data; TE and NCPC wrote the paper; all authors contributed to the final version.
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355(23):2452–66. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 2.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 3.Orazi A, Germing U. The myelodysplastic/myeloproliferative neoplasms: myeloproliferative diseases with dysplastic features. Leukemia. 2008;22(7):1308–19. doi: 10.1038/leu.2008.119. [DOI] [PubMed] [Google Scholar]
- 4.Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106(6):2162–8. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch-Ginsberg C, LeMaistre AC, Kantarjian H, Talpaz M, Cork A, Freireich EJ, et al. RAS mutations are rare events in Philadelphia chromosome-negative/bcr gene rearrangement-negative chronic myelogenous leukemia, but are prevalent in chronic myelomonocytic leukemia. Blood. 1990;76(6):1214–9. [PubMed] [Google Scholar]
- 6.Tyner JW, Erickson H, Deininger MW, Willis SG, Eide CA, Levine RL, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009;113(8):1749–55. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunbar AJ, Gondek LP, O’Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68(24):10349–57. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113(24):6182–92. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 9.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460 (7257):904–8. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 10.De Keersmaecker K, Cools J. Chronic myeloproliferative disorders: a tyrosine kinase tale. Leukemia. 2006;20(2):200–5. doi: 10.1038/sj.leu.2404064. [DOI] [PubMed] [Google Scholar]
- 11.Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348(13):1201–14. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 12.De Keersmaecker K, Graux C, Odero MD, Mentens N, Somers R, Maertens J, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32) Blood. 2005;105(12):4849–52. doi: 10.1182/blood-2004-12-4897. [DOI] [PubMed] [Google Scholar]
- 13.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36(10):1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 14.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 15.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood. 2004;104(5):1474–81. doi: 10.1182/blood-2004-02-0754. [DOI] [PubMed] [Google Scholar]
- 16.Fröhling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22(4):624–33. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 17.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 18.Virappane P, Gale R, Hills R, Kakkas I, Summers K, Stevens J, et al. Mutation of the Wilms’ tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol. 2008;26(33):5429–35. doi: 10.1200/JCO.2008.16.0333. [DOI] [PubMed] [Google Scholar]
- 19.Wouters BJ, Louwers I, Valk PJ, Löwenberg B, Delwel R. A recurrent in-frame insertion in a CEBPA transactivation domain is a polymorphism rather than a mutation that does not affect gene expression profiling-based clustering of AML. Blood. 2007;109 (1):389–90. doi: 10.1182/blood-2006-08-042325. [DOI] [PubMed] [Google Scholar]
- 20.Biggio V, Renneville A, Nibourel O, Philippe N, Terriou L, Roumier C, et al. Recurrent in-frame insertion in C/EBPalpha TAD2 region is a polymorphism without prognostic value in AML. Leukemia. 2008;22(3):655–7. doi: 10.1038/sj.leu.2404926. [DOI] [PubMed] [Google Scholar]
- 21.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5(5):376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 22.Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93(6):1817–24. [PubMed] [Google Scholar]
- 23.Preudhomme C, Warot-Loze D, Roumier C, Grardel-Duflos N, Garand R, Lai JL, et al. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2 alpha B gene in MO acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood. 2000;96(8):2862–9. [PubMed] [Google Scholar]
- 24.Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103(6):2316–24. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 25.Harada H, Harada Y, Tanaka H, Kimura A, Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101(2):673–80. doi: 10.1182/blood-2002-04-1010. [DOI] [PubMed] [Google Scholar]
- 26.Gelsi-Boyer V, Trouplin V, Adélaïde J, Aceto N, Remy V, Pinson S, et al. Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1 genes. BMC Cancer. 2008;8:299. doi: 10.1186/1471-2407-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo MC, Liang DC, Huang CF, Shih YS, Wu JH, Lin TL, et al. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23(8):1426–31. doi: 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- 28.Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104(5):755–67. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 29.Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH, et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114(26):5352–61. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 30.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27(4):619–28. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19(3):329–34. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 32.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27(3):263–70. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 33.Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26(31):5078–87. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088–91. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351(23):2403–7. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 36.Kaeferstein A, Krug U, Tiesmeier J, Aivado M, Faulhaber M, Stadler M, et al. The emergence of a C/EBPalpha mutation in the clonal evolution of MDS towards secondary AML. Leukemia. 2003;17(2):343–9. doi: 10.1038/sj.leu.2402805. [DOI] [PubMed] [Google Scholar]
- 37.Shih LY, Huang CF, Lin TL, Wu JH, Wang PN, Dunn P, et al. Heterogeneous patterns of CEBPalpha mutation status in the progression of myelodysplastic syndrome and chronic myelomonocytic leukemia to acute myelogenous leukemia. Clin Cancer Res. 2005;11(5):1821–6. doi: 10.1158/1078-0432.CCR-04-1932. [DOI] [PubMed] [Google Scholar]
- 38.Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 39.Gaidzik VI, Schlenk RF, Moschny S, Becker A, Bullinger L, Corbacioglu A, et al. Prognostic impact of WT1 mutations in cytogenetically normal acute myeloid leukemia: a study of the German-Austrian AML Study Group. Blood. 2009;113(19):4505–11. doi: 10.1182/blood-2008-10-183392. [DOI] [PubMed] [Google Scholar]
- 40.Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrózek K, Maharry K, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26(28):4595–602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabst T, Eyholzer M, Fos J, Mueller BU. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100(8):1343–6. doi: 10.1038/sj.bjc.6604977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou HA, Lin LI, Chen CY, Tien HF. Reply to ‘Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favorable prognosis’. Br J Cancer. 2009;101(4):738–40. doi: 10.1038/sj.bjc.6605207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metzgeroth G, Walz C, Erben P, Popp H, Schmitt-Graeff A, Haferlach C, et al. Safety and efficacy of imatinib in chronic eosinophilic leukaemia and hypereosinophilic syndrome: a phase-II study. Br J Haematol. 2008;143(5):707–15. doi: 10.1111/j.1365-2141.2008.07294.x. [DOI] [PubMed] [Google Scholar]
- 44.Chase A, Ernst T, Fiebig A, Collins A, Grand F, Erben P, et al. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica. 2010;95(1):20–6. doi: 10.3324/haematol.2009.011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelsi-Boyer V, Trouplin V, Adélaïde J, Bonansea J, Cervera N, Carbuccia N, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 46.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 47.Tefferi A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Patnaik MM, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23(7):1343–5. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403–10. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]