Abstract
Background
The existence of multiple subsets of chronic lymphocytic leukemia expressing ‘stereotyped’ B-cell receptors implies the involvement of antigen(s) in leukemogenesis. Studies also indicate that ‘stereotypy’ may influence the clinical course of patients with chronic lymphocytic leukemia, for example, in subsets with stereotyped IGHV3-21 and IGHV4-34 B-cell receptors; however, little is known regarding the genomic profile of patients in these subsets.
Design and Methods
We applied 250K single nucleotide polymorphism-arrays to study copy-number aberrations and copy-number neutral loss-of-heterozygosity in patients with stereotyped IGHV3-21 (subset #2, n=29), stereotyped IGHV4-34 (subset #4, n=17; subset #16, n=8) and non-subset #2 IGHV3-21 (n=13) and non-subset #4/16 IGHV4-34 (n=34) patients.
Results
Over 90% of patients in subset #2 and non-subset #2 carried copy-number aberrations, whereas 75–76% of patients in subset #4 and subset #16 showed copy-number aberrations. Subset #2 and non-subset #2 patients also displayed a higher average number of aberrations compared to patients in subset #4. Deletion of 13q was the only known recurrent aberration detected in subset #4 (35%); this aberration was even more frequent in subset #2 (79%). del(11q) was more frequent in subset #2 and non-subset #2 (31% and 23%) patients than in subset #4 and non-subset #4/16 patients. Recurrent copy-number neutral loss-of-heterozygosity was mainly detected on chromosome 13q, independently of B-cell receptor stereotypy.
Conclusions
Genomic aberrations were more common in subset #2 and non-subset #2 than in subset #4. The particularly high frequency of del(11q) in subset #2 may be linked to the adverse outcome reported for patients in this subset. Conversely, the lower prevalence of copy-number aberrations and the absence of poor-prognostic aberrations in subset #4 may reflect an inherently low-proliferative disease, which would prevent accumulation of genomic alterations.
Keywords: chronic lymphocytic leukemia, stereotyped B-cell receptors, antigens, leukemogenesis
Introduction
In chronic lymphocytic leukemia (CLL), the mutation status of the immunoglobulin heavy chain variable (IGHV) genes has emerged as one of the strongest prognostic markers, since it divides patients into two clinical subgroups, those with IGHV-mutated and those with IGHV-unmutated CLL, with different prognoses.1,2 Furthermore, CLL displays a remarkably biased IGHV repertoire with over-representation of a limited number of genes, such as IGHV1-69, IGHV4-34, IGHV3-23 and IGHV3-21.2–4 Several groups have reported multiple CLL subsets with almost identical, ‘stereotyped’ B-cell receptors (BCR) in up to about 30% of patients.3,5–11 These subsets are defined by certain criteria such as usage of similar IGHV-D-J genes and light-chain genes and an amino acid identity of 60% or more in the heavy-chain complementarity determining region 3 (CDR3), which is the main determinant of antigen specificity.6,8–10 Considering the very low probability of finding two B-cell clones with almost identical BCR by chance alone, these findings have supported the notion that the development of CLL is not stochastic and suggests a potential role for antigens in leukemogenesis through the recognition of similar epitopes within each subset.
So far, more than 100 different subsets have been defined with stereotyped BCR, with some of the subsets being more frequent than others.9,10 Interestingly, CLL subsets expressing a certain stereotyped BCR have also been indicated to share biological and clinical features.9,12 For instance, IGHV4-34/IGKV2-30 patients with stereotyped BCR (20 amino acids long, known as subset #4) have an indolent disease course compared to those with non-stereotyped IGHV4-34.9 Furthermore, subset #4 patients have a low median age at diagnosis, IgG-switched BCR6,9,13 and a potential association with persistent infection by common herpesviruses.14 Another IGHV4-34 subset, sub-set #16 (IGHV4-34/IGKV3-20, with a CDR3 that is 24 amino acids long), has been identified, although less is known about the clinical outcome for these patients. Approximately half of the patients with IGHV3-21 CLL display a stereotyped BCR (known as subset #2) with a short and highly similar CDR3 (9 amino acids long) and usage of one particular IG lambda gene, IGLV3-21.5,12,15 However, it appears that IGHV3-21 patients with stereotyped and non-stereotyped BCR share an equally poor overall survival, independently of IGHV mutational status5,15,16 although stereotypy has been associated with a shorter time to progression and the presence of other markers of a poor prognosis.9,17
No single genetic event has been found in all cases of CLL, although certain recurrent aberrations (e.g. deletions of 13q, 11q, 17p and trisomy 12) are frequently detected and can assist outcome prediction.18 Patients carrying del(13q) as the sole abnormality have a favorable prognosis,19 whereas patients with del(11q) (ATM) and del(17p) (TP53) have an inferior outcome.18 Until now, few studies have investigated genomic aberrations specifically in relation to CLL subsets with stereotyped BCR. In one study, fluorescence in situ hybridization (FISH) analysis indicated a higher frequency of del(11q) in IGHV3-21 CLL15 and, in another study, del(13q) was detected more frequently in IGHV4-34 subset #4 patients than in non-subset patients, who carried more heterogeneous aberrations.20
Since genomic profiling of stereotyped subsets might offer clues about the underlying leukemic processes, we here applied 250K single nucleotide polymorphism (SNP)-arrays to investigate the presence of whole-genome copy-number aberrations and copy-number neutral loss of heterozygosity (CNN-LOH) in CLL patients with stereotyped and non-stereotyped IGHV3-21 and IGHV4-34 BCR. These relatively common subsets were chosen for analysis as they display distinctive as well as divergent clinical and biological features.
Design and Methods
Groups of patients
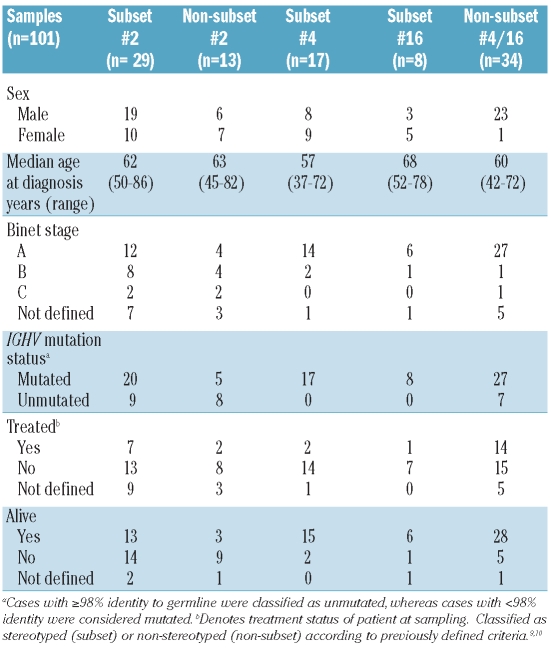
In total, 101 CLL patients from collaborating institutes in France (n=15), Greece (n=25), Denmark (n=4) and Sweden (n=57) were studied. Most samples were derived from peripheral blood (n=88), but samples from bone marrow (n=8) and spleen (n=5) were also included. All samples were diagnosed according to recently revised criteria for CLL,21 and showed a typical immunophenotype and 70% or more tumor cells. Clinical data for all patients are summarized in Table 1. Subsets were defined according to Stamatopoulos et al. and Murray et al.9,10 Twenty-nine samples were classified as subset #2 (IGHV3-21/IGLV3-21 usage, CDR3 of 9 amino acids long) and 13 samples as non-subset #2 (IGHV3-21 usage, heterogeneous CDR3 lengths). Seventeen samples were defined as subset #4 (IGHV4-34/IGKV2-30 usage, CDR3 of 20 amino acids long, IgG-switched), 8 samples as subset #16 (IGHV4-34/IGKV3-20 usage, CDR3 of 24 amino acids long) and 34 as non-subset #4/16 (IGHV4-34 usage, heterogeneous CDR3 lengths).
Table 1.
Clinical data for IGHV3-21 and IGHV4-34 chronic lymphocytic leukemia subgroups included in this study.
Single nucleotide polymorphism-array analysis
SNP-array experiments were performed according to the standard protocols for Affymetrix GeneChip® Mapping NspI-250K arrays (Gene Chip Mapping 500K Assay Manual (P/N 701930 Rev2.), Affymetrix Inc., Santa Clara, CA, USA) and the arrays were scanned using the GeneChip® Scanner 3000 7G. Genotype calling and probe level normalization were performed in the Affymetrix GeneChip® Genotyping Analysis Software (GTYPE) 4.1 using the Dynamic Model (DM) algorithm and BRLMM.22 The quality control specifies a neighbor score that represents the average of the Euclidean distances between the log2ratio of flanking SNP along the chromosomes. Low neighbor scores indicate a low noise level: a neighbor score of 0.4 or less was, therefore, applied as the cut-off for inclusion of samples. In order to produce log2ratios, copy-number normalization was performed using the Copy Number Analysis Tool (CNAT) 4.0.1. Normal samples (n=82) analyzed at the Uppsala Array Platform were used as a reference set.
Analysis of Affymetrix data for copy-number alterations
Copy-number analysis using the rank segmentation algorithm and group comparisons were performed employing BioDiscovery Nexus Copy Number 3.0 software (BioDiscovery, El Segundo CA, USA). Copy-number analysis was performed using a significance threshold (P value) of 1×10−6 and a log2ratio cut-off at ±0.2 for regions sized 200–500 kbp and ±0.15 for regions longer than 500 kbp. These settings were defined through validation experiments in a previously described sample-set including 203 CLL samples.23 Copy-number variations are genomic variants that exist in the human population and these polymorphic regions can be detected as gains and losses.24 To exclude such polymorphic regions, we compared the regions detected in the copy-number analysis to frequent regions of copy-number variations reported by Redon et al.25 and MacCarroll et al.26 Regions that overlapped by less than 50% with copy-number variations were considered as true copy-number aberrations and included in the final analysis, as previously described.23
Copy-number neutral loss-of-heterozygosity analysis
Analysis of CNN-LOH in CLL cells was performed using SNP array-data, taking into account the fraction of normal cells obtained from the flow-cytometry data, as previously described.27 CNN-LOH regions were visualized by mapping them to 200 kb segments beginning at the chromosome start site. Regions containing CNN-LOH larger than 3 Mbp with less than 50% overlap with copy-number variations were considered for further analysis.
Statistical analysis
Statistical analysis was performed using the Statistica Software 8.0 (Stat Soft Inc., Tulsa, OK, USA). A χ2 test was applied to determine any statistically significant differences in the frequency of the known recurrent aberrations between the studied groups (subsets/non-subsets and our general Swedish CLL cohort). Similarly, the one-way ANOVA test and t-test were applied to determine any statistically significant differences in the number and size of copy-number aberrations between groups of patients.
Results
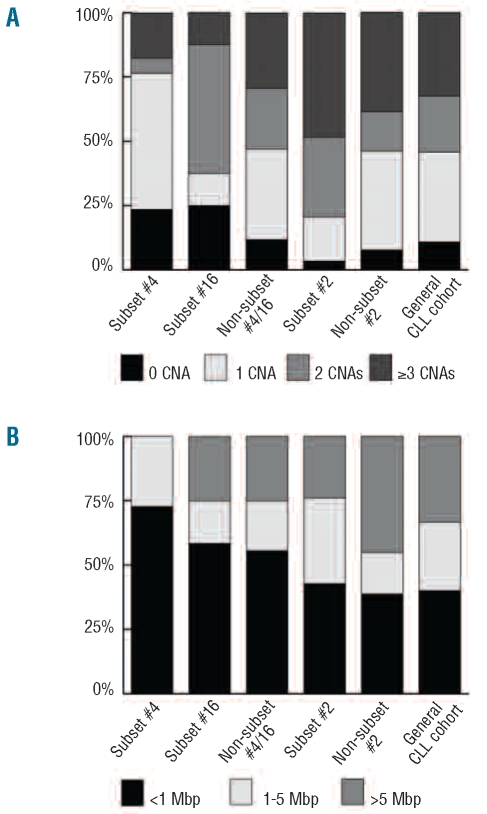
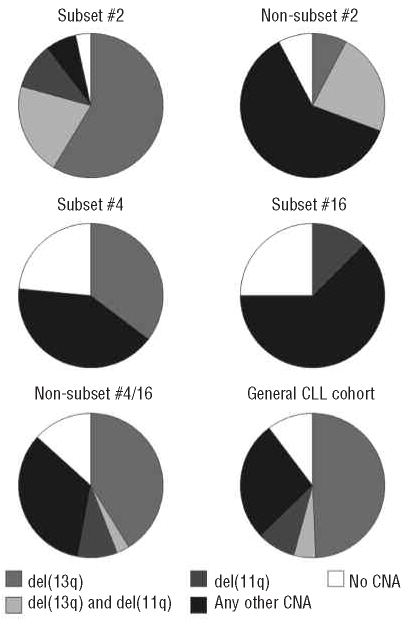
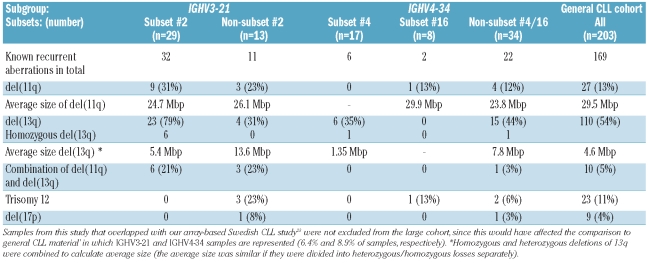
We investigated stereotyped and non-stereotyped IGHV3-21 and IGHV4-34 CLL samples using high-density 250K SNP arrays, with particular interest in detecting genomic alterations in these specific subsets. Figure 1 shows the distributions of the number and size of copy-number aberrations in the various subsets, while details concerning the known recurrent aberrations found in these subgroups are provided in Figure 2 and Table 2.
Figure 1.
Level of genomic complexity and size distribution of copy-number aberrations. The columns represent IGHV4-34 samples; subset #4 (n=17), subset #16 (n=8) and non-subset #4/16 (n=34); IGHV3-21 samples; subset #2 (n=29) and non-subset #2 (n=13) and a ‘general’ CLL cohort (n=203) from the study by Gunnarsson et al.23 Samples from the current study that overlapped with our recent array-based Swedish CLL cohort study23 were not removed from analysis, as the overlap between studies was limited. (A) Level of genomic complexity in each group is based on the frequency of samples that carry a certain number of CNAs (0, 1, 2 or ≥3). (B) The size distribution of copy-number aberrations (<1 Mbp, 1–5 Mbp or >5 Mbp) is shown as the frequency of the total number of copy-number aberrations. Note that subset #4 samples do not carry copy-number aberrations greater than 5 Mbp, whereas the non-subset #2 samples show the highest frequency of copy-number aberrations greater than 5 Mbp.
Figure 2.
Frequency of patients carrying deletions of 11q and 13q individually or in combination. Each pie represents IGHV3-21 samples in subset #2 (n=29) and non-subset #2 (n=13), IGHV4-34 samples in subset #4 (n=17), subset #16 (n=8) and non-subset #4/16 (n=34), and a ‘general’ CLL cohort (n=203) from the study by Gunnarsson et al.23 Samples that carry ‘Any CNA' also include del(17p) and trisomy 12, which were infrequently detected in this study, as indicated in Table 2.
Table 2.
Known recurrent alterations in IGHV3-21 and IGHV4-34 subsets compared to in a general chronic lymphocytic leukemia cohort.
IGHV3-21 chronic lymphocytic leukemia - subset #2 versus non-subset #2
Our copy-number analysis showed that 97% (28/29) of subset #2 and 92% (12/13) of non-subset #2 IGHV3-21 samples carried copy-number aberrations (Figure 1A). Both subgroups displayed a similar average of copy-number aberrations/sample (2.6 vs. 2.3 copy-number aberrations/sample), although non-subset #2 patients carried larger copy-number aberrations compared to subset #2 (median size, 1.8 Mbp versus 1.39 Mbp, P=0.001) (Figure 1B, Online Supplementary Table S1). The known recurrent aberrations were detected in 90% of subset #2 and 54% of non-subset #2 patients (Table 2). The majority of subset #2 samples carried del(13q) (79%), while a smaller proportion of the non-subset #2 samples (31%) displayed this aberration (P=0.0024). A particularly high frequency of del(11q) was detected in subset #2 patients (9/29 samples, 31%) compared with our previous Swedish CLL array-based report (13%) (P=0.013) (Figure 2).23 Also, a relatively high frequency of del(11q) was found in non-subset #2 cases (23%). Furthermore, of the samples with del(11q), 75% (9/12) also carried del(13q) (Figure 2–3). All 11q deletions were shown to cover the ATM gene. del(17p), covering the TP53 gene, and trisomy 12 were only detected in non-subset #2 patients.
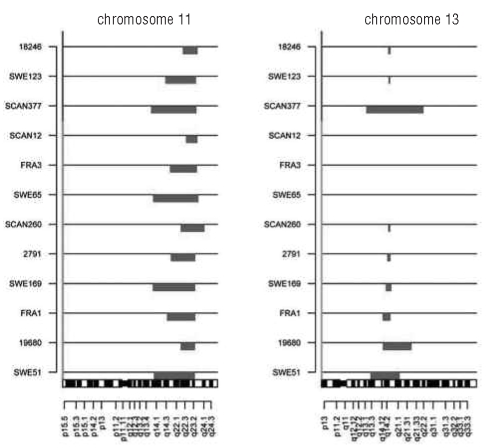
Figure 3.
A high frequency of patients with IGHV3-21 with del(11q) have a concomitant del(13q). All 12 IGHV3-21 patients who carried del(11q) are displayed. The upper three are non-subset #2 patients and the remaining belong to subset #2.
When excluding the known recurrent aberrations, losses were shown to be more common than gains in both subset #2 and non-subset #2 (Online Supplementary Table S2). When considering alterations less than or greater than 1 Mbp in size, a similar average of copy-number aberrations/sample was observed within subset #2 and non-subset #2 cases (Online Supplementary Table S1). Only two other recurrent alterations were identified on chromosomes 2q and 3p. Specifically, two subset #2 samples (6.9%) displayed a gain at 2q32.1 with an average size of 0.21 Mbp. These samples also demonstrated concurrent deletions of 11q and 13q. Furthermore, two subset #2 samples with 13q deletion demonstrated a loss at 3p21.31–3p21.1 with an average size of 2.3 Mbp. Here, one non-subset #2 patient carried del(17p), trisomy 12, a loss at 7q11.21 (0.7 Mbp) and a large novel loss at 16q11.2–16q21 (13.4 Mbp). In addition, large (>20 Mbp) non-recurring aberrations were identified at 4q, 4p, 5q, 14q and 21q, predominantly within non-subset #2 cases (Online Supplementary Table S3).
IGHV4-34 chronic lymphocytic leukemia - subset #4 versus subset 16 versus non-subset #4/16
Assessment of copy-number aberrations showed that 76% of subset #4 (13/17), 75% of subset #16 (6/8) and 88% of non-subset #4/16 (30/34) IGHV4-34 patients carried such aberrations. The average number of copy-number aberrations/sample for non-subset #4/16, subset #16 and subset #4 cases were 2.1, 1.5 and 1.3, respectively (Online Supplementary Table S1). Subset #16 and non-subset #4/16 carried larger aberrations than subset #4 samples (Figure 1B), although the median size was not statistically different (0.64 Mbp, 0.95 Mbp, 0.74 Mbp, respectively, P=0.24). The known recurrent aberrations were identified in 35% of subset #4, 25% of subset #16 and 59% of non-subset #4/16 cases (Figure 2 and Table 2). The exclusive recurrent aberration in subset #4 cases was deletion of 13q (6/6 cases with known recurrent aberrations). Deletion of 13q was also detected in 44% (15/34) of non-subset #4/16 cases, while this aberration was absent in subset #16 cases. Deletion of 11q and trisomy 12 were only detected in subset #16 and non-subset #4/16 samples, whereas del(17p) was only observed in non-subset #4/16 samples (Table 2).
Upon removal of known recurrent aberrations from the analysis, losses were more frequent than gains in subset #4, and non-subset #4/16, but not in subset #16 (Online Supplementary Table S2). Furthermore, on examination of alterations greater or less than 1 Mbp, subset #4, subset #16 and non-subset #4/16 patients showed a similar average number of copy-number aberrations/sample and a comparable median length of the aberrations (Online Supplementary Table S1). Among the IGHV4-34 cases three recurrent aberrations were revealed. A deletion on 2q37.3 (5 Mbp), covering the telomeric region, was detected in two subset #4 and two non-subset #4/16 samples. Two samples in the non-subset #4/16 group showed a gain on 7q34 covering 0.3 Mbp, while one subset #4 and one non-subset #4/16 case carried an overlapping gain of 14q23.3 with an average size of 0.6 Mbp. Finally, non-recurring, larger aberrations (>20 Mbp) including trisomy 3, losses of 6q, 14q and gains of 8q, 13q, 22q were only detected in non-subset #4/16 (Online Supplementary Table S3).
Copy-number neutral loss-of-heterozygosity in IGHV3-21 and IGHV4-34 chronic lymphocytic leukemia
The evaluation of CNN-LOH, i.e. allelic imbalances without a change in copy-number, revealed that most regions were non-recurrent between samples. For instance, when regions greater than 3 Mp were evaluated, ten non-overlapping regions were shown in subset #2/non-subset #2 IGHV3-21 samples, whereas 16 individual CNN-LOH regions were detected in subset #4 and non-subset #4/16 IGHV4-34 samples. However, a large recurrent CNN-LOH was detected on chromosome 13q in two subset #2, one subset #4 and two non-subset #4/16 samples, all of which carried a homozygous loss of 13q. Moreover, partially overlapping regions were identified on chromosome 20q11.21-q11.23 (overlapping region: 3.6 Mbp) in one subset #2 and one non-subset #2 IGHV3-21 sample. Among the IGHV4-34 patients, two non-subset #4/16 samples had an overlapping CNN-LOH of 3.2 Mbp on chromosome 6p22.1.
Discussion
Recently, several groups have reported multiple CLL subsets carrying closely homologous BCR with similar heavy and light-chain CDR3 sequences.6–10 These stereotyped BCR suggest a potential role for antigens in leukemogenesis, a hypothesis also supported by recent studies detailing the BCR specificity in CLL.28–32 Interestingly, patients in CLL subsets expressing a certain stereotyped BCR have been indicated to share biological and clinical features, for instance IGHV3-21 subset #2 patients and IGHV4-34 subset #4 patients.9,12 Despite this, limited knowledge exists about the spectrum of genomic aberrations that occur in these different subsets, although such information might give hints of important genetic events occurring during disease development and/or evolution. To gain knowledge on this issue, we used a high-density SNP array to screen for whole-genome copy-number aberrations and CNN-LOH events in stereotyped subset #2 and subset #4/#16 cases and compared them to their non-stereotyped counterparts.
Focusing on IGHV3-21 patients, our analysis revealed that a high frequency of subset #2 and non-subset #2 samples carried copy-number aberrations. In fact, both subset #2 and non-subset #2 displayed a similar average number of copy-number aberrations/sample, which was comparable to the level observed in our recent array-based study on Swedish CLL.23 Hence, when accounting for all genomic alterations, IGHV3-21 subset #2 and non-subset #2 cases do not appear to be more complex than other cases of CLL. That notwithstanding, differences were noted in the frequency of the known recurrent alterations, such as a high frequency of del(11q) among subset #2 patients (Figure 2). In fact, the frequency of del(11q) was higher in subset #2 than in either our Swedish CLL study or other previously reported FISH studies in CLL,15,18,23 although a relatively high frequency was also observed in non-subset #2 patients.
Both ATM and TP53 are important for maintaining genomic stability, with the former acting as a positive regulator of TP53. To assess the possibility that patients carrying a deletion of ATM also have TP53 mutations, which may contribute to an adverse prognosis in CLL, we sequenced all IGHV3-21 patients with del(11q) in search of TP53 mutations (exons 4–8). However, none of the patients displayed any TP53 mutation, which suggests that the deletion of ATM is sufficient to contribute to the adverse outcome observed in IGHV3-21-expressing CLL, particularly in subset #2 patients. Furthermore, the frequency of del(13q) was considerably higher in subset #2 than in non-subset #2 cases or in any other CLL material reported to date.18,23 These aberrations may represent important genetic events during the pathogenesis of IGHV3-21 CLL, particularly when considering the frequent finding of concurrent 11q and 13q deletions, especially in subset #2 cases (Figures 2 and 3). Hypothetically, these aberrations may be acquired during the phase of active stimulation by (unknown) antigens, which trigger the IGHV3-21 precursor cell to undergo rapid cell division and hence make it susceptible to clonal alterations.
When investigating subset #4 IGHV4-34 patients, a group known to have a favorable prognosis with few patients needing treatment, we observed a significantly lower average number of aberrations compared to that in non-subset #4/16, subset #2, non-subset #2 and patients in our recent array-based study (P=0.0067, Online Supplementary Table S1).23 This difference was due to the fact that subset #4 carried a higher number of samples with no genomic aberrations compared to subset #2 and non-subset #2 samples (17% versus 5%), and that a larger proportion of subset#2/non-subset #2 patients displayed a higher number of copy-number aberrations per sample (Figure 1). Furthermore, the frequency of samples carrying the known recurrent alterations was considerably lower in subset #4 (Table 2). Indeed, the only known recurrent aberration identified in subset #4 was the favorable prognostic marker del(13q) (Figure 2), albeit detected at a lower frequency (35%) compared to a recent report, in which FISH analysis detected del(13q) in 62% of subset #4 cases.20 However, this apparent discrepancy may be attributed to the fact that many of the subset #4 samples included in the aforementioned study showed a low percentage of CLL cells (7–15%) carrying del(13q) by applying FISH (K. Stamatopoulos, unpublished data), which would have remained undetected by SNP-array.
What could be the reasons why subset #4 cases acquire fewer genomic aberrations than subset #2 cases? We speculate that one important aspect could be the type of antigen stimulation that subset #4 CLL cells experience. Interestingly, IGHV4–34 antibodies are inherently autore-active due to their recognition of the carbohydrate epitopes on the surface of red blood cells.33–35 Sequence alterations by somatic hypermutation may be a means to eliminate or abrogate this autoreactivity, and thus, render IGHV4–34-expressing cells sufficiently safe to enter the peripheral repertoire.33,36 In analogy to this observation, it is perhaps relevant to note that (i) IGHV4-34-expressing CLL is most often mutated,10 and (ii) IGHV-mutated CLL cells have been shown to be less responsive than their unmutated counterparts upon IgM-crosslinking.37 Hence, subset #4 CLL cells may represent a population of cells which are relatively anergic upon BCR crosslinking.13 Additionally, we have recently demonstrated that subset #4 CLL cells show extensive intraclonal diversification, possibly due to ongoing antigenic stimulation.14,38 Following these lines of evidence, it seems plausible that continuous (super)antigenic interaction may be necessary for IGHV4-34 CLL clones to be maintained, yet the low-proliferative “anergic” state cannot be overcome, thereby preventing the accumulation of additional genomic alterations. This is supported by our findings that subset #4 cases in general showed few copy-number aberrations, carried only the good prognostic marker del(13q), and followed a very indolent disease course.
Although our high density SNP-arrays could detect some novel aberrations, few of these were overlapping in subset and non-subset groups. For example, a concurrent gain on chromosome 2q and a loss at 3p were identified in two subset #2 cases. In IGHV4-34 CLL, three recurrent aberrations were observed on chromosomes 2q, 7q and 14q but not in any particular subset. Furthermore, evaluation of CNN-LOH events showed that small non-recurring regions were frequent in all subset and non-subset groups. When focusing on larger regions (>3 Mbp), overlapping CNN-LOH events were detected on chromosome 6 in two non-subset #4/16 patients and on chromosome 20 in one subset #2 and non-subset #2 case. As described in previous CLL studies, we identified five cases with CNN-LOH on chromosome 13q.23,39,40 This recurrent event was detected in both IGHV3-21 and IGHV4-34 CLL with no bias towards any subset/non-subset group. Since CNN-LOH on 13q always harbored a homozygous del(13q), this LOH might be of specific importance in CLL biology although not specifically in relation to stereotypy.
In conclusion, high-density screening for genomic aberrations revealed differences in the genomic spectra between subset #4 and subset #2/non-subset #2. As the frequencies of del(13q) and del(11q) were particularly high in subset #2, these deletions probably represent important genetic events during the development of IGHV3-21 CLL. In contrast, subset #4 patients exclusively showed del(13q) and had a lower overall incidence of genomic aberrations. These features suggest that genomic aberrations may not play such a significant role in the pathogenesis of subset #4 CLL cases, which is reflected by their indolent disease course.
Footnotes
Funding: this work was supported by the Swedish Cancer Society, the Swedish Research Council, the Medical Faculty of Uppsala University, Uppsala University Hospital, and Lion’s Cancer Research Foundation in Uppsala, Sweden. ABBEST, Ireland, provided a stipend for NC.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
MiM, NC and RG performed research, analyzed data and wrote the paper. AI, HG and MR performed bioinformatic analyses. MJ and MaM performed laboratory work and analyzed data. FR supervised the research. KK, HOA, JJ, and FD provided samples and associated data. GJ, KS and RR supervised the research and wrote the paper.
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. [PubMed] [Google Scholar]
- 2.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–54. [PubMed] [Google Scholar]
- 3.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102(8):1515–25. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tobin G, Thunberg U, Johnson A, Thorn I, Soderberg O, Hultdin M, et al. Somatically mutated Ig V(H)3–21 genes characterize a new subset of chronic lymphocytic leukemia. Blood. 2002;99(6):2262–4. doi: 10.1182/blood.v99.6.2262. [DOI] [PubMed] [Google Scholar]
- 5.Tobin G, Thunberg U, Johnson A, Eriksson I, Soderberg O, Karlsson K, et al. Chronic lymphocytic leukemias utilizing the VH3–21 gene display highly restricted Vlambda2–14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood. 2003;101(12):4952–7. doi: 10.1182/blood-2002-11-3485. [DOI] [PubMed] [Google Scholar]
- 6.Messmer BT, Albesiano E, Efremov DG, Ghiotto F, Allen SL, Kolitz J, et al. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200(4):519–25. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Widhopf GF, 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 2004;104(8):2499–504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 8.Tobin G, Thunberg U, Karlsson K, Murray F, Laurell A, Willander K, et al. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104(9):2879–85. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
- 9.Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood. 2007;109(1):259–70. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 10.Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111(3):1524–33. doi: 10.1182/blood-2007-07-099564. [DOI] [PubMed] [Google Scholar]
- 11.Darzentas N, Hadzidimitriou A, Murray F, Hatzi K, Josefsson P, Laoutaris N, et al. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia. 2010;24(1):125–32. doi: 10.1038/leu.2009.186. [DOI] [PubMed] [Google Scholar]
- 12.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stella S, Guida G, et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3–21 gene. Blood. 2005;105(4):1678–85. doi: 10.1182/blood-2004-07-2606. [DOI] [PubMed] [Google Scholar]
- 13.Potter KN, Mockridge CI, Neville L, Wheatley I, Schenk M, Orchard J, et al. Structural and functional features of the B-cell receptor in IgG-positive chronic lymphocytic leukemia. Clin Cancer Res. 2006;12(6):1672–9. doi: 10.1158/1078-0432.CCR-05-2164. [DOI] [PubMed] [Google Scholar]
- 14.Kostareli E, Hadzidimitriou A, Stavroyianni N, Darzentas N, Athanasiadou A, Gounari M, et al. Molecular evidence for EBV and CMV persistence in a subset of patients with chronic lymphocytic leukemia expressing stereotyped IGHV4–34 B-cell receptors. Leukemia. 2009;23(5):919–24. doi: 10.1038/leu.2008.379. [DOI] [PubMed] [Google Scholar]
- 15.Thorselius M, Krober A, Murray F, Thunberg U, Tobin G, Buhler A, et al. Strikingly homologous immunoglobulin gene rearrangements and poor outcome in VH3–21-using chronic lymphocytic leukemia patients independent of geographic origin and mutational status. Blood. 2006;107(7):2889–94. doi: 10.1182/blood-2005-06-2227. [DOI] [PubMed] [Google Scholar]
- 16.Ghia EM, Jain S, Widhopf GF, 2nd, Rassenti LZ, Keating MJ, Wierda WG, et al. Use of IGHV3–21 in chronic lymphocytic leukemia is associated with high-risk disease and reflects antigen-driven, post-germinal center leukemogenic selection. Blood. 2008;111(10):5101–8. doi: 10.1182/blood-2007-12-130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bomben R, Dal Bo M, Capello D, Forconi F, Maffei R, Laurenti L, et al. Molecular and clinical features of chronic lymphocytic leukaemia with stereotyped B cell receptors: results from an Italian multicentre study. Br J Haematol. 2009;144(4):492–506. doi: 10.1111/j.1365-2141.2008.07469.x. [DOI] [PubMed] [Google Scholar]
- 18.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 19.Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720–4. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 20.Athanasiadou A, Stamatopoulos K, Gaitatzi M, Stavroyianni N, Fassas A, Anagnostopoulos A. Recurrent cytogenetic findings in subsets of patients with chronic lymphocytic leukemia expressing IgG-switched stereotyped immunoglobulins. Haematologica. 2008;93(3):473–4. doi: 10.3324/haematol.11872. [DOI] [PubMed] [Google Scholar]
- 21.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22(1):7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 23.Gunnarsson R, Isaksson A, Mansouri M, Goransson H, Jansson M, Cahill N, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2010;24(1):211–5. doi: 10.1038/leu.2009.187. [DOI] [PubMed] [Google Scholar]
- 24.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 25.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40(10):1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 27.Goransson H, Edlund K, Rydaker M, Rasmussen M, Winquist J, Ekman S, et al. Quantification of normal cell fraction and copy number neutral LOH in clinical lung cancer samples using SNP array data. PLoS One. 2009;4(6):e6057. doi: 10.1371/journal.pone.0006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different anti-body reactivity. J Clin Invest. 2005;115(6):1636–43. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiorazzi N, Hatzi K, Albesiano E. B-cell chronic lymphocytic leukemia, a clonal disease of B lymphocytes with receptors that vary in specificity for (auto)antigens. Ann N Y Acad Sci. 2005;1062:1–12. doi: 10.1196/annals.1358.002. [DOI] [PubMed] [Google Scholar]
- 30.Lanemo Myhrinder A, Hellqvist E, Sidorova E, Soderberg A, Baxendale H, Dahle C, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111(7):3838–48. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- 31.Catera R, Silverman GJ, Hatzi K, Seiler T, Didier S, Zhang L, et al. Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol Med. 2008;14(11–12):665–74. doi: 10.2119/2008-00102.Catera. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu CC, Catera R, Hatzi K, Yan XJ, Zhang L, Wang XB, et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize non-muscle myosin heavy chain IIA. Blood. 2008;112(13):5122–9. doi: 10.1182/blood-2008-06-162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat NM, Bieber MM, Chapman CJ, Stevenson FK, Teng NN. Human antilipid A monoclonal antibodies bind to human B cells and the i antigen on cord red blood cells. J Immunol. 1993;151(9):5011–21. [PubMed] [Google Scholar]
- 34.Spellerberg M, Chapman C, Hamblin T, Stevenson F. Dual recognition of lipid A and DNA by human antibodies encoded by the VH4–21 gene. A possible link between infection and lupus. Ann NY Acad Sci. 1995;764:427–32. doi: 10.1111/j.1749-6632.1995.tb55858.x. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Spellerberg MB, Stevenson FK, Capra JD, Potter KN. The I binding specificity of human VH 4–34 (VH 4–21) encoded antibodies is determined by both VH framework region 1 and complementarity determining region 3. J Mol Biol. 1996;256(3):577–89. doi: 10.1006/jmbi.1996.0110. [DOI] [PubMed] [Google Scholar]
- 36.Pugh-Bernard AE, Silverman GJ, Cappione AJ, Villano ME, Ryan DH, Insel RA, et al. Regulation of inherently autoreactive VH4–34 B cells in the maintenance of human B cell tolerance. J Clin Invest. 2001;108(7):1061–70. doi: 10.1172/JCI12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mockridge CI, Potter KN, Wheatley I, Neville LA, Packham G, Stevenson FK. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood. 2007;109(10):4424–31. doi: 10.1182/blood-2006-11-056648. [DOI] [PubMed] [Google Scholar]
- 38.Sutton LA, Kostareli E, Hadzidimitriou A, Darzentas N, Tsaftaris A, Anagnostopoulos A, et al. Extensive intraclonal diversification in a subgroup of chronic lymphocytic leukemia patients with stereotyped IGHV4–34 receptors: implications for ongoing interactions with antigen. Blood. 2009;114(20):4460–8. doi: 10.1182/blood-2009-05-221309. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, et al. Genomewide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109(3):1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, et al. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008;112(6):1296–305. doi: 10.1002/cncr.23270. [DOI] [PubMed] [Google Scholar]