Summary
Tamoxifen is a standard endocrine therapy for the prevention and treatment of steroid hormone receptor-positive breast cancer. Tamoxifen requires enzymatic activation by CYP 450 enzymes for the formation of clinically relevant metabolites, 4-OH-tamoxifen and endoxifen, which both have a greater affinity to the estrogen receptor and ability to inhibit cell proliferation when compared to the parent drug. CYP2D6 is the key enzyme in this biotransformation, and recent mechanistic, pharmacologic, and clinical pharmacogenetic evidence suggests that genetic variants and drug interaction by CYP2D6 inhibitors influence plasma concentrations of active tamoxifen metabolites and outcome of patients treated with adjuvant tamoxifen. Particularly, non-functional (poor metabolizer) and severely impaired (intermediate metabolizer) CYP2D6 variants are associated with higher recurrence rates. Accordingly, CYP2D6 genotyping prior to treatment for prediction of metabolizer status and outcome may open new avenues for the individualization of endocrine treatment choice and benefit. Moreover, strong CYP2D6 inhibitors such as the selective serotonin reuptake inhibitor paroxetine should be avoided as co-medication.
Key Words: Breast cancer, Tamoxifen, Prediction of treatment outcome, CYP2D6 metabolism, Poor metabolizer
Zusammenfassung
Tamoxifen ist ein Standard in der Prävention und Behandlung des hormonrezeptorpositiven Mammakarzinoms. Tamoxifen wird von CYP 450-Enzymen zu den klinisch wirksamen Metaboliten 4-OH-Tamoxifen und Endoxifen aktiviert, die im Vergleich zur Muttersubstanz mit höherer Affinität an den Östrogenrezeptor binden und so die Proliferation von Brustkrebszellen inhibieren. Dabei spielt das P450-Enzym CYP2D6 eine Schlüsselrolle. Im Rahmen mechanistischer, pharmakologischer und klinisch-pharmakogenetischer Studien wurde belegt, dass sowohl die Plasmakonzentrationen der aktiven Metabolite als auch der Krankheitsverlauf durch genetische CYP2D6-Varianten und Interaktion mit CYP2D6-Inhibitoren als Begleitmedikation beeinflusst werden. Insbesondere nichtfunktionelle (poor metabolizer) und in der Funktion stark eingeschränkte (intermediate metabolizer) CYP2D6-Varianten sind mit einer höheren Rückfallrate assoziiert. Die Bestimmung des CYP2D6-Genotyps zur Vorhersage des Metabolizer-Phänotyps vor Behandlungsbeginn könnte ein wichtiger Schritt hin zur individualisierten endokrinen Behandlung mit verbesserten Heilungschancen darstellen. Darüber hinaus sollte im Rahmen der adjuvanten Tamoxifen-Therapie auf die Gabe von starken CYP2D6-Inhibitoren, wie z. B. Paroxetin, einem selektiven Serotonin-Wiederaufnahme-Hemmer, verzichtet werden.
Introduction
The anti-estrogen tamoxifen is an essential part of standard adjuvant and palliative systemic therapy for patients with steroid hormone receptor-positive breast tumors [1]. Adjuvant tamoxifen significantly decreases relapse rates and mortality in pre- and postmenopausal patients, and the therapy benefit resulting from 5 years of adjuvant tamoxifen is maintained even more than 10 years after diagnosis [2]. Tamoxifen is a valid therapy option next to aromatase inhibitors (AI) in postmenopausal patients with endocrine responsive disease [3]; it is considered the standard care for premenopausal patients, for prevention of invasive breast cancer in women at high risk including those who have had ductal carcinoma in situ [4], and for the treatment of male breast cancer [5]. Tamoxifen is generally well tolerated, and menopausal symptoms including hot flashes are the most common side effects whereas severe side effects such as thromboembolic events or endometrial carcinoma are rather rare [2].
Although the clinical benefit of tamoxifen has been evident for more than 3 decades, up to 50% of patients receiving adjuvant tamoxifen relapse or die due to tumor resistance or constitutional factors. New insights into the metabolism of tamoxifen and the pharmacological activity of some of its metabolites hold promise to identify patients who will most likely benefit from tamoxifen, and distinguish them from those in need for alternative treatment options.
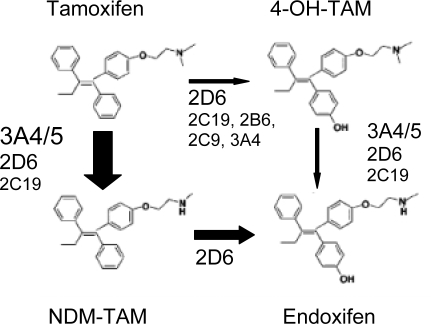
Tamoxifen is extensively metabolized and several lines of evidence suggest that the therapeutic efficacy is not mediated by the parent drug but by 4-hydroxy tamoxifen (4-OH-TAM) and 4-hydroxy-N-desmethyl tamoxifen (endoxifen) (fig. 1). Compared to tamoxifen and its major metabolite N-desmethyl tamoxifen (NDM), endoxifen and 4-OH-TAM have an up to 100-fold higher affinity for the estrogen receptor (ER) and potency to suppress breast cancer cell proliferation [6,7,8,9,10,11,12]. The key enzyme for the formation of endoxifen and 4-OH-TAM is cytochrome P450 2D6 (CYP2D6). Because CYP2D6 is subject to genetic polymorphism and the enzyme can be inhibited by co-medication, these genetic and non-genetic patient factors affecting the formation of active metabolites can impede the therapy response in breast cancer patients. This review provides an overview on the available pharmacological and pharmacogenetic evidence relevant to tamoxifen metabolism and its impact on outcome after tamoxifen treatment.
Fig 1.
Metabolic pathways leading to the formation of anti-estrogenic tamoxifen metabolites 4-OH-TAM and endoxifen as well as CYP 450 enzymes involved in these pathways. The size of the arrows symbolize the importance of the respective pathway in the tamoxifen metabolism. Adapted from [9].
CYP2D6 Genotype Phenotype Relationships
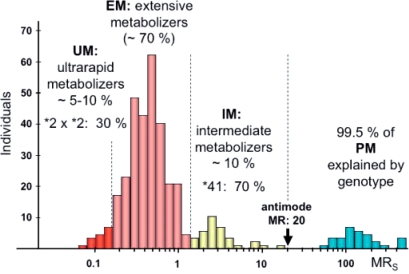
The enzymatic activity of CYP2D6 as assessed with a probe drug such as sparteine or debrisoquine shows a clear trimodal distribution indicating that the metabolism of CYP2D6 substrates is under strong genetic control giving rise to 3 distinct phenotypes in European populations: the poor metabolizer (PM), the intermediate metabolizer (IM), and the extensive metabolizer (EM) (fig. 2). The molecular mechanisms responsible for these phenotypes have been elucidated, and up to now more than 80 mutations have been identified in the gene encoding CYP2D6. The PM phenotype constituting 5–10% of the European population carries 2 non-functional (null) alleles leading to a loss of enzyme function. Of the 17 null alleles identified so far, the *3, *4, and *5 alleles are the most frequent in populations of European descent. CYP2D6 *4 is by far the most frequent null allele (20–25% in Europeans) and present in 70-90% of all PMs [13]. The 10–15% IMs have a severely impaired CYP2D6 expression und function similar to the PM due to the presence of *9, *10, and *41 alleles, and are genetically either homozygous for these IM mutations or compound heterozygous for an IM allele in combination with one null allele [13, 14]. In Orientals, the frequency of PMs is much lower (approximately 1%), but much higher IM frequencies are observed because up to 50% of the population carry the *10 allele. The predictive value of genotyping for both the PM and IM phenotype is very good provided it is not restricted to the *4 allele. The largest proportion of subjects − 60–70% of the European population – are EMs because they carry the *1 and *2 alleles which encode for an enzyme with normal expression levels and catalytic function. Based on genotypes, this phenotype can be separated into homozygous or heterozygous EMs depending on whether they carry 2 or 1 functional allele. Since heterozygous EMs who carry one *1 or *2 allele in combination with an IM or PM allele have a somewhat impaired enzyme expression and function, they have been classified as ‘IMs’ assuming a gene dose effect in such a way that heterozygous EMs would have only 50% of enzyme and catalytic activity as compared to homozygous EMs. However, this assumption is not correct, and there is substantial overlap both in enzyme content and activity between homozygous and heterozygous EMs resulting in a rather poor predictive value of the genotype. Of note, the IM is a phenotype and genotype distinct from the heterozygous EM. Since in clinical studies genotyping is used to predict the phenotype, it is absolutely essential not only to analyze all relevant mutations, but also to define the genotypes which translate into the EM, IM, and PM phenotypes.
Fig 2.
Trimodal frequency distribution of the metabolic ratio (MR) of the probe drug spartein in a German population. MR reflects the drug metabolizing enzyme activity of CYP2D6. MR refers to the amount of parent drug excreted divided by the amount of metabolite excreted. Subjects with a very low MR have a very high metabolic capacity whereas subjects with a high MR have a low metabolic capacity. The antimode of MR greater than 20 defines PMs, whereas IMs have an antimode of greater 1.2 and smaller than 20. Adapted from [14].
Although not clearly identifiable as a distinct subgroup or phenotype based on the enzymatic activity, approximately 10% of the European population account for the so called ultra rapid metabolizers (UM) (fig. 2). In 15–20% of these UMs gene duplication with up to 13 gene copies involving *1, *2, and *4 alleles have been identified as the molecular mechanism. However, most of the UMs cannot be explained by genotype, and hence its predictive value for the UM phenotype is low. The frequency of the CYP2D6 gene duplications is 1–10% in Europeans but much higher frequencies of 29 and 21% have been observed in Ethiopeans [15] and Saudi Arabians [16].
Tamoxifen Efficacy and Prediction by CYP2D6 Genotyping
Although it has been known for long time that tamoxifen requires metabolic activation to 4-OH-TAM to exert its therapeutic effect [6, 17], the knowledge about the link between tamoxifen metabolism and clinical efficacy has only recently been rapidly evolving and has been redefined based on current important pharmacological findings (fig. 3). Although endoxifen had been recognized as a secondary tamoxifen metabolite earlier on [18], only recently its pharmacologic properties have been evaluated. Stearns et al. [8] showed in 2003 that endoxifen has high ER affinity and potency to suppress breast cancer cell proliferation similar to 4-OH-TAM which by then was thought to be the principal anti-estrogenic metabolite. Moreover, both metabolites induced similar changes on global gene expression patterns in MCF7 breast cancer cells [19]. Recent work by Buck et al. [12] investigated the growth inhibitory effects of tamoxifen and its metabolites in MCF7 cells. Significant effects were observed for 4-OH-TAM and endoxifen but not for all other metabolites, and this antiproliferative activity was associated with induction of transforming growth factor beta (TGFß) and transforming growth factor ß type II receptor, which are key factors of the antiproliferative TGFß signal transduction pathway.
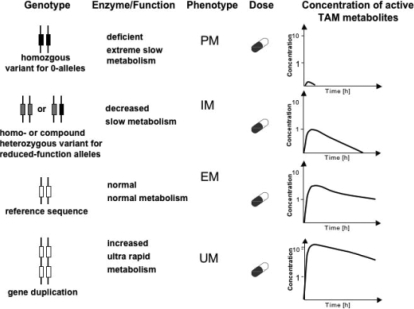
Fig 3.
Consequences of CYP2D6 genotypes for the plasma concentrations of active tamoxifen metabolites. Depending on the genotype, pronounced differences in the plasma concentration of active metabolites will be observed. PM = poor metabolizer; IM = intermediate metabolizer; EM = extensive metabolizer; UM = ultrarapid metabolizer. Frequency in Europeans: PM: 5-10%, IM: 5-10%; EM: 60-70%, and UM: 1-10%.
In plasma of tamoxifen-treated patients, endoxifen was found at 6-fold higher concentrations when compared to 4-OH-TAM, suggesting an important pharmacologic role. Comprehensive in vitro investigations and evaluation of tamoxifen's sequential biotransformation by the human CYP450 system performed by Desta et al. [9] recognized CYP3A and CYP2D6 as the most important enzymes with CYP2D6 being exclusively responsible for the formation of endoxifen from the primary metabolite NDM. Moreover, a large interindividual variability in the formation of tamoxifen metabolites was noticed and clinical implications suggested.
In a subsequent clinical study, Jin et al. [11] linked the bioavailability of active tamoxifen metabolites, particularly endoxifen, to CYP2D6 genetic variants. They performed CYP2D6 genotyping of patients treated with adjuvant tamoxifen (20 mg/day, orally) and measured metabolite plasma levels at the beginning of therapy (1 month) and at steady state (4 month). Among 80 primary breast cancer patients, 48 (60%) had a CYP2D6 wt/wt genotype, 29 (36%) a wt/variant (*3, *4, *5, *6 allele), and 3 (4%) a vt/vt genoytype. All of the 3 vt/vt patients carried a *4/*4 genotype. After 4 months of tamoxifen therapy, plasma endoxifen concentrations were significantly lower in the CYP2D6 homozygous variant genotype (20.0 nM, 95% confidence interval (CI) 11.1-28.9 nM) or the heterozygous genotype (43.1 nM, 95% CI 33.3–52.9 nM) compared to those in patients with a homozygous wild-type genotype (78.0 nM, 95% CI 65.9–90.1 nM) (p = 0.003). There were no significant associations of other candidate enzymes involved in tamoxifen biotransformation such as CYP2C9, CYP3A5, or SULT1A1 genotypes with plasma concentrations of tamoxifen or its metabolites.
While Jin et al. [11] focused on CYP2D6 PM genotypes, Borges et al. [20] investigated the quantitative relationship between CYP2D6 variants and endoxifen plasma concentrations in an updated analysis of 158 patients after 4 months treatment with 20 mg daily tamoxifen. This included variants known to cause a loss of protein (e.g. *4) or exerting decreased function (e.g. *10), i.e. PM and IM genotypes, as well as variants known to increase enzyme function, i.e. UM genotypes. The mean endoxifen/NDM ratio was low (0.04 +/− 0.02) in patients lacking any functional alleles, intermediate (0.08 +/− 0.04) in patients with 1 active allele, and high (0.15 +/− 0.09) in patients with 2 or more functional alleles. Accordingly, CYP2D6 genotypes are highly associated with endoxifen plasma concentrations and moreover, account for their variability.
With respect to NDM, the precursor of endoxifen, Serrano et al. [21] recently reported at the 2007 meeting of the American Society of Clinical Oncology (ASCO) that mutated CYP2D6 was associated with increased plasma levels. The investigators reported significantly higher plasma levels of NDM in mutation carriers compared to heterozygous or wildtype genotype carriers after 1 year of tamoxifen (n = 118; p = 0.001) indicating that conversion into the clinically relevant endoxifen may be impaired.
While CYP2D6 polymorphisms had already been linked with plasma levels of endoxifen, their impact on the clinical outcome in breast cancer patients was shown for the first time by Götz et al. [22] who investigated this relationship in patients recruited within a large prospective clinical trial of the North Central Cancer Treatment Group (NCCTG). The NCCTG 89-30-52 trial was a randomized phase III clinical trial in postmenopausal women with primary ER-positive breast cancer which evaluated the value of adding the androgen fluoxymestrone for 1 year, to the standard 5 year adjuvant tamoxifen therapy. They genotyped CYP2D6 variants *4 and *6 in DNA obtained from paraffin-embedded tissue specimens from patients who had received adjuvant tamoxifen therapy (n = 223 of 256 eligible). Of the 190 patients for whom CYP2D6 (*4) genotyping was possible, 137 (72.1%) had wt/wt, 40 (21.1%) wt/*4, and 13 (6.8%) *4/*4 genotype. The concordance rate between the genotype obtained from additional buccal cells in 17 patients, and the corresponding tumor tissue was 100%. After a median follow-up of 11.4 years, the CYP2D6 *4/*4 genotype was associated with poor patient outcome. CYP2D6 *4/*4 was associated with worse relapse-free (p = 0.023) and disease-free survival (p = 0.012). The genotype did not have an impact on overall survival (p = 0.169). Moreover, the authors looked at the CYP3A5*3 variant and did not find any association with any adverse clinical outcome.
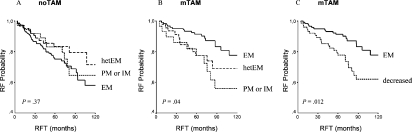
Strong evidence for the correlation between CYP2D6 genotype and outcome under tamoxifen therapy has been recently provided by Schroth et al. [23]. Their approach was based on a non-randomized cohort study including 206 breast cancer patients treated with adjuvant tamoxifen monotherapy and 280 patients without tamoxifen. Median follow-up was 71 months. Their comprehensive genotyping approach using constitutional DNA derived from formalin-fixed paraffin-embedded normal breast tissues included the CYP2D6 variants *4, *5, *10, and *41 to cover the vast majority (up to 95%) of PM genotypes and IM genotypes (up to 90%). Altogether, these analyses were aimed at the investigation of approximately 15-25% of patients expected to be carriers of PM or IM alleles and genotypes. The investigators found that carriers of CYP2D6 *4, *5 *10 and *41 alleles had significantly more breast cancer recurrences, shorter relapse-free time (hazard ratio (HR) = 2.24; 95% CI 1.16-4.33; p = 0.02), and worse event-free survival (HR = 1.89; 95% CI 1.10-3.25; p = 0.02) compared to carriers of functional alleles (fig. 4). Among 4 other tamoxifen metabolizing genes (CYP2C19, CYP2B6, CYP2C9, and CYP3A5) and variants, they also observed that the CYP2C19 high enzyme activity promoter variant *17 was associated with a more favorable clinical outcome (HR = 0.45; 95% CI 0.21-0.92; p = 0.03) than that of carriers of *1, *2, and *3 alleles. The authors appreciated that genetically determined impaired tamoxifen metabolization results in worse treatment outcome and suggested that genotyping for CYP2D6 alleles *4, *5, *10 and *41 could identify patients who will derive little benefit from adjuvant tamoxifen therapy. Moreover, they showed that the combination of functional CYP2D6 alleles and CYP2C19*17 variant identified those patients likely to benefit from tamoxifen which comprises up to 32% of tamoxifen treated patients.
Fig 4.
Kaplan-Meier probabilities of relapse-free time (RFT) for CYP2D6 metabolizer phenotypes predicted from genotypes: A Patients not treated with tamoxifen (noTAM); B Patients treated with adjuvant tamoxifen monotherapy (mTAM); C Carriers of one or two impaired CYP2D6 alleles predictive for ‘decreased’ enzyme activity were combined; EM, IM, PM, extensive, intermediate, poor metabolizer; hetEM, heterozygous extensive. (Reprinted with permission from the American Society of Clinical Oncology: Schroth et al. [23]).
Supportive evidence for a relationship between CYP2D6 genotypes and metabolite plasma concentrations as well as disease outcome has also been recently provided by a Korean study by Lim et al. [24] including 202 breast cancer patients treated with 20 mg/daily tamoxifen (either primary or metastatic breast cancer) for more than 8 weeks. As expected for Asian populations, the CYP2D6 *10 allele significantly contributed to the overall fraction of IM genotypes. The authors reported a frequency of 31.6% for CYP 2D6 wt/wt, 44% for wt/*10, and 24.2% for *10/*10 genotype. Patients with *10/*10 genotype (n = 49) had significantly lower steady-state plasma concentrations of endoxifen and 4-OH-TAM than those with other genotypes (n = 153). In a small cohort of 21 patients with metastatic breast cancer treated with tamoxifen, all patients (n = 6) with progressive disease or stable disease lasting less than 24 weeks carried the *10/*10 genotype (p = 0.0186). This significant impact of CYP2D6 genotype on time to progression was maintained in multivariate analysis. Median time to progression for CYP2D6*10/*10 patients was significantly shorter than that for all other genotypes (5.0 vs. 21.8 months, p = 0.0032).
CYP2D6 variation may not only be important for treatment outcome but also for prediction of the risk to develop breast cancer under tamoxifen prevention as recently suggested by preliminary data from an Italian chemoprevention trial including 5,408 healthy hysterectomized women aged 35-70 years, who were randomly assigned to receive 20 mg daily tamoxifen or placebo. In a nested case control study including 46 women who developed breast cancer and 136 controls, the frequency of CYP2D6 *4/*4 genotype was significantly higher in women who developed breast cancer than in those who did not: 9 vs. 1% (p = 0.015) and also within the tamoxifen treated population: 15 vs. 2% (p = 0.04) [25]. Unexpectedly, hot flashes were reported for all 3 CYP2D6 *4/*4 allele carriers who developed breast cancer under tamoxifen.
Contradictory results for the relationship of CYP2D6 genotype and outcome under tamoxifen were reported in 2 studies from the US and Sweden by Nowell et al. [26] (162 patients with tamoxifen, 175 patients without tamoxifen) and Wegmann et al. [27] (112 patients with 40 mg/daily tamoxifen, and mean follow-up of 10.7 years), respectively: While Nowell et al. reported no association between CYP2D6 *4 and tamoxifen response or breast cancer prognosis, Wegman et al. reported a decrease in the number of recurrences in tamoxifen treated patients who carried the CYP2D6 *4 variant (OR = 0.28; 95% CI 0.11-0.74; p = 0.0089). Within their recent work, Wegman et al. [28] looked at genetic variants of CYP3A5, CYP2D6, SULT1A1, or UGT2B15 in a cohort of 677 tamoxifen-treated postmenopausal patients, 238 of whom were randomized to 2 vs. 5 years of treatment. With respect to patients homozygous for CYP2D6 *4, a significantly better disease-free survival was observed compared to patients homozygous or heterozygous for the *1 allele (p = 0.05 and p = 0.04, respectively). However, this effect was not significant in multivariate Cox analysis (p = 0.055).
Tamoxifen Adverse Reaction, CYP2D6 and Drug Interactions Through Co-Medication
Hot flashes frequently occur in women using tamoxifen and are considered an indicator of drug efficacy. They are attributed to ER agonistic effects in the central nervous system due to higher tamoxifen metabolite levels in patients with functional CYP2D6. Accordingly, Götz et al. [22] showed that the incidence of hot flashes on adjuvant tamoxifen correlated with the CYP2D6 genotype. None of the patients with CYP2D6 *4/*4 genotype (0/13) reported moderate or severe hot flashes whereas in the *4/wt group 23% (9/40) and in the wt/wt group 20% (27/137) of the patients reported moderate or severe hot flashes (p = 0.064). At ASCO 2007, these data were substantiated by results from the Women's Healthy Eating and Living trial (WHEL) [29]. The WHEL trial enrolled primary breast cancer patients (n = 3,088 younger than 75 years) up to 4 years after initial diagnosis to either dietary intervention or observation alone. In this trial, 864 women received adjuvant tamoxifen. Of these patients, 78% reported hot flashes, and among those 69% also reported night sweats; only 4% reported night sweats without hot flashes, and 18% did not report either hot flashes or night sweats. A Cox proportional hazards model showed that patients reporting hot flashes had a significantly lower risk for recurrence (HR 0.51; 95% CI 0.32–0.79). For patients on adjuvant tamoxifen, hot flashes were more predictive of outcome than age, grade, hormone receptor status, or stage II cancer.
Within the context of hot flashes under adjuvant tamoxifen, a close-up look on the influence of co-medications on tamoxifen metabolism and treatment outcome is important. Women who take tamoxifen often are prescribed selective serotonin reuptake inhibitor (SSRI) antidepressants (e.g. paroxetine and fluoxetine) to treat hot flashes. SSRIs are known to inhibit CYP2D6 activity and to interfere with tamoxifen efficacy. While the effect of SSRIs on the plasma levels of endoxifen had been previously reported by Stearns et al. [8], this endoxifen lowering effect has been subsequently linked to the patients' CYP2D6 genotype by Jin et al. [11]. In their study, 24 of the 80 patients took CYP2D6 inhibitors including paroxetine, fluoxetine, sertraline, citalopram, amiodarone, and metoclopramide. Among patients who carried a homozygous wildtype genotype, the mean plasma endoxifen concentration for patients using CYP2D6 inhibitors was 58% lower than that of patients not using SSRI co-medication (38.6 vs. 91.4 nM, p = 0.0025). When the authors further investigated this effect with respect to weak and potent CYP2D6 inhibitors, they showed that women taking the weak CYP2D6 inhibitor venlafaxine (a serotonine norepinephrine reuptake inhibitor, SNRI) had slightly reduced plasma endoxifen concentrations compared to women taking the potent CYP2D6 inhibitor paroxetine. Altogether, the authors demonstrated that both pharmacogenomic effects as well as pharmacological interactions of CYP2D6 have an influence on the metabolism of tamoxifen, and therefore ultimately may affect drug efficacy.
These observations again were extended by Borges et al. [20]. In their investigation of the quantitative effects of CYP2D6 genotype and inhibitors on endoxifen plasma levels, they showed that while there were no significant differences in mean plasma concentrations of tamoxifen, NDM, and 4-OH-TAM between users and non-users of concomitant CYP2D6 inhibitors, the mean endoxifen plasma concentration was significantly lower in patients taking CYP2D6 inhibitors compared to that in patients who did not (39.6 +/− 28.4 vs. 71.5 +/−41.2 nmol/L; p < 0.01). When the authors divided the CYP2D6 inhibitors into potent (paroxetine, fluoxetine n = 19) and weak (SSRI: sertraline and citalopram (n = 14) as well as celecoxib, diphenydramine and chlorpheniramine (n = 13)), they found a more pronounced decrease in mean endoxifen plasma concentrations with potent inhibitors than with weak inhibitors. Concomitant use of venlafaxine, which is considered the least potent inhibitor, did not show any significant effect. Taking these observations to the level of CYP2D6 genotypes, the authors observed that the mean plasma endoxifen concentration was significantly lower in CYP2D6 EM patients who were taking potent CYP2D6 inhibitors compared to that in patients who were not taking CYP2D6 inhibitors (23.5 +/− 9.5 vs. 84.1 +/−39.4 nmol/L, p < 0.001). Thus, CYP2D6 genotype and concomitant potent CYP2D6 inhibitors are highly associated with plasma endoxifen concentrations and may substantially impact outcome under tamoxifen treatment by conversion of EM to PM phenotype status.
The interplay of CYP2D6 genotype and enzyme inhibitors was further explored by Götz et al. [30] in their recent followup of the NCCTG trial. They investigated the role of CYP2D6 inhibitors in 256 patients that had been randomized to the tamoxifen alone arm. Patients with CYP2D6 wt/wt genotype who did not take CYP2D6 inhibitors were classified as EM (n = 115), whereas patients with either 1 or 2 *4 alleles or those taking a CYP2D6 inhibitor were classified as IM or PM (n = 65), depending on the strength of the inhibitor. Decreased CYP2D6 metabolism was associated with poor clinical outcome in univariate and multivariate analysis: Patients with decreased metabolism had shorter time to breast recurrence (p = 0.015), relapse-free (p = 0.007), disease-free (p = 0.009), and overall survival (p = 0.082) compared to those with extensive CYP2D6 metabolism. Adjusting for tumor size and nodal status, the significant impact on time to breast recurrence, relapse-free and disease-free survival was maintained in multivariate analysis [30]. The authors concluded that CYP2D6 metabolism, as measured by genetic variation and enzyme inhibition, is an independent predictor of breast cancer outcome in postmenopausal primary breast cancer patients receiving adjuvant tamoxifen. Accordingly, outcome under tamoxifen treatment may be influenced by its pharmacogenetics as well as co-prescription of drugs interfering with the CYP2D6-mediated tamoxifen metabolism.
Clinical Relevance of CYP2D6 in Breast Cancer: Discussion
Whereas tamoxifen is still considered standard in premenopausal patients, national and international guidelines strongly recommend the use of AIs in postmenopausal women [1, 31]. Whether upfront AI use or a tamoxifen – AI sequence is the optimal treatment strategy cannot be definitely decided at this moment since trials directly comparing these treatment strategies such as BIG 1–98 or TEAM have not yet released their data regarding this question. Recent data from BIG 1-98 suggest that in patients with a high risk for early relapse, upfront AI therapy does offer a significant survival advantage [32]. However, at ASCO 2007, Punglia et al. [33] presented a biomathematical modeling exercise of estimated benefit of adjuvant tamoxifen according to CYP2D6 gene status. Using the BIG 1-98 information on relapse risk and assuming that AI metabolism was CYP2D6 independent, the authors suggested that the benefit of 5 years of adjuvant tamoxifen may even exceed that of upfront AI treatment in postmenopausal CYP2D6 wt/wt patients.
There is now strong mechanistic, pharmacological, and clinical evidence that tamoxifen efficacy and clinical outcome depend on the CYP2D6 metabolism which is subject to CYP2D6 genotype and/or pharmaco-interactions. Data from numerous international studies yielded consistent results in linking active tamoxifen metabolite plasma concentrations with genetically determined CYP2D6 metabolizer status, interference with strong CYP2D6 inhibitors as well as clinical outcome. Few conflicting data may be explained by variations in patient inclusion criteria into respective studies which may include variations in tamoxifen doses and length of treatment as well as additional chemotherapy regimens or lack of consistent ER testing. Importantly, most authors agree that genetic CYP2D6 variants as well as CYP2D6 inhibition by prescribed co-medications such as antidepressants may decrease tamoxifen metabolism, and thus negatively impact tamoxifen efficacy and treatment outcome.
There are a number of potential clinical consequences from these emerging CYP2D6-tamoxifen treatment outcome data. First of all, potent SSRIs such as paroxetine or fluoxetine should not be used for the relief of hot flashes in breast cancer patients receiving tamoxifen. Even though SSRIs are one of the few evidence-based therapy options for menopausal vasomotor symptoms [34], there is now convincing data that they may compromise tamoxifen efficacy due to their interference with CYP2D6 dependent tamoxifen metabolism. Yet, differences in tamoxifen metabolite plasma levels were seen depending on the strength of the CYP2D6 inhibitor [11, 20]. If treatment of hot flashes is indicated, an SSRI such as citalopram or escitalopram or SNRI such as venlafaxine should be used because these substances showed no significant inhibition of CYP2D6. Second, the CYP2D6 genotype-phenotype-treatment outcome relationship points to the possible benefit of upfront CYP2D6 genotyping prior to an adjuvant endocrine treatment decision. A comprehensive robust, standardized, and quality controlled CYP2D6 genotyping test will need to analyze all genetic variants that may affect tamoxifen metabolism which, based on data by Götz et al. [22, 30] and Schroth et al. [23], should include common PM alleles (*3, *4 and *5) and IM alleles depending on the individual's ethnic origin. Of note, *41 is the most frequent IM allele in Europeans, *17 is the principal IM allele in Africans, and *10 dominates in Asians (in addition *9 should also be considered). Other areas of interest with respect to clinical application are the measurement of endoxifen plasma levels as a surrogate of CYP2D6 phenotype and a possible dose increase of tamoxifen to overcome impaired CYP2D6 metabolism, the latter option may need to be evaluated further before definite conclusions can be made.
Given alternative treatment options, i.e. tamoxifen vs. AI, and considering available scientific and clinical evidence, an individualized approach for endocrine treatment of postmenopausal breast cancer patients is desirable. One might speculate that tamoxifen alone may be adequate for CYP2D6 EM/EM (wt/wt) carriers whereas postmenopausal patients with variant CYP2D6 alleles may fare better with upfront AI therapy. However, as of yet, formal recommendation on the integration of CYP2D6 genotypes in treatment decisions must await their validation in prospective clinical trials. In Germany, we are currently recruiting postmenopausal ER-positive breast cancer patients within the IKP211 observational study (www.rbk.de) dedicated to the identification of tamoxifen and AI treatment outcome predictors. The study is being performed in association with the AGO TRAFO Commission (www.ago-online.de). Patient inclusion criteria are adjuvant treatment with either tamoxifen (5 years or switch to AI after 2 years tamoxifen) or AI (5 years), respectively. It is expected that these patient cohorts will allow a first prospective confirmatory analysis of CYP2D6 metabolism, drug interaction, and treatment outcome. Whether determination of CYP2D6 genotype will be a diagnostic tool for selection of the appropriate adjuvant endocrine therapy for an individual ER-positive postmenopausal breast cancer patient awaits the validation in prospective clinical trials randomizing tamoxifen vs. AI treatment based on CYP2D6 genotypes. Such prospective clinical trials are currently being planned. Other open questions may address the clinical relevance of other cytochrome P450 isoenzymes and mutations as well as ethnic variations in the prevalence of their treatment outcome relevant genotypes.
Finally, it is important to appreciate that most cancer therapies in current use have been established empirically. The recent progress in our understanding of pharmacology and pharmacogenetics of tamoxifen, however, holds the promise for improvement through personalized medicine. Because this approach is genome-based by utilizing CYP2D6 genotyping for prediction of a patient's metabolizer phenotype, ethical issues need to be sufficiently addressed. In the light of acceptable alternatives, an informed choice about adjuvant endocrine treatment and most importantly the avoidance of a therapy that might potentially lack efficacy must be prime interests. It will therefore be important to make patients and their care-takers aware of these issues and also to initiate discussions with regulatory authorities.
References
- 1.AGO: Diagnostic and treatment of patients with primary and metastatic breast cancer: guidelines of AGO Breast Commission, Kommission Mamma-Arbeitsgemeinschaft Gynaekologische Onkologie, 2007.
- 2.EBCTCG Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol. 2007;18:1133–44. doi: 10.1093/annonc/mdm271. [DOI] [PubMed] [Google Scholar]
- 4.Fisher B, Costantino JP, Wickerham DL, Cecchini RS, Cronin WM, Robidoux A, Bevers TB, Kavanah MT, Atkins JN, Margolese RG, Runowicz CD, James JM, Ford LG, Wolmark N. Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J Natl Cancer Inst. 2005;97:1652–62. doi: 10.1093/jnci/dji372. [DOI] [PubMed] [Google Scholar]
- 5.Fentiman IS, Fourquet A, Hortobagyi GN. Male breast cancer. Lancet. 2006;367:595–604. doi: 10.1016/S0140-6736(06)68226-3. [DOI] [PubMed] [Google Scholar]
- 6.Jordan VC. Metabolites of tamoxifen in animals and man: Identification, pharmacology, and significance. Breast Cancer Res Treat. 1982;2:123–38. doi: 10.1007/BF01806449. [DOI] [PubMed] [Google Scholar]
- 7.Lien EA, Solheim E, Lea OA, Lundgren S, Kvinnsland S, Ueland PM. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–83. [PubMed] [Google Scholar]
- 8.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–64. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 9.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–75. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 10.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 11.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 12.Buck MB, Coller JK, Murdter TE, Eichelbaum M, Knabbe C. TGFbeta2 and TbetaRII are valid molecular biomarkers for the antiproliferative effects of tamoxifen and tamoxifen metabolites in breast cancer cells. Breast Cancer Res Treat. 2008;107:15–24. doi: 10.1007/s10549-007-9526-7. [DOI] [PubMed] [Google Scholar]
- 13.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 14.Toscano C, Klein K, Blievernicht J, Schaeffeler E, Saussele T, Raimundo S, Eichelbaum M, Schwab M, Zanger UM. Impaired expression of CYP2D6 in intermediate metabolizers carrying the *41 allele caused by the intronic SNP 2988G>A: evidence for modulation of splicing events. Pharmacogenet Genomics. 2006;16:755–66. doi: 10.1097/01.fpc.0000230112.96086.e0. [DOI] [PubMed] [Google Scholar]
- 15.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278:441–6. [PubMed] [Google Scholar]
- 16.McLellan RA, Oscarson M, Seidegard J, Evans DA, Ingelman-Sundberg M. Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics. 1997;7:187–91. doi: 10.1097/00008571-199706000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75:305–16. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 18.Lien EA, Solheim E, Kvinnsland S, Ueland PM. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48:2304–8. [PubMed] [Google Scholar]
- 19.Lim YC, Li L, Desta Z, Zhao Q, Rae JM, Flockhart DA, Skaar TC. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318:503–12. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 20.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, Hillman G, Hayes DF, Stearns V, Flockhart DA. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Serrano D, Macis D, Gandini S, Johansson H, Guerrieri-Gonzaga A, Mellgren G, Lien EA, Mariett F, Sandri M, Decensi A, Bonanni B: Correlation of low dose tamoxifen and its metabolites plasma level with Cyp 2D6, and SULT polymorphism; a pharmacogenetics study within a chemoprevention trial: ASCO Annual Meeting Proceedings Part I, 2007, vol 25, pp. 503.
- 22.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 23.Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, Simon W, Eichelbaum M, Brauch H. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–93. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 24.Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25:3837–45. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 25.Bonanni B, Macis D, Maisonneuve P, Johansson HA, Gucciardo G, Oliviero P, Travaglini R, Muraca MG, Rotmensz N, Veronesi U, Decensi AU. Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol. 2006;24:3708–9. doi: 10.1200/JCO.2006.06.8072. author reply 3709. [DOI] [PubMed] [Google Scholar]
- 26.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, MacLeod SL, Kadlubar FF, Ambrosone CB. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–58. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 27.Wegman P, Vainikka L, Stal O, Nordenskjold B, Skoog L, Rutqvist LE, Wingren S. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–90. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3 A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortimer JE, Flatt SW, Parker BA, Gold EB, Wasserman L, Natarajan L, Pierce JP. Tamoxifen, hot flashes and recurrence in breast cancer. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9612-x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–21. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 31.Carlson RW, Hudis CA, Pritchard KI. Adjuvant endocrine therapy in hormone receptor-positive postmenopausal breast cancer: evolution of NCCN, ASCO, and St Gallen recommendations. J Natl Compr Canc Netw. 2006;4:971–9. doi: 10.6004/jnccn.2006.0082. [DOI] [PubMed] [Google Scholar]
- 32.Mauriac L, Keshaviah A, Debled M, Mouridsen H, Forbes JF, Thurlimann B, Paridaens R, Monnier A, Lang I, Wardley A, Nogaret JM, Gelber RD, Castiglione-Gertsch M, Price KN, Coates AS, Smith I, Viale G, Rabaglio M, Zabaznyi N, Goldhirsch A. Predictors of early relapse in postmenopausal women with hormone receptor-positive breast cancer in the BIG 1-98 trial. Ann Oncol. 2007;18:859–67. doi: 10.1093/annonc/mdm001. [DOI] [PubMed] [Google Scholar]
- 33.Punglia RS, Winer EP, Weeks JC, Burstein HC: Could treatment with tamoxifen be superior to aromatase inhibitors in early-stage breast cancer after pharmacogenomic testing? A modeling analysis: ASCO Annual Meeting Proceedings Part I, 2007, vol 25, pp. 502.
- 34.Hickey M, Davis SR, Sturdee DW. Treatment of menopausal symptoms: what shall we do now? Lancet. 2005;366:409–21. doi: 10.1016/S0140-6736(05)66519-1. [DOI] [PubMed] [Google Scholar]