Abstract
Shaker-related Kv1 channels contain four channel-forming α subunits. Subfamily member Kv1.1 often occurs oligomerized with Kv1.2 α subunits in synaptic membranes, and so information was sought on the influence of their positions within tetramers on the channels’ properties. Kv1.1 and 1.2 α genes were tandem linked in various arrangements, followed by expression as single-chain proteins in mammalian cells. As some concatenations reported previously seemed not to reliably position Kv1 subunits in their assemblies, the identity of expressed channels was methodically evaluated. Surface protein, isolated by biotinylation of intact transiently transfected HEK-293 cells, gave Kv1.1/1.2 reactivity on immunoblots with electrophoretic mobilities corresponding to full-length concatenated tetramers. There was no evidence of protein degradation, indicating that concatemers were delivered intact to the plasmalemma. Constructs with like genes adjacent (Kv1.1-1.1-1.2-1.2 or Kv1.2-1.2-1.1-1.1) yielded delayed-rectifying, voltage-dependent K+ currents with activation parameters and inactivation kinetics slightly different from the diagonally positioned genes (Kv1.1-1.2-1.1-1.2 or 1.2–1.1-1.2-1.1). Pore-blocking petidergic toxins, α dendrotoxin, agitoxin-1, tityustoxin-Kα, and kaliotoxin, were unable to distinguish between the adjacent and diagonal concatamers. Unprecedentedly, external application of the pore-blocker tetraethylammonium (TEA) differentially inhibited the adjacent versus diagonal subunit arrangements, with diagonal constructs having enhanced susceptibility. Concatenation did not directly alter the sensitivities of homomeric Kv1.1 or 1.2 channels to TEA or the toxins. TEA inhibition of currents generated by channels made up from dimers (Kv1.1-1.2 and/or Kv1.2-1.1) was similar to the adjacently arranged constructs. These collective findings indicate that assembly of α subunits can be directed by this optimized concatenation, and that subunit arrangement in heteromeric Kv channels affects TEA affinity.
INTRODUCTION
Voltage-gated K+ (Kv) channels are a diverse family of proteins, in keeping with their roles in neuronal excitability, shaping action potentials, determining inter-spike intervals, and, indirectly, regulating transmitter release (Jan and Jan, 1997; Pongs, 2008). The Shaker-related Kv1 subfamily, the most intensely studied at the primary and quaternary structural levels (for review see Jan and Jan, 1997; Pongs, 2008), should prove advantageous as therapeutic targets because of their modulatory roles (Judge and Bever, 2006). Kv1 channels in the brain, purified using the selective blockers α dendrotoxin (αDTX) or dendrotoxink (DTXk), are large (Mr, ∼400 kD) sialoglycoprotein complexes (Parcej et al., 1992) consisting of four pore-forming α subunits and four cytoplasmically associated auxiliary β proteins (Scott et al., 1994a,b). When heterologously expressed alone, each of the Kv1.1-1.6 α subunits yields a unique homotetrameric channel, and further diversity in vivo results from heteropolymerization. Only a subset of the possible oligomeric combinations has been isolated from bovine, rat, and human brain (Koch et al., 1997; Shamotienko et al., 1997; Koschak et al., 1998; Coleman et al., 1999), suggesting that their synthesis and/or assembly are restricted. Kv1.2 is the most prevalent in neuronal membranes, where a fraction occurs as a homotetramer and the remainder is heteromerized with other Kv1 α subunits (Shamotienko et al., 1997; Coleman et al., 1999). Interestingly, in these preparations there is a preponderance of the less abundant Kv1.1 subunit in oligomers with Kv1.2. We aim to obtain heteromers with compositions representative of their neuronal counterparts to serve as meaningful targets for the development of therapeutics and to act as validated probes for functional characterization of Kv1 channels in situ. Toward these goals, a new and versatile platform was devised for the rapid generation of constructs encoding controlled combinations and spatial arrangements of Kv1 α subunits for expression as tetramers in mammalian cells.
This paper describes concatamers of Kv1.1 and 1.2 cDNA that yield proteins with predefined positional arrangements. As previous studies with concatemeric Kv constructs have noted aberrations of surface-expressed channels from the expected channel types (Hurst et al., 1992, 1995; McCormack et al., 1992), an extensive series of controls was undertaken to ratify the identification of the surface channels, including rearrangement of subunits, biochemical assessment of the surface-expressed polypeptide chain, biophysical characterization of channel properties, as well as pharmacological dose–response profiles assayed by radioligand displacement and inhibition of IK. Results indicate that the stoichiometry of cell surface channels is fixed by the sequence of the expression construct used.
Peptide toxins from venomous creatures interact with the external mouth of the ion pore in K+ channels and involve binding interactions from multiple Kv α subunits to a single peptide (e.g., δ-DTX) (Imredy and MacKinnon, 2000). Such subunit-spanning binding sites make the affinity of peptide toxins for heteromeric channels difficult to predict, and pore-blocking peptides have been found that bind with higher affinity to heteromeric Kv1.1/1.2 channels than to either parental homomer (Middleton et al., 2003). An original intent of studies with the constructs herein was to identify peptide toxins that might distinguish between the orderings of channel subunits; yet, an unexpected result emerged: the arrangement of Kv subunits in concatamers affected the affinity of resultant channels for external tetraethylammonium (TEA). Although previous studies of TEA blockade have concluded that only subunit stoichiometry, not subunit arrangement, influences a K+ channel’s affinity for TEA (Hurst et al., 1992; Shen et al., 1994), these results provide an example to the contrary. Dissimilarity observed in the TEA susceptibilities for adjacently and diagonally arranged oligomers indicates that the order of genes in the tetrameric constructs used determines the positioning of α subunits in assembled channels.
MATERIALS AND METHODS
Molecular biology
PCR amplification of Kv1.X constituents.
Rat Kv1.1- and 1.2-pAKS plasmids served as PCR templates, with amplification achieved by using the requisite primers and conditions that incorporated XbaI and XhoI sites at the 5′ and 3′ ends, respectively. PCR products were purified by electrophoresis on agarose gels and cloned into an “intermediate” pCRBlunt plasmid (Invitrogen); competent DH5α cells were transformed with the ligated products. DNA was prepared on a large scale from positive clones; after digestion with XhoI and XbaI, inserts were purified electrophoretically as described above.
Modification of pβUT2 plasmid and cloning of Kv1.X inserts with flanking half-linkers.
pβUT2 plasmid served as the source of an intersubunit linker (Akhtar et al., 2002); SalI, BamHI, and Bg1II sites within its multiple cloning site (MCS) were eliminated by sequential in-filling to prevent their interference with assembly of the concatemeric constructs into the expression plasmid. Purified Kv1.X inserts were ligated into the mutated pβUT2 plasmid, and positive clones were identified by digestion with XbaI and XhoI; the resultant fragments acted as templates for the subsequent PCR. The forward and reverse primers used were based on the first (up to the XbaI site) and last (downstream of XhoI site) 30 nucleotides of the untranslated regions of the Xenopus β-globin gene flanking the MCS of pβUT2. Single bands of the expected sizes were purified by agarose gel electrophoresis, cloned into pCRBlunt plasmid and assembled into the pIRES2-EGFP vector (Takara Bio Inc.); restriction sites introduced during this PCR allowed positional cloning (Fig. 1 A). All constructs were verified by DNA sequencing.
Figure 1.
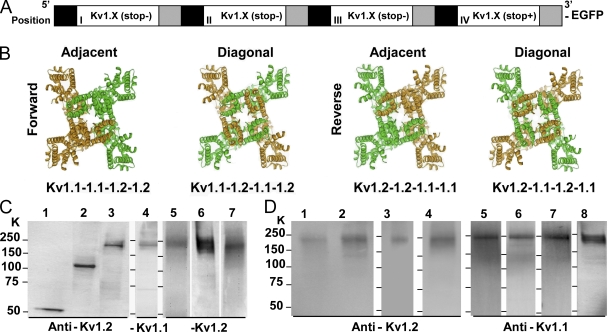
Concatenation of Kv1.X genes, expression in HEK-293 cells, and detection of complete heterotetrameric K+ channels on the plasmalemma. (A) Representation of the attachment to Kv1 genes of half-linkers and restriction sites at the 5′ (black) and 3′ (gray) end of each gene for assembly of concatemers in pIRES2-EGFP. Complete linkers form during the assembly of concatemers retaining all the sequences in the same open reading frame. (B) Adjacent and diagonal arrangements of the two α-subunit genes in the tandem-linked constructs, named in a clockwise direction (forward and reverse), used to produce the concatenated tetrameric proteins composed of Kv1.1 (green) and Kv1.2 (brown), based on a PDB file of the structure of the Kv1.2 α/β–subunit complex (Long et al., 2005). (C) HEK-293 cells transfected with pIRES2-EGFP containing the following constructs are shown in lanes: 1, Kv1.2; 2, Kv1.2-1.2; 3 and 4, Kv1.1-1.1-1.2-1.2; 5, Kv1.1-1.2-1.1-1.2; 6, Kv1.2-1.2-1.1-1.1; 7, Kv1.2-1.1-1.2-1.1. After expression for 48 h, the cells were lysed and subjected to SDS-PAGE (on 4–12% polyacrylamide gels), followed by Western blotting with the antibodies specified: Kv1.2 (lanes 1–3 and 5–7) and Kv1.1 (lane 4). (D) Intact cells expressing tetramers (as in C) were washed, biotinylated, detergent solubilized, precipitated with streptavidin-agarose beads, and analyzed using antibodies specific for Kv1.2 or Kv1.1. Lanes: 1 and 5, Kv1.1-1.1-1.2-1.2; 2 and 6, Kv1.1-1.2-1.1-1.2; 3 and 7, Kv1.2-1.2-1.1-1.1; 4 and 8, Kv1.2-1.1-1.2-1.1. Mobilities of standard proteins are indicated. Cells subjected to a mock transfection with the vector lacking any K+ channel construct, and biotinylated as before, did not show any band reactive with Kv1.1 or 1.2 IgGs. Exclusive extracellular labeling of intact HEK-293 cells by the biotinylation procedure was established from the inability to biotinylate actin, an intracellular protein, unless a cell lysate was used.
Assembly of Kv1.X tetrameric constructs and expression in mammalian cells: surface localization by biotinylation, and αDTX binding.
Paired sites for NheI/BglII, BglII/EcoRI, EcoRI/SalI, and SalI/BamHI, respectively, were used to individually subclone the genes from pCRBlunt into positions I–IV of pIRES2-EGFP (Fig. 1 A). Correct positioning of all the genes was confirmed by restriction analysis.
Biochemical characterization
The Kv1 channel constructs were expressed in HEK-293 cells for 36–96 h after transfection with Polyfect reagent (QIAGEN) and monitored by SDS-PAGE plus Western blotting with Kv1.1 and 1.2 subunit–specific antibodies (Shamotienko et al., 1997; Coleman et al., 1999). Surface biotinylation was performed on HEK-293 cells transfected for 48 h with the concatenated genes in pIRES2-EGFP, harvested, washed, resuspended at ∼2–3 × 107 cells/ml of PBS, pH 8.0, and incubated with 1 mg/ml sulfo-NHS-LC-biotin (Thermo Fisher Scientific) at room temperature for 30 min. Remaining reagent was quenched with 100 mM glycine for 30 min, samples were solubilized in 2% Triton X-100 for 1 h at 4°C, and the supernatants were incubated with streptavidin-agarose (70 µl slurry/ml; Thermo Fisher Scientific) overnight at 4°C with rotation. After washing the pelleted streptavidin-agarose with ice-cold 25 mM Tris-buffered saline, pH 8.0, containing 0.1% Tween-20, bound proteins were dissolved in SDS-PAGE sample buffer before Western blotting as described above. αDTX was radiolabeled as described previously; its saturable binding to HEK-293 cells expressing the channels and displacement by unlabeled toxins were quantified using a filtration assay (Sokolov et al., 2007). The resultant data were analyzed using GraphPad Prism software. Kaliotoxin (KTX), agitoxin-1 (AgTX1), and tityustoxin-Kα (TsTX-Kα) were purchased from Alomone Labs.
Electrophysiology
Whole cell voltage clamp was performed as before (Sokolov et al., 2007), except where specified, using an internal solution that contained (in mM): 90 KCl, 50 KF, 30 KOH, 10 EGTA, and 20 HEPES, pH 7.2. The extracellular bath solution contained (in mM): 135 choline chloride, 20 KOH, 1.8 CaCl2, 1 MgCl2, and 40 HEPES, pH 7.4, and 0.01% (wt/vol) BSA. In the conventional recording system, TEA chloride (Sigma-Aldrich) was substituted for an equivalent amount of choline chloride while it was introduced in the extracellular medium for the automated patch clamp system. Both systems gave similar Kv1 channel sensitivities to TEA. Just before use, peptide toxins were diluted from frozen aqueous stocks into external recording solution containing 0.01% (wt/vol) BSA using silanized pipette tips and tubes; test solutions were exchanged by continuous microfluidic flow with a Dynaflow-16 system (Cellectricon). Data were taken from one to six toxin concentrations to quantify inhibition, according to the relationship IC50 = fc/(1−fc)[Tx], where fc is the fractional current and [Tx] is the toxin concentration. TEA inhibition was determined by the Hill equation fit to 6–12 concentrations. Series resistance compensation was applied to restrict voltage error to <10 mV, and correction was made for a calculated liquid junction potential of +8.4 mV. Holding potential was −100 mV. P/n capacitive and leak subtraction was used to isolate K+ current (IK). Only cells with an IK of 0.8–5 nA were chosen for experimentation; this precaution avoided interference from endogenous outward currents that were found to have lower amplitude (50–200 pA at +60 mV potential). Analogue traces were filtered at 5 kHz and sampled at 50 kHz. Nonlinear fitting was performed with equations described previously (Sack and Aldrich, 2006; Sokolov et al., 2007).
For larger scale recordings, an automated patch clamp system (QPatch 16; Sophion Bioscience) was used. Its disposable Qplates contain 16 individual and parallel patch clamp positions. Cells were detached from culture plates with 0.05% trypsin/EDTA solution or Accutase (Analab) and kept in serum-free medium (CHO-S-SFM II [Invitrogen], 25 mM HEPES, pH 7.4, 0.04 µg/ml soya bean trypsin inhibitor, and 10 µg/ml penicillin-streptomycin) in an onboard stirred reservoir. Before testing, the cells were automatically transferred to an integral mini-centrifuge, pelleted, resuspended in external solution (as detailed above), and washed before being applied to the pipetting wells in the Qplate. Giga-seals were formed upon execution of a combined suction/voltage protocol; gradually increasing suction leads to the whole cell configuration. Compounds were applied, via a four-way pipetting robot, through integrated glass-coated microfluidic flow channels. Liquid flow is laminar with exchange time constants in the range of 50–100 ms. Whole cell currents were measured at a holding potential of −100 mV, and then depolarized to +60 mV for 300 ms or stepped from the holding potential in +10-mV increments. Data analysis was performed using an integrated database (Oracle) within QPatch software (Sophion Bioscience). Data are reported as mean ± SE (SEM) or SD; n values refer to number of individual cells tested. Statistical significance was evaluated by an unpaired two-tailed Student’s t test or, where indicated in the text, a Mann-Witney U test, using data obtained from at least three independent experiments. P values <0.05 were considered significant.
RESULTS
Concatenated Kv1 channels expressed in HEK cells after position-specific assembly of Kv1.1 and 1.2 gene cassettes in pIRES2-EGFP
To rapidly create a variety of constructs encoding recombinant K+ channels of predefined α-subunit stoichiometries, a novel strategy was developed for concatenating Kv1.1 and 1.2 genes (Fig. 1 A) via a linker from Xenopus β-globin gene known to be suitable (Akhtar et al., 2002). The individual genes were amplified with flanking linkers attached, and pairs of restriction sites were introduced. These allowed positional cloning of the verified constructs into any of the four positions within the MCS of the pIRES2-EGFP expression vector (Fig. 1, A and B). The positions of concatenated subunits are referred to as domains I, II, III, and IV in deference to the architecture of voltage-gated Na+ and Ca2+ channels. Sequential incorporation of Kv1.2 (+ stop codon) and/or Kv1.2 (− stop codon) into positions IV and III resulted in constructs that, upon expression in HEK-293 cells, yielded proteins of Mr of ∼56 and ∼120 kD, respectively, as revealed by SDS-PAGE and Western blotting (Fig. 1 C). As for the latter, other concatenated dimers (Kv1.1-1.2, Kv1.2-1.2, and Kv1.2-1.1) and their monomers were obtained. Likewise, the addition of Kv1.1 to positions I and II allowed for expression of the adjacent tetramer (Kv1.1-1.1-1.2-1.2), whereas the alternative insertion of Kv1.2 and 1.1 into positions II and III led to production of the diagonal tetramer (Kv1.1-1.2-1.1-1.2) (Fig. 1 B). Both gave expressed proteins with Mr of ∼240 kD, detected by subunit-specific antibodies (Fig. 1 C). In a similar manner, two additional constructs were generated by sequential incorporation of Kv1.1 and 1.1 (+ stop codon) into positions III and IV, respectively, followed by the addition of Kv1.2 to positions I and II resulting in Kv1.2-1.2-1.1-1.1 (reverse adjacent); finally, the genes were successfully added to the requisite positions to form the Kv1.2-1.1-1.2-1.1 (reverse diagonal) construct (Fig. 1 B). Again, each expressed protein gave a major band (Mr, ∼240 kD) on Western blots that proved reactive with antibodies specific for Kv1.1 or Kv1.2. For comparative purposes, concatenated homotetramers of Kv1.1 and 1.2 were generated similarly and characterized.
Intact heterotetrameric K+ channels trafficked to the cell surface in active form
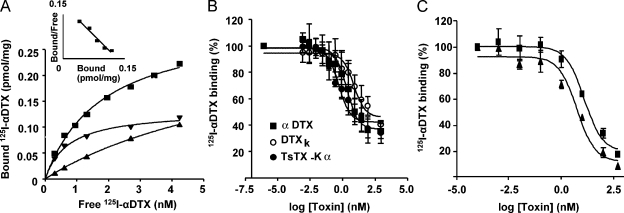
Although the cells transfected with each of these Kv1.X concatamers exhibited cell surface expression, immunofluorescent staining (unpublished data) suggested a majority of the channels were localized intracellularly. Thus, it was essential to biochemically analyze the channels on the plasmalemma by biotinylation of the surface components of HEK-293 cells transfected with the individual tandem-linked constructs. Precipitation of the solubilized biotinylated proteins with streptavidin agarose, followed by Western blotting with anti-Kv1.1 or -1.2 IgGs, revealed expression of intact channels on the surface of cells in all four cases, containing both subunit types and with Mr of ∼240 kD (Fig. 1 D). Evidence for these channels being correctly folded and assembled on the plasmalemma was provided by the avid binding of the specific blocker αDTX to Kv1.1-1.1-1.2-1.2 and Kv1.2-1.1-1.2-1.1 on intact HEK-293 cells, giving KD values of 0.74 (Fig. 2) and 1.7 nM, respectively. This high affinity corresponds to that observed for channels composed of tandem-linked Kv1.1-1.2 (Akhtar et al., 2002), suggesting that tetrameric concatenation does not affect toxin binding. Similar maximal displacement of 125I-αDTX binding to Kv1.1-1.1-1.2-1.2 (Fig. 2 B) by toxins specific for Kv1.1 (DTXk; Wang et al., 1999; Akhtar et al., 2002), Kv1.2 (TsTX-Kα; Werkman et al., 1993; Rogowski et al., 1994), or both (αDTX; Wang et al., 1999; Akhtar et al., 2002) (Table II) supports the presence of the two different subunits in assembled heterotetramers.
Figure 2.
Recombinant Kv1 channels assembled on the surface of mammalian cells in active form. (A) Saturable binding (▾) of 125I-αDTX to intact HEK-293 cells, expressing Kv1.1-1.1-1.2-1.2, quantified by a filtration assay. Relatively low values were recorded for nonsaturable (▴) binding compared with total (▪). Inset shows a Scatchard plot of the saturable binding. (B) Competition of 2.5 nM 125I-αDTX binding to cells transfected (as in A) by αDTX, DTXk, or TsTx-Kα, which gave respective mean values for Ki of 0.6, 2.0, and 0.1 nM (error bars are ± SD; n = 4). (C) Antagonism of 2.5 nM 125I-αDTX binding to cells expressing Kv1.1-1.1-1.2-1.2 (▪) or Kv1.2-1.1-1.2-1.1 (▴) channels by KTX gave respective IC50 values of 12.4 ± 0.2 and 5.6 ± 0.1 nM (mean ± SD; n = 3).
Table II.
Pharmacological properties of homomeric and heteromeric Kv1 channels
| IC50 values (± SEM) | |||||||||
| Blockers | Adjacent Kv1.1-1.1-1.2-1.2 | Reverse adjacent Kv1.2-1.2-1.1-1.1 | Diagonal Kv1.1-1.2-1.1-1.2 | Reverse diagonal Kv1.2-1.1-1.2-1.1 | Dimer Kv1.1-1.2 | Kv1.1 | Concatenated Kv(1.1)4 | Kv1.2 | Concatenated Kv(1.2)4 |
| TEA (mM) | 10 ± 0.2 (5) | 8 ± 1 (4) | 0.9 ± 0.1 (8) | 0.8 ± 0.1 (5) | 9 ± 1 (8) | 0.47 ± 0.1 (7) | 0.67 ± 0.1 (4) | 51 ± 5 (4) | 47 ± 6 (3) |
| AgTX1 (nM) | 15 ± 2 (5) [3.5 ± 0.1 (3)] | 17 ± 2 (3) | 9 ± 2 (2) | 12 ± 1 (11) [4.5 ± 0.1 (3)] | 20 ± 2 (8) | >100 (3) | >100 (3) | 35 ± 4 (3) | 26 ± 9 (2) |
| TsTx-Kα (nM) | 15 ± 2 (4) [1.1 ± 0.05(3)] | 12 ± 1 (6) | 14 ± 0.2 (2) | 17 ± 4 (9) [0.5 ± 0.2 (3)] | 11 ± 1 (7) | >100 (3) | >100 (3) | 3.1 ± 0.5 (5) | 2.6 ± 0.2 (3) |
| KTX (nM) | 4.4 ± 0.5 (5) [6.3 ± 0.1 (3)] | 6.4 ± 1 (4) | 11 ± 3 (2) | 7 ± 1 (3) [2.3 ± 0.04 (3)] | 6 ± 2 (6) | 53 ± 9 (2) | 36 ± 1 (2) | >100 (3) | >100 (2) |
Values (mean ± SEM) for activation and inactivation were derived by fitting to single- and double-exponential functions, respectively. V1/2 and slope k were calculated from Boltzmann equation fitting of the gk-V plots. IC50 values are from Hill equation fitting of Ik inhibition by TEA, AgTX1, TsTX-Kα, or KTX. Numbers in square brackets represent Ki values from competition binding experiments (in nM; see Fig. 2 D). n values in round brackets represent the number of experiments.
Adjacent or diagonal positioning of Kv1.1 and 1.2 genes in concatamers yielded K+ channels with slightly different biophysical properties
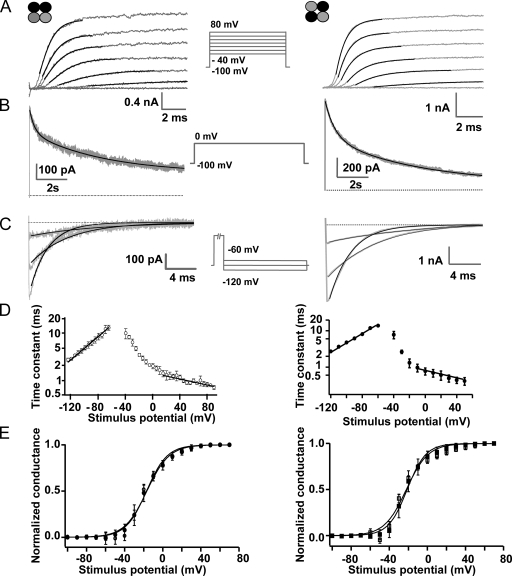
Kv1 heteromers were examined by conventional whole cell voltage clamp recording from HEK-293 cells transfected separately with each of the four tetrameric constructs. The adjacent construct, Kv1.1-1.1-1.2-1.2, gave a delayed-rectifying Kv current (IK) consistent with a single population of channels (Fig. 3, A–E). Depolarizing voltage steps triggered outward current, IK, typical of αDTX-sensitive neuronal K+ currents (Stansfeld et al., 1987). After a sigmoidal time course of activation (Fig. 3 A), the slow inactivation of IK showed two time constants from double-exponential fitting (Fig. 3 B; see τinact1 and τinact2 in Table I). Upon returning to negative voltage, IK decayed with a mono-exponential time course (Fig. 3, C and D, and Table I). Activation gating accelerated with a voltage dependence equivalent to 0.24 elementary charges at values positive to +20 mV (Fig. 3 D), giving a time constant (τact) at 0 mV of ∼2 ms (Table I). At membrane potentials <−60 mV, deactivation occurred with a voltage dependence corresponding to −0.81 elementary charges traversing the transmembrane and τ of ∼10 ms at −70 mV (Fig. 3 D). The conductance–voltage relationship (gK-V) revealed a profile well fitted by a single Boltzmann function (Fig. 3 E). Swapping the position of Kv1.1 and 1.2 subunits in the reverse adjacent expression construct yielded currents with similar kinetic and steady-state parameters (Table I) that were not statistically distinguishable in this dataset (P > 0.05; Mann-Whitney U test). Likewise, the reverse diagonal channel (Kv1.2-1.1-1.2-1.1) displayed parameters similar to its forward counterpart (Fig. 3, A–E, and Table I). Between the adjacent and diagonal heteromers, the conductance–voltage (V1/2) values and inactivation (τinact1) showed a small, yet significant difference (P < 0.05; Mann-Whitney U test). K+ currents generated from homomers were similar after concatenation with V1/2 values for Kv1.1 = −28.3 ± 4 mV (n = 3) and Kv1.1-1.1-1.1-1.1 = −25.3 ± 3 (n = 5), whereas Kv1.2 = −32 ± 1 (n = 3), Kv1.2-1.2 = −32 ± 1 (n = 2), and Kv1.2-1.2-1.2-1.2 = −29 ± 2 (n = 2), indicating that the voltage dependence of activation is not greatly altered by concatenating subunits.
Figure 3.
Kv1.1-1.1-1.2-1.2 and Kv1.2-1.1-1.2-1.1 yield functionally uniform K+ currents when expressed in HEK-293 cells. In A–E, data for the adjacent and diagonal channels are presented in the left and right panels, respectively. (A) K+ currents (IK), recorded by conventional patch clamp, in response to depolarizing steps (gray traces), from −40 to 80 mV in 20-mV increments (see protocol below), fit with a power of an exponential function (black lines). (B) Inactivation of IK (gray trace) during a pulse to 0 mV for 10 s, fitted with a double-exponential function (black line). (C) Deactivation of IK (gray traces) at −60, −90, or −120 mV after a 50-ms depolarization at +60 mV, fitted with a mono-exponential function (black lines). (D) Time constants associated with activation (>−50 mV) or deactivation (<−50 mV); lines are fits of exponential functions. Error bars represent ± SEM; n = 3. (E) Conductance–voltage relationship of IK currents after 100 ms at the indicated voltages from the forward adjacent (○; Kv1.1-1.1-1.2-1.2), reverse adjacent (•; Kv1.2-1.2-1.1-1.1), the forward diagonal (□; Kv1.1-1.2-1.1-1.2), and the reverse adjacent (▪; Kv1.2-1.1-1.2-1.1), calculated using −55 mV as reversal potential. Line is a Boltzmann fit, and parameters are shown in Table I. Error bars represent ± SD; some error bars fall within the data points; n ≥ 7.
Table I.
Biophysical properties of heteromeric Kv1 channels
| Parameters | Adjacent Kv1.1-1.1-1.2-1.2 | Reverse adjacent Kv1.2-1.2-1.1-1.1 | Diagonal Kv1.1-1.2-1.1-1.2 | Reverse diagonal Kv1.2-1.1-1.2-1.1 |
| τact at 0 mV (ms) | 1.9 ± 0.1 (3) | 2.3 ± 0.6 (3) | 1.4 ± 0.1 (3) | 1.3 ± 0.1 (4) |
| τinact1 at 0 mV (ms) | 540 ± 2 (5) | 529 ± 50 (4) | 306 ± 34 (3)a | 341 ± 17 (4)a |
| τinact2 at 0 mV | 6,200 ± 500 (5) | 5,053 ± 596 (4) | 3,586 ± 485 (3) | 4,372 ± 404 (4) |
| V 1/2 (mV) | −17 ± 1 (9) | −18 ± 1 (14) | −22 ± 1 (7)a | −21 ± 1 (10)a |
| Slope (k) | 11 ± 1 (9) | 12 ± 1 (14) | 11 ± 1 (7) | 10 ± 1 (10) |
Values (mean ± SEM) for activation and inactivation were derived by fitting to single- and double-exponential functions, respectively. V1/2 and slope k were calculated from Boltzmann equation fitting of the gk-V plots.
Significant difference from adjacent constructs (P < 0.05; Mann-Whitney U test).
Switching of Kv1.1 and 1.2 genes from adjacent to diagonal positions in the concatamers altered channel sensitivity to TEA, but not peptide toxins
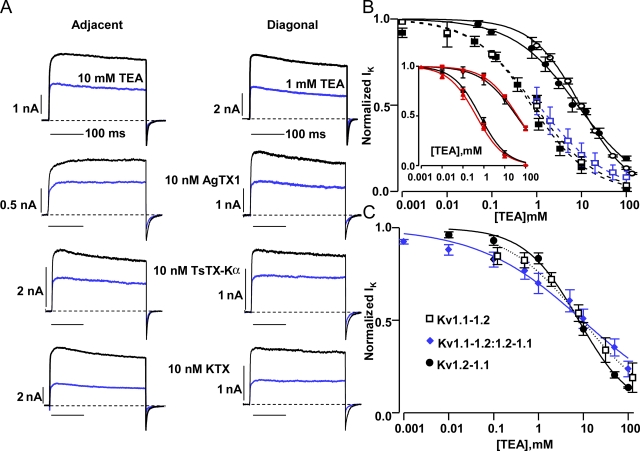
K+ currents from the forward and reverse versions of the adjacently and diagonally arranged channels showed similar susceptibility to AgTX1, TsTX-Kα, or KTX (Fig. 4 A and Table II). Toxins also showed similar Ki values for 125I-αDTX displacement from adjacent and diagonal constructs (Fig. 2 C and Table II). Concatamers of Kv1.1 and 1.2 homomeric channels showed almost identical pharmacological profiles as their homotetrameric counterparts, indicating that concatenation itself does not affect binding (Table II). Kv1.1 homomers were inhibited by KTX but displayed little susceptibility to AgTX1 or TsTX-Kα, indicating that in heteromers, Kv1.1 subunits should be permissive to the binding of AgTX1 or TsTX-Kα. In contrast, concatenated and non-concatenated Kv1.2 exhibited appreciable sensitivity to AgTX1 and TsTX-Kα, but not KTX. Thus, inclusion of Kv1.2 in tetramers allows, even enhances, KTX binding. The uniform sensitivity of the adjacent and diagonal channels to toxins is consistent with all of the expressed channels maintaining the same subunit stoichiometry.
Figure 4.
Adjacently and diagonally arranged Kv1.1 and 1.2 tandem-linked gene constructs gave channels that were distinguished by TEA, but not AgTX1, TsTX-Kα, or KTX. (A) Current traces recorded using QPatch from an adjacent (Kv1.1-1.1-1.2-1.2) and diagonal (Kv1.2-1.1-1.2-1.1) channel in the absence (black) and presence of TEA, AgTX1, TsTX-Kα, or KTX (blue). (B) Dose–response curves for the forward and reverse adjacent channels show an ∼10-fold lower affinity for TEA than the corresponding diagonals. ○, Kv1.1-1.1-1.2-1.2; n = 5 cells for each concentration, manual patch clamp; Hill equation fit IC50 = 9.6 mM, slope = 0.8. •, Kv1.2-1.2-1.1-1.1; Qpatch, n = 2–3; 8.2 mM, 0.6. □, Kv1.1-1.2-1.1-1.2; n = 4, Qpatch; 0.9 mM, 0.6. (blue) □, Kv1.1-1.2-1.1-1.2; n = 6–8, manual patch clamp; 1.1 mM, 0.6. ▪, Kv1.2-1.1-1.2-1.1; n = 3–5, Qpatch; 0.8 mM, 0.7. (Inset) Non-concatenated (red ▴; n = 6–7; 0.4 mM, 0.6) and concatenated (black ▴; n = 2–4; 0.7 mM, 0.7) homotetrameric Kv1.1 channels showed almost identical sensitivity to TEA, as with the less sensitive homotetrameric Kv1.2 (red ▾; n = 2–4; 41 mM, 0.6) channel and the tandem-linked homotetramer (black ▾; n = 3; 36 mM, 0.5). All homomer data were from Qpatch. (C) Channel dimers showed TEA susceptibilities similar to that for the pair of adjacently arranged tetrameric concatamers. □, Kv1.1-1.2; n = 8; 9.3 mM, 1.0. •, Kv1.2-1.1; n = 5–6; 7.8 mM, 0.7. ◆, Kv1.1-1.2 + Kv1.2-1.1; n = 9–11; 9.5 mM, 0.4. Data were from Qpatch. Error bars represent ± SEM.
K+ currents produced by the forward and reverse adjacent constructs were found to be inhibited by TEA, which is known to bind all four sensitive subunits just external to the K+ conduction pore (Hurst et al., 1992). Representative traces from cells expressing the Kv1.1-1.1-1.2-1.2 channel showed that 10 mM TEA was required to block ∼50% of the K+ current (Fig. 4, A and B). Averages of full datasets for the forward and reverse adjacent arrangements revealed that the reverse adjacent construct is similarly sensitive (Fig. 4 B and Table II). The levels of TEA inhibition seen with these adjacent Kv1.1/1.2 heteromers were intermediate between those of Kv1.1 and Kv1.2 parental homomers, with concatenation having no discernable effect on TEA inhibition (Fig. 4 B, inset, red lines, and Table II). In marked contrast, the IK produced by channels constructed with diagonal positioning of the Kv1 genes proved to be more sensitive to TEA, with 50% block at ∼1 mM (Fig. 4, A and B, and Table II). This surprising observation, made with both the forward and reverse diagonal arrangements, provides evidence that interspersed ordering of the two Kv1.1 subunits results in structurally distinct channels that allow more intimate interaction of TEA with the channel mouth. The TEA sensitivity of the diagonal constructs is similar to Kv1.1. Also, dose responses obtained from the conventional patch clamp recordings are in agreement with the above-noted values observed with the automated system. It is noteworthy that channels obtained from Kv1.1-1.2 or Kv1.2-1.1 dimeric constructs displayed TEA susceptibilities not distinguishable from both of the adjacent (P > 0.05; t test) but significantly different from the diagonal tetramers (P < 0.0001). Likewise, similar susceptibility to TEA was observed (IC50 = 10 ± 2 mM; n = 11; Fig. 4 C) for channels formed when Kv1.1-1.2 and Kv1.2-1.1 dimers were coexpressed using equal amounts of their constructs.
DISCUSSION
Demonstrably uniform populations of concatamerized Kv channels have yielded novel biophysical insights into channel function and pharmacology (Kavanaugh et al., 1991; Hurst et al., 1992; Liman et al., 1992; Gagnon and Bezanilla, 2009). Although such desirable channel assemblies can be engineered, expression products may not resemble the intended design, and each concatamerized construct needs to be validated. Failures of concatemeric Kv constructs have been thoughtfully documented in earlier studies (McCormack et al., 1992; Hurst et al., 1995), necessitating careful controls to assess the composition and uniformity of the expressed channel population (see Sack et al., 2008). In the current study, the expressed channels were tested for uniformity by a variety of means. The fact that the respective conductance–voltage relationships obtained for all the various assemblies of K+ channels expressed (Kv1.1 or 1.2 homomers and their concatenated forms between the pairs of adjacently and diagonally arranged heterotetramers) were similar, and the absence of channels with voltage sensitivity deviating from the norm, is indicative of changes not being introduced by concatenation. The forward and reverse versions of each construct were readily obtainable. Forward constructs acted as internal controls for reverse counterparts. Positions of subunits were completely swapped for each construct and gave no indication of a position-dependent overrepresentation of subunits, a problem noted with Kv1 concatamers expressed in Xenopus oocytes (McCormack et al., 1992; Hurst et al., 1995). Because the turret extracellular region between Kv1.1 and 1.2 subunits is different (see Scheme 1), and C-type inactivation involves rearrangements at the selectivity filter and extracellular entrance to the pore, positioning of these subunits in diagonal or adjacent format might explain the difference in their susceptibilities to TEA and τinact1 values. The dose–response profiles of TEA inhibition revealed Hill slopes consistently less than unity (Fig. 4, B and C), which is troubling, as such behavior would be expected of mixed populations of channels with different TEA affinities. However, the Hill slopes of TEA inhibition of homomeric channels were similar, suggesting that these shallow slopes are an artifact of the experimental conditions, likely from the variable contribution of endogenous channels and/or leak currents to each cell. Importantly, although TEA affinities differed between adjacent and diagonal constructs, sensitivities to αDTX, AgTX1, TsTX-Kα, and KTX were indistinguishable in channels expressed from all constructs bearing two Kv1.1 and two Kv1.2 subunits. Although possible, the fortuitous manifestation of these ligand-binding properties in both the expected channels and ones composed of aberrant stoichiometries would be remarkable. Thus, the most logical explanation of the data appears to be that tandem linkage of the genes determines the arrangement of subunits in channels containing two copies of Kv1.1 and Kv1.2. Previous efforts using tandem-linked constructs of Kv1.1 and Kv1.2 sequences have resulted in channels with ligand-binding properties indicative of a heterogenous population of channel assemblies (Middleton et al., 2003). The distinct TEA sensitivities observed herein for adjacent versus diagonal arrangements offer evidence that the optimized concatenation technique overcomes this limitation of past constructs and fixes the rotational ordering of K+ channel subunits. The linkages created with the present methodology appear to ensure the desired combination of α subunits in the recombinant tetramers and, also, allow these to be arranged in adjacent or diagonal positions. The finding that the adjacently ordered tetramers and tandem-linked dimers have a similar TEA affinity leads one to the supposition that Kv1.1/1.2 heterodimers preferentially assemble into a structure similar to adjacently ordered tetramers. Presently, it is unclear how this surprising experimental observation relates to the in vivo assembly of hetero-oligomeric Kv1 channels.
Thanks to extensive research and especially mutant cycle screens of agitoxin-2 binding to the Shaker Kv1 channel, the binding site for pore-blocking scorpion peptides is well understood. These toxins occlude the outer mouth of the channel pore by interacting with all four Kv1 α subunits (Hidalgo and MacKinnon, 1995; Long et al., 2005). Many residues in the S5–S6 segment of Kv1 channels interact with these toxins, and residues that, when mutated, caused a 10-fold or greater change in affinity (Gross et al., 1994; Ranganathan et al., 1996) are indicated in Scheme 1.
Sequence alignment. The most critical TEA-binding residue is circled, and those that interact with scorpion toxins are red.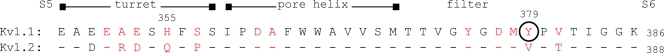
|
SCHEME 1. |
The differential sensitivity of Kv1.1 and Kv1.2 channels to the AgTX2-related scorpion toxins tested in this study is likely to result from variations in these toxin-interacting residues (Garcia et al., 1998). This variability also creates a unique binding interface for toxins in each distinct heteromer of the two channels. Interestingly, KTX, TsTX-Kα, and AgTX1 all bound avidly to channels that contained two subunits insensitive to each toxin, yet none of the toxins could distinguish between adjacent and diagonal subunit arrangements. This suggests that the ability of heteromers to bind these peptides is determined by a high affinity binding site on only a portion of the outer pore, with toxin-insensitive Kv1 subunits being permissive to toxin binding. The postulate of permissive subunits is supported by the ability of other scorpion toxins to bind to Kv1 heteromultimers from the brain that contain toxin-insensitive subunits (Koschak et al., 1998), and is reminiscent of the ability of snake-derived dendrotoxins to bind channels with toxin-insensitive subunits (Shamotienko et al., 1997; Coleman et al., 1999; Akhtar et al., 2002). It is worth noting that not all mutations to the toxin-binding site of Kv1 channels are permissive in heteromultimers and will abolish toxin binding in channels with two insensitive subunits (Gross et al., 1994). Thus, the sensitivity of Kv channels to pore-blocking peptide toxins cannot be predicted a priori by the presence of sensitive subunits, and empirical testing of heteromers is required to establish ligand affinity.
It remains a challenge to decipher a mechanism for the most intriguing outcome from this investigation, namely, that K+ channel concatamers with a diagonal arrangement display a higher susceptibility to TEA than the adjacent configuration. All four K+ channel subunits are known to make an energetic contribution to the binding of a single TEA molecule at the external mouth, where an aromatic residue in the pore-forming region is required for high sensitivity (MacKinnon et al., 1990; Kavanaugh et al., 1991; Heginbotham and MacKinnon, 1992; Hurst et al., 1992). The Kv1.1 homotetramer, which possesses Y379 at that position, proved much more susceptible to TEA in the present and past reports than that composed of Kv1.2, which has a valine at the equivalent location (see Scheme 1). Studies in which this aromatic residue alone was mutated found that the arrangement of TEA-sensitive subunits does not affect the channels’ affinity (Hurst et al., 1992; see also Shen et al., 1994). However, these observations do not preclude the possibility that arrangement of other sensitive and insensitive subunits may affect TEA binding. The key aromatic residue is not a sole determinant of the affinity for this blocker, as mutation of others also alters its binding (Kavanaugh et al., 1991; Heginbotham and MacKinnon, 1992; Andalib et al., 2004; Ahern et al., 2009). These investigations all suggest that ligand affinity can be influenced by residues that do not directly contact TEA. Dissimilarities between Kv1.1 and 1.2, other than the tyrosine to valine change, may contribute to the difference in TEA sensitivities of adjacent and diagonal heteromers. For example, the charged H355 at the external mouth of Kv1.1 (when protonated at lower pH) can repel the cationic TEA (Bretschneider et al., 1999). Moreover, affinity for external TEA can be modulated by allosteric means. For example, the affinity of Kv2.1 channel for external TEA is dynamic and radically altered by conformational changes within the channel pore (Ikeda and Korn, 1995; Immke and Korn, 2000). A reorganization of residues in the TEA-binding region within the pore of adjacent concatamers might also result in unexpected binding. In the absence of detailed structure of the TEA-binding site in these heteromeric channels, it is difficult to discern what can account for the differential sensitivity. One final, intriguing hypothesis we present is that the increased sensitivity of the diagonal tetramers is due to the tetramers matching the symmetry of the TEA molecule itself. In a structure of TEA binding to the external pore of the KcsA channel, TEA is positioned with a twofold rotational symmetry (Lenaeus et al., 2005). That is, it makes equivalent contacts with each diagonally positioned subunit, via cation–π interactions with the critical aromatic tyrosine (Ahern et al., 2009). Perhaps one set of these diagonal contacts provides more binding energy than the other, and the diagonally positioned tyrosines of Kv1.1 provide these high affinity contacts, making the diagonal heteromers more sensitive to TEA.
Acknowledgments
We thank Professor Olaf Pongs for the kind gift of Kv1 α subunit genes, Patrick Jennings for the cell culture, and Alison Byrne for manuscript preparation.
This work was supported by a Research Professorship award from Science Foundation Ireland (to J.O. Dolly), a PRTLI 4 grant from the Irish Higher Education Authority for the Neuroscience section of “Target-driven therapeutics and theranostics,” and an IRCSET/Marine Institute postgraduate studentship (to N. Muniyappa).
Lawrence G. Palmer served as editor.
Footnotes
Abbreviations used in this paper:
- αDTX
- α dendrotoxin
- AgTX1
- agitoxin-1
- DTXk
- dendrotoxink
- KTX
- kaliotoxin
- Kv
- voltage-gated K+
- MCS
- multiple cloning site
- TEA
- tetraethylammonium
- TsTX-Kα
- tityustoxin-Kα
References
- Ahern C.A., Eastwood A.L., Dougherty D.A., Horn R. 2009. An electrostatic interaction between TEA and an introduced pore aromatic drives spring-in-the-door inactivation in Shaker potassium channels. J. Gen. Physiol. 134:461–469 10.1085/jgp.200910260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar S., Shamotienko O., Papakosta M., Ali F., Dolly J.O. 2002. Characteristics of brain Kv1 channels tailored to mimic native counterparts by tandem linkage of alpha subunits: implications for K+ channelopathies. J. Biol. Chem. 277:16376–16382 10.1074/jbc.M109698200 [DOI] [PubMed] [Google Scholar]
- Andalib P., Consiglio J.F., Trapani J.G., Korn S.J. 2004. The external TEA binding site and C-type inactivation in voltage-gated potassium channels. Biophys. J. 87:3148–3161 10.1529/biophysj.104.046664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider F., Wrisch A., Lehmann-Horn F., Grissmer S. 1999. External tetraethylammonium as a molecular caliper for sensing the shape of the outer vestibule of potassium channels. Biophys. J. 76:2351–2360 10.1016/S0006-3495(99)77392-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman S.K., Newcombe J., Pryke J., Dolly J.O. 1999. Subunit composition of Kv1 channels in human CNS. J. Neurochem. 73:849–858 10.1046/j.1471-4159.1999.0730849.x [DOI] [PubMed] [Google Scholar]
- Gagnon D.G., Bezanilla F. 2009. A single charged voltage sensor is capable of gating the Shaker K+ channel. J. Gen. Physiol. 133:467–483 10.1085/jgp.200810082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.L., Hanner M., Kaczorowski G.J. 1998. Scorpion toxins: tools for studying K+ channels. Toxicon. 36:1641–1650 10.1016/S0041-0101(98)00157-3 [DOI] [PubMed] [Google Scholar]
- Gross A., Abramson T., MacKinnon R. 1994. Transfer of the scorpion toxin receptor to an insensitive potassium channel. Neuron. 13:961–966 10.1016/0896-6273(94)90261-5 [DOI] [PubMed] [Google Scholar]
- Heginbotham L., MacKinnon R. 1992. The aromatic binding site for tetraethylammonium ion on potassium channels. Neuron. 8:483–491 10.1016/0896-6273(92)90276-J [DOI] [PubMed] [Google Scholar]
- Hidalgo P., MacKinnon R. 1995. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 268:307–310 10.1126/science.7716527 [DOI] [PubMed] [Google Scholar]
- Hurst R.S., Kavanaugh M.P., Yakel J., Adelman J.P., North R.A. 1992. Cooperative interactions among subunits of a voltage-dependent potassium channel. Evidence from expression of concatenated cDNAs. J. Biol. Chem. 267:23742–23745 [PubMed] [Google Scholar]
- Hurst R.S., North R.A., Adelman J.P. 1995. Potassium channel assembly from concatenated subunits: effects of proline substitutions in S4 segments. Receptors Channels. 3:263–272 [PubMed] [Google Scholar]
- Ikeda S.R., Korn S.J. 1995. Influence of permeating ions on potassium channel block by external tetraethylammonium. J. Physiol. 486:267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immke D., Korn S.J. 2000. Ion–ion interactions at the selectivity filter. Evidence from K+-dependent modulation of tetraethylammonium efficacy in Kv2.1 potassium channels. J. Gen. Physiol. 115:509–518 10.1085/jgp.115.4.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imredy J.P., MacKinnon R. 2000. Energetic and structural interactions between delta-dendrotoxin and a voltage-gated potassium channel. J. Mol. Biol. 296:1283–1294 10.1006/jmbi.2000.3522 [DOI] [PubMed] [Google Scholar]
- Jan L.Y., Jan Y.N. 1997. Cloned potassium channels from eukaryotes and prokaryotes. Annu. Rev. Neurosci. 20:91–123 10.1146/annurev.neuro.20.1.91 [DOI] [PubMed] [Google Scholar]
- Judge S.I., Bever C.T., Jr 2006. Potassium channel blockers in multiple sclerosis: neuronal Kv channels and effects of symptomatic treatment. Pharmacol. Ther. 111:224–259 10.1016/j.pharmthera.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Kavanaugh M.P., Varnum M.D., Osborne P.B., Christie M.J., Busch A.E., Adelman J.P., North R.A. 1991. Interaction between tetraethylammonium and amino acid residues in the pore of cloned voltage-dependent potassium channels. J. Biol. Chem. 266:7583–7587 [PubMed] [Google Scholar]
- Koch R.O., Wanner S.G., Koschak A., Hanner M., Schwarzer C., Kaczorowski G.J., Slaughter R.S., Garcia M.L., Knaus H.G. 1997. Complex subunit assembly of neuronal voltage-gated K+ channels. Basis for high-affinity toxin interactions and pharmacology. J. Biol. Chem. 272:27577–27581 10.1074/jbc.272.44.27577 [DOI] [PubMed] [Google Scholar]
- Koschak A., Bugianesi R.M., Mitterdorfer J., Kaczorowski G.J., Garcia M.L., Knaus H.G. 1998. Subunit composition of brain voltage-gated potassium channels determined by hongotoxin-1, a novel peptide derived from Centruroides limbatus venom. J. Biol. Chem. 273:2639–2644 10.1074/jbc.273.5.2639 [DOI] [PubMed] [Google Scholar]
- Lenaeus M.J., Vamvouka M., Focia P.J., Gross A. 2005. Structural basis of TEA blockade in a model potassium channel. Nat. Struct. Mol. Biol. 12:454–459 10.1038/nsmb929 [DOI] [PubMed] [Google Scholar]
- Liman E.R., Tytgat J., Hess P. 1992. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 9:861–871 10.1016/0896-6273(92)90239-A [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- MacKinnon R., Heginbotham L., Abramson T. 1990. Mapping the receptor site for charybdotoxin, a pore-blocking potassium channel inhibitor. Neuron. 5:767–771 10.1016/0896-6273(90)90335-D [DOI] [PubMed] [Google Scholar]
- McCormack K., Lin L., Iverson L.E., Tanouye M.A., Sigworth F.J. 1992. Tandem linkage of Shaker K+ channel subunits does not ensure the stoichiometry of expressed channels. Biophys. J. 63:1406–1411 10.1016/S0006-3495(92)81703-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R.E., Sanchez M., Linde A.R., Bugianesi R.M., Dai G., Felix J.P., Koprak S.L., Staruch M.J., Bruguera M., Cox R., et al. 2003. Substitution of a single residue in Stichodactyla helianthus peptide, ShK-Dap22, reveals a novel pharmacological profile. Biochemistry. 42:13698–13707 10.1021/bi035209e [DOI] [PubMed] [Google Scholar]
- Parcej D.N., Scott V.E., Dolly J.O. 1992. Oligomeric properties of alpha-dendrotoxin-sensitive potassium ion channels purified from bovine brain. Biochemistry. 31:11084–11088 10.1021/bi00160a018 [DOI] [PubMed] [Google Scholar]
- Pongs O. 2008. Regulation of excitability by potassium channels. Results Probl. Cell Differ. 44:145–161 10.1007/400_2007_032 [DOI] [PubMed] [Google Scholar]
- Ranganathan R., Lewis J.H., MacKinnon R. 1996. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 16:131–139 10.1016/S0896-6273(00)80030-6 [DOI] [PubMed] [Google Scholar]
- Rogowski R.S., Krueger B.K., Collins J.H., Blaustein M.P. 1994. Tityustoxin K alpha blocks voltage-gated noninactivating K+ channels and unblocks inactivating K+ channels blocked by alpha-dendrotoxin in synaptosomes. Proc. Natl. Acad. Sci. USA. 91:1475–1479 10.1073/pnas.91.4.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack J.T., Aldrich R.W. 2006. Binding of a gating modifier toxin induces intersubunit cooperativity early in the Shaker K channel’s activation pathway. J. Gen. Physiol. 128:119–132 10.1085/jgp.200609492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack J.T., Shamotienko O., Dolly J.O. 2008. How to validate a heteromeric ion channel drug target: assessing proper expression of concatenated subunits. J. Gen. Physiol. 131:415–420 10.1085/jgp.200709939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott V.E., Muniz Z.M., Sewing S., Lichtinghagen R., Parcej D.N., Pongs O., Dolly J.O. 1994a. Antibodies specific for distinct Kv subunits unveil a heterooligomeric basis for subtypes of alpha-dendrotoxin-sensitive K+ channels in bovine brain. Biochemistry. 33:1617–1623 10.1021/bi00173a001 [DOI] [PubMed] [Google Scholar]
- Scott V.E., Rettig J., Parcej D.N., Keen J.N., Findlay J.B., Pongs O., Dolly J.O. 1994b. Primary structure of a beta subunit of alpha-dendrotoxin-sensitive K+ channels from bovine brain. Proc. Natl. Acad. Sci. USA. 91:1637–1641 10.1073/pnas.91.5.1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamotienko O.G., Parcej D.N., Dolly J.O. 1997. Subunit combinations defined for K+ channel Kv1 subtypes in synaptic membranes from bovine brain. Biochemistry. 36:8195–8201 10.1021/bi970237g [DOI] [PubMed] [Google Scholar]
- Shen K.Z., Lagrutta A., Davies N.W., Standen N.B., Adelman J.P., North R.A. 1994. Tetraethylammonium block of Slowpoke calcium-activated potassium channels expressed in Xenopus oocytes: evidence for tetrameric channel formation. Pflugers Arch. 426:440–445 10.1007/BF00388308 [DOI] [PubMed] [Google Scholar]
- Sokolov M.V., Shamotienko O., Dhochartaigh S.N., Sack J.T., Dolly J.O. 2007. Concatemers of brain Kv1 channel alpha subunits that give similar K+ currents yield pharmacologically distinguishable heteromers. Neuropharmacology. 53:272–282 10.1016/j.neuropharm.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Stansfeld C.E., Marsh S.J., Parcej D.N., Dolly J.O., Brown D.A. 1987. Mast cell degranulating peptide and dendrotoxin selectively inhibit a fast-activating potassium current and bind to common neuronal proteins. Neuroscience. 23:893–902 10.1016/0306-4522(87)90166-7 [DOI] [PubMed] [Google Scholar]
- Wang F.C., Bell N., Reid P., Smith L.A., McIntosh P., Robertson B., Dolly J.O. 1999. Identification of residues in dendrotoxin K responsible for its discrimination between neuronal K+ channels containing Kv1.1 and 1.2 alpha subunits. Eur. J. Biochem. 263:222–229 10.1046/j.1432-1327.1999.00494.x [DOI] [PubMed] [Google Scholar]
- Werkman T.R., Gustafson T.A., Rogowski R.S., Blaustein M.P., Rogawski M.A. 1993. Tityustoxin-K alpha, a structurally novel and highly potent K+ channel peptide toxin, interacts with the alpha-dendrotoxin binding site on the cloned Kv1.2 K+ channel. Mol. Pharmacol. 44:430–436 [PubMed] [Google Scholar]