Abstract
End-stage liver disease caused by chronic hepatitis C virus (HCV) infection is a leading cause for liver transplantation (LT). Due to viral evasion from host immune responses and the absence of preventive antiviral strategies, reinfection of the graft is universal. The mechanisms by which the virus evades host immunity to reinfect the liver graft are unknown. In a longitudinal analysis of six HCV-infected patients undergoing LT, we demonstrate that HCV variants reinfecting the liver graft were characterized by efficient entry and poor neutralization by antibodies present in pretransplant serum compared with variants not detected after transplantation. Monoclonal antibodies directed against HCV envelope glycoproteins or a cellular entry factor efficiently cross-neutralized infection of human hepatocytes by patient-derived viral isolates that were resistant to autologous host-neutralizing responses. These findings provide significant insights into the molecular mechanisms of viral evasion during HCV reinfection and suggest that viral entry is a viable target for prevention of HCV reinfection of the liver graft.
Hepatitis C virus (HCV)–related cirrhosis and hepatocellular carcinoma are leading indications for liver transplantation (LT). A major limitation is the universal HCV reinfection of the graft followed by an accelerated course of virus-induced liver disease (Brown, 2005). A prophylactic strategy for prevention of reinfection is lacking, and interferon-based antiviral therapies have limited efficacy and tolerability in LT recipients (Brown, 2005). Thus, recurrent liver disease with poor outcome has become an increasing problem facing hepatologists and transplant surgeons, underlying the urgent need for novel strategies for prevention of reinfection.
The development of preventive antiviral strategies has been hampered by a limited understanding of the mechanisms leading to HCV reinfection. Reinfection occurs within a few hours of graft reperfusion despite the presence of anti-HCV antibodies (Brown, 2005). Evolution of viral quasispecies rapidly changes after transplantation, and only a small fraction of viral variants present before transplantation is selected after LT (Moreno Garcia et al., 2003; Feliu et al., 2004; Brown, 2005; Schvoerer et al., 2007). These observations suggest that HCV has developed efficient strategies to evade host immunity and adapt rapidly to the new host environment. The mechanisms by which viral variants are selected and HCV evades host immunity to establish persistence in transplanted patients are not understood.
HCV has a very high replication rate, and the highly error-prone viral polymerase allows for rapid production of minor viral variants that may outpace humoral and cellular immune responses (Bowen and Walker, 2005; Ray et al., 2005; von Hahn et al., 2007; Uebelhoer et al., 2008; Aurora et al., 2009; Dazert et al., 2009). These variants are under constant immune pressure in the infected host, and selection processes lead to domination of the viral quasispecies by the most fit virus that can also evade immune recognition (Uebelhoer et al., 2008).
Both viral and host factors are potential determinants for evasion from host responses and adaptation of the virus after transplantation. Viral entry is the very first step of HCV infection (Evans et al., 2007; Zeisel et al., 2007, 2008; von Hahn and Rice, 2008; Ploss et al., 2009) and is thus an important factor for initiation of infection of the naive liver graft. Moreover, viral entry is a major target of neutralizing antibodies, a first-line host defense inhibiting viral spread. Indeed, the rapid induction of cross-neutralizing antibodies in the very early phase of infection has been suggested to contribute to control of HCV infection (Lavillette et al., 2005; Pestka et al., 2007).
In this study, we aimed to investigate whether viral entry and escape from neutralizing antibodies are determinants for viral evasion and persistence during the very early phase of graft infection. Infectious retroviral HCV pseudoparticles (HCVpps) have been shown to represent a robust and valid system for the study of HCV entry and antibody-mediated neutralization in clinical cohorts (Lavillette et al., 2005; Dreux et al., 2006; Pestka et al., 2007; Grove et al., 2008; Haberstroh et al., 2008; Zeisel et al., 2008; Witteveldt et al., 2009). Using HCVpps bearing viral envelope glycoproteins derived from patients undergoing LT, we show that efficient viral entry and escape from antibody-mediated neutralization are key determinants for selection of viral variants reinfecting the liver graft. Furthermore, we demonstrate that mAbs directed against viral or host entry factors efficiently cross-neutralized infection of human hepatocytes by patient-derived viral isolates that were resistant to autologous host-neutralizing responses. These results define the molecular mechanisms of viral evasion during HCV reinfection and suggest that viral entry is a viable target for the prevention of HCV reinfection of the liver graft.
RESULTS
Composition and diversity of HCV variants before and after LT
To study the impact of viral entry and neutralization on viral evasion during LT, we investigated viral quasispecies evolution in six patients infected with HCV genotype 1b undergoing LT (Table I). A total of 439 clones (mean, 24 per time point and patient; range, 20–31) was obtained by RT-PCR from serum before and 7 d and 1 mo after transplantation. To investigate the evolutionary dynamics of the envelope quasispecies evolution, distribution of viral quasispecies was analyzed as described previously for other cohorts (Farci et al., 2000; Feliu et al., 2004; Schvoerer et al., 2007).
Table I.
Clinical and virological features of HCV-infected patients undergoing LT
| Patient number | Age R/D | Gender | Indication of transplantation and MELD | HCV genotype | Viral load | IS treatment | ||
| BT | D7 | M1 | ||||||
| yr | log10 IU/ml | log10 IU/ml | log10 IU/ml | |||||
| P01 | 60/73 | Male | Cirrhosis + HCC MELD 9.32 | 1b | 5. 94 | 5.20 | 5.72 | tac/cor |
| P02 | 34/65 | Male | Cirrhosis MELD 10.37 | 1b | 5.14 | 4.18 | 5.14 | tac/rap/cor |
| P03 | 64/74 | Male | Cirrhosis + HCC MELD 9.53 | 1b | 5. 61 | 3.65 | 6.83 | tac/rap/cor |
| P04 | 69/15 | Female | Cirrhosis + HCC MELD 23.59 | 1b | 6.42 | 4.94 | 6.90 | tac/rap/cor |
| P05 | 51/22 | Male | Cirrhosis MELD 14.25 | 1b | 5.39 | 4.85 | 5.22 | tac/cor |
| P06 | 65/68 | Female | Cirrhosis + HCC MELD 11.99 | 1b | 5.96 | 6.19 | 6.90 | tac/rap/cor |
HCC, hepatocellular carcinoma; tac, tacrolimus; rap, rapamycin; cor, corticosteroids. Patient number, age of recipient (R) and donor (D), recipient gender, indication of transplantation, model of end-stage liver disease (MELD) score before transplantation, HCV genotype, viral load (before [BT] and day 7 [D7] and month 1 [M1] after transplantation), and immunosuppressive (IS) treatment are shown.
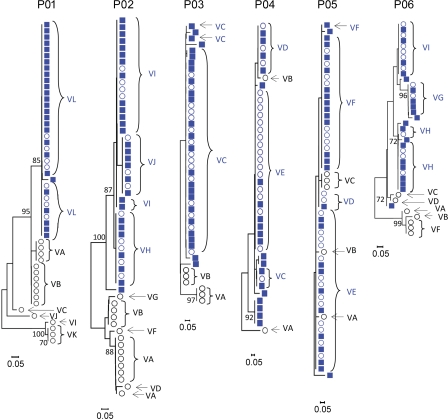
Sequence analysis demonstrated the presence of different viral variants in the serum before and after transplantation (Figs. 1 and 2 and Figs. S1 and S2). Several variants (e.g., VA to VK in patient 01) were not detected after transplantation (termed nonselected variants), whereas other variants (termed selected variants) were detected both before and after transplantation and were the predominant viral isolates after transplantation (e.g., variant VL in patient 01; Fig. 2). Phylogenetic analyses of viral sequences detected before and 7 d after transplantation revealed a marked and abrupt decrease of diversity (bottleneck effect) at day 7 in four out of six patients (P01, P02, P03, and P06), with selected variants clustering in a single branch (Fig. 1). The viral diversity in the other two patients (P04 and P05), which was low before transplantation, remained stable before and after transplantation (Fig. 1).
Figure 1.
Phylogenetic analyses of viral quasispecies evolution before and 7 d after LT. Evolution of HCV variants before and 7 d after LT. Rooted neighbor-joining trees of HCV HVR1 amino acid sequences from the six patients depicted in Table I are shown. Bootstrap values are expressed as percentages per 1,000 replicates. Only bootstrap proportions >70 are indicated. Sequences of variants isolated before LT and not detected on day 7 after transplantation are depicted as open black circles, sequences of variants isolated before LT and reinfecting the graft are depicted as open blue circles, and sequences isolated 7 d after LT are depicted as blue squares. Viral isolates used for functional analysis are indicated with capital letters.
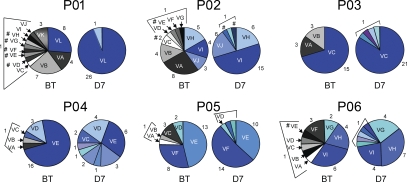
Figure 2.
Evolution of HCV variants before and 7 d after LT. Distribution of full-length E1E2 variants (mean number of clones per patient, 24; range, 20–31) depicted in Fig. 1 is shown for each patient. Circle graphs represent the percentage of each clone detected. The number of each clone is indicated. Viral isolates are indicated by an individual color and capital letters. Variants containing stop codons, insertions, or deletions altering the HCV open reading frame are depicted with a number sign (#) and were not further analyzed in HCVpp assays. Variants reinfecting the liver graft are depicted in blue, and nonselected variants not detected after transplantation are depicted in white, gray, or black. BT, before LT; D7, 7 d after LT
Compared with the marked decrease in diversity of variants in the majority of patients immediately after transplantation (Fig. 1), the virus population remained homogeneous 1 mo after transplantation (Fig. S1). The mean genetic distances between the sequences obtained at day 7 and those obtained at month 1 were 1%, 1%, <0.1%, 1.1%, 1.3%, and 4% for P01, P02, P03, P04, P05, and P06, respectively. Sequences isolated at day 7 and month 1 after LT were clustered in the same branch of phylogenetic trees (Fig. S1), and variants selected at day 7 remained predominant 1 mo after transplantation in all patients (Fig. S1 and not depicted). Sequence analysis of serum samples at month 6 and 12 after transplantation available from three patients demonstrated that variants selected during the early phase of transplantation (P01VL, P04VE, and P06VI) had the same or similar envelope glycoprotein sequence as predominant variants present at later time points after transplantation (unpublished data). These data indicate that the new host environment leads to an abrupt and marked change in the composition and diversity of the viral quasispecies in the majority of patients, which remains relatively stable after transplantation.
Viral entry is important for selection of viral variants after LT
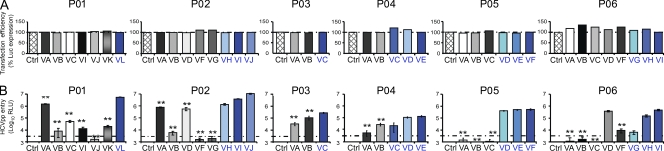
To determine the mechanism of selection and evasion of viral variants during reinfection, we produced HCVpps harboring full-length E1E2 glycoproteins of all detected and grouped variants before transplantation (nonselected variants; Figs. 1 and 2) as well as the most prevalent variants reinfecting the liver graft (selected variants; Fig. 2). Sequence analysis of viral isolates showed that 13 out of 63 isolates contained stop codons or insertions altering the open reading frame. These variants were not further analyzed for HCVpp production and are indicated by a number sign (#) in the respective pie charts of Fig. 2. Expression of envelope glycoproteins from 33 out of 50 isolates (66%) resulted in the production of infectious HCVpps (Fig. 3). Complete sequences of isolates characterized in functional experiments are shown in Fig. S2. The strains reinfecting the graft always produced functional E1E2 (Fig. 3), whereas a large fraction of nonselected variants failed to produce infectious HCVpps (Fig. 3). The percentage of patient-derived viral isolates resulting in infectious HCVpps was higher than in a recent longitudinal study of a single HCV-infected patient (von Hahn et al., 2007). This difference is most likely caused by virus-specific factors allowing different use of entry factors because we observed a large variation in the percentage of infectious isolates within the six patients (37–100%). Alternatively, technical factors such as the use of more permissive Huh7 cells may have resulted in the isolation of more variants with more easily detectable HCVpp infection in this study.
Figure 3.
Viral entry is important for selection of viral variants after LT. (A) Transfection efficiency was analyzed for each variant and control (Ctrl; empty vector without E1E2) by quantifying luciferase expression (expressed as normalized percentage of transfection efficiency based on the predominant selected variant). (B) Comparative analysis of viral entry of HCVpps containing full-length E1E2 functional envelope proteins from pretransplant variants in Huh7 cells depicted in Fig. 2. HCVpps bearing patient-derived HCV envelope glycoproteins were added to Huh7 cells, and infection was analyzed by luciferase reporter gene expression. Results are expressed in relative light units (RLU) plotted in a logarithmic scale. The threshold for a detectable infection in this system is indicated by dashed lines. The detection limit for positive luciferase reporter protein expression was 3 × 103 RLU/assay, corresponding to the mean ± 3 SD of background levels, i.e., luciferase activity of naive noninfected cells or cells infected with pseudotypes without HCV envelopes (Dimitrova et al., 2008). Background levels of the assay were determined in each experiment. Means ± SD from at least four independent experiments (performed in triplicate) are shown. Statistically significant differences (repeated measures ANOVA) in HCVpp entry between the predominant selected strain and nonselected strains are indicated by asterisks (**, P < 0.001). Variants reinfecting the liver graft are depicted in blue, and nonselected variants not detected after transplantation are depicted in white, gray, or black.
To address whether the efficiency of infection was different in strains reinfecting the liver graft (selected variants) and strains not detected during the first days of LT (nonselected variants), we performed a comprehensive analysis of viral entry of all patient-derived HCVpps containing functional full-length HCV envelope glycoproteins. As shown in Fig. 3, HCVpps generated from variants reinfecting the graft were characterized by more efficient entry into Huh7 cells compared with HCVpps generated from nonselected variants that were not detected after transplantation (mean enhancement 152-fold; P < 0.0001). To confirm the results obtained with Huh7 hepatoma cell lines, we infected primary human hepatocytes obtained from four different donors with patient-derived HCVpps from selected and nonselected variants. Infection efficiency was 10–100-fold lower in human hepatocytes, as indicated by the level of reporter gene expression in cells infected with the same HCVpp preparations. Similar to results obtained with hepatoma cells, HCVpps of the selected variants showed more efficient entry than HCVpps produced from the nonselected variants (Fig. S3). The difference in viral entry observed between selected and nonselected variants did not result from variations in transfection efficiency (Fig. 3), nor was it caused by impaired HCVpp assembly or release because the levels of envelope glycoprotein incorporation into different pseudoparticles were similar (Fig. S4). These results demonstrate that viral isolates that are selected during transplantation are characterized by more efficient entry compared with isolates not detected after transplantation. Collectively, these data suggest that the efficiency of viral entry into naive hepatocytes of the liver graft is important for selection of viral variants after LT.
HCV variants reinfecting the liver graft escape autologous neutralizing responses
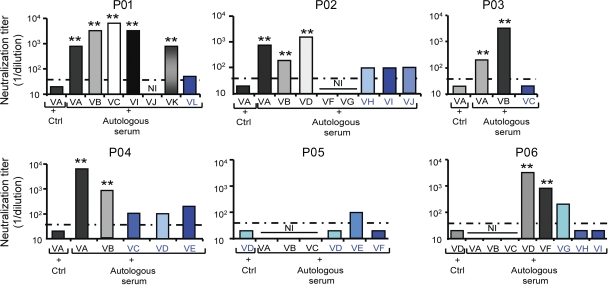
Because viral entry is a major target of neutralizing antibodies and HCV reinfection of the liver graft occurs efficiently despite the presence of high titer of anti-HCV antibodies, we investigated whether neutralizing antibodies present in pretransplantation serum were able to neutralize infection of patient-derived HCVpps. Patient-derived HCVpps were preincubated with autologous pretransplantation serum, and their subsequent infection levels were quantified. As shown in Fig. 4, HCVpps derived from selected variants were only poorly neutralized by antibodies present in pretransplant serum (mean neutralizing titer, 1:69; range, 1:20 to 1:200). In contrast, HCVpps derived from HCV variants that were not detected after transplantation were efficiently neutralized by autologous host-neutralizing antibodies (mean neutralizing titer, 1:2,257; range, 1:200 to 1:6,400). The differences between neutralization of selected and nonselected variants were highly significant (P < 0.001). Furthermore, nonselected isolates showing high infectivity (e.g., strains P01VA, P02VA, P02VD, and P06VD) were efficiently neutralized by autologous pretransplant serum as shown in Fig. 4. These data demonstrate that escape from antibody-mediated neutralization represents an important mechanism for viral evasion from host immunity during LT.
Figure 4.
Escape from antibody-mediated neutralization is a key determinant for selection of viral variants after LT. HCVpps were incubated with pretransplant anti-HCV–positive serum or control anti-HCV–negative serum (Ctrl) in serial dilutions for 1 h at 37°C. HCVpp–antibody complexes were then added to Huh7 cells, and infection was analyzed as described in Fig. 3. Calculation of neutralization and determination of background and thresholds for neutralization are described in Materials and methods. End point dilution titers are indicated for each variant. Dashed lines indicate the threshold for a positive neutralization titer corresponding to 1:40. Variants reinfecting the liver graft are depicted in blue, and nonselected variants not detected after transplantation are depicted in white, gray, or black. Means from at least four independent experiments (performed in triplicate) are shown. Statistically significant differences (repeated measures ANOVA) in neutralization between the predominant selected strain and nonselected strains are indicated by asterisks (**, P < 0.001). NI, noninfectious.
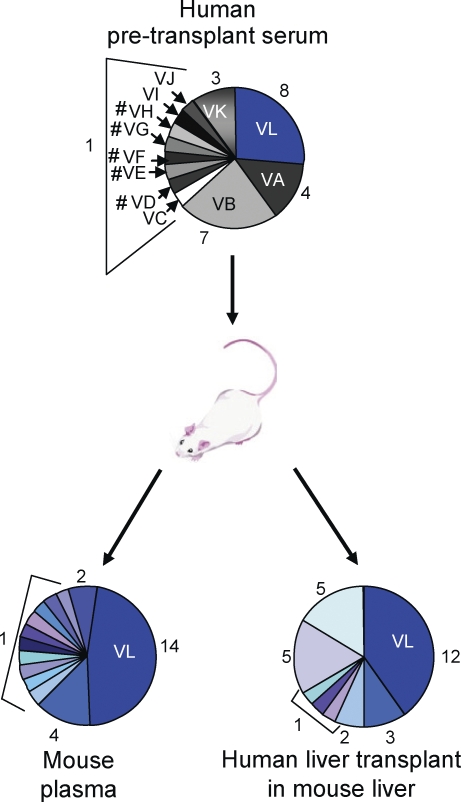
To ensure that the selected variants emerge because of a clear competitive advantage and not just by chance, we analyzed HCV infection in the chimeric liver uPA-SCID (urokinase-type plasminogen activator/severe combined immunodeficient) mouse model after injection with pretransplant serum of patient P01 (Meuleman et al., 2008). As shown in Fig. 5, variant P01VL (which was selected after LT in the respective patient) was the most prevalent variant in both plasma and liver of the infected uPA-SCID mouse. This result corroborates the presumed selective advantage of the variant reinfecting the graft and demonstrates the relevance of our analysis and validity of the HCVpp model system for HCV infection in vivo.
Figure 5.
Evolution of HCV variants in the uPA-SCID mouse model infected with patient-derived pretransplant serum. A human liver–chimeric mouse was challenged by intraperitoneal injection of pretransplant serum of patient P01 (infectious dose administered, 3.3 × 105 IU) and sacrificed at week 3 after infection. HCV RNA was isolated and amplified by RT-PCR from liver and plasma as described previously (Schvoerer et al., 2007). HCV isolates in plasma and liver samples were cloned and sequenced as described in Materials and methods (30 clones per sample in one animal). Circle graphs represent the percentage of each clone detected, and numbers of clones are shown. Variant VL, which was selected after LT in patient P01, was the most prevalent variant in both plasma and liver of the infected uPA-SCID mouse. Variants containing stop codons, insertions, or deletions altering the HCV open reading frame are depicted with a number sign (#).
Functional analysis of HVR1 (hypervariable region 1) subvariants confirms that phylogenetic grouping allows a representative functional analysis of full-length variants
Grouping of full-length E1E2 quasispecies (Figs. 1 and 2) was performed on HVR1 variation because quasispecies grouping based on HVR1 variation has been shown to be the most accurate approach to study the evolutionary dynamics of the HCV quasispecies population (Farci et al., 2000; Feliu et al., 2004; Schvoerer et al., 2007). To address whether grouping of full-length E1E2 genomes by HVR1 analysis results in selection of clones that are representative of the functional properties of the entire quasispecies population, we performed a functional analysis of all detected E1E2 subvariants (VA, VB, VK, and VL) containing the same HVR1 of patient P01. This approach allowed us to address the question whether all the subvariants harboring the same HVR1 have the same level of infectivity and if not, what is the ratio of highly infectious subvariants compared with the less infectious ones. As shown in Fig. S5, the ratio of highly infectious subvariants compared with the less infectious ones was high (5:8) in HVR1 variants with a high infectivity phenotype (VL) and low in HVR1 variants with a low infectivity phenotype (the ratio of high to low infectivity subvariants was 2:4 for VA, 1:7 for VB, and 0:3 for VK subvariants). Most importantly, we demonstrate that all of the nonselected E1E2 subvariants exhibiting high infectivity were efficiently neutralized by autologous patient serum (e.g., subvariants VAs1 and VBs1 in Fig. S5). Finally, grouping of viral quasispecies on complete envelope glycoprotein sequences with subsequent functional analysis of all individual full-length clones in patient 01 again demonstrated that selected strains were characterized by efficient viral entry and escape from neutralizing responses, whereas nonselected strains exhibited a poor entry and neutralization phenotype (unpublished data). These analyses corroborate that phylogenetic grouping of full-length E1E2 genomes by HVR1 analysis is valid and allows a functional analysis of full-length clones whose function is representative of the entire E1E2 quasispecies population. Collectively, these analyses confirm that entry and neutralization are important factors for selection of viral variants during HCV reinfection in LT independent of the method of quasispecies grouping.
Mutations in CD81 binding domains of E2 confer enhanced viral entry
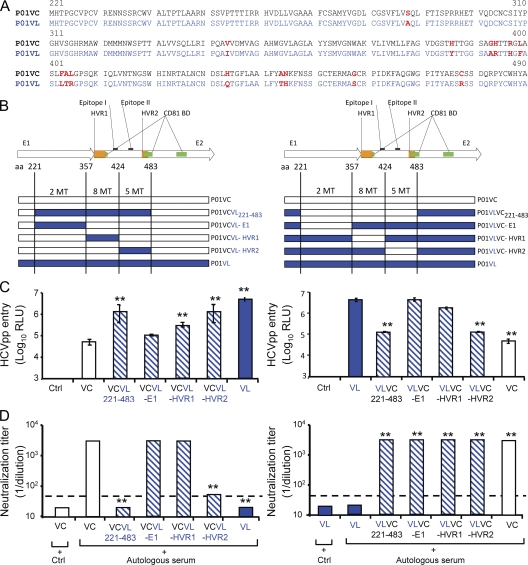
To map envelope regions mediating enhanced entry and viral escape, we focused on an individual patient (P01) and exchanged four regions spanning aa 221–483 (C-terminal E1 and N-terminal E2), 221–357 (E1), 358–424 (E2-HVR1), and 425–483 (E2-HVR2) of the envelope glycoproteins of selected variant P01VL and nonselected variant P01VC of patient P01 (Fig. 6, A and B). The exchange of HVR1 of selected and nonselected variants resulted in chimeric HCVpps (P01VCVL-HVR1) with higher infectivity than HCVpps derived from the P01VC parental strain (Fig. 6 C). The exchange of the E2 region comprising aa 425–483 resulted in chimeric HCVpps (P01VCVL-HVR2) with the highest infectivity and poor neutralization by the autologous pretransplant serum, the phenotype of the selected variant (Fig. 6, C and D). These data indicate that mutations in the HCV envelope region E2425–483 predominantly mediate enhanced viral entry and escape from neutralizing antibodies during reinfection of the liver graft in this patient. Furthermore, these data confirm that the observed differences in entry and neutralization were indeed mediated by mutations in the viral envelope glycoproteins and were not caused by any technical artifacts or by impairment in HCVpp production. Sequence analysis of P01VC and P01VL showed five mutations in the E425–483 region (H434Q, A444T, N445H, G458S, and C478R), including one mutation in CD81 BDII (C478R; Fig. 6, Fig. S2, and Table S1). Analysis of the sequences encompassing the same region (aa 425–483) in the other patients showed mutations in the majority of strains of all patients but one (P05; Fig. S2 and Table S1). This finding suggests that mutations in this region are also involved in enhanced entry and sensitivity to neutralization in the other patients. Interestingly, nonselected variants exhibiting a low entry phenotype very frequently showed mutations in CD81 binding domains I, II, and III (Table II and Fig. S2). Collectively, these data support a model whereby acquired mutations alter envelope glycoprotein–CD81 binding, which subsequently modulates the entry phenotype.
Figure 6.
Mutations in envelope region E2425–483 mediate enhanced viral entry and escape from neutralizing antibodies. To map envelope regions mediating enhanced entry and viral escape, we exchanged four regions spanning C-terminal E1 and N-terminal E2, E1, E2- HVR1, and E2-HVR2 of the envelope glycoproteins of selected variant VL and nonselected variant VC of patient P01 (see Figs. 2–4). These regions include aa 221–483, aa 221–357, aa 358–424, and aa 425–483, respectively. (A) Deduced amino acid sequences of the exchanged region between P01VC (black) and P01VL (blue). Amino acid changes are indicated in red bold letters. (B) Construction of recombinant chimeric HCVpps P01VCVL221–483 and P01VLVC221–483, P01VCVL-E1 and P01VLVC-E1, P01VCVL-HVR1 and P01VLVC-HVR1, and P01VCVL-HVR2 and P01VLVC-HVR2 by exchanging E1E2 envelope domain aa 221–483, aa 221–357, aa 358–424, and aa 425–483, respectively, of nonselected variant VC (patient 01) depicted in white and escape isolate VL (patient 01) depicted in blue (see Figs. 2–4). HVR1 and HVR2 are shown in orange, and CD81 binding domains (CD81 BD) are shown in green. Positions of E2 epitopes I and II are indicated (Zhang et al., 2007, 2009). The number of mutations within each region is shown. (C) Viral entry of HCVpps containing chimeric envelope proteins in Huh7 cells. HCVpps were incubated with Huh7 cells, and infection was analyzed as described in Fig. 3. Results are expressed in relative light units (RLU) plotted in a logarithmic scale. The threshold for a detectable infection is 3 × 103 RLU and was determined as described in Fig. 3. (D) Neutralization of HCVpps by autologous pretransplant serum was performed as described in Fig. 4. End point dilution titers are indicated for each variant. Dashed lines indicate the threshold for a positive neutralization titer corresponding to 1:40. Calculation of neutralization and determination of threshold titers are described in Materials and methods. Chimeric HCVpps are depicted in dashed blue. Statistically significant differences (repeated measures ANOVA) in HCVpp entry or neutralization between VC and VL wild-type and mutant variants are indicated by asterisks (**, P < 0.001). Ctrl, negative control; MT, mutation.
Table II.
Mutations within CD81 binding domains of viral variants
| Viral variants | CD81 BDI amino acid positions | CD81 BDII amino acid positions | CD81 BDIII amino acid positions | ||||||||||||||||||
| 412 | 414 | 416 | 420 | 474 | 475 | 476 | 477 | 478 | 479 | 480 | 481 | 492 | 521 | 522 | 524 | 528 | 531 | 533 | 537 | 538 | |
| P01VLa | Q | V | T | W | Y | A | E | S | R | S | S | D | Q | R | S | V | T | E | K | L | L |
| P01VAa | |||||||||||||||||||||
| P01VBb | |||||||||||||||||||||
| P01VCb | C | K | |||||||||||||||||||
| P01VIb | L | ||||||||||||||||||||
| P01VKb | P | L | F | A | E | ||||||||||||||||
| P01VJc | I | P | R | L | |||||||||||||||||
| P02VIa | Q | I | T | W | Y | A | E | P | H | D | L | G | Q | R | S | A | N | E | E | L | I |
| P02VJa | Q | D | R | ||||||||||||||||||
| P02VHa | Q | D | R | ||||||||||||||||||
| P02VAa | V | D | |||||||||||||||||||
| P02VDa | V | D | R | ||||||||||||||||||
| P02VBb | L | A | D | ||||||||||||||||||
| P02VFc | S | R | S | D | R | ||||||||||||||||
| P02VGc | V | D | R | T | |||||||||||||||||
| P03VCa | K | V | T | W | Y | A | K | P | N | S | S | D | R | R | F | A | S | E | E | L | L |
| P03VAa | Q | E | T | ||||||||||||||||||
| P03VBb | E | ||||||||||||||||||||
| P04VEa | Q | I | T | W | Y | A | E | P | R | S | L | D | Q | R | F | V | N | E | E | L | L |
| P04VDa | |||||||||||||||||||||
| P04VCb | |||||||||||||||||||||
| P04VAb | N | K | |||||||||||||||||||
| P04VBb | |||||||||||||||||||||
| P05VEa | Q | V | T | W | Y | T | E | P | E | A | L | D | Q | R | F | V | S | E | E | L | L |
| P05VFa | |||||||||||||||||||||
| P05VDa | |||||||||||||||||||||
| P05VAc | |||||||||||||||||||||
| P05VBc | |||||||||||||||||||||
| P05VCc | |||||||||||||||||||||
| P06VHa | Q | I | T | W | H | V | V | P | H | G | L | D | R | R | F | A | T | E | E | F | I |
| P06VIa | R | ||||||||||||||||||||
| P06VDa | Q | ||||||||||||||||||||
| P06VGb | A | R | |||||||||||||||||||
| P06VFb | Y | A | E | S | R | S | V | N | L | L | |||||||||||
| P06VAc | Y | A | E | R | S | ||||||||||||||||
| P06VBc | Y | A | E | R | S | V | N | L | L | ||||||||||||
| P06VCc | |||||||||||||||||||||
BD, binding domain; RLU, relative light unit. For each patient, amino acid changes in the CD81 BDI, BDII, and BDIII (Owsianka et al., 2006) relative to the most prevalent variant showing efficient viral entry are indicated. Amino acid positions where at least one mutation was observed in the six patients are shown; conserved amino acid positions are not depicted.
Variants showing high infectivity (RLU ≥ 5 log10).
Variants showing low infectivity (5 log 10 ≥ RLU ≤ 3.5 log10).
Noninfectious variants (RLU ≤ 3 log10).
Sequence analysis of P01VB and P05 variants did not reveal any amino acid mutations within aa 425–483 or the three CD81 binding domains. The absence of mutations within these regions suggests that in these variants, other envelope regions are responsible for the alteration of the entry and neutralization phenotype (Table II, Fig. S2, and Table S1). These could include SR-BI (scavenger receptor B I) binding sites within HVR1 (Fig. S2) or functional regions that contribute to viral entry steps mediated by claudin-1 or occludin. Interestingly, a further analysis of the full-length E1E2 sequences of low entry variants P05VB and P05VC variants identified two mutations of cysteine residues at positions C229R and C564R, respectively. Structural modeling suggests that these mutations may alter disulfide bond formation, resulting in altered envelope glycoprotein folding (Krey et al., 2010).
Cross-neutralizing mAbs efficiently inhibit entry of escape variants resistant to host responses
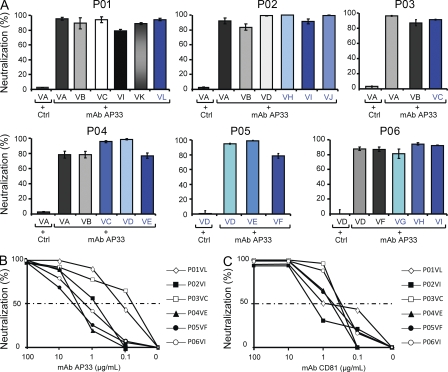
To investigate whether targeting entry of viral isolates infecting the liver graft by cross-neutralizing mAbs is a suitable approach for prevention of HCV reinfection, we incubated patient-derived HCVpps with mAb AP33 directed against an epitope within the HCV E2 protein and studied its ability to neutralize patient-derived HCVpps. As shown in Fig. 7 (A and B), anti-E2 mAb AP33 efficiently inhibited infection of HCVpps containing envelope glycoproteins from both selected and nonselected viral variants in Huh7 cells and primary human hepatocytes (IC50 0.1–3.3 µg/ml). Similar results were obtained by preincubating hepatocytes with an mAb directed against the large extracellular loop of HCV entry factor CD81 (Fig. 7 C; Pileri et al., 1998). Efficient neutralization of HCV isolates having escaped autologous neutralizing responses by cross-neutralizing antibodies suggests that targeting viral entry by mAbs is a promising strategy for preventing HCV reinfection of the liver graft.
Figure 7.
Cross-neutralization of escape variants infecting the liver graft by antienvelope and antireceptor mAbs. (A) Neutralization of HCVpps from viral isolates by cross-neutralizing anti-E2 mAb AP33. HCVpps derived from different isolates were incubated with 10 µg/ml anti-E2 AP33 or isotype monoclonal control IgG, and neutralization of viral entry in Huh7 cells was determined as described in Fig. 4 (entry in the presence of isotype monoclonal control IgG = 100%). Neutralization was calculated as described in Materials and methods. Means ± SD from three independent experiments (performed in triplicate) are shown. (B and C) Neutralization of HCV isolates having escaped patients’ neutralizing responses by anti-E2 and anti-CD81 mAbs in primary human hepatocytes. HCVpps derived from viral isolates selected after LT (P01VL, P02VI, P03VC, P04VE, P05VF, and P06VI) were incubated with serial dilutions of anti-E2 AP33 (B) or control IgG from mouse (Ctrl) as described in Fig. 4 and then added to primary human hepatocytes. For analysis of neutralization using anti-CD81, primary human hepatocytes were preincubated with anti-CD81 or isotype control IgG for 1 h at 37° before incubation with patient-derived HCVpps (C). Means from one representative experiment (performed in triplicate) out of two independent experiments are shown. 50% neutralization of HCVpp entry is indicated by a dashed line.
DISCUSSION
In this study, we demonstrate for the first time that viral entry into hepatocytes and escape from host-neutralizing antibodies represent important mechanisms for selection of HCV variants during HCV reinfection of the liver graft. Furthermore, we show that infection of strains selected during transplantation can be efficiently neutralized by broadly neutralizing mAbs. These findings define the molecular mechanisms of viral evasion during HCV reinfection and pave the way for novel antiviral strategies for prevention of HCV reinfection of the liver graft.
Both viral and host factors may contribute to HCV reinfection during LT. Viral factors include efficiency of entry, replication, and production of progeny virus. Host factors include humoral and cellular responses, graft- or donor-related factors, and immunosuppressive therapy. The pretransplantation environment is characterized by chronic HCV infection, where viral population is subjected to selective pressures from both humoral and cellular immunity. Viral variants evade immune response by continuous generation of B and T cell escape variants (von Hahn et al., 2007), resulting in slow and progressive change in composition of viral quasispecies with coexistence of highly neutralized and escape viral variants (von Hahn et al., 2007). In contrast, the early phase of the posttransplantation period is characterized by rapid de novo infection of naive hepatocytes in a host with impaired cellular immune responses. As shown by phylogenetic analyses (Fig. 1), this change of host environment results in an abrupt and marked change in the composition of viral quasispecies population. Phylogenetic analyses demonstrated a decrease in the quasispecies diversity after LT, with selection of only a fraction of relatively homogeneous viral variants in the majority of patients (Fig. 1). Thus, it is likely that the implantation of the new liver and start of immunosuppressive therapy generate a bottleneck effect by selecting variants that are able to infect the liver graft more efficiently. Because viral entry is a key determinant for the efficient and rapid initiation of viral infection and neutralizing antibodies are a first-line host defense in the transplanted liver, the ability of a viral isolate to efficiently infect naive hepatocytes and escape host-neutralizing responses may be an important advantage for the initiation of graft infection, allowing rapid dissemination of the virus. Indeed, the functional investigation of entry and neutralization of patient-derived viral particles before and after transplantation demonstrates that viral variants selected during transplantation were characterized by more efficient viral entry and escape from neutralizing antibodies compared with viral variants not detected after transplantation. This suggests that entry and evasion from antibody-mediated neutralization play a key role for selection of viral variants in the early phase of transplantation. The concept that the selected variants emerge because of a clear competitive advantage and not just by chance is further supported by the analysis of HCV infection in the chimeric liver uPA-SCID mouse model (Fig. 5): the variant which was selected after LT in the respective patient was the most prevalent variant in both plasma and liver of the infected uPA-SCID mouse. This result corroborates the presumed selective advantage of the variant reinfecting the graft and confirms that the abrupt change of the host environment leads to selection of viral variants with an efficient entry phenotype.
The key impact of escape from neutralization is underlined by the finding that all nonselected variants, including isolates showing high infectivity (e.g., strains P01VA, P02VA, P02VD, and P06VD), were efficiently neutralized by autologous pretransplant serum and that all selected variants, including isolates with a lower infectivity phenotype (P02VH, P04VC, P06VG, P06VH, and P06VI), escaped neutralization (Figs. 3 and 4).
Interestingly, in all patients undergoing LT, the strains having escaped host immune responses after transplantation were also among the most prevalent strains before transplantation (Figs. 2–4). These findings indicate that efficient entry and evasion from neutralizing responses result already in a survival advantage of the virus in the patient with end-stage liver disease and suggest that preselection of strains occurs already before transplantation. Nevertheless, the marked and abrupt homogenization of quasispecies observed in phylogenetic analyses (Fig. 1), the stability of variants after transplantation (Fig. S1), and the relative advantage of selected variants in the uPA-SCID mouse model (Fig. 5) suggest that the identified shifts in the viral populations with selection of neutralization-resistant and entry-efficient variants are related to the reinfection situation and do not simply represent processes that occur continuously during chronic infection.
Grouping of full-length E1E2 quasispecies (Figs. 1 and 2) was performed on HVR1 variation because quasispecies grouping based on HVR1 variation has been shown to be the most accurate approach to study the evolutionary dynamics of the HCV quasispecies population (Farci et al., 2000; Feliu et al., 2004; Schvoerer et al., 2007). The validity of this approach for our cohort was confirmed by a side by side comparison of phylogenetic analysis of HVR1 and full-length E1E2 sequences: when HVR1 was excluded, the phylogenetic reconstructions of the rest of the E1E2 region showed a monophyletic population with absent discrimination between selected and nonselected variants similar to previous observations by Farci et al. (2000; and unpublished data). A functional analysis of entry and neutralization of variants containing the same HVR1 further corroborated that grouping of full-length E1E2 genomes by HVR1 analysis with subsequent functional analysis of full-length clones is valid and represents the functional properties of the quasispecies population (Fig. S5).
It is conceivable that cell-mediated immune responses also contribute to the differences observed in variants before and after transplantation. Indeed, binding of envelope glycoprotein E2 to CD81 has been suggested to induce functional changes of natural killer or dendritic cells (Nattermann et al., 2006; Crotta et al., 2010). Because of the profound immunosuppression with severely impaired cell-mediated immune response after transplantation, efficient viral entry most likely becomes the predominant determinant of selection of viral variants in the transplanted host. The pretransplantation environment may allow nonselected variants to compete with the selected variants despite disadvantages in terms of neutralizing antibody response and viral entry. In contrast, with immunosuppression after transplantation, which primarily impacts cell-mediated immunity, selected variants take over because efficient viral entry and escape from neutralizing responses give them a relative advantage. This hypothesis is also supported by the experiments in the uPA-SCID mouse model: when the cell-mediated immune system is taken out of the equation, then viral entry becomes an important determinant of relative fitness in the host (Fig. 5). Other factors may also contribute to relative viral fitness, including different replication abilities among HCV variants (Uebelhoer et al., 2008; Dazert et al., 2009) or the ability to spread by cell–cell transmission (Timpe et al., 2008; Witteveldt et al., 2009). Finally, we cannot exclude the idea that mutations that improve virus entry will affect other steps of the viral life cycle.
The association of evasion of host-neutralizing responses and efficient entry may be explained by the fact that viral entry and neutralization are closely linked by the interference of neutralizing antibodies with envelope–host entry factor interactions. Indeed, we have previously shown that purified antiviral antibodies derived from HCV-infected patients inhibit HCV infection at an entry step closely linked to CD81 and SR-BI interaction (Haberstroh et al., 2008). Nevertheless, escape from neutralization could not be explained simply by efficient viral entry because several variants with a high infectivity phenotype were also efficiently neutralized as shown in Figs. 3 and 4 (P01VA, P02VA, P02VD, and P06VD). Moreover, differences in neutralization remained similar when the amount of HCVpps for neutralization experiments was adjusted for relative infectivity or quantity of pseudoparticles (unpublished data).
Exchange of envelope domains of selected and nonselected HCV variants demonstrated that mutations within the region E2425–483 mediate enhanced viral entry and escape from neutralizing antibodies. This region contains an important CD81 binding site (Owsianka et al., 2006) and is a target of patient-derived neutralizing antibodies (Haberstroh et al., 2008; Law et al., 2008). Thus, it is likely that the mutations present in this region enhance viral entry by modulating the affinity of E2 to CD81 and induce a conformational change allowing resistance to binding of neutralizing antibodies. Because the exchange of HVR1 of selected and nonselected HCV variants also resulted in a partial enhancement of viral entry (Fig. 6), it is likely that mutations in this region contribute to enhanced entry, e.g., by modulating SR-BI dependency (Bartosch et al., 2005).
It is of interest to note that the mapped E2 region conferring escape from neutralizing antibodies (E2425–483) contains an epitope (epitope II, aa 434–446) recognized by host nonneutralizing antibodies that have been proposed to disrupt virus neutralization by antibodies that target epitope I, a highly conserved glycoprotein segment located upstream (aa 412–419; Zhang et al., 2007, 2009). Thus, the mutations identified in this epitope (e.g., H434Q, A444T, and N445H in variant VL; Fig. 6, Fig. S2, and Table S1) may enhance the binding of nonneutralizing antibodies and contribute to attenuation or escape from neutralization of selected variants by antibodies binding to epitope I. However, it is noteworthy that neutralization of the selected variants by our broadly neutralizing mAb AP33 (Fig. 7 B), which recognizes amino acid residues located in epitope I (Owsianka et al., 2005; Tarr et al., 2006), was not affected.
Collectively, our data suggest that adaptive mutations within HVR1, CD81 binding domains, and neutralizing epitopes appear during HCV quasispecies evolution after LT, leading to the emergence of the most fit or best adapted virus that is resistant to host autologous neutralizing responses and that is capable of efficient de novo infection of the liver graft. The use of state of the art model systems, including patient-derived HCVpps and naive human hepatocytes from different donors, suggests that the findings obtained indeed mimic virus–host interactions occurring in the initiation of graft infection in the HCV-infected patient. This is further supported by our in vivo experiments investigating viral evolution in the uPA-SCID mouse model repopulated with human hepatocytes and infected with pretransplant serum. The predominant variant which was selected after LT in the respective patient was the most prevalent variant in both plasma and liver of the infected uPA-SCID mouse (Fig. 5). These results demonstrate the relevance of our analysis and validity of the HCVpp model system for HCV infection in vivo. This is in line with the fact that in vitro neutralization in the HCVpp model system has been shown to reflect neutralization of infectious recombinant cell culture–derived HCV in vitro (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005; Haberstroh et al., 2008; von Hahn and Rice, 2008; Krieger et al., 2010) and in human hepatocytes in vivo (Law et al., 2008; Vanwolleghem et al., 2008). Furthermore, the HCVpp system is characterized by high robustness (von Hahn and Rice, 2008; Zeisel et al., 2008) and efficient infection of human hepatocytes (Krieger et al., 2010) and allows the production of a large number of patient-derived viruses. Nevertheless, we cannot exclude the idea that the HCVpp system may or may not mimic all of the complexities of an authentic virus when it comes to neutralization and entry.
Finally, our findings have important implications for the development of novel preventive strategies and management of the HCV-infected patient undergoing LT. It is of interest to note that first generation anti-HCV Ig or antibodies failed to prevent HCV recurrence after LT in clinical trials (Davis et al., 2005; Schiano et al., 2006). The reason why these antibodies failed was most likely their poor neutralizing capacity. Indeed, in vitro and in vivo characterization of anti-E2 HCV-AbXTL68 in preclinical experiments had shown an inefficient neutralization in the HCVpp model system and HCV-Trimera mouse model (Eren et al., 2006). Very recent studies in an HCV animal model have shown that new generation cross-neutralizing anti-E2 or anti-CD81 antibodies are capable of neutralizing genetically diverse HCV isolates and protect against heterologous HCV quasispecies challenge (Law et al., 2008; Meuleman et al., 2008). Our study demonstrates for the first time that cross-reactive mAbs neutralize infection of the liver graft by viral variants that are resistant to host-neutralizing responses and as such paves the way for the development of novel preventive strategies and improved management of the HCV-infected patient undergoing LT. Efficient inhibition of infection of diverse HCV escape variants studied here suggests that antireceptor or antienvelope mAbs will have sufficient cross-reactivity to neutralize highly variable quasispecies variants. Combination of antienvelope and antireceptor antibodies may further increase the genetic barrier for resistance and may offer a viable and promising strategy to prevent HCV reinfection of the liver graft.
MATERIALS AND METHODS
Patients.
Clinical and virological features of patients followed at the Strasbourg University Hospitals are shown in Table I. Blood samples were collected before and 7 d and 1 mo after LT with approval of the Strasbourg University Hospital Institutional Review Board (http://clinicaltrials.gov identifier: NCT00213707). Anti-HCV antibodies and HCV RNA were analyzed using AxSYM (Abbott) and VERSANT HCV RNA 3.0 (Bayer).
Cells and reagents.
293T cells and Huh7 cells were isolated and cultured as described previously (Pestka et al., 2007; Haberstroh et al., 2008). Primary human hepatocytes from liver resections were provided by P. Bachellier and D. Jaeck (Hôpitaux Universitaires de Strasbourg, Strasbourg, France) and isolated by C. Royer (Institut National de la Santé et de la Recherche Médicale, Unité 748, Strasbourg, France) as described previously (Krieger et al., 2010). Anti-E1 (11B7) and anti-E2 (AP33; 3E5-1) mAbs and human anti-HCV IgG have been described previously (Owsianka et al., 2005; Pestka et al., 2007; Haberstroh et al., 2008). Anti-CD81 (JS81) was obtained from BD.
Amplification and cloning of HCV E1E2.
Total RNA was purified from 280 µl of serum using the RNeasy kit (QIAGEN). cDNA was synthesized using superscript II reverse transcription (Invitrogen) with random primers (Invitrogen). The cDNA was used as the template for PCR amplification of the sequence encoding the C-terminal core and full-length E1E2 proteins using HotStar HiFidelity PCR Taq polymerase (QIAGEN) and primers MLVextS (5′-CGTAGGTCGCGTAACTTGGGTAA-3′) and MLVextAS (5′-GTGCGCCTCGGCTCTGGTGATAAA-3′), corresponding to nt 681–703 and 2886–2909, respectively. A nested PCR was performed using primers AD78-1 and -2 containing an EcoRV restriction site as described previously (Pestka et al., 2007). Full-length E1E2 PCR products were cloned into PCR 2.0-TOPO (Invitrogen). After sequencing (Beckman Coulter), cDNA containing the full-length E1E2 coding sequence was digested by EcoRV and subcloned into expression vector pCMV-IRES for production of HCVpps (Pestka et al., 2007). Phylogenetic analysis of viral quasispecies was performed as described previously (Schvoerer et al., 2007). Neighbor-joining trees based on Kimura two-parameter distance matrices (1,000 bootstrap resampling replications) were constructed using the MEGA 4 software package (Tamura et al., 2007; Schramm et al., 2008). Amino acid genetic diversity was based on the pairwise analysis of all the sequences within each time point.
HCVpp production, infection, and neutralization.
HCVpps were generated by transfection of 293T cells as described previously (Bartosch et al., 2003; Pestka et al., 2007), using the CMV-Gag-Pol MLV (mouse leukemia virus) packaging construct, an MLV-Luc reporter plasmid, and the expression plasmids encoding the HCV envelope glycoproteins of HCV variants. To study HCV entry, HCVpps were added to Huh7 cells or primary human hepatocytes in triplicate and incubated for 72 h at 37°C. HCV entry was determined by analysis of luciferase reporter gene expression as described previously (von Hahn et al., 2007; Haberstroh et al., 2008). The detection limit for positive luciferase reporter protein expression was 3 × 103 RLU/assay corresponding to the mean ± 3 SD of background levels, i.e., luciferase activity of naive noninfected cells or cells infected with pseudotypes without HCV envelopes (Dimitrova et al., 2008). Background levels of the assay were determined in each individual experiment. For the study of antibody-mediated neutralization, HCVpps were mixed with autologous anti-HCV, anti-E2 mAb AP33, control serum (consisting of a pool of three anti-HCV–negative sera), or irrelevant isotype control IgG and preincubated for 1 h at 37°C and added to Huh7 cells or primary human hepatocytes in triplicate for 72 h at 37°C (von Hahn et al., 2007). Anti-CD81 mAb was preincubated with primary human hepatocytes for 1 h at 37°C, and then HCVpps were added. Antibodies were serially diluted to determine the IC50, the concentration of antibody that gave a 50% inhibition of HCVpps. The specific neutralization was determined according to the following equation: specific neutralization = 100 − [100 × (infectivity of HCVpps in the presence of autologous serum or mAb/infectivity of HCVpps in the presence of anti-HCV–negative control sera or irrelevant isotype control IgG)]. Background levels of HCVpp infection were taken into account by subtracting infectivity in the presence of control serum used at the same dilution as for autologous serum. The neutralization titer was defined as the last dilution of the sample that conveyed a ≥50% reduction of HCVpp entry compared with an equivalent dilution of control serum, as described previously (Pestka et al., 2007). Background of neutralization of HCVpp infection was determined by analysis of a large reference panel of anti-HCV–negative control sera exhibiting a neutralization titer of ≤1:20. Thus, a neutralization titer of 1:40 was defined as the threshold of positive antibody-mediated neutralization.
HCVpp infection of primary human hepatocytes.
Primary human hepatocytes were isolated as described previously (Krieger et al., 2010). Primary hepatocytes were infected with HCVpps expressing envelope glycoproteins of predominant viral isolates described in Fig. 2. 1 d after hepatocyte isolation and plating, hepatocytes were washed, and HCVpps were added for 3 h at 37°C. After infection, the supernatant was removed and replaced by fresh William’s E medium. HCVpp infection was assessed by measurement of luciferase activity 72 h after infection, as described previously (Krieger et al., 2010).
Analysis of HCVpp envelope glycoprotein expression.
Expression of HCV glycoproteins of predominant strains was characterized in 293T producer cells, and HCVpps were purified by sucrose gradient ultracentrifugation as described previously (Bartosch et al., 2003; Barth et al., 2006; Haberstroh et al., 2008). Immunoblots of HCV glycoproteins were performed using antienvelope antibodies (anti-E1 mAb 11B7 and anti-E2 mAb AP33 or 3E5-1) or anti-HCV IgG as described previously (Pestka et al., 2007).
Mapping of envelope domains mediating enhanced entry and escape from neutralization.
For mapping experiments, four major fragments comprising the entire E1E2-coding region were exchanged between the selected variant P01VL and the nonselected variant P01VC of patient P01. These regions included aa 221–483, aa 221–357, aa 358–424, and aa 425–483 and correspond to the functional regions E1, HVR1, HVR2, and CD81 binding domain II, respectively (Fig. 6, A and B). To allow the exchange, new restriction sites were generated by introducing silent mutations using QuikChange II XL (Agilent Technologies). The resulting expression constructs encoding for P01VC/VL chimeric envelope glycoproteins were used to generate HCVpps as described previously (Pestka et al., 2007).
Infection of uPA-SCID mice transplanted with human hepatocytes.
Human liver–uPA-SCID mice were infected with patient serum (P01), and viral quasispecies were analyzed as described previously (Law et al., 2008; Meuleman et al., 2008).
Statistical analysis.
Statistical analysis (repeated measures analysis of variance [ANOVA]) was performed with the SPSS 16.0 software for Windows (SPSS Inc.).
Online supplemental material.
Fig. S1 shows phylogenetic analyses of viral quasispecies evolution in the early phase posttransplantation (day 7 and month 1). Fig. S2 shows comparative alignment of amino acid sequences of full-length E1E2 variants isolated from the six transplanted patients and analyzed in functional experiments. Fig. S3 shows infection of primary human hepatocytes by HCVpps bearing envelope glycoproteins from the most prevalent selected and nonselected strains. Fig. S4 shows expression of envelope glycoproteins in 293T producer cells and HCVpps. Fig. S5 shows functional analysis of subvariants containing the same HVR1 in an individual patient. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090766/DC1.
Acknowledgments
We thank M. Bastien–Valle for excellent technical assistance, P. Bachellier, D. Jaeck, and C. Royer for providing and isolating primary human hepatocytes, N. Meyer for support in statistical analysis, GenImmune, Novartis Vaccines, and Genentech for the gift of antibodies, C. Combet and F. Penin for analyses of envelope sequences, and R. Bartenschlager, J.A. McKeating, M.H. Heim, M.B. Zeisel, E. Schvoerer, C. Rice, D. Samuel, and H.E. Blum for helpful discussions.
This work was supported by Institut National de la Santé et de la Recherche Médicale, the European Union (ERC-2008-AdG-233130-HEPCENT and Interreg IV FEDER-Hepato-Regio-Net 2009), the Agence Nationale de la Recherche chair of excellence program (ANR-05-CEXC-008), Agence Nationale des Recherches sur le SIDA et les Hépatites Virales (2007/306 and 2008/354), the Région d’Alsace (2007/09), the Else Kröner-Fresenius Foundation (EKFS P17//07//A83/06), the Ligue Contre le Cancer (CA 06/12), Institut National de la Santé et de la Recherche Médicale, the University of Strasbourg Hospitals, the Ghent University concerted action grant 01G00507, the Belgian state (Interuniversity Attraction Poles Program P6/36–HEPRO), and a fellowship of the Research Foundation Flanders (FWO–Vlaanderen to P. Meuleman).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ANOVA
- analysis of variance
- HCV
- hepatitis C virus
- HCVpp
- HCV pseudoparticle
- LT
- liver transplantation
- uPA-SCID
- urokinase-type plasminogen activator/severe combined immunodeficient
References
- Aurora R., Donlin M.J., Cannon N.A., Tavis J.E. 2009. Genome-wide hepatitis C virus amino acid covariance networks can predict response to antiviral therapy in humans. J. Clin. Invest. 119:225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H., Schnober E.K., Zhang F., Linhardt R.J., Depla E., Boson B., Cosset F.L., Patel A.H., Blum H.E., Baumert T.F. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579–10590 10.1128/JVI.00941-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B., Dubuisson J., Cosset F.L. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J. Exp. Med. 197:633–642 10.1084/jem.20021756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B., Verney G., Dreux M., Donot P., Morice Y., Penin F., Pawlotsky J.M., Lavillette D., Cosset F.L. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217–8229 10.1128/JVI.79.13.8217-8229.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen D.G., Walker C.M. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 436:946–952 10.1038/nature04079 [DOI] [PubMed] [Google Scholar]
- Brown R.S. 2005. Hepatitis C and liver transplantation. Nature. 436:973–978 10.1038/nature04083 [DOI] [PubMed] [Google Scholar]
- Crotta S., Brazzoli M., Piccioli D., Valiante N.M., Wack A. 2010. Hepatitis C virions subvert natural killer cell activation to generate a cytokine environment permissive for infection. J. Hepatol. 52:183–190 10.1016/j.jhep.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Davis G.L., Nelson D.R., Terrault N., Pruett T.L., Schiano T.D., Fletcher C.V., Sapan C.V., Riser L.N., Li Y., Whitley R.J., Gnann J.W., Jr 2005. A randomized, open-label study to evaluate the safety and pharmacokinetics of human hepatitis C immune globulin (Civacir) in liver transplant recipients. Liver Transpl. 11:941–949 10.1002/lt.20405 [DOI] [PubMed] [Google Scholar]
- Dazert E., Neumann-Haefelin C., Bressanelli S., Fitzmaurice K., Kort J., Timm J., McKiernan S., Kelleher D., Gruener N., Tavis J.E., et al. 2009. Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J. Clin. Invest. 119:376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova M., Affolter C., Meyer F., Nguyen I., Richard D.G., Schuster C., Bartenschlager R., Voegel J.C., Ogier J., Baumert T.F. 2008. Sustained delivery of siRNAs targeting viral infection by cell-degradable multilayered polyelectrolyte films. Proc. Natl. Acad. Sci. USA. 105:16320–16325 10.1073/pnas.0800156105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M., Pietschmann T., Granier C., Voisset C., Ricard-Blum S., Mangeot P.E., Keck Z., Foung S., Vu-Dac N., Dubuisson J., et al. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285–18295 10.1074/jbc.M602706200 [DOI] [PubMed] [Google Scholar]
- Eren R., Landstein D., Terkieltaub D., Nussbaum O., Zauberman A., Ben-Porath J., Gopher J., Buchnick R., Kovjazin R., Rosenthal-Galili Z., et al. 2006. Preclinical evaluation of two neutralizing human monoclonal antibodies against hepatitis C virus (HCV): a potential treatment to prevent HCV reinfection in liver transplant patients. J. Virol. 80:2654–2664 10.1128/JVI.80.6.2654-2664.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., von Hahn T., Tscherne D.M., Syder A.J., Panis M., Wölk B., Hatziioannou T., McKeating J.A., Bieniasz P.D., Rice C.M. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 446:801–805 10.1038/nature05654 [DOI] [PubMed] [Google Scholar]
- Farci P., Shimoda A., Coiana A., Diaz G., Peddis G., Melpolder J.C., Strazzera A., Chien D.Y., Munoz S.J., Balestrieri A., et al. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 288:339–344 10.1126/science.288.5464.339 [DOI] [PubMed] [Google Scholar]
- Feliu A., Gay E., García-Retortillo M., Saiz J.C., Forns X. 2004. Evolution of hepatitis C virus quasispecies immediately following liver transplantation. Liver Transpl. 10:1131–1139 10.1002/lt.20206 [DOI] [PubMed] [Google Scholar]
- Grove J., Nielsen S., Zhong J., Bassendine M.F., Drummer H.E., Balfe P., McKeating J.A. 2008. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J. Virol. 82:12020–12029 10.1128/JVI.01569-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberstroh A., Schnober E.K., Zeisel M.B., Carolla P., Barth H., Blum H.E., Cosset F.L., Koutsoudakis G., Bartenschlager R., Union A., et al. 2008. Neutralizing host responses in hepatitis C virus infection target viral entry at postbinding steps and membrane fusion. Gastroenterology. 135:1719–1728: e1 10.1053/j.gastro.2008.07.018 [DOI] [PubMed] [Google Scholar]
- Krey T., d’Alayer J., Kikuti C.M., Saulnier A., Damier-Piolle L., Petitpas I., Johansson D.X., Tawar R.G., Baron B., Robert B., et al. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6:e1000762 10.1371/journal.ppat.1000762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger S.E., Zeisel M.B., Davis C., Thumann C., Harris H.J., Schnober E.K., Mee C., Soulier E., Royer C., Lambotin M., et al. 2010. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 51:1144–1157 [DOI] [PubMed] [Google Scholar]
- Lavillette D., Morice Y., Germanidis G., Donot P., Soulier A., Pagkalos E., Sakellariou G., Intrator L., Bartosch B., Pawlotsky J.M., Cosset F.L. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J. Virol. 79:6023–6034 10.1128/JVI.79.10.6023-6034.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M., Maruyama T., Lewis J., Giang E., Tarr A.W., Stamataki Z., Gastaminza P., Chisari F.V., Jones I.M., Fox R.I., et al. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27 10.1038/nm1698 [DOI] [PubMed] [Google Scholar]
- Lindenbach B.D., Evans M.J., Syder A.J., Wölk B., Tellinghuisen T.L., Liu C.C., Maruyama T., Hynes R.O., Burton D.R., McKeating J.A., Rice C.M. 2005. Complete replication of hepatitis C virus in cell culture. Science. 309:623–626 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]
- Meuleman P., Hesselgesser J., Paulson M., Vanwolleghem T., Desombere I., Reiser H., Leroux-Roels G. 2008. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 48:1761–1768 10.1002/hep.22547 [DOI] [PubMed] [Google Scholar]
- Moreno Garcia J., del Campo Terron S., Moraleda Garcia G., Garcia Gonzalez M., de Vicente Lopez E., Nuño Vazquez-Garza J., Fortun Abete J., Martin P., Barcena Marugan R. 2003. Analysis of hepatitis C viral quasispecies in liver transplantation. Transplant. Proc. 35:1838–1840 10.1016/S0041-1345(03)00632-8 [DOI] [PubMed] [Google Scholar]
- Nattermann J., Zimmermann H., Iwan A., von Lilienfeld-Toal M., Leifeld L., Nischalke H.D., Langhans B., Sauerbruch T., Spengler U. 2006. Hepatitis C virus E2 and CD81 interaction may be associated with altered trafficking of dendritic cells in chronic hepatitis C. Hepatology. 44:945–954 10.1002/hep.21350 [DOI] [PubMed] [Google Scholar]
- Owsianka A., Tarr A.W., Juttla V.S., Lavillette D., Bartosch B., Cosset F.L., Ball J.K., Patel A.H. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104 10.1128/JVI.79.17.11095-11104.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka A.M., Timms J.M., Tarr A.W., Brown R.J., Hickling T.P., Szwejk A., Bienkowska-Szewczyk K., Thomson B.J., Patel A.H., Ball J.K. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704 10.1128/JVI.00271-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka J.M., Zeisel M.B., Bläser E., Schürmann P., Bartosch B., Cosset F.L., Patel A.H., Meisel H., Baumert J., Viazov S., et al. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. USA. 104:6025–6030 10.1073/pnas.0607026104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A.J., Houghton M., Rosa D., Grandi G., Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science. 282:938–941 10.1126/science.282.5390.938 [DOI] [PubMed] [Google Scholar]
- Ploss A., Evans M.J., Gaysinskaya V.A., Panis M., You H., de Jong Y.P., Rice C.M. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 457:882–886 10.1038/nature07684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S.C., Fanning L., Wang X.H., Netski D.M., Kenny-Walsh E., Thomas D.L. 2005. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J. Exp. Med. 201:1753–1759 10.1084/jem.20050122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiano T.D., Charlton M., Younossi Z., Galun E., Pruett T., Tur-Kaspa R., Eren R., Dagan S., Graham N., Williams P.V., Andrews J. 2006. Monoclonal antibody HCV-AbXTL68 in patients undergoing liver transplantation for HCV: results of a phase 2 randomized study. Liver Transpl. 12:1381–1389 10.1002/lt.20876 [DOI] [PubMed] [Google Scholar]
- Schramm F., Soulier E., Royer C., Weitten T., Fafi-Kremer S., Brignon N., Meyer N., Ellero B., Woehl-Jaegle M.L., Meyer C., et al. 2008. Frequent compartmentalization of hepatitis C virus with leukocyte-related amino acids in the setting of liver transplantation. J. Infect. Dis. 198:1656–1666 10.1086/592986 [DOI] [PubMed] [Google Scholar]
- Schvoerer E., Soulier E., Royer C., Renaudin A.C., Thumann C., Fafi-Kremer S., Brignon N., Doridot S., Meyer N., Pinson P., et al. 2007. Early evolution of hepatitis C virus (HCV) quasispecies after liver transplant for HCV-related disease. J. Infect. Dis. 196:528–536 10.1086/519691 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tarr A.W., Owsianka A.M., Timms J.M., McClure C.P., Brown R.J., Hickling T.P., Pietschmann T., Bartenschlager R., Patel A.H., Ball J.K. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology. 43:592–601 10.1002/hep.21088 [DOI] [PubMed] [Google Scholar]
- Timpe J.M., Stamataki Z., Jennings A., Hu K., Farquhar M.J., Harris H.J., Schwarz A., Desombere I., Roels G.L., Balfe P., McKeating J.A. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 47:17–24 10.1002/hep.21959 [DOI] [PubMed] [Google Scholar]
- Uebelhoer L., Han J.H., Callendret B., Mateu G., Shoukry N.H., Hanson H.L., Rice C.M., Walker C.M., Grakoui A. 2008. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog. 4:e1000143 10.1371/journal.ppat.1000143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwolleghem T., Bukh J., Meuleman P., Desombere I., Meunier J.C., Alter H., Purcell R.H., Leroux-Roels G. 2008. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology. 47:1846–1855 10.1002/hep.22244 [DOI] [PubMed] [Google Scholar]
- von Hahn T., Rice C.M. 2008. Hepatitis C virus entry. J. Biol. Chem. 283:3689–3693 10.1074/jbc.R700024200 [DOI] [PubMed] [Google Scholar]
- von Hahn T., Yoon J.C., Alter H., Rice C.M., Rehermann B., Balfe P., McKeating J.A. 2007. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 132:667–678 10.1053/j.gastro.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H.G., Mizokami M., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteveldt J., Evans M.J., Bitzegeio J., Koutsoudakis G., Owsianka A.M., Angus A.G., Keck Z.Y., Foung S.K., Pietschmann T., Rice C.M., Patel A.H. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90:48–58 10.1099/vir.0.006700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel M.B., Koutsoudakis G., Schnober E.K., Haberstroh A., Blum H.E., Cosset F.L., Wakita T., Jaeck D., Doffoel M., Royer C., et al. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 46:1722–1731 10.1002/hep.21994 [DOI] [PubMed] [Google Scholar]
- Zeisel M.B., Cosset F.L., Baumert T.F. 2008. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology. 48:299–307 10.1002/hep.22307 [DOI] [PubMed] [Google Scholar]
- Zhang P., Wu C.G., Mihalik K., Virata-Theimer M.L., Yu M.Y., Alter H.J., Feinstone S.M. 2007. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc. Natl. Acad. Sci. USA. 104:8449–8454 10.1073/pnas.0703039104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhong L., Struble E.B., Watanabe H., Kachko A., Mihalik K., Virata-Theimer M.L., Alter H.J., Feinstone S., Major M. 2009. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc. Natl. Acad. Sci. USA. 106:7537–7541 10.1073/pnas.0902749106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D.R., Wieland S.F., Uprichard S.L., Wakita T., Chisari F.V. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA. 102:9294–9299 10.1073/pnas.0503596102 [DOI] [PMC free article] [PubMed] [Google Scholar]