Abstract
Studies have suggested that antigen receptor loci adopt contracted conformations to promote long-distance interactions between gene segments during V(D)J recombination. The Tcra/Tcrd locus is unique because it undergoes highly divergent Tcrd and Tcra recombination programs in CD4−CD8− double negative (DN) and CD4+CD8+ double positive (DP) thymocytes, respectively. Using three-dimensional fluorescence in situ hybridization, we asked whether these divergent recombination programs are supported by distinct conformational states of the Tcra/Tcrd locus. We found that the 3′ portion of the locus is contracted in DN and DP thymocytes but not in B cells. Remarkably, the 5′ portion of the locus is contracted in DN thymocytes but is decontracted in DP thymocytes. We propose that the fully contracted conformation in DN thymocytes allows Tcrd rearrangements involving Vδ gene segments distributed over 1 Mb, whereas the unique 3′-contracted, 5′-decontracted conformation in DP thymocytes biases initial Tcra rearrangements to the most 3′ of the available Vα gene segments. This would maintain a large pool of distal 5′ Vα gene segments for subsequent rounds of recombination. Thus, distinct contracted conformations of the Tcra/Tcrd locus may facilitate a transition from a Tcrd to a Tcra mode of recombination during thymocyte development.
The diversity of antigen-specific receptors on T and B lymphocytes is generated by the assembly of variable (V), diversity (D), and joining (J) gene segments in a process termed V(D)J recombination (Schatz and Spanopoulou, 2005). This process is catalyzed by the lymphocyte-specific recombinase proteins RAG1 and RAG2 (recombination-activating gene 1 and 2), which initiate the recombination reaction by generating double-strand breaks between V, D, and J gene segments and their flanking recombination signal sequences (RSSs). V(D)J recombination is regulated in a lineage- and developmental stage–specific fashion during T lymphocyte development in the thymus, with four TCR genes, Tcra, Tcrb, Tcrg, and Tcrd, rearranging at two distinct developmental stages to generate αβ or γδ T lymphocytes (Cobb et al., 2006; Krangel, 2009). The Tcrb, Tcrg, and Tcrd genes rearrange first, in CD4−CD8− double negative (DN) thymocytes, and a small subset of DN thymocytes with in-frame Tcrg and Tcrd rearrangements may become γδ T lymphocytes. A larger subset of DN thymocytes with in-frame Tcrb rearrangements differentiate to the CD4+CD8+ double positive (DP) stage, in which Tcra gene recombination occurs. In-frame Tcra rearrangement leads to cell surface expression of αβ TCRs, and those TCRs that can support positive selection promote maturation of DP thymocytes to either the CD4+CD8− or CD4−CD8+ single positive stage.
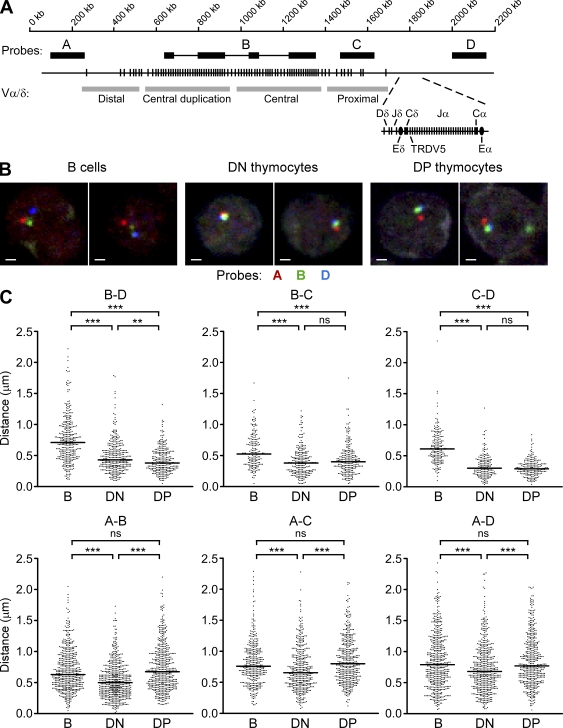
Tcrd and Tcra gene segments are organized together in a complex genetic locus (Krangel et al., 2004; Krangel, 2009). The mouse Tcra/Tcrd locus spans 1.6 Mb and contains about 100 Vα/δ gene segments that occupy the largest portion of the locus (Fig. 1 A). The 3′ 100 kb of the locus contains two Dδ, two Jδ, and a Cδ gene segment, followed by 61 Jα and a Cα gene segment. Tcrd and Tcra recombination events are distinct in their developmental timing. However, they are distinct in other important respects as well. For example, only a small subset of V gene segments commonly rearrange to Dδ and Jδ segments in DN thymocytes, whereas essentially all V gene segments rearrange to Jα segments in DP thymocytes (Krangel et al., 2004; Krangel, 2009). The V segments that rearrange in DN thymocytes include several that are relatively close to Dδ and Jδ (e.g., TRDV1, TRDV2-2, TRDV4, and TRDV5) and several that are as far as 1 Mb away (e.g., TRAV15/DV6 family). Tcrd and Tcra recombination events also differ in the following way: because germline Dδ gene segments are eliminated by Vδ to Dδ to Jδ rearrangement, Tcrd has only a single opportunity for recombination on an allele. In contrast, Tcra typically undergoes multiple rounds of Vα to Jα rearrangement (Krangel et al., 2004; Krangel, 2009). Tcra recombination initiates with the use of relatively 3′ Vα and 5′ Jα gene segments, and these initial rearrangements can be replaced by multiple cycles of Vα to Jα rearrangement involving progressively more 5′ Vα and more 3′ Jα gene segments as thymocytes search for a TCR that can mediate positive selection (Pasqual et al., 2002).
Figure 1.
Subregion-specific contraction of the Tcra/Tcrd locus during thymocyte development. (A) Organization of Tcra/Tcrd locus and relative positions of four BAC probes (A, B, C, and D). V segments are categorized as proximal, central, central duplication, or distal. Probe B (C57BL/6 origin) hybridizes discontinuously to the central and central duplication regions of the strain 129 Tcra/Tcrd locus. (B) 3D-FISH analysis of strain 129 Tcra/Tcrd locus conformation in splenic B cells and recombinase-deficient DN and DP thymocytes using probes A, B, and D. Each image represents a single z-plane with representative loci. Bars, 1 µm. (C) Scatter plots display pairwise distance measurements between the centers of probe hybridization compiled from use of the A–B–D, A–C–D, and A–B–C probe combinations. Data were accumulated from two to five independent experiments for each cell type (184–592 total alleles) with 78–126 alleles per experiment. Median values are indicated by horizontal lines. **, P < 0.01; ***, P < 0.001; ns, not statistically significant.
The distinct dynamics of Tcrd and Tcra recombination events can be partly understood based on the known properties of locus cis-acting regulatory elements. The Tcrd enhancer (Eδ) is activated in DN thymocytes and is thought to locally regulate Dδ and Jδ chromatin structure and accessibility to support Vδ to Dδ to Jδ rearrangement (Monroe et al., 1999; Bassing et al., 2003). The Tcra enhancer is activated in DP thymocytes and activates the TEA (T early α) promoter at the 5′ end of the Jα array (Sleckman et al., 1997); TEA germline transcription is essential to modify 5′ Jα chromatin and target 5′ Jα segments for initial recombination events (Villey et al., 1996; Hawwari et al., 2005; Abarrategui and Krangel, 2007). During subsequent rounds of recombination, promoters of the rearranged Vα gene segments target additional Vα rearrangements to progressively more 3′ Jα gene segments (Hawwari and Krangel, 2007). Less is understood about the parameters that bias initial Tcra rearrangements to relatively 3′ Vα gene segments and thereby result in a 3′ to 5′ progression of Vα gene usage. One idea is that the 3′ Vα gene segments are preferred initial targets for the recombinase because these Vα gene segments are selectively activated by Eα, which is known to function across the 3′ one third of the Tcra/Tcrd locus in DP thymocytes (Hawwari and Krangel, 2005).
A variety of studies have implicated conformational changes in antigen receptor loci as important determinants of long-distance V(D)J recombination events (for review see Jhunjhunwala et al., 2009). Three-dimensional (3D) fluorescence in situ hybridization (FISH) analyses have demonstrated developmental stage-specific contraction of Ig and TCR loci that is thought to promote synapsis of widely separated RSSs (Kosak et al., 2002; Roldán et al., 2005; Skok et al., 2007). For example, the well-studied Igh locus contracts in pro-B cells to prepare for VH to DHJH rearrangement and then decontracts in the subsequent pre-B stage (Roldán et al., 2005). Igh contraction depends on transcription factors Pax5, Ikaros, and YY1 (Fuxa et al., 2004; Liu et al., 2007; Reynaud et al., 2008); depleting any of these factors reduces distal VH rearrangement even though distal VH chromatin accessibility may be normal (Hesslein et al., 2003; Fuxa et al., 2004; Liu et al., 2007). A recent elegant study has provided a detailed picture of Igh locus contraction in pro-B cells (Jhunjhunwala et al., 2008). A notable feature of the contracted conformation is that all VH gene segments are brought into proximity to the DHJH cluster in 3D space. This is thought to provide all VH gene segments a similar opportunity for recombination (Jhunjhunwala et al., 2008), which may be particularly important because individual Igh alleles can support only a single round of VH to DHJH rearrangement.
In DN thymocytes, the Tcra/Tcrd locus behaves similarly to the Igh locus in that it undergoes a single round of Vδ to Dδ to Jδ rearrangement and must allow rearrangement of both Dδ-proximal and -distal Vδ gene segments. However, in DP thymocytes, the locus behaves differently, with multiple rounds of Vα to Jα recombination starting with proximal Vα gene segments. This raises the question of whether these divergent modes of recombination are supported by distinct conformational states during thymocyte development. In this study, we provide a detailed characterization of Tcra/Tcrd locus conformation in DN and DP thymocytes. We find that the entire locus is contracted in DN thymocytes, but that the 5′ portion is selectively decontracted in DP thymocytes. We suggest that decontraction of the 5′ region facilitates a transition of the locus from a Tcrd to a Tcra mode of recombination during thymocyte development.
RESULTS AND DISCUSSION
To determine the spatial conformation of the Tcra/Tcrd locus during thymocyte development, we used 3D-FISH to track the relative positions of four distinct segments of the locus using bacterial artificial chromosome (BAC) probes (A, B, C, and D; Fig. 1 A). Tcra/Tcrd locus V gene segments can be classified as proximal, central, and distal according to their positioning relative to Dδ, Jδ, and Jα gene segments. Sets of unique V gene segments are distributed across the proximal and distal 300 kb of the V array (identified by probes C and A, respectively); in contrast, the central V gene segments are found in a 400-kb region that is duplicated in strain 129 mice (identified by probe B, which hybridizes discontinuously with an extended region covering almost the entire duplicated region). We used these and probe D (which hybridizes downstream of Cα) to analyze Tcra/Tcrd locus conformation in three cell types: DN thymocytes from Rag2−/− mice, DP thymocytes from Rag2−/− mice injected with anti-CD3 antibody to drive the DN to DP transition, and control splenic B lymphocytes of wild-type mice. Use of these cell populations allowed us to uniformly analyze unrearranged Tcra/Tcrd loci; this approach was valid because previous studies had shown locus contraction to occur independent of recombinase expression (Fuxa et al., 2004; Roldán et al., 2005; Skok et al., 2007; Jhunjhunwala et al., 2008).
The four probes were used in various three-way combinations (A–B–D, A–C–D, and A–B–C) in three-color 3D-FISH followed by confocal microscopy. Selected images obtained using the A–B–D probe strategy are shown (Fig. 1 B). We then evaluated all pairwise distances between the centers of mass of the hybridization signals on large numbers of alleles for each cell type (Fig. 1 C). We found that all median distances measured across the 3′ portion of the locus (B–D, B–C, and C–D) were substantially reduced in both DN and DP thymocytes as compared with splenic B cells (Fig. 1 C, top row). The B–C and C–D distances were indistinguishable between DN and DP thymocytes; however, there was a small but significant difference in the B–D distance. From these data, we conclude that the 3′ half of the Tcra/Tcrd locus (B–C–D) is extended in B cells and is similarly, although not identically, contracted in both thymocyte subsets. In contrast, all median center to center distances measured across the 5′ end of the locus (A–B, A–C, and A–D) were substantially reduced in DN thymocytes but then increased in DP thymocytes (Fig. 1 C, bottom row). The measured increases in the A–C and A–D distances likely reflect a selective extension of the A–B segment because the B–C, C–D, and B–D distances are all similar in DN and DP thymocytes. Thus, the 5′ end of the locus (A–B) displays a strikingly different behavior than the 3′ end; it contracts in DN thymocytes but then decontracts in DP thymocytes. The results indicate that the Tcra/Tcrd locus possesses different contracted configurations in DN and DP thymocytes: in the former, it is contracted along its entire length, whereas in the latter, the 3′ half is contracted, but the 5′ half is decontracted.
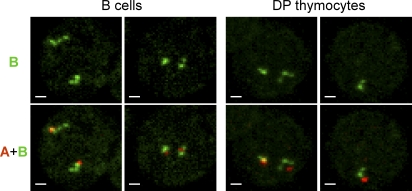
We noticed that on a fraction of alleles, probe B appeared to detect two or three foci rather than a single focus (Fig. 2). In each instance, this complex hybridization pattern could be attributed to a single allele because the second allele was well separated, as judged by simultaneous hybridization with additional probes. Alleles with multiple foci did not result from DNA replication because additional probes always detected single foci on the same allele (Fig. 2). Because probe B hybridizes discontinuously to four segments spanning ∼700 kb of the duplicated central portion of the V array (Fig. 1 A), the detection of multiple foci by probe B is likely to reflect an extended conformation of this portion of the Tcra/Tcrd locus. Remarkably, examples of multiple probe B foci were frequent in B cells and DP thymocytes (38% and 30% of alleles, respectively) but were rare in DN thymocytes (5% of alleles; Table I). These data indicate that the 5′ end extension of the Tcra/Tcrd locus includes both distal and central V gene segments.
Figure 2.
Conformation of central V segments as defined by probe B hybridization. Representative examples of alleles with multiple foci detected by probe B in B cells and DP thymocytes. Tcra/Tcrd alleles were detected using a combination of probes A, B, and C. The top row of images shows probe B hybridization; the bottom row shows a combination of probes A and B. Probe C identified a single focus in each instance (not depicted). Similar results were observed in two independent experiments using the same probe combination. Bars, 1 µm.
Table I.
Enumeration of alleles with one, two, and three foci detected by probe B
| Cell type | Probe B foci per allele | ||
| One | Two | Three | |
| % | % | % | |
| B cellsa | 62 | 35 | 3 |
| DN thymocytesb | 95 | 5 | 0 |
| DP thymocytesc | 70 | 24 | 6 |
The proportion of alleles with multiple probe B foci in DN thymocytes was significantly different from B cells and DP thymocytes (P < 0.0001 by Fisher’s exact test). Data were from one experiment for each cell type.
158 total alleles.
140 total alleles.
166 total alleles.
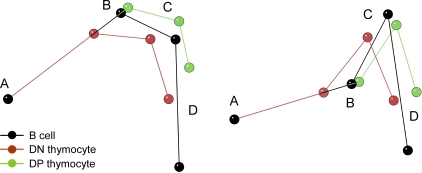
To evaluate the overall organization of the locus in 3D space, we calculated the coordinates of sites A, B, C, and D from the full set of median distance measurements. The resulting models (Fig. 3, which presents two different views of the superimposed models) depict the mean conformation of the locus in nuclei of all three cell types. The range of conformations adopted by the locus is obviously much more complex, as indicated by the heterogeneous pattern of probe B hybridization and the broadly distributed probe to probe distance measurements on the entire population of alleles (Figs. 1 and 2). Nevertheless, the models highlight a nonlinear mean configuration of the locus in nuclei of all three cell types. Furthermore, the models emphasize that the conformational difference between B cells and DN thymocytes affects the entire locus, whereas the conformational difference between DN and DP thymocytes preferentially impacts the 5′ half of the locus: there is a striking change in the A–B distance between DN and DP thymocytes, with relative conservation of spatial relationships among B, C, and D.
Figure 3.
3D models of Tcra/Tcrd locus conformation. Median distances for the three cell types, determined from the entire dataset in Fig. 2, were used to generate 3D models that are aligned so that they share point A and the A–B axis. Two views are shown from different angles.
Previous studies have linked locus contraction with V(D)J recombination at the Igh, Igk, Tcra, and Tcrb loci (for review see Jhunjhunwala et al., 2009). In this study, we showed that the Tcra/Tcrd locus is contracted in both DN and DP thymocytes but that the contracted configurations in the two developmental compartments are distinct. We suggest that the two Tcra/Tcrd locus conformations support different modes of V gene segment usage during Tcrd and Tcra recombination. We think that the globally contracted configuration adopted in DN thymocytes provides widely distributed Vδ gene segments the opportunity to undergo Vδ to Dδ to Jδ rearrangement and that this is important because there is only one chance for Vδ to Dδ to Jδ rearrangement per allele. In these ways, the Tcra/Tcrd locus in DN thymocytes resembles the Igh locus in pro-B cells. In contrast, we believe that the 5′-decontracted, 3′-contracted conformation of Tcra/Tcrd locus in DP thymocytes insures that primary Vα to Jα rearrangement initiates with proximal Vα gene segments, thereby maintaining a large pool of distal Vα gene segments for subsequent rounds of recombination. Because of the extended 5′ region, use of distal Vα gene segments would only be favored once they are brought into proximity of Jα gene segments by prior rounds of Vα to Jα recombination. A limitation of our study is that we have only determined the conformation of unrearranged Tcra/Tcrd alleles in DP thymocytes, even though many Tcra/Tcrd alleles would normally have undergone prior Vδ to Dδ to Jδ rearrangement in DN thymocytes. We presume that the remaining upstream Vα gene segments on previously rearranged alleles would adopt an extended configuration in DP thymocytes, thereby promoting ordered Vα usage on these alleles as well.
Previous studies have implicated decontraction of the Igh locus in pre-B cells and the Tcrb locus in DP thymocytes in feedback inhibition of V segment recombination that is associated with allelic exclusion (Roldán et al., 2005; Skok et al., 2007). Notably, analyses of decontracted Igh alleles in µ transgenic mice and in Pax5- or YY1-deficient mice have demonstrated residual recombination that involves proximal VH segments only (Fuxa et al., 2004; Liu et al., 2007). However, in none of these cases does restricted recombination of proximal VH segments represent a significant physiological process. In contrast, our data suggest a potential novel physiological role for locus decontraction as a means to direct initial Tcra recombination events to relatively proximal Vα gene segments, thus allowing a robust progression of primary and secondary Vα to Jα recombination events.
We note that our results conflict with those of a previously published study, which claimed that the Tcra/Tcrd locus does not contract until the DP stage (Skok et al., 2007). We cannot easily explain the apparent absence of contraction in DN thymocytes in that study because both studies analyzed 3′ end contraction in Rag2−/− DN thymocytes using probes B and D. One potential difference is that we focused on 129 mice, which have an internal duplication in the Tcra/Tcrd locus, whereas Skok et al. (2007) analyzed C57BL/6 mice, which have an internal triplication. However, neither this nor other background differences account for the discrepant results because we also detected contraction when we compared C57BL/6 Rag2−/− DN thymocytes to C57BL/6 B cells (Fig. S1). It remains possible that distinct methodologies for distance measurements may account for the different conclusions of the two studies. We suggest that a contracted configuration of the locus in DN thymocytes provides a logical basis for long-distance Tcrd gene recombination events that are known to occur in DN thymocytes.
Although several factors have been shown to be required for Igh locus contraction in pro-B cells, the molecular mechanisms regulating locus contraction and decontraction are still unknown. Prior work has indicated that Eα can regulate histone modifications and transcription of unrearranged V gene segment promoters at a distance of nearly 500 kb (Hawwari and Krangel, 2005). Because enhancer–promoter communication is thought to occur through direct long-distance interactions, we asked whether Eα plays a role in 3′ end contraction of the Tcra/Tcrd locus in DP thymocytes. However, comparison of wild-type and Eα-deficient alleles in DP thymocytes revealed no significant difference in 3′ end contraction (Fig. S2 A). We also observed no significant difference in locus contraction between wild-type and Eδ- or Eα-deficient alleles in DN thymocytes (Fig. S2 B). Chromatin-organizing proteins such as SATB1, CTCF, and cohesin have been shown to promote long-distance looping interactions at nonantigen receptor loci (Cai et al., 2006; Splinter et al., 2006; Nativio et al., 2009). Whether they play similar roles at antigen receptor loci remains to be established.
It is intriguing that Eα has been documented to regulate Tcra/Tcrd locus chromatin across a remarkably long 500-kb region that corresponds to the contracted portion of the locus in DP thymocytes (Hawwari and Krangel, 2005). Because 3′ end contraction appears to occur in an Eα-independent fashion, we speculate that when Eα is activated in DP thymocytes, this preexisting conformation may promote interactions between Eα and distant promoters, much as it may promote interactions between distant RSSs for Vα to Jα recombination. If the 3′ end contracted conformation of the locus in DP thymocytes dictates the region that is influenced by Eα, this conformation may be viewed as promoting not only RSS synapsis but RSS accessibility to RAG proteins as well. The same general principal may hold at other antigen receptor loci; enhancers and promoters may establish direct molecular contacts over long distances (Liu and Garrard, 2005; Oestreich et al., 2006), but they may do so only after having been brought into proximity by an enhancer- and promoter-independent conformational change.
In conclusion, our data demonstrate a Tcra/Tcrd locus conformational change during thymocyte development that we propose facilitates a transition from a Tcrd mode to a Tcra mode of V gene segment usage. The unique 5′ end extended and 3′ end contracted configuration in DP thymocytes may provide a novel conformation-based mechanism to direct Vα to Jα recombination events to the most proximal of the available Vα gene segments. The molecular basis for locus subregion-specific conformational changes will be important to unravel in future studies.
MATERIALS AND METHODS
Mice.
DN thymocytes were obtained from 2–3-wk-old Rag2−/− mice (129 or C57BL/6 background). DP thymocytes were obtained from Rag2−/− mice (129 background) 10 d after a single i.p. injection of 150 µg anti-CD3 antibody (145-2C11; BioLegend). B lymphocytes were isolated from splenocytes of wild-type 129 or C57BL/6 mice by depleting non-B cells on a MACS CS column using biotin-conjugated anti-CD11b, anti–Gr-1, anti-Thy1.1, anti-Thy1.2, anti-CD5 (eBioscience), and anti-CD43 (BD) antibodies together with streptavidin-conjugated magnetic beads. B cell purity was ∼90% with no apparent contaminating T cells. Eα−/− Rag2−/− and Eδ−/− Rag2−/− mice were of mixed background but carried the strain 129 Tcra/Tcrd locus. All mice were used in accordance with protocols approved by the Duke University Animal Care and Use Committee.
FISH probes and antibodies.
BAC clones RP23-304L21 (probe A, distal V), RP24-334B8 (probe B, central V), bMQ-440L6 (probe C, proximal V), and RP23-10K20 (probe D, 3′ end) were used to generate DNA probes. BACs were directly labeled with Alexa Fluor 488–5-dUTP (Invitrogen) or Alexa Fluor 586–5-dUTP (Invitrogen) using a nick translation kit (Roche) or were labeled with digoxigenin–11-dUTP using a DIG-nick translation kit (Roche). Digoxigenin-conjugated DNA probes were detected by indodicarbocyanine (Cy5)-conjugated antidigoxigenin antibody (Jackson ImmunoResearch Laboratories, Inc.).
3D DNA immuno-FISH.
Cells were fixed and hybridized as described previously (Schlimgen et al., 2008). In brief, cells attached to poly-l-lysine–coated slides were fixed in 4% (vol/vol) paraformaldehyde for 10 min, followed by permeabilization with 0.5% (wt/vol) saponin and 0.5% (vol/vol) Triton X-100 for 20 min and incubation with 0.1 N HCl for 10 min. After four cycles of freeze/thaw in 20% (wt/vol) glycerol, the slides were stored at −80°C. For hybridization, the cells were denatured by incubation at 78°C for 3 min in 70% (vol/vol) formamide and 2× SSC, followed by 1 min in 50% (vol/vol) formamide and 2× SSC. The DNA probe cocktail consisted of 1 µg of each DNA probe per slide and blocking DNAs (mixture of mouse C0t DNA, human placental DNA, and salmon sperm DNA) in HYBRISOL VII (MP Biomedicals). Denatured slides were then hybridized for 24–48 h at 37°C with boiled and preannealed probe cocktails. Excess probes were removed by three incubations for 5–7 min at 42°C in 50% (vol/vol) formamide and 2× SSC, followed by three incubations at 63°C in 0.2× SSC. The slides were then blocked by incubation for 30 min in 4% BSA and 2× SSC and were then incubated for 1 h with Cy5-conjugated antidigoxigenin antibody in 4% BSA and 2× SSC. Excess antibodies were removed by three 5-min incubations in 0.1% (vol/vol) Triton X-100 and 2× SSC. The slides were mounted in Vectashield (Vector Laboratories).
Confocal imaging, distance calculations, and statistical analysis.
Slides were imaged on a confocal microscope (SP5; Leica) using a 100× NA 1.4 objective lens and a 2× optical zoom. ImageJ (National Institutes of Health) software was used to process images using a Kalman stack filter and to determine the coordinates (x, y, z) of focus centers. Distances between pairs of foci (d, in micrometers) were calculated using the formula d2 = [(x′ − x) × 0.151]2 + [(y′ − y) × 0.151]2 + [(z′ − z) × 0.126]2, where 0.151 µm is the size of each pixel and 0.126 µm is the z-plane separation. Only nuclei with distinguishable signals from two alleles were analyzed. Mann-Whitney statistical tests were performed using Prism 3.0 (GraphPad Software, Inc.). The 3D models were built using the KiNG program (Richardson and Richardson, 1992; Chen et al., 2009) with the coordinates calculated as follows: A(0, 0, 0), B(xb, 0, 0), C(xc, yc, 0), and D(xd, yd, zd) where
and
Online supplemental material.
Fig. S1 shows an analysis of 3′ end contraction in DN thymocytes of C57BL/6 mice. Fig. S2 shows analyses of locus contraction on Eα- and Eδ-deficient alleles. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100772/DC1.
Acknowledgments
We would like to thank R. Schlimgen, M. Kuraoka, and Y. Chen for technical advice, S. Johnson and Y. Gao of the Duke Comprehensive Cancer Center Light Microscopy Facility for imaging support, and H. Kondilis, E. Chan, Q.-J. Li, and S. Unniraman for their helpful comments on the manuscript.
This work was supported by National Institutes of Health grant R37 GM41052.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- 3D
- three-dimensional
- BAC
- bacterial artificial chromosome
- DN
- double negative
- DP
- double positive
- FISH
- fluorescence in situ hybridization
- RSS
- recombination signal sequence
References
- Abarrategui I., Krangel M.S. 2007. Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO J. 26:4380–4390 10.1038/sj.emboj.7601866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing C.H., Tillman R.E., Woodman B.B., Canty D., Monroe R.J., Sleckman B.P., Alt F.W. 2003. T cell receptor (TCR) α/δ locus enhancer identity and position are critical for the assembly of TCR δ and α variable region genes. Proc. Natl. Acad. Sci. USA. 100:2598–2603 10.1073/pnas.0437943100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Lee C.C., Kohwi-Shigematsu T. 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38:1278–1288 10.1038/ng1913 [DOI] [PubMed] [Google Scholar]
- Chen V.B., Davis I.W., Richardson D.C. 2009. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 18:2403–2409 10.1002/pro.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R.M., Oestreich K.J., Osipovich O.A., Oltz E.M. 2006. Accessibility control of V(D)J recombination. Adv. Immunol. 91:45–109 10.1016/S0065-2776(06)91002-5 [DOI] [PubMed] [Google Scholar]
- Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422 10.1101/gad.291504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Krangel M.S. 2005. Regulation of TCR δ and α repertoires by local and long-distance control of variable gene segment chromatin structure. J. Exp. Med. 202:467–472 10.1084/jem.20050680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Krangel M.S. 2007. Role for rearranged variable gene segments in directing secondary T cell receptor α recombination. Proc. Natl. Acad. Sci. USA. 104:903–907 10.1073/pnas.0608248104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Bock C., Krangel M.S. 2005. Regulation of T cell receptor α gene assembly by a complex hierarchy of germline Jalpha promoters. Nat. Immunol. 6:481–489 10.1038/ni1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein D.G., Pflugh D.L., Chowdhury D., Bothwell A.L., Sen R., Schatz D.G. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42 10.1101/gad.1031403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Cutchin S., Riblet R., van Dongen J.J., Grosveld F.G., Knoch T.A., Murre C. 2008. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 133:265–279 10.1016/j.cell.2008.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Murre C. 2009. Chromatin architecture and the generation of antigen receptor diversity. Cell. 138:435–448 10.1016/j.cell.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- Krangel M.S. 2009. Mechanics of T cell receptor gene rearrangement. Curr. Opin. Immunol. 21:133–139 10.1016/j.coi.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel M.S., Carabana J., Abbarategui I., Schlimgen R., Hawwari A. 2004. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ locus. Immunol. Rev. 200:224–232 10.1111/j.0105-2896.2004.00155.x [DOI] [PubMed] [Google Scholar]
- Liu Z., Garrard W.T. 2005. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 25:3220–3231 10.1128/MCB.25.8.3220-3231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Schmidt-Supprian M., Shi Y., Hobeika E., Barteneva N., Jumaa H., Pelanda R., Reth M., Skok J., Rajewsky K., Shi Y. 2007. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 21:1179–1189 10.1101/gad.1529307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe R.J., Sleckman B.P., Monroe B.C., Khor B., Claypool S., Ferrini R., Davidson L., Alt F.W. 1999. Developmental regulation of TCR δ locus accessibility and expression by the TCR δ enhancer. Immunity. 10:503–513 10.1016/S1074-7613(00)80050-3 [DOI] [PubMed] [Google Scholar]
- Nativio R., Wendt K.S., Ito Y., Huddleston J.E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J.M., Murrell A. 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5:e1000739 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Cobb R.M., Pierce S., Chen J., Ferrier P., Oltz E.M. 2006. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 24:381–391 10.1016/j.immuni.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Pasqual N., Gallagher M., Aude-Garcia C., Loiodice M., Thuderoz F., Demongeot J., Ceredig R., Marche P.N., Jouvin-Marche E. 2002. Quantitative and qualitative changes in V-J α rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J. Exp. Med. 196:1163–1173 10.1084/jem.20021074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z., Smale S.T., Winandy S., Singh H. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 9:927–936 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D.C., Richardson J.S. 1992. The kinemage: a tool for scientific communication. Protein Sci. 1:3–9 10.1002/pro.5560010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán E., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A. 2005. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 6:31–41 10.1038/ni1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., Spanopoulou E. 2005. Biochemistry of V(D)J recombination. Curr. Top. Microbiol. Immunol. 290:49–85 10.1007/3-540-26363-2_4 [DOI] [PubMed] [Google Scholar]
- Schlimgen R.J., Reddy K.L., Singh H., Krangel M.S. 2008. Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nat. Immunol. 9:802–809 10.1038/ni.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok J.A., Gisler R., Novatchkova M., Farmer D., de Laat W., Busslinger M. 2007. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat. Immunol. 8:378–387 10.1038/ni1448 [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon C.G., Ferrini R., Davidson L., Alt F.W. 1997. Function of the TCR α enhancer in alphabeta and gammadelta T cells. Immunity. 7:505–515 10.1016/S1074-7613(00)80372-6 [DOI] [PubMed] [Google Scholar]
- Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20:2349–2354 10.1101/gad.399506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villey I., Caillol D., Selz F., Ferrier P., de Villartay J.P. 1996. Defect in rearrangement of the most 5′ TCR-J α following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity. 5:331–342 10.1016/S1074-7613(00)80259-9 [DOI] [PubMed] [Google Scholar]