Abstract
Neutrophils trap and kill bacteria by forming highly decondensed chromatin structures, termed neutrophil extracellular traps (NETs). We previously reported that histone hypercitrullination catalyzed by peptidylarginine deiminase 4 (PAD4) correlates with chromatin decondensation during NET formation. However, the role of PAD4 in NET-mediated bacterial trapping and killing has not been tested. Here, we use PAD4 knockout mice to show that PAD4 is essential for NET-mediated antibacterial function. Unlike PAD4+/+ neutrophils, PAD4−/− neutrophils cannot form NETs after stimulation with chemokines or incubation with bacteria, and are deficient in bacterial killing by NETs. In a mouse infectious disease model of necrotizing fasciitis, PAD4−/− mice are more susceptible to bacterial infection than PAD4+/+ mice due to a lack of NET formation. Moreover, we found that citrullination decreased the bacterial killing activity of histones and nucleosomes, which suggests that PAD4 mainly plays a role in chromatin decondensation to form NETs instead of increasing histone-mediated bacterial killing. Our results define a role for histone hypercitrullination in innate immunity during bacterial infection.
Peripheral blood neutrophils are the first line of defense after bacterial infection (Segal, 2005; Nathan, 2006). Neutrophils can migrate to sites of infection in response to inflammatory signals, where they engulf and kill bacteria by phagocytosis. Recently, a novel mechanism of extracellular bacterial killing mediated by highly decondensed chromatin structures, termed neutrophil extracellular traps (NETs), was identified (Brinkmann et al., 2004). NET formation is induced by PMA, LPS, and bacteria (Brinkmann et al., 2004). NETs trap and kill pathogenic bacteria, such as Shigella flexneri (Brinkmann et al., 2004) and group A Streptococcus pyogenes (GAS; Buchanan et al., 2006). The role of NETs in innate immunity has kindled much attention due to its involvement in human health and diseases and its therapeutic implications (Brinkmann and Zychlinsky, 2007; Nizet, 2007; Wartha and Henriques-Normark, 2008; von Köckritz-Blickwede and Nizet, 2009). For example, impaired NET formation predisposes newborn infants to bacterial infection (Yost et al., 2009). Further, in chronic granulomatous disease patients with impaired nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activity and reactive oxygen species (ROS) production, neutrophils cannot generate NETs and possess poor antimicrobial activity (Fuchs et al., 2007; Bianchi et al., 2009). Conversely, gene therapy with the NADPH oxidase gene in a chronic granulomatous disease patient to restore both NET formation and antimicrobial functions has provided an effective treatment for this disease (Bianchi et al., 2009).
In the eukaryotic nucleus, 147 bp of DNA is wrapped around a core histone octamer, which includes two of each histone H3, H2B, H2A, and H4, to form a nucleosome. Nucleosomes are ∼11 nm in diameter and are the basic structural units of chromatin (Richmond and Davey, 2003). The association of linker histone H1 with linker DNA further organizes nucleosomes to form higher-order chromatin structures (Brown et al., 2006). During NET formation, chromatin is extremely decondensed to form 15–25-nm chromatin fibers (Brinkmann et al., 2004). However, the mechanisms regulating this extreme chromatin decondensation are less clear. Posttranslational histone modifications play an important role in regulating chromatin structure and function (Jenuwein and Allis, 2001; Kouzarides, 2007), including chromatin decondensation/condensation and transcription regulated by citrullination (Cuthbert et al., 2004; Wang et al., 2004, 2009; Neeli et al., 2008). We and others have recently found that an increase in histone citrullination is associated with chromatin decondensation during NET formation (Neeli et al., 2008; Wang et al., 2009). The conversion of histone Arg or monomethyl-Arg to citrulline residues is catalyzed by peptidylarginine deiminase 4 (PAD4; also called PADI4 or PADV; Wang et al., 2004), a neutrophil enriched nuclear enzyme (Nakashima et al., 2002). Excessive PAD4 function has been related to rheumatoid arthritis (Yamada and Yamamoto, 2007) and cancer (Chang and Han, 2006; Chang et al., 2009). However, whether PAD4 is an important factor for NET-mediated innate immune functions has not been investigated. Here, using the PAD4 knockout mice, we show that PAD4 is required for bacterial killing by NETs.
RESULTS AND DISCUSSION
Effects of PAD4 knockout on mouse early development and neutrophil differentiation
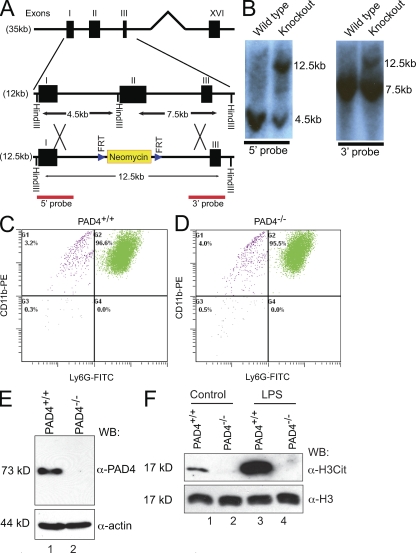
To analyze the function of PAD4 in NET formation and bacterial killing, we generated PAD4−/− mice by deleting PAD4 exon II (Fig. 1, A and B). Exon II deletion generates a frame shift close to the 5′ end of PAD4 coding domain sequence, thereby terminating PAD4 translation prematurely. PAD4 mutant mice were genotyped using wild-type and mutant-specific PCR primers (Fig. S1, A–D). The PCR product of the mutant PAD4 locus was sequenced to confirm the replacement of exon II by a residual flippase recognition target (FRT) sequence (Fig. S1 E). PAD4 homozygous mice survived to adulthood and demonstrated no detectable physical abnormality (unpublished data). However, PAD4 heterozygous and homozygous mice were born at a rate lower than predicted by the Mendelian ratio (Fig. S1 F), which suggests that the loss of PAD4 affects embryonic development.
Figure 1.
PAD4 is essential for histone citrullination in mouse neutrophils. (A) Schematic illustration of the replacement of exon II in the PAD4 gene with the FRT site–flanked neomycin cassette. A Hind III restriction site adjacent to exon II was also replaced. (B) Replacement of exon II leads to an appearance of a 12.5-kb fragment in addition to the 4.5-kb fragment detected by the 5′ probe (left) or the 7.5-kb fragment detected by the 3′ probe (right) in the Southern blot after digestion of ES cell genomic DNA with Hind III (representative results from three independent experiments are shown). (C and D) Flow cytometry analyses of peripheral blood neutrophils in PAD4+/+ (C) and PAD4−/− (D) mice using antibodies against neutrophil surface markers CD11b and Ly6G. Peripheral blood neutrophils were purified from five PAD4+/+ or five PAD4−/− paired mouse siblings in each experiment. Representative results from three independent experiments are shown. (E) Western blotting of PAD4 protein in PAD4+/+ and PAD4−/− neutrophils. Mouse PAD4 was detected as an ∼73-kD protein in SDS-PAGE (five mice per genotype, representative results of two independent experiments). (F) Western blotting of histone citrullination in PAD4+/+ and PAD4−/− neutrophils were performed before and after LPS treatment. General H3 antibody was used to ensure equal loading (five mice per genotype, three independent experiments).
PAD4 expression increases when human leukemia HL-60 cells differentiate along the granulocytic pathway (Nakashima et al., 1999), and PAD4 is abundant in the nucleus of peripheral blood neutrophils (Nakashima et al., 2002). To investigate whether PAD4 depletion affects neutrophil differentiation, we used antibodies against neutrophil cell surface markers CD11b and Ly6G to perform flow cytometry analyses. Comparable amounts of neutrophils were detected in the peripheral blood of PAD4+/+ and PAD4−/− mice, respectively (Fig. 1, C and D). Western blotting showed the expression of PAD4 in PAD4+/+ but not PAD4−/− neutrophils (Fig. 1 E). Electron microscopy analyses found that both PAD4+/+ and PAD4−/− neutrophils possess characteristic neutrophil morphology, including lobular nucleus and condensed chromatin regions under the nuclear envelope (Fig. S1 G). The above results indicate that PAD4 deletion did not affect the generation of mouse neutrophils during hematopoiesis.
PAD4 is required for histone citrullination and chromatin decondensation
Next, we analyzed whether PAD4 is important for histone citrullination. First, we found that recombinant mouse PAD4 citrullinated histone H3 in biochemical assays (Fig. S2, A–C). Second, endogenous histone H3 citrullination was detected in neutrophils from PAD4+/+ but not PAD4−/− neutrophils in Western blotting (Fig. 1 F), which indicates that the basal level of histone H3 citrullination is PAD4 dependent.
NET formation is induced after LPS and PMA treatment in human neutrophils (Brinkmann et al., 2004). To test whether PAD4 is important for chromatin decondensation and NET formation upon LPS treatment, immunostaining experiments were performed. Before LPS treatment, ∼3.69% of PAD4+/+ neutrophils are positively stained for histone H3 citrullination, but NET formation was not detected (Fig. 2 A and Table I), whereas histone H3 citrullination or NET formation was not observed in PAD4−/− neutrophils (Fig. 2 A and Table I). After 3 h of LPS treatment, an increase in histone H3 citrullination was detected in PAD4+/+ but not in PAD4−/− neutrophils by Western blotting (Fig. 1 F) or immunostaining analyses (Fig. 2 B and Table I). DNA staining (pseudo-colored green) identified chromatin decondensation to various degrees after LPS treatment, including swelling nuclei (Fig. 2 B; Nuclear area quantification with ImageJ in Fig. S3 A showed an ∼3.5-fold increase of these swelling nuclei) and elongated chromatin in PAD4+/+ neutrophils (Fig. 2 B and Table I). NETs are composed of highly decondensed chromatin and anti-bacterial granular proteins (Brinkmann et al., 2004). The decondensed chromatin induced by LPS showed positive staining by histone H3 citrullination and neutrophil elastase antibodies (Fig. 2 C), which indicates that these structures are indeed NETs. The PAD inhibitor Cl-amidine was previously found to repress the formation of a NET-like structure in human HL-60 granulocytic cells (Wang et al., 2009). After pretreatment of PAD4+/+ mouse neutrophils with Cl-amidine for 30 min, histone H3 citrullination and chromatin decondensation induced by LPS were much more decreased than untreated neutrophils (Table I and Fig. S3 B), which indicates that PAD activity is important for NET formation. Furthermore, chromatin decondensation and histone citrullination were detected in PAD4+/+ but not in PAD4−/− neutrophils after 3 h of PMA treatment (Table I and Fig. S3 C). These results indicate that PAD4−/− mouse neutrophils lack the ability to citrullinate histones or form NETs after LPS and PMA treatment.
Figure 2.
PAD4 is required for chromatin decondensation and NET formation after LPS and H2O2 treatment. (A) Histone citrullination and nuclear morphology in untreated neutrophils. A small number (∼3.69%) of PAD4+/+ neutrophils showed robust histone H3 citrullination (H3Cit) staining before stimulation. DNA dye Hoechst staining was pseudo-colored green. Decondensed chromatin was not observed before stimulation. Histone H3Cit or chromatin decondensation were not observed in PAD4−/− neutrophils before stimulation. (B) LPS treatment induced histone citrullination and chromatin structural changes in PAD4+/+ neutrophils. DNA was pseudo-colored green. Notice the swelling nucleus (top) and chromatin elongation (middle). In contrast, LPS treatment did not induce histone citrullination or chromatin structural changes in PAD4−/− neutrophils (bottom). (C) Histone citrullination and neutrophil elastase staining colocalized with decondensed chromatin stained by DNA dye in PAD4+/+ neutrophils after LPS treatment. (D) H2O2 treatment induced histone citrullination and chromatin structural changes in PAD4+/+ neutrophils but not in PAD4−/− neutrophils. For assays in A–D, peripheral blood neutrophils were purified from five PAD4+/+ or five PAD4−/− paired mouse siblings in each experiment, and at least three independent experiments for each treatment condition were performed.
Table I.
Percentages of mouse neutrophils with positive histone citrullination, enlarged nucleus, and NET formation after treatment with LPS, PMA, and H2O2
| Treatment | PAD4+/+ | PAD4−/− | ||||
| Histone citrullination | Enlarged nucleusab | NET formationcd | Histone citrullination | Enlarged nucleus | NET formation | |
| Control | 3.7 ± 0.3%e | UDf | UD | UD | UD | UD |
| LPS | 42.3 ± 3.9%e | 23.4 ± 2.7%e | 9.5 ± 0.5%e | UD | UD | UD |
| Cl-amidinef → LPS | 8.1 ± 1.3%e | 1.2 ± 0.2%e | 0.32 ± 0.3%e | UD | UD | UD |
| PMA | 48.5 ± 3%e | 10.9 ± 0.3%e | 2.7 ± 0.5%e | UD | UD | UD |
| H2O2 | 48.3 ± 5.4%e | 13.4 ± 1.4%e | 3 ± 0.3%e | UD | UD | UD |
U.D., undetectable.
The nucleus is scored as an enlarged nucleus if its nuclear diameter is >1.5-fold larger than a regular neutrophil, which is ∼10 µm in diameter.
Enlarged nuclei were found to be histone citrullination positive.
NET formation is scored if chromatin has extruded and elongated from the nucleus to extracellular space and is confirmed by neutrophil elastase staining after LPS treatment.
NETs were found to be histone citrullination positive.
Averages and standard deviations were shown (three independent experiments, >500 cells were counted from each experiment).
200 µM Cl-amidine was used to treat neutrophils for 30 min before LPS treatment for 3 h.
ROS, such as H2O2, efficiently induce the formation of NETs in adult neutrophils (Fuchs et al., 2007; Neeli et al., 2008) but not in newborn infant neutrophils (Yost et al., 2009), which suggests that other cellular mechanisms downstream of ROS are involved in regulating NET formation. Histone H3 citrullination and NET formation were detected in PAD4+/+ but not in PAD4−/− neutrophils after 3 h of H2O2 treatment (Fig. 2 D and Table I), which indicates that PAD4 functions downstream of H2O2 stimulus during NET formation. Table I summarizes the efficacy of LPS, PMA, and H2O2 in inducing chromatin structure changes in neutrophils. Although each of the treatments elicited nuclear morphology changes and NET formation to a certain extent, LPS was the most potent inducer for NET formation under current treatment conditions (Table I). Furthermore, RT-PCR experiments showed that the depletion of PAD4 in neutrophils did not affect the expression of other active PAD family members, including PAD1, -2, and -3 (Fig. S3 D). Collectively, the above results indicate that PAD4 is important for NET formation in mouse neutrophils upon treatment with proinflammatory stimuli.
PAD4 is required for S. flexneri killing by NETs
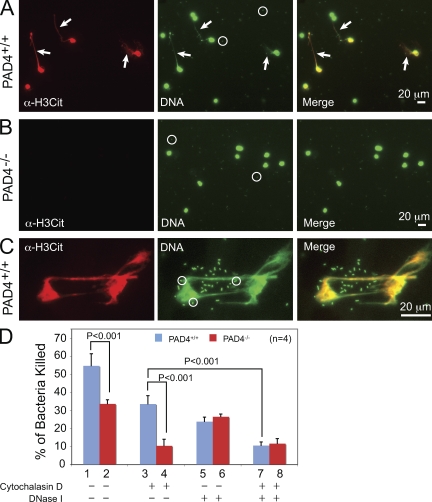
To analyze whether PAD4 is essential for NET formation after incubation with bacteria, we chose the pathogenic bacteria strain S. flexneri, which was previously shown to induce NET formation (Brinkmann et al., 2004). After priming neutrophils with IL-8 for 30 min and then incubation with S. flexneri for 2 h after a previously described procedure (Brinkmann et al., 2004), histone citrullination was detected in 28.1 ± 4.7% of PAD4+/+ neutrophils, and formation of NETs was detected in 13.9 ± 1.8% of PAD4+/+ neutrophils (Fig. 3 A, arrows denote NETs and open circles denote bacteria), which indicates that a fraction of neutrophils commit NET formation to kill extracellular bacteria. In contrast, histone citrullination or NET formation was not detected in PAD4−/− neutrophils (Fig. 3 B). Immunostaining images at higher magnification showed that histone H3 citrullination antibody stained NETs that are in close contact with bacteria (Fig. 3 C, circles).
Figure 3.
PAD4 is required for bacterial killing mediated by NETs. (A and B) Preincubation of neutrophils with IL-8 followed by incubation with S. flexneri bacteria induced histone citrullination and NET formation in PAD4+/+ (A) but not in PAD4−/− neutrophils (B). Arrows in A denote decondensed chromatin forming NETs. Circles in A and B highlight bacteria stained by the DNA dye. (C) Higher magnification images showing histone H3 citrullination and DNA staining of decondensed chromatin associated with bacteria. Circles indicate bacteria stained by DNA dye. (D) Percentages of S. flexneri bacteria killed by PAD4+/+ or PAD4−/− neutrophils, or neutrophils treated with cytochalasin D, DNase I, or both cytochalasin D and DNase I before incubation with bacteria. P-values (n = 4) were determined by a Student’s t test. Error bars indicate standard deviation. For assays in A–D, peripheral blood neutrophils were purified from five PAD4+/+ or five PAD4−/− paired mouse siblings in each experiment, and at least three independent experiments were performed.
To test whether PAD4-mediated NET formation is important for bacterial killing, S. flexneri were incubated with IL-8–primed PAD4+/+ and PAD4−/− neutrophils for 30 min, and bacterial killing was analyzed by colony formation assays. Under conditions when both phagocytosis and NET formation were permitted, 54.6 ± 6.6% and 33.6 ± 2.2% of bacteria were killed by PAD4+/+ and PAD4−/− neutrophils, respectively (Fig. 3 D). When phagocytosis was inhibited by cytochalasin D, 33.6 ± 4.5% and 10.5 ± 3.5% of bacteria were killed by the PAD4+/+ and the PAD4−/− neutrophils, respectively (Fig. 3 D), demonstrating a significant decrease (n = 4, P < 0.001, Student’s t test) in NET-mediated bacterial killing by PAD4−/− neutrophils compared with PAD4+/+ neutrophils. In contrast, when DNase I was added to disrupt NETs, PAD4+/+ and PAD4−/− neutrophils showed similar bacterial killing efficacy (Fig. 3 D), which suggests that the phagocytosis pathway is not affected by PAD4 deletion. When both cytochalasin D and DNase I were used simultaneously, bacteria killing by both the PAD4+/+ and the PAD4−/− neutrophils was decreased to low and comparable levels (Fig. 3 D). Collectively, these data indicate that PAD4 is important for S. flexneri killing mediated by NETs. Further, PAD4+/+ and PAD4−/− neutrophils uptake fluorescent bioparticles at comparable efficacy in phagocytosis analyses (Fig. S4, A and B), which indicates that the loss of PAD4 does not affect phagocytosis.
Killing M1 GAS or M1 ΔSda1 GAS bacteria by neutrophils
Certain serotypes of GAS, such as M1 GAS, cause an invasive infection called necrotizing fasciitis (Buchanan et al., 2006). M1 GAS secretes an extracellular DNase (Sda1) that helps bacteria to evade trapping and killing by NETs (Buchanan et al., 2006). We postulated that the M1 GAS and M1 ΔSda1 GAS strains offer a unique system to test the role of PAD4 in mediating NET formation and bacterial killing in vitro and in vivo. Similarly as previously reported (Buchanan et al., 2006), DNase activity was detected from the culture supernatant of M1 GAS but not from M1 ΔSda1 GAS (Fig. 4 A, compare lanes 2 and 3). Consistent with the notion that Sda1 helps M1 GAS disrupt the DNA backbone of NETs, M1 GAS induced histone H3 citrullination in PAD4+/+ neutrophils, but NETs were rarely observed (Fig. 4 B, top; and Fig. 4 D, light gray bars). In contrast, M1 ΔSda1 GAS induced both histone citrullination and NET formation in PAD4+/+ neutrophils (Fig. 4 C, top; and Fig. 4 D, dark gray bars). Higher-magnification images showed NET structures associated with bacteria after incubation of PAD4+/+ neutrophils with M1 ΔSda1 GAS (Fig. 4 E). Moreover, neither M1 GAS nor M1 ΔSda1 GAS induced histone citrullination or NET formation in PAD4−/− neutrophils (Fig. 4, B and C, bottom), which indicates that PAD4 is important for histone citrullination and NET formation induced by GAS bacteria.
Figure 4.
Antagonism of PAD4-mediated NET formation and bacterial extracellular DNase-mediated NET destruction. (A) Genomic DNA (untreated in lane 1) was incubated with cell culture supernatant from wild-type M1 GAS (lane 2) or M1 ΔSda1 GAS (lane 3). DNA degradation by DNase Sda1 was observed (lane 2; representative results of three independent experiments). (B) Upon incubation of M1 GAS with PAD4+/+ neutrophils, histone H3 citrullination was detected but NETs were rarely observed by immunostaining (top). In contrast, histone H3 citrullination or NETs were not detected after incubation of M1 GAS with PAD4−/− neutrophils (bottom). (C) Both histone H3 citrullination and NETs were detected after incubation of M1 ΔSda1 GAS with PAD4+/+ neutrophils (top, arrows denote NETs). In contrast, histone H3 citrullination or NETs were not detected after incubation of M1 ΔSda1 GAS with PAD4−/− neutrophils (bottom). (D) Percentages of PAD4+/+ neutrophils with H3 citrullination staining or with both H3 citrullination and NET formation after incubation with M1 GAS or M1 ΔSda1 GAS were analyzed. The presence of Sda1 decreased NET formation by ∼4.2-fold (P < 0.003 by a Student’s t test). All NETs were positive for the H3 citrullination antibody staining. Error bars indicate standard deviation. (E) Higher-magnification images show NETs formed in PAD4+/+ neutrophils after incubation with M1 ΔSda1 GAS (arrows denote NETs). For assays in B-E, peripheral blood neutrophils were purified from five PAD4+/+ or five PAD4−/− paired mouse siblings in each experiment, and three independent experiments were performed.
Next, we tested the bacterial killing efficacy of M1 GAS or M1 ΔSda1 GAS by PAD4+/+ or PAD4−/− neutrophils (Fig. S4 C). Wild-type M1 GAS bacteria were killed by PAD4+/+ and PAD4−/− neutrophils at low but comparable efficacy, whereas M1 ΔSda1 GAS was killed more efficiently by PAD4+/+ neutrophils than by PAD4−/− neutrophils (Fig. S4C, 1–4), which suggests that (a) NET-mediated bacterial killing by PAD4+/+ neutrophils was inhibited by the DNase Sda1, and (b) that PAD4−/− neutrophils have a decreased ability in killing M1 ΔSda1 GAS due to a lack of NET formation. Furthermore, when phagocytosis was inhibited by cytochalasin D, PAD4+/+ neutrophils killed M1 ΔSda1 GAS more effectively, but not more M1 GAS than PAD4−/− neutrophils (Fig. S4 C, 5–8). In contrast, when NET formation was inhibited by DNase I treatment, PAD4+/+ neutrophils were somewhat less efficient in killing bacteria than PAD4−/− neutrophils (Fig. S4 C, 9–12). These results underscore the importance of PAD4 in NET-mediated killing of GAS and a dynamic contribution of NETs and phagocytosis in bacterial killing.
PAD4−/− mice are more susceptible to infection by M1 ΔSda1 GAS
To analyze the susceptibility of PAD4−/− mice to bacterial infection, we applied the mouse model of necrotizing fasciitis (Buchanan et al., 2006), in which M1 GAS or M1 ΔSda1 GAS bacteria were subcutaneously injected into paired PAD4+/+ and PAD4−/− mouse siblings. In agreement with the in vitro bacterial killing results, PAD4+/+ and PAD4−/− mice developed large lesions at 3 d after injection of 5 × 106 M1 GAS bacteria (Fig. 5 A, mouse left flanks). In contrast, PAD4+/+ mice developed small lesions, whereas PAD4−/− mice developed much larger lesions at 3 d after injection of 5 × 106 M1 ΔSda1 GAS bacteria (Fig. 5 A, mouse right flanks). The lesion areas were measured using ImageJ. Compared with that in PAD4+/+ mice, M1 ΔSda1 GAS formed ∼4.03-fold larger lesions in PAD4−/− mice (P < 0.001; Fig. 5 B, dark gray bars). Colony formation assays of bacteria recovered from necrotic lesion sites found that M1 ΔSda1 GAS proliferated ∼3.97-fold more in PAD4−/− mice than in PAD4+/+ mice (P < 0.001; Fig. 5 C). Skin lesions were also taken at 2 d after infection and prepared for histology analyses. The lesion severity was scored from 0 (normal) to 4 (severe) based on the presence of neutrophils, edema, necrosis, and skin ulceration according to a previously described method (Mann et al., 2004). The difference was most pronounced with the M1 ΔSda1 GAS infections where the average pathology score for PAD4−/− mice was approximately four times greater than that of the PAD4+/+ mice (unpublished data). Representative photomicrographs of lesions from each group were shown (Fig. 5 D). These histopathological findings are in close agreement with the overall findings and colony formation assays.
Figure 5.
PAD4 is important in immune defense against GAS in a mouse model of necrotizing fasciitis. (A) In necrotizing fasciitis assays, M1 GAS but not M1 ΔSda1 GAS induced large lesions in PAD4+/+ mice (two representative PAD4+/+ mice are shown on the left panels). In contrast, both M1 GAS and M1 ΔSda1 GAS formed large lesions in PAD4−/− mice (right). Arrows indicate lesion sites. (B) Lesion size was measured using ImageJ. The size of lesion formed by M1 ΔSda1 GAS in PAD4−/− mice increased ∼4.03-fold compared with PAD4+/+ mice (P < 0.001 by a Student’s t test). (C) The number of bacteria recovered from the lesion formation site was analyzed by colony formation assays. The number of M1 ΔSda1 GAS bacteria recovered from PAD4−/− mice increased ∼3.97-fold compared with PAD4+/+ mice (P < 0.001 by a Student’s t test). For A-C, three independent experiments were performed, with two pairs of PAD4+/+ and PAD4−/− mouse siblings used for each experiment. (D) Representative photomicrographs of skin lesions from PAD4+/+ and PAD4−/− mice infected with M1 GAS and M1 ΔSda1 GAS. Arrows indicate neutrophilic infiltrates and the star highlights intact epithelium that is absent in the other sections. Magnification is 100×. Three independent experiments, with one pair of PAD4+/+ and PAD4−/− mouse siblings, were formed for each experiment. (E) GST-PAD4 expressed and purified from Escherichia coli was used to treat histone H3 and mononucleosomes. The citrullination of histone H3 was tested by using histone H3 citrullination antibody (H3Cit) in Western blotting. Histone H3 was probed to show the amount of histone H3 in each lane (representative results of three independent experiments). (F) Analyses of the percentages of S. flexneri bacteria killed by histone H3, citrullinated histone H3, mononucleosomes, and citrullinated mononucleosomes at concentrations indicated. Standard deviations (indicated by error bars) are calculated from three independent repeat experiments.
Histones are highly enriched for positively charged Lys and Arg residues and have been found to effectively kill bacteria at low concentrations (Hirsch, 1958). To test the effects of Arg-Cit conversion on bacterial killing by histones, we treated free histone H3 or mononucleosomes with PAD4 and confirmed the histone citrullination by Western blotting (Fig. 5 E). In bacterial killing assays, we found that the citrullination of Arg residues of free histone H3 or nucleosomal histones decreased their respective efficacy of bacterial killing (Fig. 5 F). Collectively with the bacterial killing results by neutrophils, our results indicate that histone citrullination contributes to antibacterial function mainly by facilitating NET formation instead of increasing histone’s antibacterial activity.
Although the role of histone hypercitrullination in NET formation has been implicated in previous studies, it is now clear that PAD4 is an important immune factor required for NET-mediated anti-bacterial innate immunity. We have shown here that the deletion of PAD4 in mice abolishes both basal and inducible histone citrullination in peripheral blood neutrophils. This lack of histone citrullination is likely caused by the absence of PAD4 because the depletion of PAD4 does not affect the expression of other active PAD family members. We have also found that PAD4 is necessary for the formation of NETs induced by LPS, PMA, H2O2, and bacteria in cell culture, and that PAD4−/− neutrophils posses significantly decreased activity in killing bacteria in vitro due to a lack of NET formation.
Interestingly, Buchanan et al. (2006) showed that certain bacterial pathogens have evolved a mechanism in which they secrete an extracellular DNase to escape killing and trapping by NETs to enhance their survival within the host. Using the necrotizing fasciitis model with S. pyogenes strains of M1 GAS and M1 ΔSda1 GAS in PAD4+/+ and PAD4−/− mice, we demonstrated that PAD4−/− mice are more susceptible to bacterial infection than PAD4+/+ mice due to a lack of NET function.
Many pathological and genetic studies linked PAD4 and protein citrullination in synovial fluid with rheumatoid arthritis (Yamada and Yamamoto, 2007). We envision that under chronic inflammatory conditions, PAD4 is released from neutrophils during NET formation inside joints, leading to citrullination of self-proteins and thereby generating autoimmune antigens. As such, the decision for neutrophils to form NET must be deliberately controlled. It has been observed that neutrophils in newborns lack the ability to form NETs (Yost et al., 2009), and only a certain percentage of neutrophils engage NET formation in mice and human neutrophils (Fuchs et al., 2007). Further study of how NET formation is regulated will help us understand more about this fascinating process of bacterial killing.
MATERIALS AND METHODS
Generation of PAD4 knockout mice.
To generate the PAD4 knockout construct, a 3.1-kb PAD4 intron I fragment and a 2.7-kb PAD4 intron II fragment were PCR amplified and cloned before and after the neomycin cassette of the pGK-Neo-FRT vector (a gift from B. Wang, Weill Medical College, Cornell University, New York, NY). The knockout construct was sequence confirmed and linearized before electroporation to transfect embryonic stem (ES) J1 cells. ES cell colony screening was performed as described previously (Xiong et al., 2002). ES cells were microinjected into the early embryos to generate chimeric mice at the Transgenic Mouse Facility at Pennsylvania State University. To remove the neomycin cassette inserted in the PAD4 locus, neomycin-containing PAD4 homozygous mice were crossed with flippase-expressing 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J mice purchased from the Jackson Laboratory. The F1 generation was sibling crossed, and the F2 generation was PCR genotyped to identify PAD4 mice with the neomycin cassette removed from its insertion site at PAD4 exon II. All mice were maintained in an authorized animal facility, and mouse experiments were performed in accordance with the approved guideline of the Institutional Animal Care and Use Committee of Pennsylvania State University.
PCR genotyping of the PAD4+/+, PAD4+/−, and PAD4−/− mice.
Tail clipping and mouse genomic DNA extraction was performed essentially as described previously (Laird et al., 1991). Primers for PAD4 wild type were 5′-CACCGGGATTAAATACCCAATA-3′ and 5′-TGTGAACGTGACTGCATTTAGA-3′. Primers for PAD4 knockout were 5′-AGACCTGAGGAAGCTCAGACCTC-3′ and 5′-AGTGGTTTAGTCACACTGTGGATGT-3′.
Mouse peripheral blood neutrophil purification.
Mice peripheral blood was collected from the mandibular vein. Blood collected from five paired PAD4+/+ and PAD4−/− mouse siblings was pooled for neutrophil purification using Histopaque-1077 and -1119 density gradients according to the manufacturer’s instructions (Sigma-Aldrich). Contaminating erythrocytes were removed by hypotonic treatment. Neutrophils were finally resuspended in ice-cold PBS containing 10 mM d-glucose before further experiments.
Immunostaining, flow cytometry, and TEM analyses.
Cell staining was performed essentially as described previously (Wang et al., 2004). Antibodies used for immunostaining included α-H3Cit (Abcam) and α-neutrophil elastase (Santa Cruz Biotechnology, Inc.). DNA was stained by the DNA dye Hoechst. For flow cytometry analysis, 2 × 105 cells purified using Histopaque were resuspended in 200 µl PBS supplemented with 3% FBS. Biotinized CD11b antibody (BD) was added and incubated for 1 h on ice. After washing with PBS, streptavidin-conjugated PE (SA-PE; eBioscience) and fluorescein isothiocyanate-conjugated Ly6G antibody (BD) were added and incubated for 30 min in the dark. At least 104 positive stained cells were counted using a flow cytometer (FC500; Beckman Coulter) and analyzed with the CXP software at the Penn State Flow Cytometry Facility. Transmission electron microscopy analyses of PAD4+/+ and PAD4−/− neutrophil morphology were performed using the service of the Penn State Electron Microscopy Facility.
Western blotting and RT-PCR.
Western blotting using the α-PAD4 antibody was performed essentially as described previously (Wang et al., 2004). For RT-PCR, total RNA was extracted from neutrophils purified from peripheral blood of paired PAD4+/+ and PAD4−/− siblings using the RNeasy minikit (QIAGEN). RNA concentrations were measured using a spectrophotometer and further normalized using the amount of 18S rRNA. Equal amounts of RNA (0.3 µg) were used to perform RT-PCR using the Superscript One-Step RT-PCR kit (10928–042; Invitrogen). RT-PCR primers were mPAD1 (5′-ATGACTTCAAGGTGAAGGTG-3′ and 5′-TGTAGTTGGAGAGGGATGTC-3′), mPAD2 (5′-CTACATCTCCATGTCGGACT-3′ and 5′-GTGCCTCAATATAGCCAAAC-3′), mPAD3 (5′-GACTGTGACATGAACTGTGC-3′ and 5′-TCGGAGAAGTCCTCATTAGA-3′), and mPAD4 (5′-CTCTCCAGGAGTCATCGTAG-3′ and 5′-CCAACACCAGCTGATACTTT-3′).
Bacterial strains and growth conditions.
The S. flexneri strain (provided by the E. coli Reference Center at Penn State University) was grown in Luria-Bertani media. The M1 serotype of the GAS strain and the M1 ΔSda1 GAS strain were provided by V. Nizet (University of California, San Diego, San Diego, CA) and were grown in Todd-Hewitt broth (THB) or agar (THA).
Treatment of neutrophils with LPS, PMA, H2O2, IL-8, and the bacterial strains S. flexneri and GAS.
Mouse neutrophils were stimulated for 3 h with 1 µg/ml LPS, 100 µM H2O2, and 25 nM PMA, in the presence of 2 mM calcium at 37°C and 5% CO2 in PBS supplemented with 10 mM d-glucose before an immunostaining assay to determine NET formation. Enlarged neutrophil nuclei after 3 h of LPS treatment were measured using ImageJ (National Institutes of Health) to determine nuclear area changes. For PAD inhibitor treatment, neutrophils were incubated with 200 µM Cl-amidine for 30 min before LPS treatment. For IL-8 and bacteria treatment, 2 × 105 neutrophils were first incubated with 100 ng/ml IL-8 at 37°C and 5% CO2 for 30 min. 2 × 107 exponential phase S. flexneri or GAS were then added, and samples were centrifuged at 200 g for 10 min and further incubated with neutrophils for 2 h before immunostaining.
Phagocytosis assay.
Phagocytosis assays were performed using a Vybrant Phagocytosis Assay kit (Invitrogen) with modifications for use with flow cytometry essentially as described previously (Siemsen et al., 2007).
DNase activity assay.
M1 and M1 ΔSda1 GAS bacteria were grown overnight in THB, and the supernatant was collected. 1 µg of U2OS cell genomic DNA was incubated with 3 µl of bacteria supernatant for 10 min at 37°C in a total volume of 50 µl of buffer (300 mM Tris-HCl, pH 8.0, 3 mM CaCl2, and 3 mM MgCl2). EDTA was then added to a final concentration of 60 mM to stop the reaction. DNA was visualized in 1.2% agarose gel electrophoresis.
Bacterial killing assays using neutrophils.
Neutrophils purified from the peripheral blood of paired PAD4+/+ and PAD4−/− mouse siblings were resuspended in Locke’s solution (10 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM KCl, 2 mM CaCl2, and 10 mM d-glucose) at 2 × 106 cells/ml and incubated with 100 ng/ml IL-8 at 37°C in a 5% CO2 incubator for 30 min. 10 µg/ml cytochalasin D and/or 100 U/ml DNase I were then added to each individual group and further incubated for 15 min. Exponential phase S. flexneri, M1 GAS, or M1 ΔSda1 GAS bacteria were incubated with neutrophils to a multiplicity of infection (bacteria/cells) of 1:100. Cultures were centrifuged at 700 g for 10 min and further incubated for 30 min at 37°C. Samples were plated in lysogeny broth agar or THA at a series of 1:10 dilution, and CFUs were analyzed. Bacterial killing efficacy was calculated as the percentage of control values (bacteria incubated alone without neutrophils).
Mouse infection, necrotizing fasciitis, and histology assays.
Necrotizing fasciitis experiments were performed essentially as described previously (Datta et al., 2005; Buchanan et al., 2006). In brief, 5 × 106 M1 GAS or M1 ΔSda1 GAS bacteria at exponential growth phase were pelleted, washed, resuspended in 50 µl PBS, diluted 1:1 with 2.5 mg/ml Cytodex beads (Sigma-Aldrich), and subcutaneously injected into the shaved left and right flanks of 8–10-wk-old paired PAD4+/+ and PAD4−/− mouse siblings, respectively. Two pairs of PAD4+/+ and PAD4−/− mouse siblings were simultaneously injected for each independent experiment. Three independent experiments were performed. At 3 d after infection, mice were euthanized with CO2 and the lesion areas were pictured alongside with a scale bar. Lesion size was measured using ImageJ. The lesion areas were removed by skin biopsy and added to PBS. Bacteria extracted into PBS were plated in THA plates at a series of 1:10 dilution to determine CFUs. For histological evaluation, the skin lesions were excised at 2 d after bacterial infection and fixed with 10% neutral buffered formalin, embedded in paraffin, and sectioned. Slides were routinely prepared and stained with hematoxylin and eosin. An assessment of microscopic lesions was made by one of the authors (M.J. Kennett), a veterinarian with training and experience in rodent pathology and blinded to experimental treatment. The presence of neutrophils, edema, necrosis, and ulceration in each lesion were graded subjectively, on a relative scale of 0 to 4, with 0 being normal, 1 being mild, 2 being moderate, 3 being marked, and 4 being severe. The pathology score was calculated as the mean of the four categories for each infection.
Bacteria killing using histones and nucleosomes with or without citrullination.
Histone H3 and mononucleosomes were citrullinated by PAD4 as described previously (Wang et al., 2004). 106 exponential growing phase S. flexneri were incubated with unmodified or citrullinated histone H3 and mononucleosomes at various concentrations in a total volume of 100 µl at 37°C, with shaking in Hanks’ balanced salt solution buffer supplemented with 10 mM Hepes, pH 7.4. Samples were plated in lysogeny broth agar at a series of 1:10 dilution to determine CFUs. Bacterial killing efficacy was calculated as the percentage of control values (bacteria incubated alone without histone H3 or mononucleosomes).
Online supplemental material
Fig. S1 shows the generation of PAD4 knockout mice and the characterization of mutant mice and neutrophils. Fig. S2 shows that mouse GST-PAD4 fusion protein citrullinated histone H3. Fig. S3 shows that PAD4 activity is required for histone citrullination and NET formation after LPS and PMA treatment. Fig. S4 shows that PAD4−/− neutrophils are normal in phagocytosis but lack the ability to kill GAS bacteria by NETs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100239/DC1.
Acknowledgments
We are grateful to the Transgenic Mouse, the Flow Cytometry, and the Electron Microscopy Facilities at Pennsylvania State University for their technical help. We thank Dr. V. Nizet (University of California, San Diego) for GAS bacteria strains, Dr. B. Wang (Weill Medical College, Cornell University) for reagents, and Drs. J.C. Reese, D.S. Gilmour, B.F. Pugh, S.A. Coonrod (Weill Medical College, Cornell University), and C.D. Allis (Rockefeller University) for suggestions and critical reading of the manuscript.
Research is supported in part by a Pennsylvania State University start-up fund and a National Institutes of Health grant (R01 CA136856) to Y. Wang. M. Li generated the knockout ES cells and the chimera mice. The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- Cit
- citrullination
- ES
- embryonic stem
- FRT
- flippase recognition target
- GAS
- group A Streptococcus pyogenes
- NET
- neutrophil extracellular trap
- PAD4
- peptidylarginine deiminase 4
- ROS
- reactive oxygen species
References
- Bianchi M., Hakkim A., Brinkmann V., Siler U., Seger R.A., Zychlinsky A., Reichenbach J. 2009. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 114:2619–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V., Zychlinsky A. 2007. Beneficial suicide: why neutrophils die to make NETs. Nat. Rev. Microbiol. 5:577–582 10.1038/nrmicro1710 [DOI] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Brown D.T., Izard T., Misteli T. 2006. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 13:250–255 10.1038/nsmb1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan J.T., Simpson A.J., Aziz R.K., Liu G.Y., Kristian S.A., Kotb M., Feramisco J., Nizet V. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16:396–400 10.1016/j.cub.2005.12.039 [DOI] [PubMed] [Google Scholar]
- Chang X., Han J. 2006. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol. Carcinog. 45:183–196 10.1002/mc.20169 [DOI] [PubMed] [Google Scholar]
- Chang X., Han J., Pang L., Zhao Y., Yang Y., Shen Z. 2009. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer. 9:40 10.1186/1471-2407-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert G.L., Daujat S., Snowden A.W., Erdjument-Bromage H., Hagiwara T., Yamada M., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Kouzarides T. 2004. Histone deimination antagonizes arginine methylation. Cell. 118:545–553 10.1016/j.cell.2004.08.020 [DOI] [PubMed] [Google Scholar]
- Datta V., Myskowski S.M., Kwinn L.A., Chiem D.N., Varki N., Kansal R.G., Kotb M., Nizet V. 2005. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol. Microbiol. 56:681–695 10.1111/j.1365-2958.2005.04583.x [DOI] [PubMed] [Google Scholar]
- Fuchs T.A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., Zychlinsky A. 2007. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176:231–241 10.1083/jcb.200606027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J.G. 1958. Bactericidal action of histone. J. Exp. Med. 108:925–944 10.1084/jem.108.6.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. 2001. Translating the histone code. Science. 293:1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128:693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Laird P.W., Zijderveld A., Linders K., Rudnicki M.A., Jaenisch R., Berns A. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293 10.1093/nar/19.15.4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann P.B., Elder K.D., Kennett M.J., Harvill E.T. 2004. Toll-like receptor 4-dependent early elicited tumor necrosis factor alpha expression is critical for innate host defense against Bordetella bronchiseptica. Infect. Immun. 72:6650–6658 10.1128/IAI.72.11.6650-6658.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Hagiwara T., Ishigami A., Nagata S., Asaga H., Kuramoto M., Senshu T., Yamada M. 1999. Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1alpha,25-dihydroxyvitamin D(3). J. Biol. Chem. 274:27786–27792 10.1074/jbc.274.39.27786 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Hagiwara T., Yamada M. 2002. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 277:49562–49568 10.1074/jbc.M208795200 [DOI] [PubMed] [Google Scholar]
- Nathan C. 2006. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6:173–182 10.1038/nri1785 [DOI] [PubMed] [Google Scholar]
- Neeli I., Khan S.N., Radic M. 2008. Histone deimination as a response to inflammatory stimuli in neutrophils. J. Immunol. 180:1895–1902 [DOI] [PubMed] [Google Scholar]
- Nizet V. 2007. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J. Allergy Clin. Immunol. 120:13–22 10.1016/j.jaci.2007.06.005 [DOI] [PubMed] [Google Scholar]
- Richmond T.J., Davey C.A. 2003. The structure of DNA in the nucleosome core. Nature. 423:145–150 10.1038/nature01595 [DOI] [PubMed] [Google Scholar]
- Segal A.W. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197–223 10.1146/annurev.immunol.23.021704.115653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemsen D.W., Schepetkin I.A., Kirpotina L.N., Lei B., Quinn M.T. 2007. Neutrophil isolation from nonhuman species. Methods Mol. Biol. 412:21–34 10.1007/978-1-59745-467-4_3 [DOI] [PubMed] [Google Scholar]
- von Köckritz-Blickwede M., Nizet V. 2009. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. 87:775–783 10.1007/s00109-009-0481-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wysocka J., Sayegh J., Lee Y.H., Perlin J.R., Leonelli L., Sonbuchner L.S., McDonald C.H., Cook R.G., Dou Y., et al. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 306:279–283 10.1126/science.1101400 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li M., Stadler S., Correll S., Li P., Wang D., Hayama R., Leonelli L., Han H., Grigoryev S.A., et al. 2009. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184:205–213 10.1083/jcb.200806072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartha F., Henriques-Normark B. 2008. ETosis: a novel cell death pathway. Sci. Signal. 1:pe25 10.1126/stke.121pe25 [DOI] [PubMed] [Google Scholar]
- Xiong N., Kang C., Raulet D.H. 2002. Redundant and unique roles of two enhancer elements in the TCRgamma locus in gene regulation and gammadelta T cell development. Immunity. 16:453–463 10.1016/S1074-7613(02)00285-6 [DOI] [PubMed] [Google Scholar]
- Yamada R., Yamamoto K. 2007. Mechanisms of disease: genetics of rheumatoid arthritis—ethnic differences in disease-associated genes. Nat. Clin. Pract. Rheumatol. 3:644–650 10.1038/ncprheum0592 [DOI] [PubMed] [Google Scholar]
- Yost C.C., Cody M.J., Harris E.S., Thornton N.L., McInturff A.M., Martinez M.L., Chandler N.B., Rodesch C.K., Albertine K.H., Petti C.A., et al. 2009. Impaired neutrophil extracellular trap (NET) formation: a novel innate immune deficiency of human neonates. Blood. 113:6419–6427 10.1182/blood-2008-07-171629 [DOI] [PMC free article] [PubMed] [Google Scholar]