Abstract
Numerous genetic studies associated the Neuregulin 1 (NRG1) Icelandic haplotype (HAPice), and its single nucleotide polymorphism SNP8NRG243177 [T/T], with schizophrenia. Because SNP8NRG243177 [T/T] has characteristics of a functional polymorphism that maps close to NRG1 type IV coding sequences, our initial goal was to map precisely the human type IV transcription initiation site. We determined that the initiation site is 23bp upstream of the previously reported type IV exon, and that no other transcripts map to the SNP8NRG243177 region. Because NRG1 type IV transcripts are specific to human, we isolated full-length NRG1 type IV cDNAs from human hippocampi and expressed them in non-neural cells and dissociated rat hippocampal neurons to study protein expression, processing and function. Using an antiserum we generated against the NRG1 type IV-specific N-terminus, we found that the protein is targeted to the cell surface where PKC activation promotes its cleavage and release of the extracellular domain. Conditioned medium derived from type IV expressing cells stimulates ErbB receptor phosphorylation, as well as downstream Akt and Erk signaling, demonstrating that NRG1 type IV possesses biological activity similar to other releasable NRG1 isoforms. To study the subcellular targeting of distinct isoforms, neurons were transfected with the Ig-domain-containing NRG1 types I and IV, or the cysteine-rich domain (CRD) type III isoform. Three dimensional confocal images from transfected neurons indicate that, whereas all isoforms are expressed on somato-dendritic membranes, only the type III-CRD isoform is detectable in distal axons. These results suggest that NRG1 type IV expression levels associated with SNP8NRG243177 [T/T] can selectively modify signaling of NRG1 released from somato-dendritic compartments, in contrast to the type III NRG1 that is also associated with axons.
Keywords: Neuregulin, erbB4 receptor, neuregulin-erbB signaling, schizophrenia, polymorphism, subcellular localization
Introduction
NRG1 is the most extensively studied member of the Neuregulin family of growth and differentiation factors (NRG1–4) that signal through ErbB receptor tyrosine kinases (ErbB2–4) to regulate various intracellular signaling cascades. NRGs are part of the epidermal growth factor (EGF) superfamily and characterized by a conserved EGF-like domain that is both necessary and sufficient to elicit the autophosphorylation of ErbB receptors (Buonanno and Fischbach 2001; Mei and Xiong 2008). NRG1 isoforms are synthesized as transmembrane precursors that are cleaved at their extracellular juxtamembrane domain by metalloproteases in an activity and PKC-dependent fashion (Loeb et al. 1998; Wang et al. 2001); recently, neurotrophin-induced NRG1 release was shown to specifically require PKC delta activity (Esper and Loeb 2009). As discussed below, differential promoter usage and alternative splicing give rise to a large repertoire of NRG1 isotypes that can subserve distinct functions attributable to the functional domains encoded by the different exons (Buonanno and Fischbach 2001; Falls 2003).
Three promoters were originally identified in the NRG1 gene that generate the type I (ARIA, NDF, heregulin), type II (GGF) and type III (SMDF, CRD) pro-NRG1 isoforms. A recent primer extension and RT-PCR study, using internal primers corresponding to human sequences encoding the EGF-like domain, identified an additional putative transcription initiation site that encoded a 5' 187 base pair exon. The novel putative transcript was denoted NRG1 type IV (Steinthorsdottir et al. 2004), and was later shown to encode a full-length NRG1 transcript that is only expressed in the human brain (Tan et al. 2007). The major functional consequence of alternative promoter usage is differences in trans-membrane topology and types of extracellular domains. Whereas type I, II and IV NRG1 isoforms are single-pass transmembrane proteins, type III NRG1 harbors an additional transmembrane domain (TM) as well as a cysteine-rich domain (CRD), encoded by the first exon. NRG1 transcripts are also differentially spliced, giving rise to transcripts that contain different number of immunoglobulin-like (Ig) domains found in type I, II and IV NRG1 isotypes, but absent in the CRD-type isoform. The topography and presence of Ig-domains influence the function of distinct NRG1 isoforms because the Ig domains can localize released NRG1 to cell surface proteoglycans (Loeb and Fischbach 1995; Loeb et al. 1999), whereas the additional TM encoded by the type III exon retains cleaved NRG1 on the cell surface. Hence, NRG1 isotypes can signal through ErbB receptors either in a paracrine or direct cell-to-cell fashion. The different patterns of NRG1 isoform expression and processing suggested distinct functions (Sandrock et al. 1995; Yang et al. 1998). Consistent with this view, NRG1 type III locally regulates GABAergic interneuron development in the medial ganglionic emminence, whereas the type I/II isoforms can signal from a distance to promote interneuron migration from the medial ganglionic emminence into the neocortex (Flames et al. 2004).
In addition to its well-established role in glial development (Chen et al. 2006), the NRG1/ErbB pathway has more recently been implicated in the regulation of glutamatergic function and plasticity in the adult hippocampus (Huang et al. 2000; Kwon et al. 2005; Bjarnadottir et al. 2007; Kwon et al. 2008)} and prefrontal cortex (Gu et al. 2005). NRG1 can inhibit LTP induction and reverse its expression (depotentiate) at hippocampal Schaeffer collateral to CA1 pyramidal neuron synapses, and loss of ErbB4 results in increased LTP levels (Pitcher et al. 2008). We recently demonstrated that NRG1 can acutely (less than 2 min) enhances dopamine release in the hippocampus, and that its activation of D4 dopamine receptors reverses LTP by promoting the internalization of AMPA receptors (Kwon et al. 2008). Based on the depotentiating effects of acute NRG1 treatment on early-phase LTP at hippocampal glutamatergic synapses, we proposed that NRG1 contributes to the homeostatic regulation of glutamatergic and dopaminergic transmission that have critically been associated with schizophrenia (Kwon et al. 2005; Kwon et al. 2008).
There is accumulating genetic and functional evidence for an involvement of NRG1 and its ErbB receptors in the etiology of schizophrenia. Genetic studies in different populations have identified a NRG1 “schizophrenia at-risk” haplotype denoted HAPice” (Stefansson et al. 2002; Stefansson et al. 2003; Williams et al. 2003), and a recent meta-analysis confirmed the association of this haplotype with schizophrenia (Li et al. 2006; Munafo et al. 2006). A single nucleotide polymorphism (SNP8NRG243177 [T/T]) within the HAPice is associated with endophenotypes related to schizophrenia (Hall et al. 2006; Stefanis et al. 2007; Sprooten et al. 2009). Interestingly, SNP8NRG243177 maps upstream of the NRG1 type IV 5'-exon (Steinthorsdottir et al. 2004; Law et al. 2006; Tan et al. 2007)} and the [T/T] allele has been associated with higher type IV transcript levels in postmortem tissue from control individuals and persons with schizophrenia (Law et al. 2006). Moreover, in the initial studies that identified the type IV exon (Steinthorsdottir et al. 2004), as well as in this study, an open reading frame extends the entire length of the exon, but it is not clear if the type IV sequences identified encode a functional protein. This raises two important questions: 1) whether the sequences identified to encode the type IV exon might extend further upstream and harbor additional putative start codons, and 2) whether type IV transcripts could extend further upstream to include the SNP8NRG243177 polymorphism. Taken together these studies suggest that variations in NRG1 type IV expression have important implications for schizophrenia and therefore there is a need to map the imitation site of type IV and to study it biological function.
To this end, we have used a variety of approaches to isolate and map the bona fide NRG1 type IV transcription initiation site. Here we report that type IV exon is longer than initially reported. In addition, we have isolated two full-length polyadenylated type IV transcripts from adult human brain. To study the expression and processing of the type IV protein, we raised antibodies against the unique N-terminal 13 amino acid sequence of NRG1 type IV and analyzed expression in transfected heterologous cells because NRG1 type IV is uniquely expressed in human brain and is not detected in other tissues (Steinthorsdottir et al. 2004; Tan et al. 2007). Our experiments show that full-length pro-NRG1 type IV is targeted to the cell surface, its glycosylated extracellular domain is released in a PKC-dependent fashion, and it is biologically functional. Interestingly, the subcellular distribution of NRG1 type III (CRD) and type IV proteins, which vary only in N-terminal sequences encoded by the unique upstream exons, differentially accumulate in distinct neuronal compartments suggesting that these isoforms subserve distinct functions in the brain.
Experimental procedures
Materials
HEK-293 and OVCAR-3 cells were obtained from ATCC (Manassas, VA). Human NRG1β1 peptide, encompassing the EGF-domain peptide between amino acids 176–246, was from R&D Systems (Minneapolis, MN). PD158780 was from Calbiochem (LaJolla, CA). Antibodies: mouse monoclonal anti–V5 epitope (Serotec, Kidington, UK), rabbit polyclonal anti-NRG1 CTa (SC348, Santa Cruz Biotechnology, CA), rabbit polyclonal anti-Akt (pan), rabbit polyclonal anti-phospho-Akt (s473), rabbit polyclonal anti-p44/42 MAP Kinase and rabbit polyclonal anti -phospho-p44/42 MAP Kinase (Cell Signaling Technology, MA), rabbit polyclonal anti-phohpo-erbB2 (Tyr 1248, Millipore, MA), rabbit polyclonal anti-erbB2 (Thermo, MA) mouse monoclonal anti-phospho-tyrosine (Upstate Biotechnology, NY) and mouse monoclonal-microtubule associated protein 2 (MAP2, Sigma. MO).
Generation of antibodies against NRG1 type IV
A polyclonal antiserum against a peptide comprising the NRG1 type IV-specific amino-terminal 13 residues (N-Met-Gly-Lys-Gly-Arg-Ala-Gly-Arg-Val-Gly-Thr-Thr-Ala) was raised in New Zealand white rabbits (Covance, Denver, PA). A Cys was added to the peptide at the C-terminus end for conjugation. Two rabbits were injected (HL5792 and HL5793). NRG1 type IV-specific antibodies from HL5792 were affinity-purified against the immunogenic peptide, and tested for specificity in Western blots and by immunocytochemistry of HEK-293 cells transfected with NRG1 type IV or type I expression vectors.
5' and 3'-rapid amplification of cDNA ends (RACE)
5' and 3'-RACE experiments were performed using First Choice RACE-Ready cDNA from Ambion (Austin, TX) according to the manufacturer's instructions. We chose this kit because it selected for full-length mRNAs by treatment with tobacco acid pyrophosphatase (TAP) to remove the cap structure from the full-length mRNA leaving a 5'-monophosphate. A synthetic RNA adapter is ligated to the RNA population using T4 RNA ligase so only molecules containing a 5'-phosphate - de-capped, full-length mRNAs - will accept the adapter. This approach also removes ribosomal RNA, fragmented mRNA, tRNA, and contaminating genomic DNA. In brief, for 5' RACE a gene specific reverse primer that binds close to the start codon of the type IV exon (5' CTG TGG TGC CAA CTC GGC CCG C 3') and the 5' RACE outer primer (corresponding to the 5' RACE adapter sequence) were used to amplified the type IV 5' end region from human brain RACE Ready cDNA. PCR products were subcloned into pGEM-T easy vector (Promega, Madison, WI) for sequencing.
For 3' RACE, the cDNA was amplified by nested PCR. The first 3' RACE PCR reaction was performed using a gene-specific forward primer that binds close to the 3'-end of the type IV exon-E187 (5' CGA GTT GGC ACC ACA GCC TTG 3') and the 3' RACE outer primer. A primer corresponding to the EGF-like domain (5' TCA AAC CCC TCG AGA TAC TTG TGC A 3') and the 3' RACE inner prime were used in the second PCR reaction.
Isolation of full-length NRG1 type IV transcripts
Total RNA from human hippocampi and frontal cortex was purchased from Ambion (Austin, TX). First-strand cDNA synthesis was carried out using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) with a gene specific reverse primer for the cytoplasmic tail `a' (CTa; 5' TTA TAC AGC AAT AGG GTC TTG GTT AGC 3') or for cytoplasmic tail `b' (CTb; 5' TTA GAG AAT GAA GCC CAA ATG GGG 3'); see (Falls 2003) for conventions on NRG1 domain nomenclature. The Expand Long Template PCR system (Roche, Indianapolis, IN) was used to amplify NRG1 type IV cDNAs using a gene-specific forward primer corresponding to sequences that map close to the 3'-end of the type IV exon-E187 (5' CGA GTT GGC ACC ACA GCC TTG 3') and reverse primers corresponding either to the C-terminal “a” (CTa), “b” (CTb) or “c” (CTc) tails. PCR conditions were as follows: an initial denaturating step at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 10 seconds, annealing at 58°C for 30 seconds and extension at 68°C for 3 min. Extension steps were increased by 20 seconds for each of the last 20 cycles. Resultant PCR products were cloned into pGEM-T easy vector (Promega, Madison, WI) and screened by colony hybridization using a [32P]-labeled oligonucleotide probe specific to the sequence encoding the NRG1 TM region (E103). Positive clones harboring NRG1 type IV cDNAs were confirmed by sequencing and subcloned into mammalian expression vector pcDNA3.1/V5-HIS TOPO (Invitrogen, Carlsbad, CA) using PCR. Primers were: CTa and a forward primer harboring the sequence for the V5 epitope tag (5' AGC AGC ATG GGT AAG CCT ATC CCT AAC CCT CTC CTC GGT CTC GAT TCT ACG GGG AAA GGA CGC GCG GGC CGA GTT GGC ACC ACA GAC TTG 3'). In addition, a cDNA corresponding to the β3 splice variant of NRG1 type IV was isolated by RT-PCR of human hippocampal RNA (Ambion,TX) using a β3-specific reverse primer (5' AGA CAG AAA GGG AGT GGA CGT A 3') and a NRG1 type IV specific forward primer (5' GGC AGC AGC ATG GGG AAA GGA 3'). PCR products cloned into the pcDNA3.1/V5-HIS TOPO vector as described above and verified by sequencing.
Analysis of NRG1 type IV expression in heterologous cells and stimulation with Phorbol ester
HEK-293 cells were cultured in MEM/10% FBS/ 2% L-glutamine. Cells were transfected at 60–80% confluence with NRG1 type IV expression constructs using FuGene6 transfection reagent (Roche, Indianapolis, IN). Forty-eight hours after transfection, cells were briefly washed with PBS and lysed in 10 mM Tris-HCl pH-7.5, 150 mM NaCl, 0.1% Na-Deoxycholate, 1% Triton-X 100 and 0.1% SDS to prepare whole-cell extracts for Western blotting analyses. For phorbol ester stimulation experiments the standard medium was replaced with OPTI-MEM I medium 48 hours after transfection. 12 hours later this medium was replace with a fresh OPTI-MEM medium with or without 0.5 μM Phorbol 12 Myristate 13 Acetate (Calbiochem, La Jolla, CA). The cells were incubated at 37°C for 30 min. The conditioned medium was collected, filtered through a 0.22 μm filter (Millipore, Billerica, MA) and concentrated using Amicon Ultra-15 centrifugal filters (Millipore, Billerica, MA).
NRG1/ErbB signaling assay
OVCAR-3 cells were maintained in DMEM / 10% FBS and transfected with NRG1 type IV as described above. Twenty-four hours after transfection, cells were starved for an additional 24 hours in DMEM containing 10 μg/ml transferrin. Conditioned medium was collected and used to stimulate 24 hours serum-starved untransfected OVCAR-3 cells in the present or the absent of 10 μM PD158780 for 5 min at room temperature. Conditioned medium from OVCAR-3 cells transfected with NRG1 type I and direct stimulation with NRG1-β1 peptide (0.5–5 nM) were included as controls. Cells were lysed immediately in buffer containing 100 mM Tris pH-6.8, 2% SDS, 10% glycerol and phosphatase / protease inhibitors. Lysates were incubated for 5 min at room temperature, sonicated, and heated for 5 min at 60°C.
Immunoblot analysis
Total protein from lysates of transfected HEK-293 cells (10μg), or from stimulated OVCAR-3 cells (5μg) were size fractionated on 4–12% gradient SDS-PAGE gels (Invitrogen, Carlsbad, CA) and transferred onto nitrocellulose membranes. Blots were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T), and incubated overnight at 4°C with primary antibodies in TBS-T containing 3% bovine serum albumin (BSA). Following incubation with horseradish peroxidaseconjugated goat-anti-mouse or sheep-anti-rabbit secondary antibodies, blots were developed using chemiluminescence detection (ECL plus system, GE Healthcare / Amersham).
Immunocytochemical analysis of cell lines
Transfected HEK-293 cells on glass coverslips were washed with phosphate-buffered saline (PBS) and fixed for 10 min in PBS containing 4% paraformaldehyde (PFA). Cells were then blocked in 5% normal goat serum (NGS) with or without 0.1% Triton X-100 in PBS for 1 hour at room temperature. Primary antibodies against NRG1 type IV (HL5792) and the V5 epitope in PBS containing 5% normal goat serum were used either under non-permeabilizing conditions or in the presence of 0.1% Triton X-100. Cells were incubated with antibodies overnight at 4°C, and visualized with either goat-anti-mouse Alexa 488 or goat-anti-rabbit Cy3 secondary antibodies. Fluorescence signals were analyzed in a Zeiss 510 Meta confocal microscope at 40× magnification using oil immersive objective.
Immunocytochemical analysis of transfected primary hippocampal neurons
Dissociated hippocampal cultures were prepared from embryonic day 19 Sprague-Dawley rats as previously described (Longart et al. 2004) using Neurobasal medium supplemented with B27 (Invitrogen, Carlsbad, CA) (Brewer et al. 1993). Neurons were co-transfected at 15–17 days in vitro (DIV) with CsCl-purified plasmids expressing NRG1-type IV and SNPH-GFP (Kang et al. 2008) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Forty-eight hours later, cells were briefly washed with artificial cerebrospinal fluid (aCSF; in mM: 25 HEPES pH7.4, 119 NaCl, 2.5 KCl, 1 NaH2PO4, 2 CaCl2, 30 Glucose) and live-labeled for 15 min at 37°C with affinity-purified anti-NRG1 type IV (0.15 μg/ml), followed by incubation for 15 min at 37°C with a goat-anti-rabbit antibody conjugated to Cy3. Cells were then fixed in 4% PFA and blocked with 5% NGS in PBS containing 0.1% Triton X-100 for 1 hour at room temperature. In another set of experiments, cells were co-transfected as described above with NRG1 type IV, I or III and SNPH-GFP. Forty–eight hours later, cell were fixed and labeled with a NRG1 cytoplasmic tail a antibody (sc-348) and visualized with goat-anti-rabbit Cy3 secondary antibodies. In all experiments, dendrites were labeled using a monoclonal antibody against MAP2 and visualized with a goat-anti-mouse Alexa 405 secondary antibody. Cells were imaged as described above using a 63× oil immersion objective. A three-dimensional configuration of the axonal segment was build using the Zeiss Image program base on z-stack images.
Results
Identification of the NRG1 type IV transcription initiation site
The initial studies that reported the existence of a novel human NRG1 type IV exon were performed using PCR reactions from a brain cDNA library with a downstream non-coding primer anchored close to the NRG1 TM. An exonic region of 187 base pairs (E187) was identified that encodes an open reading frame along its entire length and harbors a potential in-frame ATG that was proposed to correspond to the type IV translation initiation site (Steinthorsdottir et al. 2004). Our analysis of genomic sequences upstream of the presumed type IV start codon using the Six Frame Translation program (http://searchlauncher.bcm.tmc.edu/cgi-bin/seq-util/seq-util.pl) revealed a putative 358 bp open reading frame (ORF) extending well beyond the 5'-end of E187. Because experimental approaches that directly identify the transcription initiation site were not used in the original study (Steinthorsdottir et al. 2004), we performed experiments to determine if type IV transcripts initiate from sequence further upstream.
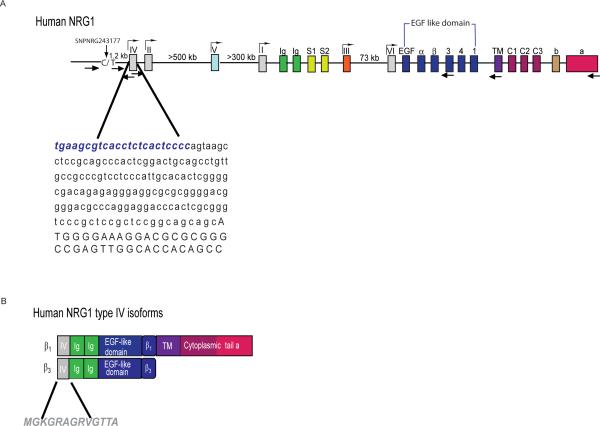
A reverse transcription protocol that selects specifically for full-length capped mRNAs (see Methods), combined with RT-PCR using type IV-specific primers mapping to the 3'-end of E187, was used to identify type IV cDNAs that extended further upstream into the mRNA CAP site (Fig. 1A). After screening for the longest cDNAs using radiolabeled type IV oligonucleotides to probe Southern blots of the RT-PCR-amplified material and DNA sequencing, we determined that the type IV exon is actually 210 bp and the transcription initiation site resides 23 bp upstream of E187. The cDNA we obtained is contiguous with genomic DNA, indicating that no additional 5' exons are present in type IV transcripts and that these transcripts do not extend into the SNP8NRG243177 polymorphic site. Given the association of SNP8NRG243177 with schizophrenia endophenotypes and data suggesting that it could be a functional SNP, we investigated the existence of transcripts extending into this region by using RT-PCR of human brain RNA and different primer sets flanking SNP8NRG243177 (see Fig. 1A). No PCR products were amplified encompassing SNP8NRG243177 (data not shown). In conclusion, NRG1 type IV transcripts have a unique 210 bp exon with a single potential initiator methionine.
Fig 1. Mapping the NRG1 type IV promoter and isolation of type IV transcripts.
A) Genomic organization of NRG1 showing putative promoters and exons encoding distinct functional domains. The exons are represented by different color boxes, and putative promoters (exons I through VI) are indicated (gray boxes). Exons encoding the different domains are color-coded and represent: Ig-like domain (green), spacers (cyan), α and β EGF-like domains splice variants with extensions 1–4 (blue), TM (purple), cytoplasmic domains 1–3 (bourdeaux), and the distinct C-termini a and b (orange). Sites and orientations of primers used in this study are indicated by the arrows below. The sequence of the type IV exon is shown, including the novel upstream 23 bp sequence identified in this study (bold blue). Location of the 8NRGSNP243177 polymorphism is indicated. (B) Schematic representation of the human type IV isoforms encoded by the cDNAs isolated in this study (β1and β3). The domains contained in each protein, as defined in the diagram above, are shown. The unique 13 amino acid type IV human sequence used to generate a type IV specific antibody is shown below.
Isolation of full-length NRG1 type IV cDNAs
Differential splicing of downstream exons gives rise to NRG1 isotypes that harbor distinct cytoplasmic carboxy-terminal (CT) tails denoted CTa, CTb and CTc (Falls 2003). In order to characterize the expression, processing and function of NRG1 type IV, we isolated cDNAs containing the entire open reading frame using RT-PCR and RNA isolated from adult human hippocampus and frontal cortex. We used a type IV-specific forward primer, mapping upstream of the initiator methionine, and reverse primers corresponding to the different CTa-c tails. We obtained a 1773 bp cDNA that corresponds to a full length NRG1 type IV transcript (accession # GQ983557) that encodes an Ig-like domain, an EGF-like (β1 isotype) motif, the TM and the CTa tail (shown schematically in Fig. 1B). No PCR products were obtained using reverse primers for either the CTb or CTc tails, suggesting that most NRG1 type IV transcripts in the adult cortex and hippocampus encode the CTa tail.
Isolation of NRG1 type IV β3 transcript by 3'-RACE
In an attempt to isolate NRG1 type IV transcripts in an unbiased fashion, we also employed 3'-RACE using an anchored oligo(dT) reverse primer (see Methods). We isolated transcripts that correspond to β3 isoforms (accession # GQ983558) and confirmed the result by RT-PCR of hippocampal RNA using a β3-specific reverse primer. This transcript encodes an Ig-like domain and EGFβ1-like motif, but it lacks sequences for the TM and is therefore presumed to give rise to soluble NRG1 protein (see also Fig. 1)
Analysis of NRG1 type IV protein glycosylation
We raised an NRG1 type IV-specific antiserum using a peptide corresponding to the unique 13 amino acids encoded by the type IV exon, in order to study the expression, processing and subcellular distribution of the protein. Antibodies were affinity-purified against the immunogenic peptide. The specificity of the antiserum was confirmed using a NRG1 type IV β1 expression construct tagged with the V5 epitope at the N-terminus. Using protein extracts from transfected HEK293 cells on Western blots (Fig 2A), the type IV antibody (rabbit HL5792) identified a major immunoreactive band of 90 kDa corresponding to the unprocessed pro-form of NRG1 type IV β1a; very minor non-specific 75 and 45 kD bands were detected with extracts from both the control (empty expression vector) and type IV transfected cells. The band at 90 kD is specific to NRG1 type IV, because it was selectively identified by the anti-V5 epitope tag antibody (Fig. 2A, middle panel). A commercial antiserum raised against the NRG1 CTa tail (SC-348) also identified the 90 kD type IV band, and it recognized a number of other products in protein extracts prepared from transfected HEK293 cells including a band at ~65 kDa that may represent the proteolytically processed intracellular domain (ICD). On the other hand, the absence of the lower molecular mass band with the type IV-specific antiserum suggests shedding of the extracellular domain (ECD). The origin of the other immunoreactive bands detected by SC-348 is unclear; nevertheless, neither the NRG1 type IV or V5 antibodies detected other major bands supporting the specificity of the rabbit type IV antiserum.
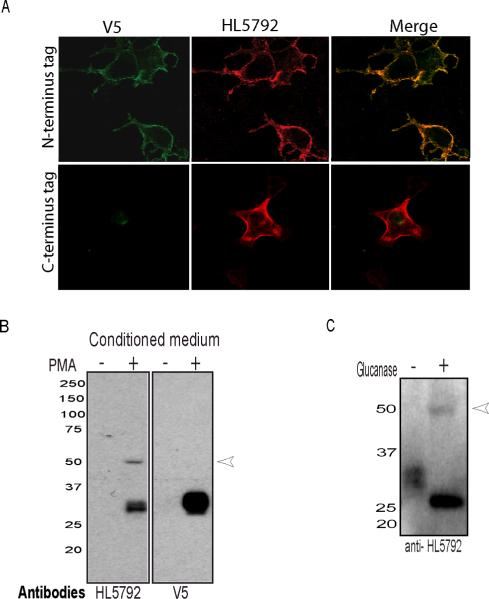
Fig 2. The NRG1 type IV antibody HL5792 identifies a glycosylated NRG1 type IVβ1a protein.
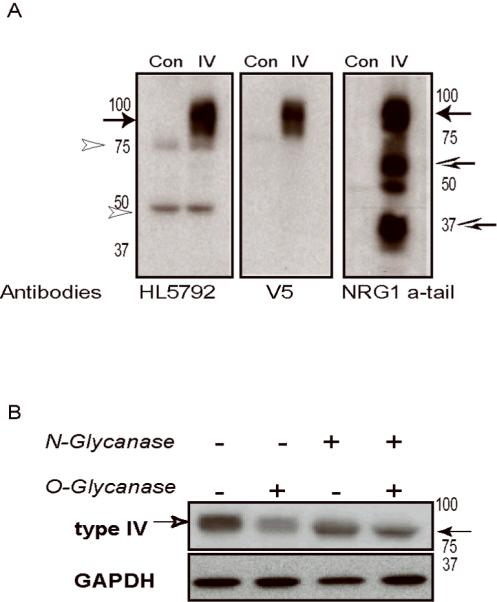
NRG1 type IV constructs expressing V5-tagged proteins were transfected into HEK293 cells. A) Western blots prepared with 10 μg protein from HEK293 whole cell extracts probed with HL5792 rabbit polyclonal antibody raised against the type IV N-terminus (left panel). A monoclonal antibody to the V5 epitope (middle) and a commercial antibody raised against the a-tail C-terminus (right) were used to confirm expression of full-length NRG1. Extracts from untransfected HEK293 cells were included as negative controls (Con). The 3 antibodies identified a major band at 90 kDa corresponding to pro-NRG1 type IV (arrow), while the C-terminal antibody also detected a putative processed ICD of ~ 65 kDa and additional truncated products (open arrows). The light bands observed at 75 and 50 kDa using HL5792 are nonspecific cross-reacting bands (open arrowheads), also seen in untransfected control. (B) Cell lysates were treated with O- or N-glycanase to determine extent of NRG1 type IV glycosylation. Western blots were probed with the V5 epitope tag antibody, stripped, and re-probed with anti-GAPDH. A shift in NRG1 type IV apparent molecular weight from 90 kD (open arrowhead, left) to 80 kD (closed arrowhead, right) was observed after N- and O- glycanase treatment; note that migration of GAPDH remains unchanged.
The predicted molecular mass of full length NRG1 type IV calculated from the deduced animo acid sequence is 66.8 kDa. However, the apparent molecular weight of type IV β1a protein in SDS-PAGE is about 90 kD; approximately 25% more than predicted. In order to determine if this discrepancy is attributable to glycosylation, lysates from transfected HEK293 cells were treated with either O-or N-Glycanase, or both, and the effects of these treatments were analyzed by Western blotting. Removal of O-linked sugars had no noticeable effect on migration of the apparent 90 kDa NRG1 type IV β1a band, while removal of N-linked sugars produced a ~10 kDa downward shift (Fig. 2B). Therefore, N-glycosylation partially contributes to the reduced mobility of NRG1 type IV β1a protein observed in SDS-PAGE. Additional post-translational modifications or protein properties must account for the aberrant migration observed in our experiments, as reported previously by other groups studying NRG1 type I (Peles et al. 1992; Wen et al. 1994). Taken together, these data demonstrate that the NRG1 type IV β1a transcript we isolated from human hippocampus is translated to yield a full-length, glycosylated pro-protein that is specifically recognized at its N-terminus by our rabbit type IV and V5-epitope tag antibodies, and recognized at its C-terminus by a NRG1 CTa antiserum.
Next we studied the expression of type IV in postmortem human brain (Clontech, Mountain View, CA). We observed a reproducible pattern of bands on Westerns (bands at 29, 35 and 50kD) that migrate in the range consistent with processed NRG1 type IV (data not shown). Interestingly, a similar banding pattern was reported for a NRG1 antiserum targeting the extracellular domain (ECD) of the protein (Pankonin et al. 2009). However, because of the differences between predicted and apparent molecular weights observed on Western blots using the NRG1 type IV-specific (HL5792) and pan intracellular domain (SC-348) antibodies, at this time we cannot conclusively designate them as processed NRG1 type IV products
PKC promotes shedding of the NRG1 type IV ECD from the cell surface
NRG1 type IV transcripts were reported to be specific for human brain and were not detected in several neuronal cancer origin cells lines (Tan et al. 2007), which imposed an experimental limitation to study its endogenous expression in human brain. We therefore resorted to studying its processing in transfected cultured cells. To examine if NRG1 type IV is targeted to the cell surface and processed, expression was analyzed by immunofluorescence in transfected cells using antibodies directed against the ECD. First, HEK-293 cells transfected with the cDNA encoding NRG1 type IVβ1a tagged at its N-terminus with the V5 epitope were fixed, and incubated with either type IV- or V5-specific antibodies using non-permeabilizing conditions. Under these conditions, both antibodies detected the fusion protein, indicating that their respective epitopes were exposed at the cell surface (Fig. 3A, top panel). In control experiments, the same antibodies were applied under permeabilizing conditions, and the resulting labeling pattern of internal and surface type IV β1a protein closely resembled that of the surface IV β1a alone (supplement Fig1A, top panel). Moreover, the fluorescence signals neatly outlined the cells, suggesting that the majority of immunoreactivity against the ECD is localized at the plasma membrane. To confirm that the immunofluorescence obtained under non-permeabilizing conditions was not contaminated by intracellular signals, we transfected cells with a NRG1 type IVβ1a construct in which the V5 epitope tag was fused to the carboxyl terminus of the protein. As shown in Fig. 3A (bottom panel), no V5-epitope immunoreactivity was detected in non-permeabilized cells, but was evident after permeabilization (supplement Fig1A, bottom panel). In contrast, the type IV-specific antibody detected the ECD in both conditions (Fig. 3A, bottom panel, supplement fig1A, bottom panel). The specificity of the immunofluorescence signals were confirmed in transfected cells treated only with secondary antibodies, or mock-transfected cells (Supplemental Fig. 1B). These results confirm the specificity of antiserum HL5792, and show that NRG1 type IV transcripts are translated and the protein targeted to the plasma membrane.
Fig 3. NRG1 type IV is targeted to the plasma membrane, proteolytically processed and released into the medium.
(A) HEK-293 cells expressing a cDNA encoding NRG1 type IV tagged at its N-terminus (top panel) or C-terminus (bottom panel) with the V5 epitope were fixed, and incubated with either type IV- or V5-specific antibodies using non-permeabilizing conditions. Type IV protein is expressed at the cell surface. (B) Type IV ECD is released into the medium in the presence of phorbol ester. HEK293 cells expressing V5 tagged-type IV were incubated with or without 0.5 μM PMA for 30 min. The condition medium was collected and concentrated by filtration. Concentrated conditioned medium was probed by Western blotting with HL5792 and V5 antibodies. Both antibodies detect a 29 kDa band corresponding to the type IV ECD. (C) Type IV ECD concentrated medium treated with O- or N- Glycanase to the determine extent of NRG1 type IV glycosylation, and analyzed by Western blotting using anti-HL5792 antibodies. A downward shift in mobility of the deglycosylated type IV ECD was observed.
Previous studies have demonstrated that shedding of the NRG1 ECD is promoted by treatment with phorbol esters (Loeb et al. 1998; Wang et al. 2001). To determine whether PKC activation promotes the release of the type IV ECD into the medium, cells expressing N-terminally tagged type IV β1a were treated with PMA (0.5 μM) at concentrations and time reported to activate PKC (Loeb et al. 1998). As shown in Fig. 3B, Western blot analysis of conditioned media collected from cells treated with either vehicle or PMA indicated that PKC activation promotes the accumulation of a ~29 kDa ECD peptide in media. Treatment of the ~29 kDa ECD with a mixture of N- and O-Glucanase enzyme caused an electrophoretic mobility shift (Fig 3C) that is consistent with the reported glycosylation of NRG1 type I, the other NRG1 isoform containing both the Ig-like and spacer domains (Peles et al. 1992; Lu et al. 1995). In conclusion, these combined results demonstrate that NRG1 type IV is a glycoprotein located on the cell surface that can be processed, in a PKC dependent fashion, to release its ECD.
NRG1 type IV ECD conditioned medium activates AKT and ERK pathways in an ErbB dependent fashion
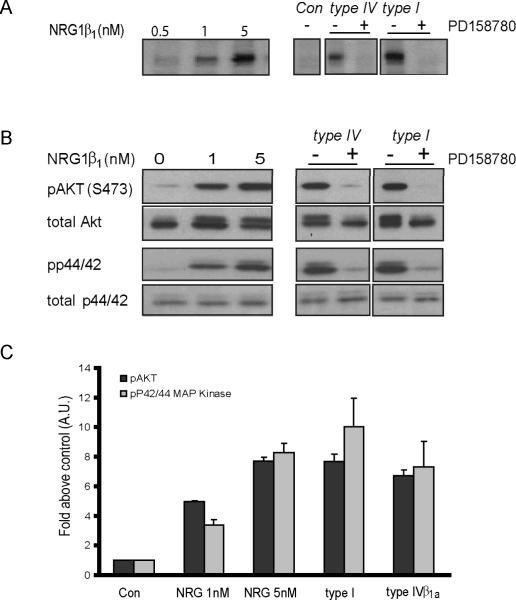
Next, we asked whether conditioned media from cells transfected with NRG1 type IV β1a stimulates ErbB receptor autophosphorylation. First, in preliminary experiments, conditioned medium from COS-7, HEK-293 and OVCAR-3 cells were tested for low ErbB receptor autophosphorylation activity in the absence of heterologously expressed NRG1. Of these three cell lines, OVCAR-3 cells exhibited the lowest levels of background phosphorylation (not shown) and were therefore chosen both to generate conditioned medium and to test its effects on ErbB receptor autophosphorylation (Fig. 4A). The dose-response curve of receptor autophosphorylation, assayed as an increase in phosphotyrosine signal levels at ~180 kDa (representing ErbB2 protein, supplementary Fig 2) upon stimulation with a purified NRG1β1 peptide encompassing the receptor-activating EGF domain of human NRG1, showed a nearlinear increase in the tested range of 0.5 – 5 nM NRG1β1 peptide (Fig 4A, left panel). Serum-starved cells were incubated with conditioned medium derived from cells expressing NRG1 type IV proteins in the absence or presence of the specific pan-ErbB receptor inhibitor PD158780. Inhibitor-sensitive increases of phosphotyrosine signals over baseline were apparent for NRG1 type IV β1a conditioned medium (Fig. 4A, right panel). As expected, cells stimulated with NRG1 type Iβ2 conditioned medium showed robust ErbB receptor phosphorylation. Taken together with our findings on glycosylation and processing of NRG1 type IV β1a protein, these results demonstrate that this NRG1 isoform gives rise to a released, biologically active ErbB ligand.
Fig 4. NRG1 type IV protein stimulates ErbB receptor and downstream signaling pathways.
A) Induction of tyrosine phosphorylation by NRG β1 peptide and NRG1 isoforms. A dose-response curve of receptor autophosphorylation, assayed as an increase in phosphotyrosine signal levels at ~180 kDa upon stimulation with a EGF-like peptide (left panel). Conditioned medium form OVCAR-3 cell expressing different NRG1 isoforms was collected used to stimulate serum starved OVCAR-3 cells. Western blots of cell lysates were analyzed using the pan-phopho-tyrosine antibody. A 180 kDa band corresponding to pErbB was absent in parallel research with or without 10 μM PD158780 was included, (right panel). (B) Activation of pAkt and p42/44, downstream molecules of the NRG1/ErbB signaling pathway, by NRG1 isoforms. Immunoblot analysis was performed on total cell lysate using antibodies against phospho-Akt or phospho-Erk p44/42. Blots were striped and re-probe for total Akt and p44/p42 protein. (C) Quantitative analysis Western blots shown in B (Data mean+SD from 3 independent experiments).
Next, we investigated whether NRG1 type IV isoforms stimulate intracellular signaling pathways downstream of activated ErbB receptors. In particular, we analyzed phosphorylation of Akt, a major target of the activated PI3 kinase pathway, at serine 473 (S473), and of P42/44 MAP kinase at threonine 202 and tyrosine 204 (Thr202/Tyr204) (Collier and Li 2003; Liu et al. 2007). OVCAR-3 cells were treated with different concentrations of purified NRG1β1 peptide (Fig. 4B), or NRG1 -conditioned medium as described above, and phosphorylated Akt and P42/44 proteins were detected by Western blotting of cell lysates using the corresponding phosphorylation state-specific antibodies. The NRG1 type IV and type I isoforms stimulated phosphorylation of both Akt and P42/44 in a PD158780-sensitive manner, indicating that both isoforms exhibit biological activity. Densitometric analysis of autoradiograms revealed that NRG1 type IV β1a stimulated Akt and p42/44 phosphorylation by 7-fold over controls (Fig 4C), and that the extent of stimulation was similar to that observed with 5nM EGF-like peptide (see Fig 4A). In conclusion, type IV signaling via ErbB receptors and activation of downstream phosphorylation cascades are similar to those observed with NRG1 type I.
Subcellular localization of NRG1 type IV in cultured hippocampal neurons
NRG1 transcripts are expressed in neurons of the hippocampus and other brain areas. Pro-NRG1 protein has been reported in motor and sensory neuron cell bodies and axons (Loeb et al. 1999), as well as presynatpic synaptophysin-positive puncta in cultured hippocampal neurons (Longart et al. 2004) However, studies of expression patterns of NRG1 isoforms propose that types I and type III have distinct functions (Sandrock et al. 1995; Yang et al. 1998; Hancock et al. 2008), suggesting that NRG1 isoforms can be distributed differently in neurons. To compare the subcellular distribution of Ig-like domain harboring NRG1 types I and IV with NRG1 type III, dissociated rat hippocampal neurons were transfected with cDNA expression constructs for each of the NRG1 isotypes. All cDNAs were cloned into the same expression vector to drive similar levels of expression and the isotypes used varied only in exons encoding the unique N-termini; splice sites in the EGF-domain (beta) and C-terminus (type “c”) were the same. Protein levels between isotypes appeared similar by immunohistochemistry. To differentiate between dendritic and axonic cell processes, we co-transfected neurons with an expression vector for GFP-tagged syntaphilin (SNPH-GFP) that labels fine axonic processes and we visualized the somoto-dendtritic compartment by labeling neurons with a monoclonal MAP2 antibody; SNPH-GFP also labels to a lower extent the somato-dendritic compartment (Kang et al. 2008).
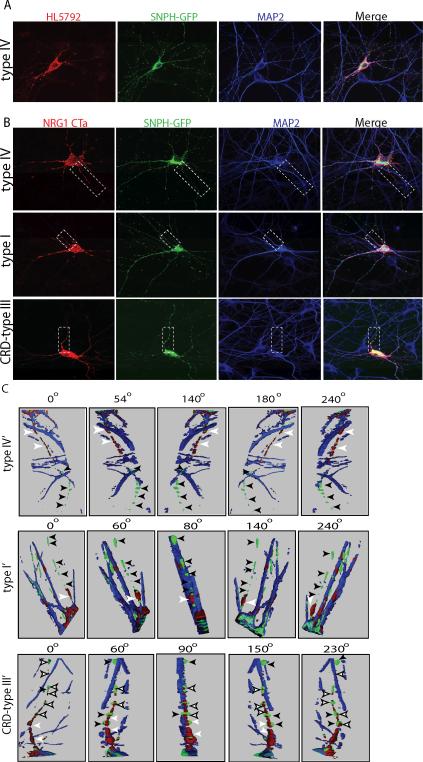
In initial experiments we analyzed the surface expression of NRG1 type IV by live-labeling transfected, unpermeabilized neurons with our HL5792 antibody. After fixation and Triton X-100 permeabilization, neurons were treated with the MAP2 antibody. As shown in Fig. 5A, NRG1 type IV accumulates at its highest levels on the somatic membrane and in proximal dendrites, and distributes in small puncta along the MAP2-positive distal dendrites. While SNPH-positive and MAP2-negative processes from the transfected and neighboring neurons can be observed (green), these are not labeled by HL5792, indicating that NRG1type IV protein does not accumulate in axons. Because commercial antibodies selective for either NRG1 type I or type III are non-existent, in a second set of experiments a NRG1 CTa tail antibody (SC-348) was used to label the C-termini of the three NRG1 isoforms in transfected neurons. Similar to the live-labeling results for NRG1 type IV, we found that immunolabeling of the three NRG1 isoforms is highest in the soma and proximal dendrites, and that these distinct NRG1 isotypes are expressed in MAP2-positive dendrites (Fig. 5B). However, in stark contrast to NRG1 type I and IV isoforms, we found that CRD-NRG1 (type III) also is found in SNPH-positive/MAP2 negative axonal processes. Given the convoluted growth of fine axonal processes, the overlay of CRD-NRG1 in SNPH-positive processes is best observed in three dimensional reconstructions of stacked confocal images rotated using the Zeiss Image Browser program (Fig. 5C); axons are marked by black arrowheads (Fig. 5C) and the reconstructed part of the cell is marked by a dashed box in Fig. 5B. It should be emphasized that immunoreactivity for the three NRG1 isotypes (red) is detected in the initial axonal segment (white arrowheads), but only in the case of CRD-NRG1 staining extends along the entire axon (white filled arrowheads). Axons of neurons transfected with either NRG1-type I, -III (CRD) or -IV that could be unambiguously identified were quantified and summarized in Table 1. These experiments showed that while 100% of axons are positive for NRG1 in neurons transfected with CRD-NRG1, only 0 and 12.5 % of neurons expressing type I or type IV showed NRG1 immunoreactivity along their axons, respectively. In summary, these results indicate that the Ig domain-containing NRG1 type I and IV fail to accumulate or be transported down distal axons, and suggest that the first N-terminal cytoplasmic domain specific to CRD-NRG1 (type III) could play an important role in trafficking this NRG1 isoform selectively into distal axons.
Fig 5. Subcellular localization of NRG1 type IV in hippocampal neurons.
A) Dissociated hippocampal neurons co-transfected with NRG1 type IV and SNPH-GFP, and live labeled with type IV antibodies. Cells were also stained for the dendritic marker MAP2 after permeabilization. Type IV immunolabeling (red) colocalizes with the dendritic marker MAP2 (blue) and is strongest in the proximal region of dendrites. (B) Hippocampal neurons were co-transfected with NRG1 type I, type IV or type III and SNPH-GFP (green), permeabilized, and incubated with antibodies against NRG1 (red) and MAP2 (blue). Type I and IV immunoreactivity is restricted to neuronal somas and dendrites, and absent from SNPH positive axons. In contrast, NRG1 CRD-type III is localized to the cell somata and dendrites but it also was targeted to the axon (green). (C) Three dimension reconstruction of the area obtained by the dash box in B. Axons (green) are indicated by black arrows. Type IV and I immunolabeling can be observed in the proximal region of the axon (white arrowheads) but not in more distal region. Unlike type IV and I, CRD-type III is expressed in the axon (white filled arrowheads).
Table 1.
Differential subcellular distribution of NRG1 isoforms in transfected neurons
| NRG1 Isoform | Transfected neuronsa | Neurons with identified axonsb | NRG1-positive distal axonsc | % of labeled distal axons |
|---|---|---|---|---|
| type IV | 22 | 8 | 1 | 12.5 |
| type I | 18 | 6 | 0 | 0 |
| type III | 26 | 10 | 10 | 100 |
Number of neurons expressing NRG1 in somato-dendritic compartments.
Number of neurons with clearly identified axons labeled by SNPH-GFP. Axonal GFP labeling had to be unambiguously traced to the cell body of origin to be counted.
Number of NRG1 and SNPH co-immunoreactive distal axons.
Discussion
In the present study we have mapped the type IV transcription initiation site and have demonstrated for the first time that type IV mRNAs are translated, and the protein processed and released in a PKC dependent fashion. The mature processed protein is active because it triggers ErbB autophosphorylation and activates downstream MAP and PI3 kinase pathways. In addition, we have shown that in hippocampal neurons NRG1 type IV protein is targeted to soma, dendrites and initial axonal segments, but unlike CRD-NRG1 (type III), it does not accumulate or is transported to distal axons. The differential localization of NRG1 types I and IV, relative to type III (CRD), has important implications for the distinct roles different NRG1 isotypes may play during development and contribute to disease.
SNP8NRG243177 [T/T] has been associated with impairment of frontal and temporal lobe activation, deficits in cognitive function and predisposition to the development of psychotic symptoms in individuals at high risk for developing schizophrenia (Hall et al. 2006; Keri et al. 2009) (However, see Corwley et al (Crowley et al. 2008)). In addition, in normal individuals the [T/T] at-risk haplotype was associated with reduced spatial working memory (Stefanis et al. 2007) and white matter in the anterior limb of the internal capsule (McIntosh et al. 2007). These studies suggest that SNP8NRG243177 is a functional polymorphism, since no other SNPs or microsatellites from this haplotype were associated with these behavioral or functional deficits. The fact that there was an open reading throughout the entire Exon 187 DNA sequence (Steinthorsdottir et al. 2004), raised the possibility that type IV transcription initiates further upstream to generate transcripts that potentially encode a larger protein or harbor sequences that regulate mRNA stability. This issue is critically important considering that SNP8NRG243177 has the properties of a functional polymorphism, and a longer sequence conceivably could extend into the SNP8NRG243177 polymorphic site. For these reasons, a rigorous Cap-trap approach was taken to map the precise type IV transcription initiation site and to identify transcripts that conceivably extend into the SNP8NRG243177 polymorphic site. Our results indicate that the type IV transcription initiation site is 23 bp upstream of the previously published 5'-end of exon E187 (Steinthorsdottir et al. 2004), and that there are no other potential translation start sites in this extended sequence. Therefore, our results confirmed that human type IV is translated from a methionine located at the 3'-end of E210 (includes the extra sequence) and that SNP8NRG243177 is not part of a NRG1 coding region. These findings are not inconsistent with the proposal that SNP8NRG243177 encompasses a serum response element that regulates transcription from the type IV promoter (Law et al. 2006; Tan et al. 2007).
In these studies we isolated a 1.8 kb cDNA that encodes a full-length NRG1 type IV protein containing an Ig-like domain, an EGF-like β1 motif, a TM domain and the CTa tail. Interestingly, we found that while the NRG1 type IVβ1a cDNA is predict to encode a 590 amino acid protein with a molecular mass of 66.8 kDa, the pro-form migrates as an apparent 90 kDa protein in Western blots of lysates from transfected HEK293 cells. The size discrepancy appears to result from the extracellular domain because the processed ICD migrates with an apparent molecular weight of 65 KDa, instead of 43 kDa. The inconsistencies between the predicted and observed apparent molecular weight has been reported previously for NRG1 type I (Longart et al. 2004) and type III (Frenzel and Falls 2001), and therefore is not unique to NRG1 type IV. The disparity between the predicted and the SDS-PAGE apparent molecular weight of pro-NRG1 was suggested to result, at least in part, from O- and N-glycosylation of the extracellular domain (Peles et al. 1992; Wen et al. 1994; Loeb et al. 1998; Wang et al. 2001). We found that glycosidases reduce the apparent mobility of NRG1 type IV on SDS-PAGE by approximately 10 kDa, thus accounting for some, but not all, of the observed discrepancy between the deduced and apparent molecular weight of processed NRG1. Therefore, other as yet unknown factors must contribute to the reduced mobility of NRG1 isoforms in SDS-PAGE.
In these studies we also investigated the processing of NRG1 type IV. Ig-containing NRG1 isoforms are expressed on the cell surface prior to cleavage and release of the soluble ectodomain (Loeb et al. 1998; Ozaki et al. 2000) by ADAM17 (TACE) (Montero et al. 2002) and ADAM19 (Shirakabe et al. 2001). We found by antibody labeling of non-permeabilized and permeabilized transfected HEK 293 cells that the majority of type IV protein is expressed at the plasma membrane. Because the cleaved extracellular domain of NRG1 type I has been shown to remain associated with the cell surface via interactions with heparin-sulfate proteoglycans (Loeb and Fischbach 1995; Loeb et al. 1999), our immunohistochemical experiments were not sufficient to determine if cell surface NRG1 type IV was unprocessed or cleaved. Using Western blotting with antibodies targeting the N- and C-terminal domains of NRG1 type IV we could determine that NRG1 type IV is partially processed, and that PKC activation by phorbol ester treatment further promoted the release of the type IV ectodomain into the medium. Taken together, these results confirm that NRG1 type IV is translated as a transmembrane glycoprotein, shuttled through the secretory machinery to the membrane surface and released to the medium through a PKC dependent mechanism.
NRG1 binds to ErbB3 and ErbB4 receptors, which upon receptor dimerization, trigger tyrosine cross-phosphorylation and the activation of intracellular downstream pathways such as Ras-MAPK and PI3K–Akt pathways (see (Linggi and Carpenter 2006) for review). Our biochemical data show that conditioned medium from OVCAR-3 expressing type IV β1a stimulated p44/42 MAP kinase and Akt phosphorylation. Activation of PI3K and Erk have been shown to be required for NRG1-dependent induction of neurite outgrowth (Gerecke et al. 2004), Schwann cell survival (Li et al. 2001) and regulation of glutamatergic neurotransmission (Gu et al. 2005). Further studies will be needed to understand the role type IV proteins play in ErbB signaling during development and normal adult remodeling.
We previously reported that NRG1 targets to axons, as well as to somatic and dendritic compartments (Longart et al. 2004). ARIA is transported down the axons of motor neurons and released at motor terminals (Goodearl et al. 1995; Sandrock et al. 1995). NDF is an axon associated survival signal for developing oligodendrocyte (Fernandez et al. 2000), and the levels of NRG1-type III in axons determine the extent of myelination in the peripheral nervous system (Taveggia et al. 2005). Here we compared in dissociated hippocampal neurons the subcellular distribution of NRG1 type I and type IV, isoforms belonging to the single-pass Ig domain-containing family, to NRG1 CRD-type III that transverses the membrane twice and whose N-terminus is cytoplasmic. The three cDNAs were cloned into the same expression vector and the same splice variants were used, as to allow direct comparison of the effects of the first unique exons. We found that type I and IV immunoreactivity is targeted to neuronal soma and dendrites but is mostly absent from axonal processes. This distribution contrasts with that of NRG1 type III (CRD) that accumulates in axons, soma and dendrites. Interestingly, the neuronal subcellular distribution of NRG1 types I and IV is similar to that of NRG2 (Longart et al. 2004), the first NRG reported to be enriched in the somato-dendritic compartment of neurons and be undetectable in axons. The similar subcellular distribution between these different NRGs is not totally unexpected. Sequence similarity between NRG1 type I and NRG2 is high, and the domain organization between NRG1 type I, type IV and NRG2 is similar - each harboring extracellular Ig-like and spacer domains and one TM domain. These results suggest that sequences in the first cytoplasmic and/or TM of type III (CRD) NRG1 may interact with intracellular proteins that selectively traffic this isoform to axons. The differentlly distribution of NRG1 Ig family members (type I and IV) and type III suggests that the non-redundant functions for these isoforms may result, at least in part, from their distinct subcellular distributions. Importantly, dendritic release of Ig-type NRG1 isoforms into the neuropil could serve as a means of regulating network activity. For example, dendritic release of brain-derived neurotrophic factor (BDNF) by dissociated hippocampal neurons affect synaptic plasticity and development (Horch and Katz 2002; Kuczewski et al. 2008). Dendritic release of oxytocin can attenuate the effect of GABA inputs (de Kock et al. 2004). We suggest that NRG1 type IV released from dendrites could modify neuronal function locally in a paracrine fashion, as we previously suggested for NRG2 (Longart et al. 2004), because ErbB4 receptors are abundantly expressed at postsynaptic glutamatergic synapses (Garcia et al. 2000; Longart et al. 2007) in selective types of GABAergic interneurons (Neddens and Buonanno 2009; Vullhorst et al. 2009) and there is no evidence for NRG1 expression in GABAergic interneurons. Another plausible target of dendritically released NRG1 type IV are glial cells. Astrocytes express ErbB-3 receptors (Gerecke et al. 2001; Gerecke et al. 2004), and are known to regulate neuronal properties and synaptic plasticity (see (Slezak et al. 2006)).
In summary, this study provides evidence for the first time that type IV is translated, proteolytically processed and released from cells to activate ErbB receptors and downstream signaling cascades. Since type IV involvement in schizophrenia was suggested based on association with SNP8NRG243177, further studies will be needed to determine if type IV may be associated with increased risk to develop schizophrenia or it may be link to the disease per se.
Supplementary Material
Acknowledgments
The authors thank Dr. Vullhorst for contributing to the characterization of the HL5792 antibody, Dr. Karavanova for preparation of neuronal cultures, and Dr Schram from the NICHD Microscopy Imaging Core for helping with the confocal microscope and assembly of 3D images. We also thank Drs. Vullhorst and Neddens for critical reading of the manuscript. We are grateful for the financial support from the Eunice Shriver Kennedy NICHD Intramural Research Program.
REFENCES
- Bjarnadottir M, Misner DL, Haverfield-Gross S, Bruun S, Helgason VG, Stefansson H, Sigmundsson A, Firth DR, Nielsen B, Stefansdottir R, Novak TJ, Stefansson K, Gurney ME, Andresson T. Neuregulin1 (NRG1) signaling through Fyn modulates NMDA receptor phosphorylation: differential synaptic function in NRG1+/− knock-outs compared with wild-type mice. J Neurosci. 2007;27:4519–4529. doi: 10.1523/JNEUROSCI.4314-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. Journal of neuroscience research. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11:287–296. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- Chen S, Velardez MO, Warot X, Yu ZX, Miller SJ, Cros D, Corfas G. Neuregulin 1-erbB signaling is necessary for normal myelination and sensory function. J Neurosci. 2006;26:3079–3086. doi: 10.1523/JNEUROSCI.3785-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DA, Li T. The genetics of schizophrenia: glutamate not dopamine? European journal of pharmacology. 2003;480:177–184. doi: 10.1016/j.ejphar.2003.08.105. [DOI] [PubMed] [Google Scholar]
- Crowley JJ, Keefe RS, Perkins DO, Stroup TS, Lieberman JA, Sullivan PF. The neuregulin 1 promoter polymorphism rs6994992 is not associated with chronic schizophrenia or neurocognition. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1298–1300. doi: 10.1002/ajmg.b.30727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kock CP, Burnashev N, Lodder JC, Mansvelder HD, Brussaard AB. NMDA receptors induce somatodendritic secretion in hypothalamic neurones of lactating female rats. The Journal of physiology. 2004;561:53–64. doi: 10.1113/jphysiol.2004.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper RM, Loeb JA. Neurotrophins induce neuregulin release through PKC delta activation. J Biol Chem. 2009 doi: 10.1074/jbc.M109.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Experimental cell research. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- Fernandez PA, Tang DG, Cheng L, Prochiantz A, Mudge AW, Raff MC. Evidence that axon-derived neuregulin promotes oligodendrocyte survival in the developing rat optic nerve. Neuron. 2000;28:81–90. doi: 10.1016/s0896-6273(00)00087-8. [DOI] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marin O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Frenzel KE, Falls DL. Neuregulin-1 proteins in rat brain and transfected cells are localized to lipid rafts. Journal of neurochemistry. 2001;77:1–12. doi: 10.1046/j.1471-4159.2001.t01-1-00132.x. [DOI] [PubMed] [Google Scholar]
- Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke KM, Wyss JM, Carroll SL. Neuregulin-1beta induces neurite extension and arborization in cultured hippocampal neurons. Molecular and cellular neurosciences. 2004;27:379–393. doi: 10.1016/j.mcn.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Gerecke KM, Wyss JM, Karavanova I, Buonanno A, Carroll SL. ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. The Journal of comparative neurology. 2001;433:86–100. doi: 10.1002/cne.1127. [DOI] [PubMed] [Google Scholar]
- Goodearl AD, Yee AG, Sandrock AW, Jr., Corfas G, Fischbach GD. ARIA is concentrated in the synaptic basal lamina of the developing chick neuromuscular junction. The Journal of cell biology. 1995;130:1423–1434. doi: 10.1083/jcb.130.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Whalley HC, Job DE, Baig BJ, McIntosh AM, Evans KL, Thomson PA, Porteous DJ, Cunningham-Owens DG, Johnstone EC, Lawrie SM. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nature neuroscience. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- Hancock ML, Canetta SE, Role LW, Talmage DA. Presynaptic type III neuregulin1-ErbB signaling targets {alpha}7 nicotinic acetylcholine receptors to axons. The Journal of cell biology. 2008;181:511–521. doi: 10.1083/jcb.200710037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horch HW, Katz LC. BDNF release from single cells elicits local dendritic growth in nearby neurons. Nature neuroscience. 2002;5:1177–1184. doi: 10.1038/nn927. [DOI] [PubMed] [Google Scholar]
- Huang YZ, Won S, Ali DW, Wang Q, Tanowitz M, Du QS, Pelkey KA, Yang DJ, Xiong WC, Salter MW, Mei L. Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron. 2000;26:443–455. doi: 10.1016/s0896-6273(00)81176-9. [DOI] [PubMed] [Google Scholar]
- Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, Sheng ZH. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keri S, Kiss I, Kelemen O. Effects of a neuregulin 1 variant on conversion to schizophrenia and schizophreniform disorder in people at high risk for psychosis. Mol Psychiatry. 2009;14:118–119. doi: 10.1038/mp.2008.1. [DOI] [PubMed] [Google Scholar]
- Kuczewski N, Porcher C, Ferrand N, Fiorentino H, Pellegrino C, Kolarow R, Lessmann V, Medina I, Gaiarsa JL. Backpropagating action potentials trigger dendritic release of BDNF during spontaneous network activity. J Neurosci. 2008;28:7013–7023. doi: 10.1523/JNEUROSCI.1673-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OB, Longart M, Vullhorst D, Hoffman DA, Buonanno A. Neuregulin-1 reverses long-term potentiation at CA1 hippocampal synapses. J Neurosci. 2005;25:9378–9383. doi: 10.1523/JNEUROSCI.2100-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OB, Paredes D, Gonzalez CM, Neddens J, Hernandez L, Vullhorst D, Buonanno A. Neuregulin-1 regulates LTP at CA1 hippocampal synapses through activation of dopamine D4 receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15587–15592. doi: 10.1073/pnas.0805722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5' SNPs associated with the disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Human molecular genetics. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- Li Y, Tennekoon GI, Birnbaum M, Marchionni MA, Rutkowski JL. Neuregulin signaling through a PI3K/Akt/Bad pathway in Schwann cell survival. Molecular and cellular neurosciences. 2001;17:761–767. doi: 10.1006/mcne.2000.0967. [DOI] [PubMed] [Google Scholar]
- Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tao YM, Woo RS, Xiong WC, Mei L. Stimulated ErbB4 internalization is necessary for neuregulin signaling in neurons. Biochem Biophys Res Commun. 2007;354:505–510. doi: 10.1016/j.bbrc.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JA, Fischbach GD. ARIA can be released from extracellular matrix through cleavage of a heparin-binding domain. The Journal of cell biology. 1995;130:127–135. doi: 10.1083/jcb.130.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JA, Susanto ET, Fischbach GD. The neuregulin precursor proARIA is processed to ARIA after expression on the cell surface by a protein kinase C-enhanced mechanism. Molecular and cellular neurosciences. 1998;11:77–91. doi: 10.1006/mcne.1998.0676. [DOI] [PubMed] [Google Scholar]
- Loeb JA, Khurana TS, Robbins JT, Yee AG, Fischbach GD. Expression patterns of transmembrane and released forms of neuregulin during spinal cord and neuromuscular synapse development. Development (Cambridge, England) 1999;126:781–791. doi: 10.1242/dev.126.4.781. [DOI] [PubMed] [Google Scholar]
- Longart M, Liu Y, Karavanova I, Buonanno A. Neuregulin-2 is developmentally regulated and targeted to dendrites of central neurons. The Journal of comparative neurology. 2004;472:156–172. doi: 10.1002/cne.20016. [DOI] [PubMed] [Google Scholar]
- Longart M, Chatani-Hinze M, Gonzalez CM, Vullhorst D, Buonanno A. Regulation of ErbB-4 endocytosis by neuregulin in GABAergic hippocampal interneurons. Brain research bulletin. 2007;73:210–219. doi: 10.1016/j.brainresbull.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HS, Hara S, Wong LW, Jones MD, Katta V, Trail G, Zou A, Brankow D, Cole S, Hu S, et al. Post-translational processing of membrane-associated neu differentiation factor proisoforms expressed in mammalian cells. J Biol Chem. 1995;270:4775–4783. doi: 10.1074/jbc.270.9.4775. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Moorhead TW, Job D, Lymer GK, Munoz Maniega S, McKirdy J, Sussmann JE, Baig BJ, Bastin ME, Porteous D, Evans KL, Johnstone EC, Lawrie SM, Hall J. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nature reviews. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero JC, Yuste L, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A. Mitogen-activated protein kinase-dependent and -independent routes control shedding of transmembrane growth factors through multiple secretases. The Biochemical journal. 2002;363:211–221. doi: 10.1042/0264-6021:3630211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- Neddens J, Buonanno A. Selective populations of hippocampal interneurons express ErbB4 and their number and distribution is altered in ErbB4 knockout mice. Hippocampus. 2009 doi: 10.1002/hipo.20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki M, Tohyama K, Kishida H, Buonanno A, Yano R, Hashikawa T. Roles of neuregulin in synaptogenesis between mossy fibers and cerebellar granule cells. Journal of neuroscience research. 2000;59:612–623. doi: 10.1002/(SICI)1097-4547(20000301)59:5<612::AID-JNR4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Pankonin MS, Sohi J, Kamholz J, Loeb JA. Differential distribution of neuregulin in human brain and spinal fluid. Brain research. 2009;1258:1–11. doi: 10.1016/j.brainres.2008.12.047. [DOI] [PubMed] [Google Scholar]
- Peles E, Bacus SS, Koski RA, Lu HS, Wen D, Ogden SG, Levy RB, Yarden Y. Isolation of the neu/HER-2 stimulatory ligand: a 44 kd glycoprotein that induces differentiation of mammary tumor cells. Cell. 1992;69:205–216. doi: 10.1016/0092-8674(92)90131-u. [DOI] [PubMed] [Google Scholar]
- Pitcher GM, Beggs S, Woo RS, Mei L, Salter MW. ErbB4 is a suppressor of long-term potentiation in the adult hippocampus. Neuroreport. 2008;19:139–143. doi: 10.1097/WNR.0b013e3282f3da10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrock AW, Jr., Goodearl AD, Yin QW, Chang D, Fischbach GD. ARIA is concentrated in nerve terminals at neuromuscular junctions and at other synapses. J Neurosci. 1995;15:6124–6136. doi: 10.1523/JNEUROSCI.15-09-06124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K, Wakatsuki S, Kurisaki T, Fujisawa-Sehara A. Roles of Meltrin beta /ADAM19 in the processing of neuregulin. J Biol Chem. 2001;276:9352–9358. doi: 10.1074/jbc.M007913200. [DOI] [PubMed] [Google Scholar]
- Slezak M, Pfrieger FW, Soltys Z. Synaptic plasticity, astrocytes and morphological homeostasis. Journal of physiology, Paris. 2006;99:84–91. doi: 10.1016/j.jphysparis.2005.12.082. [DOI] [PubMed] [Google Scholar]
- Sprooten E, Lymer GK, Maniega SM, McKirdy J, Clayden JD, Bastin ME, Porteous D, Johnstone EC, Lawrie SM, Hall J, McIntosh AM. The relationship of anterior thalamic radiation integrity to psychosis risk associated neuregulin-1 variants. Mol Psychiatry. 2009;14:237–238. 233. doi: 10.1038/mp.2008.136. [DOI] [PubMed] [Google Scholar]
- Stefanis NC, Trikalinos TA, Avramopoulos D, Smyrnis N, Evdokimidis I, Ntzani EE, Ioannidis JP, Stefanis CN. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62:784–792. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, Ingason A, Gulcher JR, Stefansson K, St Clair D. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. American journal of human genetics. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. American journal of human genetics. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, Olafsson O, Stefansson K, Gulcher JR. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Tan W, Wang Y, Gold B, Chen J, Dean M, Harrison PJ, Weinberger DR, Law AJ. Molecular cloning of a brain-specific, developmentally regulated Neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007 doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, Yano H, Rosenbluth J, Einheber S, Xu X, Esper RM, Loeb JA, Shrager P, Chao MV, Falls DL, Role L, Salzer JL. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullhorst D, Neddens J, Karavanova I, Tricoire L, Petralia RS, McBain CJ, Buonanno A. Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J Neurosci. 2009;29:12255–12264. doi: 10.1523/JNEUROSCI.2454-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Miller SJ, Falls DL. The N-terminal region of neuregulin isoforms determines the accumulation of cell surface and released neuregulin ectodomain. J Biol Chem. 2001;276:2841–2851. doi: 10.1074/jbc.M005700200. [DOI] [PubMed] [Google Scholar]
- Wen D, Suggs SV, Karunagaran D, Liu N, Cupples RL, Luo Y, Janssen AM, Ben-Baruch N, Trollinger DB, Jacobsen VL, et al. Structural and functional aspects of the multiplicity of Neu differentiation factors. Molecular and cellular biology. 1994;14:1909–1919. doi: 10.1128/mcb.14.3.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Preece A, Spurlock G, Norton N, Williams HJ, Zammit S, O'Donovan MC, Owen MJ. Support for genetic variation in neuregulin 1 and susceptibility to schizophrenia. Mol Psychiatry. 2003;8:485–487. doi: 10.1038/sj.mp.4001348. [DOI] [PubMed] [Google Scholar]
- Yang X, Kuo Y, Devay P, Yu C, Role L. A cysteine-rich isoform of neuregulin controls the level of expression of neuronal nicotinic receptor channels during synaptogenesis. Neuron. 1998;20:255–270. doi: 10.1016/s0896-6273(00)80454-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.