Abstract
Natural RNA sensors of small molecules (a.k.a. riboswitches) regulate numerous metabolic genes. In bacteria, these RNA elements control transcription termination and translation initiation by changing the folding pathway of nascent RNA upon direct binding of a metabolite. To identify and study riboswitches we used in vitro reconstituted solid-phase transcription elongation/termination system. This approach allows for direct monitoring ligand binding and riboswitch functioning, establishing the working concentration of a ligand as a function of RNA polymerase speed, and also probing RNA structure of the riboswitch. Using this system we have been able to identify and characterize first several riboswitches including those involved in vitamin biosynthesis and sulfur metabolism. The system can be utilized to facilitate biochemical studies of riboswitches in general, i.e. to simplify analysis of riboswitches that are not necessarily involved in transcriptional control.
Keywords: riboswitch, RNA polymerase, transcription elongation, termination, attenuation, metabolites
1. Introduction
In the last several years, riboswitches have become a paradigm of the RNA-driven regulation of gene expression (1–5). These non-coding stretches of RNA adopt an alternative conformation upon direct binding of a small molecular ligand to activate or repress cognate genes. In bacteria, riboswitches are ubiquitous and control over 3% of all metabolic genes. They usually reside in the 5′ untranslated regions (UTR) and consist of two coupled elements: evolutionary conserved sensor domain, which directly binds the ligand, and a variable expression platform that transmits the signal to the gene expression machinery such as RNA polymerase or a ribosome (1, 2). Depending on its design, a riboswitch either prevents or permits the formation of an RNA attenuator that in turn dictates whether a gene is expressed or not. The attenuator functions as sequester of the ribosome-binding site or intrinsic terminator positioned in front of an operon or both. A few riboswitches that have been found in fungi and plants work at the level of alternative RNA splicing (6–8). Since the sensor domain is highly conserved within each family of riboswitches, it can be placed within a context of a standard transcription termination expression platform to benefit from a simple and quantitative in vitro readout system. Such a strategy also takes full advantage of the solid-phase reconstituted transcription system for thorough biochemical analysis of riboswitch functioning. The system utilizes “walking” technology to obtain individual homogenous elongation complexes (EC) stalled at any desired position within the transcription unit (9). This allows for various types of RNA structural analysis (e.g. chemical and enzymatic probing, crosslinking, and real-time spectrometry) to be performed during transcription thus reflecting a natural situation of co-transcriptional RNA folding. The system also takes into account the kinetics of transcription elongation.
2. Materials
2.1. Protein Purification
Escherichia coli and Bacillus subtilis strains recommended for this procedure should express a chromosome-encoded β′ subunit of RNA polymerase with a His6-tag at the C-terminus (10). We used the E. coli strain RL 721 (F-l-thr-1 leu-6 pro-A2 his-4 thi-1 argE3 lacY1 galK2 ara-14 syl-5 mtl-1 tsx-33 rpsL31 supE37 recB21 recC22 sbcB15 sbcC201 rpoC3531(His6) zja::kan).
The E. coli strains M5248 (λ bio275 c1857 ΔH1) and JC4588 (F− endoI− recA56 gal his322 thi) and the plasmid pEcoRQ111 (11), which is a derivative of the pSCC2 (ApR), are kindly provided by Dr. Paul Modrich (Duke University). The pEcoRQ111 plasmid was purified from the JC4588 strain and was introduced into the M5248 expression strain just prior to preparation of the enzyme.
E. coli or Bacillus culture medium LBD (Gibco/BRL, Bethesda, MD): 0.5% (w/v) Bactotryptone, 1.0% (w/v) Bacto-yeast extract, 0.5% (w/v) NaCl, and 1% (w/v) dextrose.
Lysis buffer: 300 mM NaCl, 50 mM Tris-HCl, pH 7.9, 5% (v/v) glycerol, 10 mM ethylenediaminetetraacetic acid (EDTA), 0.1% (w/v) phenylmethanesulphonylfluoride (PMSF), 10 mM mercaptoethanol, 133 μg/mL lysozyme (Sigma).
10% (w/v) sodium deoxycholate (Sigma).
10% (v/v) polymine P solution (Sigma).
Tris-glycerol-EDTA-DTT (TGED) buffer: 50 mM Tris–HCl, pH 7.9, 5 % (v/v) glycerol, 0.1 mM EDTA, and 0.1 mM DTT.
TGED50, TGED200, TGED300, TGED500, TGED600, TGED1000 buffer: TGED buffer supplemented with 50, 200, 300, 500, 600, and 1,000 mM NaCl, respectively.
Sonic Dismembrator 60 (Fisher Scientific).
Solid NH2SO4.
Centricon-50 filter cartridges (Amicon/Millipor, Billerica, MA).
Chromatography columns: Superose 6 HR 10/30; Mono Q HR 5/5; 10 mL Heparin-Sepharose (GE Healthcare, Piscataway, NJ).
Template-dependent forward and reverse oligonucleotides: 50 pmol/μL stock. As an example: 5′-CCAGATCCCGAAAATTTATC (forward) and 5′-CACTGACCCTTTTGGGACCGC (reverse).
2.2. DNA templates and NTP substrates preparation
Peristaltic pump, gradient mixer, and UV detector (GE Healthcare).
dNTPs mixture (1 mM each nucleotide) (Promega, Madison, WI). Store at −20°C.
Tris-EDTA (TE) buffer: 10 mM Tris-HCl, pH 7.9, and 1 mM EDTA.
SeaKem low-melting agarose (Cambrex, East Rutherford, NJ).
Phusion DNA polymerase and 10x Phusion reaction buffer (Fermentas, Glen Burnie, MD) as provided by a manufacturer.
2.3. Solid-Phase in vitro Transcription
Ultra pure ribonucleotide triphosphates (GE Healthcare), prepared as 10 mM individual solutions and 1 mM (each) mixture (10X NTPs solution). Store at −20°C.
Transcription buffer (TB): 10 mM Tris-HCl, pH 7.9, 10 mM MgCl2, and 100 mM KCl.
“High salt” TB buffer: TB supplemented with 1 M KCl.
ApUpC RNA primer (Oligos Etc., Wilsonville, OR).
10X starting mix: 100 μM ApUpC RNA primer, 250 μM ATP and GTP.
[α-32P] CTP, 3,000 Ci/mmol, 0.5 μM (NEN/PerkinElmer, Waltham, MA)
Loading mix: 12 M urea, 20 mM EDTA, 10 mM Tris-HCl, pH 7.5, 0.0125 % (w/v) each of bromophenol blue (BPB) and xylene cyanol (XC).
Resins: Ni++-NTA (Qiagen, Valencia, CA); TALON (Clontech, Mountain View, CA); NutrAvidin (Pierce, Biotechnology, Rockford, IL).
1 mg/mL rifampicin (Sigma) dissolved in water (10X solution).
Restriction enzyme EcoRQ111, isolated as described in (15) and stored at ~1 mg/mL at −20°C in a buffer containing 20 mM KPO4, pH 7.4, 0.4 M KCl, 1 mM EDTA, 0.2 mM DTT, 50% (v/v) glycerol.
Sequencing gel: 8 M urea, 10% acrylamide:bisacrylamide solution (29:1).
RNAse H (Roche, Basel, Switzerland).
Microcuvettes for fluorescent studies (StarnaCells, Atascadero, CA).
Template-dependent antisense oligonucleotides.
3. Methods
3.1. Preparation of the DNA Template
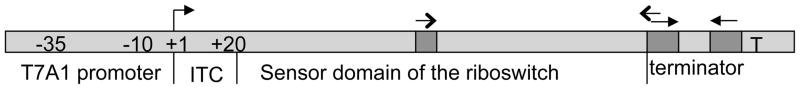
The common structure of the template for riboswitch studies is shown in Fig. 1. It consists of the A1 promoter of bacteriophage T7, adapter sequence, the sensor domain of a riboswitch, and an intrinsic transcription terminator to be coupled with a riboswitch of choice. T7A1 is one of the strongest known promoters to support in vitro transcription by RNA polymerase (RNAP) from Gram-negative or Gram-positive bacteria, including E.coli and B.subtilis. It works within a broad range of temperatures and salt concentrations and supports multi-round transcription in vitro without additional factors. The adapter sequence is a common initial transcribed sequence (ITS), which allows for one step preparation of the homogenous highly radiolabeled elongation complex stalled at position +20°C (EC20). The terminator hairpin sequence should be designed to match the structure of the sensor domain of a riboswitch. The 3′ proximal part of the terminator hairpin stem is variable and should be complementary to the antiterminator part of the expression platform. It is followed by a stretch of 8–9 thymidines (T-stretch) and GC-rich “front element” of the terminator. The latter part improves the termination efficiency (12). The following procedure exemplifies the walking technique for the T7 A1 promoter template with the initial transcribed sequence: ATCGAGAGGG10 ACACGGCGAA20 TAGCCATCCC30 AAT33.
Fig. 1.
Schematics of the unified riboswitch template. The bar represents a PCR fragment containing T7A1 promoter (~40 bp), the initial transcribed sequence (ITC; 20 bp), the sensor domain of the riboswitch under study; and the intrinsic terminator (solid arrows indicate inverted repeat of the termination hairpin followed by the U-stretch). The terminator is designed so that the left shoulder of the hairpin stem is complementary to potential antiterminator sequence of the sensor domain (open arrows).
T7A1 promoter containing linear DNA templates can be easily produced by PCR and purified in a relatively large concentration (e.g. 1 pmol/μL). At least two fold molar excess of DNA over RNAP is recommended to achieve the maximum yield of the initial EC.
Prepare an amplification reaction (50 μL) that includes 41.5 μL water, 5 μL 10x Phusion reaction buffer, 1 μL dNTPs mixture, 1 μL of each forward and reverse oligonucleotides and 0.5 μL Phusion DNA polymerase.
Run the PCR reaction as follows: 95°C 45 sec, 55°C 1 min, 72°C 45 sec (29 cycles). Master mixture is usually prepared for twenty PCR reactions and then distributed in 50-μL aliquots into 250 μL thin-wall plastic tubes.
Resolve the amplification products in a 1.2% (w/v) agarose gel, cut the desired fragment, extract it by electroelution, followed by phenol-chloroform treatment and precipitation with ethanol (see Note 1).
Dilute the DNA pellet in 50 μL TE. The final concentration of DNA is expected to be ~2 pmol/μL as determined by UV spectrometry.
3.2. Preparation of the RNA Polymerase
The procedure described below is optimized for E. coli RL 721 strain (10), a KmR derivative of JC7623 strain (13). This bacterial strain carries a chromosomal copy of the rpoC gene encoding a β′ subunit of RNA polymerase with the His-tag at its COOH-terminus. The use of this strain guarantees that all purified RNAP would be His-tagged, although essentially the same procedure could be applied to conventional E.coli and B.subtilis strains. The purification method described here yields ~1 mg of RNA polymerase σ70 (RNAPσ70) holoenzyme per 10 g of cells. The enzymatic activity of the holoenzyme is ~2–3 fold higher than that of a commercially available preparations of E.coli RNAP.
Dilute an overnight culture (50 mL) of RL 721 strain in 4 L of LBD broth containing 50 μg/mL kanamycine.
Grow cells with aeration to OD600~1.0 at 37°C (usually 3–4 h).
Harvest cells by centrifugation at 3,000× g for 7 min. The cell pellet can be stored at −70°C.
Resuspend the cell pellet in 100 mL of lysis buffer and incubate on ice for 20 min. Add sodium deoxycholate to the final concentration of ~0.2 % (v/v) constantly mixing the cell suspension.
Sonicate 20 mL aliquots of cells at the maximum output power by 30 sec pulses interrupted by 30 sec pauses (5–7 rounds). Completion of cell lysis can be monitored by reduction of viscosity in the mixture.
Clarify the lysate by centrifugation at 10,000× g for 30 min.
Slowly add polymine P to the supernatant to the final concentration of ~0.35 % (v/v) constantly stirring the mixture. Continue stirring for another 10 min. Centrifuge the suspension at 8,000× g for 5 min.
Resuspend the pellet thoroughly by a glass homogenizer in 100 mL of TGED300 buffer. Centrifuge the suspension at 8,000× g for 5 min. Discard the supernatant.
Resuspend the pellet in 100 mL of TGED500 buffer, homogenize, and centrifuge as above. Discard the supernatant.
To extract RNAP from polymine P, resuspend the pellet in 100 mL of TGED1000 buffer for one hour. Clarify the suspension at 8,000× g for 5 min.
Slowly add solid NH2SO4 to the supernatant to 65 % (39.8 g/100 mL) and stir for 30 min. The precipitate can be left overnight without loss of RNAP activity. Centrifuge the suspension at 10,000× g for 45 min. Dissolve the pellet in 50 mL of TGED buffer and centrifuge again to remove the insoluble material. The final protein concentration should not exceed 5 mg/mL as determined by UV spectrometry using an extinction coefficient of 6.2 mL/cm/mg at 280 nm. If protein concentration is too high, sedimentation may be compromised. In this case, dilute the solution with water until it becomes colorless.
Load the protein solution onto 10 mL Heparin-Sepharose column pre-equilibrated with TGED50 buffer. Loading proceeds in 15–20 mL portions using 50 mL superloop at 0.5 mL/min.
Wash the column with 20 mL of the same buffer. Change the flow rate to 1 mL/min and wash the column by 20 mL TGED300 buffer. Elute RNAP from the column with 20 mL TGED600 buffer. Fractionate the protein into 1-mL aliquots.
Concentrate the eluate from several protein-containing fractions using Centricon-50 filter cartridge at 1,000× g up to 0.5–1 mL so that the protein concentration would not exceed 10 mg/mL.
Load concentrated RNAP sample onto a Superose-6 10/30 HR column in 0.5 mL portions (usually twice) at 0.3 mL/min in TGED200 buffer.
Carry out elution at the same rate in 1 mL fractions for 90 min. RNAP is eluted as a separate peak after ~35 min. Combine protein-containing fractions, concentrate them up to 2 mL by Centricon-50, and dilute to 10 mL by TGED50.
Load RNAP sample onto Mono Q 5/5 HR column in TGED50 buffer using the 50 mL superloop at 0.2 mL/min flow rate. After washing the column with 5 mL of the same buffer, apply the 0.05–1 M NaCl gradient as follows: 0–20% - 2 mL, 21–80% - 60 mL, 81–100% - 2 mL, and collect 1 mL fractions. The RNAP core enzyme and holoenzyme are eluted as separate peaks between 30% and 45% of salt.
Concentrate the fractions containing RNAP holoenzyme with Centricon-50, mix with equal volume of cold glycerol, and store the protein at −20°C.
3.4. Transcription in Solid-Phase (Walking)
The principle of the walking reaction is that the initial EC immobilized onto a solid support undergoes rounds of washing to remove the unincorporated NTP substrates, followed by addition of the incomplete set of NTPs that allow transcription to proceed to the next DNA position corresponding to the first missing NTP.
Each component of the EC (RNAP, DNA, or RNA) can be tagged for walking. Biotin tag can be put at the 5′ terminus of the DNA strand (see below). For the His-tag, two types of metal-chelating beads are available: Ni++-NTA-agarose and TALON Co2+-Sepharose beads. The latter resin has less non-specific binding and requires less time and concentration of imidazole to elute the EC off the beads, if necessary. For the biotin tag, the best resin for walking is NeutrAvidine UltraLink from Pierce. A representative walking protocol, which is described here for the His-tagged β′ subunit of RNA polymerase, can be utilized with minimal changes for other tags (see Note 2).
3.4.1. Preparation of the Initial EC and Walking
To prepare a standard reaction (for 10–15 samples) incubate 1 pmol (~0.5 μL) of His-tagged RNAP and 2 pmol (~1 μL) of T7A1 promoter DNA fragment (~200 bp) in 8 μL of TB at 37°C for 5 min. Add 1.5 μL of the starting mix and 5 μL of TB-pre-equilibrated Ni++-NTA or TALON resin. Incubate the mixture at 37°C with slight agitation for 8 min.
Wash the beads four times with 1.5 mL TB and resuspend in 10 μL TB containing 0.3 μM [α-32P] CTP with or without ATP and GTP (25 μM). Incubate the reaction at room temperature for 8 min and perform two rounds of washing with “high salt” TB and two rounds of standard TB washing. Without ATP and GTP, the initial [α-32P] EC is stalled at position +11 (EC11); with ATP and GTP, the initial EC is stalled at position +20 (EC20). It is expected that the washed EC11 and EC20 count 1,000–2,000 and 3,000–5,000 cpm, respectively, if the bottom of the tube is placed next to the membrane of the Geiger counter.
Add the appropriate composition of NTP (1 μL) to the washed initial EC (~20 μL) and incubate for 3–5 min. Wash with TB as described above. Repeat this procedure as many times as needed to move the EC to a desired position (Fig. 2). All walking steps are usually done at room temperature. The concentration of NTPs for each step has to be selected empirically to minimize the read-through as well as incomplete chase (see Note 3). If purified NTPs are used, 20 μM of each NTP is generally recommended. If non-purified NTPs are used, 3–5 μM NTP and longer elongation time (~5–8 min) are more appropriate. At the last walking step, TB can be changed for any other buffer that suits the purpose of the experiment.
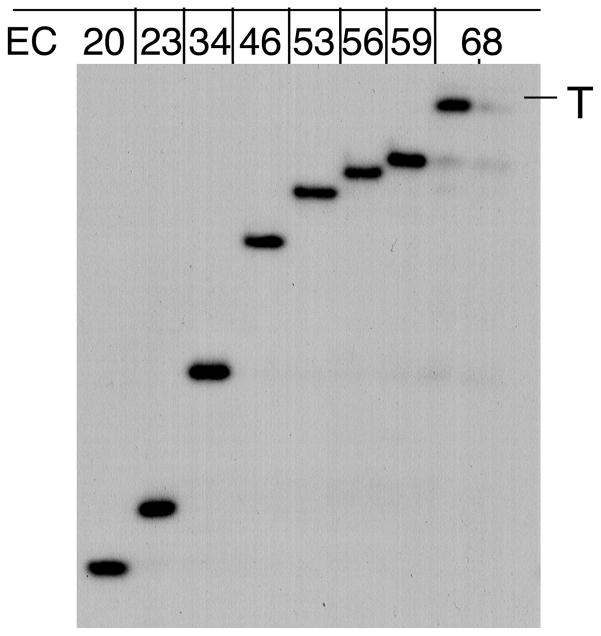
Fig. 2.
RNAP walking. The autoradiogram shows [32P]-labeled RNA transcripts isolated from the indicated ECs. The panel shows walking using His6-tagged RNAP. “T” corresponds to the termination point (position +68) of the λ tR2 terminator. The two lanes of EC68 show RNA recovery before and after washing the beads with TB.
3.4.2. Roadblocking
Walking the EC far from the promoter (e.g. over 200 bp) may be laborious and time consuming. The yield of the final EC in this case can be decreased substantially due to partial loss of material (beads) during multiple washing steps and also due to partial loss of activity at certain (arrest) positions. To avoid these complications, a transcriptional roadblock can be used. The roadblock is a site-specific DNA binding protein that is able to stop the EC completely without termination at any distance from the promoter. The roadblock, which worked well in our hands is the mutant form of EcoRI restriction endonuclease, EcoRQ111 (14). EcoRQ111 can be removed from DNA without interfering with the EC by the high salt buffers (e.g. 500 mM KCl). The use of EcoRQ111 has an advantage over other site-specific DNA binding proteins in that it requires small changes in DNA for generating its binding site.
Engineer EcoRI site by PCR a few base pairs upstream of the site of interest.
Dilute an aliquot of EcoRQ111 in TB to prepare 10X stock (~0.02 mg/mL), which can be stored at +4°C for a few days. Add 1 μL of EcoRQ111 from 10X stock to the initial EC for 2 min, followed by addition of all four NTPs (0.5 mM). The working concentration of EcoRQ111 is about 8 pmol per 1 pmol of the EcoRI site. The chase reaction proceeds for 1–3 min. More than 90% of the EC is expected to be blocked by EcoRQ111 under these conditions.
3.5. Applications for Riboswitch Studies
3.5.1. Single-Round (anti) Termination Assay
The following assay is designed for a quick assessment of the intrinsic termination efficiency as a function of riboswitch activity (Fig. 3, last two lanes).
Fig. 3.

A solid-phase transcription assay to probe riboswitch functioning. A representative radiogram shows RNA products after a single-round transcription reaction performed in solid phase using T7A1-thi-box riboswitch template (last two lanes). The initial EC20 carrying [32P]– labeled transcript was chased through the terminator in the presence or absence of 10 μM TPP. The first four lanes demonstrate walking reaction using the same template. EC20 was walked four steps to reach the thi-box domain.
Prepare the initial [α-32P]-labeled EC20 as described in Subheading 3.4.1. steps 1–2. All following reactions are performed at room temperature.
Add rifampicin to the final concentration 10 μM to prevent reinitiation by RNAP.
Distribute EC20 in 10-μl aliquots in 10 Eppendorf tubes. Aliquots can be stored at +4°C for 12 h.
Add small molecular ligands (usually 1 μL) followed by the chase reaction induced by adding 1 μL of 10X NTP mix for 5 min.
Stop the reaction by two volumes of the hot loading mix. Heat the samples at 100°C for 30 sec and immediately load on hot 10% denaturing sequencing gel that had been pre-run for at least 10 min. Freeze the rest of the samples and store at −70°C.
To calculate the efficiency of termination (%T), divide the amount of radioactivity in a termination band by the total radioactivity present in the termination and all read-through bands. %T depends on monovalent and divalent salt concentrations, NTP concentration, and temperature. Variation of the ligand concentration and amount of NTPs added to the washed EC20 allows estimation of the riboswitch termination efficiency as a function of the elongation rate.
To study the process in more detail, the assay can be modified to include walking and roadblocking steps. Once the initial immobilized EC is prepared as described above, it can be walked to any desired position along the riboswitch sequence. For example, to reach the sensor domain of the TPP-sensing riboswitch (thi-box) (16) at least four walking steps are needed (Fig. 3). Alternatively, the EcoRI site can be engineered to halt the EC at any distance within the riboswitch sequence. Using such walking techniques one can study the effect of co-transcriptional RNA folding on riboswitch functioning and also probe the RNA structure directly during elongation.
3.5.2. Riboswitch Structure Probing
Many types of chemical and enzymatic RNA probing can be performed without releasing the EC from the solid support. The following methods of RNA probing have been used to monitor riboswitch conformational changes during transcription in response to ligand binding.
Major conformational changes in RNA can be readily detected by ribonuclease H (RNAse H) following the annealing of short (8–10 nt) antisense DNA oligos to the different part of the riboswitch nascent transcript (16, 17). RNAse H specifically recognizes RNA:DNA heteroduplexes and works in a standard TB.
Add antisense oligonucleotide at 50 fold molar excess over the DNA template.
Add 0.5 U of RNAse H per 10 μL reaction. Incubate for 5 min at room temperature. Stop the reaction by the hot loading mix and analyze the products on the sequencing gel.
Walking can be used for generating specifically modified RNA transcripts for structural analysis (12). Many different NTP analogs can be utilized by E. coli RNAP and incorporated into the nascent RNA at specific positions during walking, for example, within the sensor domain of the riboswitch. The cross-linkable NTP analogs (such as 4-thio-UTP and 6-thio-GTP) and fluorescent analogs (e.g. 2-aminopurinetriphosphate (2AP); 5-(fur-2-yl)UTP; or pyrollo-C) are particular useful to probe the local RNA conformational changes and interactions directly in the elongation complexes. For example, stacking interactions with the neighboring bases quench the 2AP fluorescence. Many NTP analogs and conditions for their incorporation are available from our laboratory upon request.
3.5.3. Spectroscopic Detection of the RNA-Ligand Interaction
Direct RNA binding of a small molecule, which is intrinsically fluorescent (e.g. FMN) or carries a fluorescent tag that can be detected in real time during transcription by scanning spectrofluoremetry (16). The change of fluorescence during steady-state transcription is monitored on a Perkin-Elmer LS50B scanning fluorometer equipped with the quartz “submicrocuvette” with the 10 mm pathlength (see Note 4). For example, the formation of the riboswitch-FMN complex quenches FMN fluorescence due to photoinduced electron transfer from FMN to the aromatic rings of RNA bases.
To measure FMN binding, prepare the start-up transcription complex in 25 μL as described above in Subheading 3.4.1. steps 1–2, except that 20 μM CTP is used instead of [32P] CTP. Add FMN to 4 μM.
Transfer the mixture to the submicrocuvette and take the initial control spectrum (λ = 519em/450ex nm, 5-nm slit widths). Record spectra at various time intervals after addition of NTPs to 100 μM.
Acknowledgments
This work was supported by the NIH grants R01 GM58750 and GM72814 (E.N.)
Footnotes
Phenol and high temperature (>70°C) treatments affect the quality of DNA templates. Avoid phenol extraction or keep phenol treatment as brief as possible. To obtain DNA for biotin-tag-based walking, use for PCR the PAGE-purified forward DNA oligonucleotide carrying the 5′-biotin.
Perform all reactions in siliconized Eppendorf plastic tubes. The standard round of washing, i.e. pelleting and resuspending the beads, includes 3–5 sec centrifugation in a table-top microcentrifuge, removal of supernatant leaving ~50 μL above the pellet, and resuspension in 1.5 mL of the appropriate transcription buffer.
Commercially available NTPs are not pure enough to support walking on every DNA sequences. A read-through of certain positions due to small contamination in the NTP stocks is common. To avoid this problem, it is strongly recommended to purify original NTP stocks. The purification procedure has been described in detail (9).
The method detects the effect of various conditions (e.g. the rate of elongation, salt concentration, etc.) on ligand binding during transcription. Spectroscopic detection can be coupled with the fast kinetic and walking techniques.
References
- 1.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 4.Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 7.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 8.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nudler E, Gusarov I, Bar-Nahum G. Methods of walking with the RNA polymerase. Methods Enzymol. 2003;371:160–169. doi: 10.1016/S0076-6879(03)71011-8. [DOI] [PubMed] [Google Scholar]
- 10.Anthony LC, Artsimovitch I, Svetlov V, Landick R, Burgess RR. Rapid purification of His(6)-tagged Bacillus subtilis core RNA polymerase. Protein Expr Purif. 2000;19:350–354. doi: 10.1006/prep.2000.1272. [DOI] [PubMed] [Google Scholar]
- 11.Wright DJ, King K, Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989;264:11816–11821. [PubMed] [Google Scholar]
- 12.Epshtein V, Cardinale C, Ruckenstein AE, Borukhov S, Nudler E. Allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Kushner SR, Nagaishi H, Templin A, Clark AJ. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng SC, Kim R, King K, Kim SH, Modrich P. Isolation of gram quantities of EcoRI restriction and modification enzymes from an overproducing strain. J Biol Chem. 1984;259:11571–11575. [PubMed] [Google Scholar]
- 16.Mironov AS, Gusarov I, Rafikov R, Errais-Lopez L, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 17.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci U S A. 2003;100:5052–5026. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]