Abstract
Elucidating dynamic morphological changes of gene-carrying vectors and their nucleic acid release under varying intracellular conditions has been a technical challenge. Atomic force microscopy (AFM) was employed to observe nucleic acid/polymer polyplexes under endosomal and reducible cytosolic conditions. Both ketalized (acid-degradable) and unmodified (non-degradable) polyethylenimine (PEI) in linear and branched forms were used to prepare plasmid DNA- or siRNA-complexing polyplexes. Then, the polyplexes’ complexation and disassembly were observed by in situ AFM in various differentially changing buffers that represent intracellular conditions. Results demonstrated obvious morphological destruction of DNA/ketalized linear PEI (KL-PEI) polyplexes under mildly acidic endosomal conditions, while no morphological changes were observed by DNA/ketalized branched PEI (KB-PEI) under the same conditions. In addition, siRNA was more efficiently dissociated from KL-PEI than KB-PEI under the same conditions. Non-degradable PEI did not show any evidence that DNA or siRNA was released. Anionic biomacromolecules (e.g., heparan sulfate), which was hypothesized to dissociate nucleic acids from cationic polymers, did not successfully disassemble polyplexes but appeared to be adsorbed on cationic polymers. The in situ AFM results combined with in vitro cellular transfection and gene silencing indicated that efficient endosomal escape of plasmid DNA in a compact polyplex form is required for efficient gene expression. On the contrary, rapid dissociation of siRNA from its cationic carrier is crucial for efficient gene silencing. The findings of this study may provide new insightful information for designing stimuli-responsive nonviral gene vectors as well as expanding tools for investigating nonviral vectors in nano scales under biologically inspired conditions.
Keywords: In situ AFM, Mimicked intracellular trafficking, Nonviral gene delivery, Plasmid DNA and siRNA, Polyethylenimine
INTRODUCTION
Use of nonviral vectors, complexed nucleic acids mostly with cationic polymers, has been remarkably increased recently due to their advantages over viral vectors such as unlimited genome size to be delivered, low immunogenicity, and facile preparation at large scales and desired quality (Han et al., 2000; Read et al., 2005). Although most nonviral vectors mimic extracellular and intracellular gene delivery pathways of viral vectors, which offer substantially higher gene delivery efficiency than nonviral vectors, the mechanisms of complexation and disassembly dynamics of nonviral vectors in a cell are not fully elucidated (Schaffer et al., 2000). In order to improve the design of nonviral vectors, an increased understanding of intracellular nonviral gene delivery is needed. One of the challenges associated with understanding complexation and disassembly dynamics is technical difficulty to observe interactions of nucleic acids with cationic polymers at a molecular level under differentially varying conditions in a cell.
Over the last few decades, atomic force microscopy (AFM) has become pivotal in imaging morphologies of nanomaterials as a non-destructive technique that provides nanometer scale resolution. When it is employed under aqueous environments and physiological conditions, AFM allows for visualizing biological materials such as DNA and living cells (Fotiadis et al., 2002; Gadegaard, 2009; Hörber et al., 2003; Kada et al., 2008). Imaging native biomolecules by AFM directly within a three-dimensional aqueous volume requires little or no sample preparation (Fotiadis et al., 2002; Hörber et al., 2003; You and Lowe, 1996), and in situ AFM in the fluid-tapping mode greatly reduces frictional force between the AFM tip and sample surface, allowing minimal artifacts during imaging of soft biological materials (You and Lowe, 1996). Moreover, in situ and real-time monitoring of dynamic changes of the nucleic acid/polymer polyplexes is achievable by facilely and timely exchanging buffers via a fluid cell during the observation. Although AFM has been used to study nonviral vectors (Bettinger et al., 2001; Chim et al., 2005; Dunlap et al., 1997; Kircheis et al., 2001; Wiethoff et al., 2003), there have been limited studies to observe the dynamic morphological changes of nucleic acid/polymer polyplexes in liquid. Understanding of molecular dynamics of nonviral vectors under differentially changing conditions, which mimic intracellular gene delivery pathways, is expected to generate key information for designing efficient gene carriers.
Recently, we reported that acid-degradable ketalized PEI greatly improved delivery of both plasmid DNA and siRNA via enhanced dissociation of the nucleic acids in a cell (Shim and Kwon, 2008; Shim and Kwon, 2009a; Shim and Kwon, 2009b). It was also demonstrated that ketalization ratios, molecular weights of ketalized PEI, and types of nucleic acids (DNA vs. siRNA) played important roles in condensation of nucleic acids and intracellular localization of nucleic acids/ketalized PEI polyplexes, which consequently affected DNA transfection and RNA interference efficiencies (Shim and Kwon, 2008; Shim and Kwon, 2009a). To further comprehend 1) the reasons for such controlled intracellular localizations and gene expression/silencing by nucleic acids/ketalized PEI polyplexes and 2) nucleic acid release dynamics via polyplex disassembly under different intracellular conditions, the complexation and disassembly of the various polyplexes (DNA vs. siRNA and acid-degradable vs. nondegradable polymers) under a mildly acidic endosomal (e.g., pH 5.0) and a reducible cytosolic (e.g., presence of anionic biomacromolecules) conditions, were investigated. The polyplexes under differentially changing conditions in a fluid cell were continuously observed by in situ AFM. Cleavage of ketal linkages in ketalized PEI under the endosomal condition resulted in reduced interaction with nucleic acids, leading to facilitated release of the nucleic acids, while the polyplexes prepared with non-degradable PEI did not show noticeable morphological changes associated with nucleic acid release. It was also observed that the structural difference of ketalized PEI (linear vs. branched) significantly affected release dynamics of DNA and morphological transitions of polyplexes. The findings of this study may provide insightful information about how stimuli-responsive and non-responsive polyplexes are processed under different intracellular conditions.
MATERIALS AND METHODS
Materials
Linear polyethylenimine (2.5 kDa) and branched polyethylenimine (1.8 kDa) were supplied from Polyscience, Inc. (Warrington, PA). V-1 quality mica was supplied from Ted Pella, Inc. (Redding, CA). Enhanced green fluorescent protein (eGFP)-encoding plasmid DNA (Shim and Kwon, 2008) and Silencer GFP siRNA purchased from Ambion (Austin, TX) were used. NIH 3T3 cells (ATCC, Rockville, MD) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (MediaTech, Herndon, VA) with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT), unless otherwise noted.
Syntheses and Acid-hydrolysis Rates of Ketalized Linear PEI and Ketalized Branched PEI
Acid-degradable acrylated ketal monomers (compound 1) were grafted to linear PEI (2.5 kDa) or branched PEI (1.8 kDa) via Michael addition conjugation (Scheme 1). Ketalized linear PEI (KL-PEI) was synthesized in the same manner as reported in the previous paper (Shim and Kwon, 2009b). For the synthesis of ketalized branched PEI (KB-PEI), branched PEI (0.1 g, 2.33 mmol of amine groups, 1 equiv) dissolved in 10 mL of dichloromethane was reacted with 0.73 g of compound 1 (2.33 mmol, 1 equiv) in the presence of triethylamine (0.23 g, 2.33 mmol, 1 equiv). After 120 h of reaction at 45 °C, the polymer was precipitated into anhydrous diethyl ether and dried under vacuum, followed by trifluoroacetate group-deprotection using 1:9 MeOH/6N NaOH solution. Then the polymer was purified by dialysis against DI water at 4 °C (Mw cutoff of dialysis membrane = 500), followed by lyophilization to obtain the product as a powder with a hint of yellow color. Degrees of ketalization of KL-PEI and KB-PEI were calculated to be 34% and 37%, respectively, by using 1H NMR spectroscopy in the same manner as reported previously (Shim and Kwon, 2008; Shim and Kwon, 2009b). Hydrolysis rates of KL-PEI and KB-PEI at different pHs and temperatures were investigated by incubating the polymers in acetate (pH 5.0) and Tris-HCl (pH 7.4) buffers in D2O at 25 °C and 37 °C, respectively, for various periods of time. The disappearance of the ketal linkage peak at 1.41 ppm relative to the constant buffer salt peak at different incubation time points was quantified by 1H NMR spectroscopy in order to calculate the hydrolysis rates of the polymers.
Scheme 1.
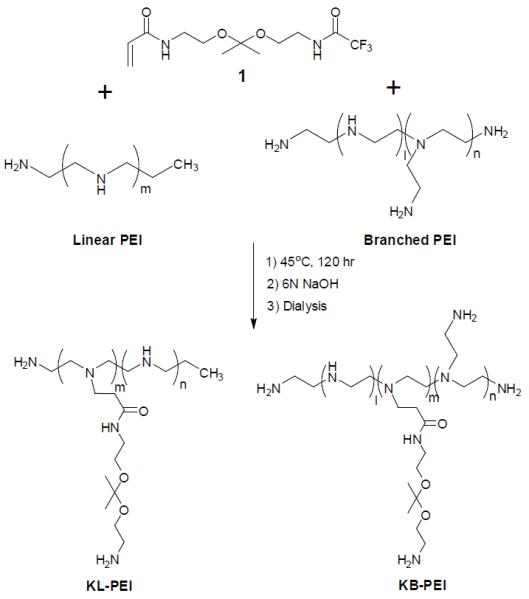
Synthesis of Ketalized PEI (KL-PEI).
Polyplex Preparation and Characterization
Various amounts of polymers dissolved in phosphate buffered saline (PBS) (2 mg/mL) were mixed with 5 μg of nucleic acid (DNA or siRNA) in 100 μL of DI water at various N/P ratios (e.g., N/P ratio of 120 for KB-PEI and N/P ratio of 160 for KL-PEI), followed by incubation for 30 min at room temperature. N/P ratio refers molecular ratio of amines of PEI and phosphates of nucleic acid. Unmodified L-PEI and B-PEI were used to complex DNA and siRNA at N/P ratios of 40 and 80, respectively. For dynamic light scattering (DLS) analysis, the resulting polyplex solution was diluted with additional 800 μL of DI water. Measurements of mean particle size for various polyplexes were performed with a ZEN3600 Zetasizer (Malvern Instruments, Worcestershire, UK) at 25 °C. The optimum N/P ratios were obtained by nucleic acid condensation determined by DLS as well as cellular transfection and gene silencing efficiencies.
AFM Characterization
AFM imaging was conducted using a MFP-3D atomic force microscopy (Asylum Research, Santa Barbara, CA). Commercial pyramidal silicon nitride tips (Asylum Research) with cantilever length of about 100 μm, a nominal resonance frequency of 34 kHz, and a spring constant of 0.08 N/m were used for tapping mode imaging in fluid. Samples for AFM imaging were prepared by depositing a 60 μL aliquot of the polyplex solution containing 3 μg of nucleic acids on freshly cleaved V-1 quality scratch-free ruby mica and incubating for 10 min. For in situ imaging of fully hydrated samples, the mica substrate was transferred into a standard closed fluid cell (Asylum Research) where 5 mL of Milli-Q water (≥ 18.2 MΩ/cm) was subsequently injected. To monitor the morphological changes of polyplexes upon hydrolysis, 5 mL of pH 5.0 acetate buffer (100 mM acetic acid adjusted with NaOH, 150 mM NaCl) was flowed into the fluid cell by using a syringe pump with a flow rate of 0.3 mL/min. To simulate post-endosomal escape conditions, 150 mM NaCl and heparan sulfate (30 μg/mL) in DI water was used to exchange buffers. All AFM imaging was performed at room temperature with a scan speed of 1.0 Hz. All acquired images contained 512 × 512 data points and were rendered using Igor Pro software 6.0 with only background slopes corrected.
Cell Transfection
NIH 3T3 cells were inoculated at a density of 3 × 104 cells/well in a 24-well plate, 24 h prior to transfection. DNA/polymer polyplexes were prepared by mixing 0.5 μg of DNA and various amounts of ketalized PEI, achieving various N/P ratios in the final volume of 50 μL. After 30 min of incubation, the prepared polyplex solution was added to wells along with 250 μL of DMEM. After 4 h of incubation, the medium was replaced with 0.5 mL of fresh DMEM containing 10% FBS, and the cells were further incubated for an additional 20 h at 37 °C. After the cells were harvested by trypsinization, the relative eGFP expression was analyzed using a Guava EasyCyte Plus cytometer (Guava Technologies, Inc., Hayward, CA).
RNA Interference
eGFP-expressing NIH 3T3 cells were acquired by retroviral transduction, followed by G418 selection for 10 days (Kwon et al., 2001). A day prior to siRNA treatment, cells were seeded in a 24-well plate at a density of 2 × 104 cells/well. siRNA/polymer polyplexes were prepared by mixing various amounts of ketalized PEI with anti-eGFP siRNA solution (75 pmol) in DI water to obtain various N/P ratios in the final volume of 75 μL. After 30 min of incubation at room temperature, the resulting polyplex solution was added to cells with 300 μL of DMEM to obtain a final siRNA concentration of 200 nM. After 4 h of incubation, the medium in the well was replaced with 0.5 mL of fresh DMEM containing 10% FBS, and the cells were further cultured for 72 h and harvested by trypsinization. eGFP fluorescence of the cells was analyzed by using a Guava EasyCyte Plus flow cytometer (Guava Technologies, Hayward, CA). The silencing of eGFP gene expression was quantified by comparing the mean fluorescence intensities of the cells incubated with various polyplexes and the ones of the cells incubated without polyplexes.
RESULTS
Studying Intracellular Fates of Nucleic Acid/Polymer Polyplexes Using In Situ AFM
While extracellular gene delivery pathways are relatively understood experimentally, intracellular behaviors of nucleic acid/polymer polyplexes are still unclear (Fig. 1) and frequently found to be controversial in literature. For example, whether nucleic acids are dissociated in the cytoplasm or in the nucleus and what happens to the polyplexes that have escaped from the endosome, which significantly affects the final intracellular destination of polyplexes (cytoplasm vs. nucleus), are unclear (Fig. 1). Confocal laser-scanning microscopy (CLSM) is the most commonly used technique to find answers to those questions. However, due to limited resolution that is at best on the submicron scale, behaviors of individual polyplexes less than 100 nm are difficult to observe using confocal microscopy. CLSM also requires fluorescence labeling of polyplexes, which may significantly alter polyplexes’ overall physical properties and, more importantly, interactions of nucleic acids and polymers. Although AFM is an excellent tool for imaging at nanometer scales, it has been seldom employed to investigate intracellular events. In this study, we used a flow cell, which enable us to create a series of intracellular conditions (e.g., ordered mildly acidic pH and presence of heparan sulfate), without disturbing polyplexes. The experimental setup also enabled a time evolution study of the same polyplexes to be observed under differentially varying conditions.
Fig. 1.
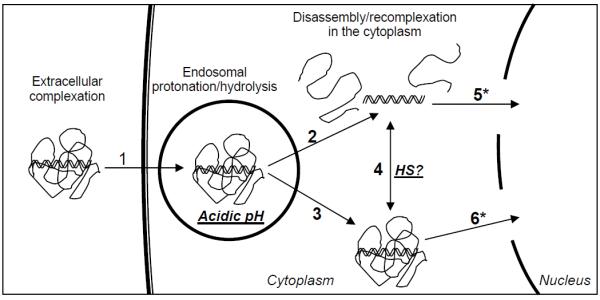
Gene delivery pathways for nucleic acid/polymer polyplexes. Complexed form of nucleic acids are internalized (1) and should survive in the mildly acidic endosome before being released into the cytoplasm after complete dissociation via hydrolysis (2) or without dissociation (3) of nonviral vectors. Reducible molecules in the cytoplasm such as negatively charged heparan sulfate (HS) may disassemble polyplexes or disassembled nucleic acid may be re-complexed in the cytoplasm due to neutral pH (4). Whether dissociated or complexed form of nucleic acids is advantageous for transnuclear localization is another key question to be answered.
Acid-Degradability of Ketalized PEI (K-PEI)
As shown in Table 1, the half-lives of ketalized linear PEI (KL-PEI) at 25 °C were measured to be 3.8 and 126 h at pH 5.0 and 7.4, respectively, using the previously used methods (Shim and Kwon, 2008; Shim and Kwon 2009b). The half-lives of ketalized branched PEI (KB-PEI) at the same temperature were 4.7 and 172 h at pH 5.0 and 7.4, respectively. It was found that hydrolysis rate of KL-PEI was slightly higher than that of KB-PEI. Regardless of structural difference (i.e., linear vs. branched), consistent acid-degradability for both KL-PEI and KB-PEI was obtained. For example, 33 and 37 times faster hydrolysis rates at pH 5.0 than at pH 7.4 for KL-PEI and KB-PEI, respectively, were obtained at 25°C. At 37 °C, hydrolysis at pH 5.0 was consistently faster (13 and 12 times for KL-PEI and KB-PEI, respectively) than at pH 7.4 regardless of structural difference of the polymers.
Table 1.
Hydrolysis kinetics of various ketalized PEI at different temperatures and pHs
| Polymer | Degree of ketalization (%) |
Half-lives (h) |
|||
|---|---|---|---|---|---|
| 37 °C | 25 °C | ||||
| pH 5.0 | pH 7.4 | pH 5.0 | pH 7.4 | ||
| KL-PEI | 34 | 1.5 | 19.8 | 3.8 | 126 |
| KB-PEI | 37 | 2.8 | 32.1 | 4.7 | 172 |
Complexation of Nucleic Acids with Ketalized PEI
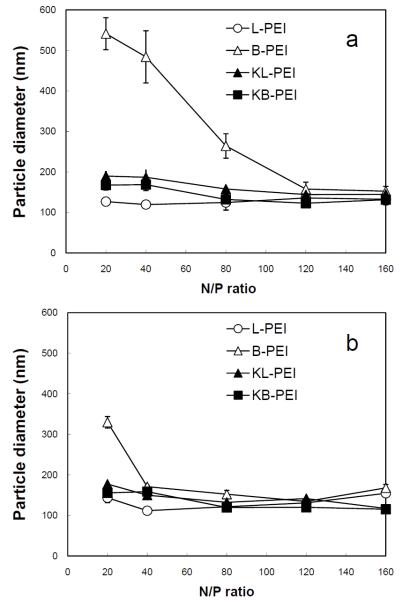
As shown in Fig. 2a, the sizes of DNA/PEI polyplexes at the N/P ratios of 20 - 160 were measured to be in the range of 120 to 180 nm except the polyplexes prepared with unmodified branched PEI (B-PEI), as determined by dynamic light scattering (DLS) particle analysis. The most compact form of the polyplexes were obtained by complexing DNA with unmodified linear PEI (L-PEI) at the N/P ratio of 40, while B-PEI formed much less compact polyplexes over the entire range of N/P ratios. Considering the fact that the molecular weights of L-PEI (2.5 kDa) and B-PEI (1.8 kDa) were relatively similar (difference of ~30%), the length of B-PEI was too short to complex the much longer plasmid DNA. This limited DNA complexation by B-PEI was greatly improved by ketalization that is possibly due to enhanced interactions of elongated side chains with DNA, as reported previously (Shim and Kwon, 2008; Shim and Kwon, 2009a). On the other hand, ketalization adds relatively bulky and short ketal branches to the linear backbone of L-PEI, and may result in reduced flexibility of interaction with DNA. Difference in siRNA complexation by both unmodified and ketalized PEI appeared significantly smaller than DNA complexation based on the measured sizes (Fig. 2b).
Fig. 2.
Particle sizes of (a) DNA/PEI and (b) siRNA/PEI polyplexes in DI water, determined by DLS.
Dynamic Interactions of DNA with Acid-degradable and Non-degradable PEI under Endosomal and Cytosolic Conditions
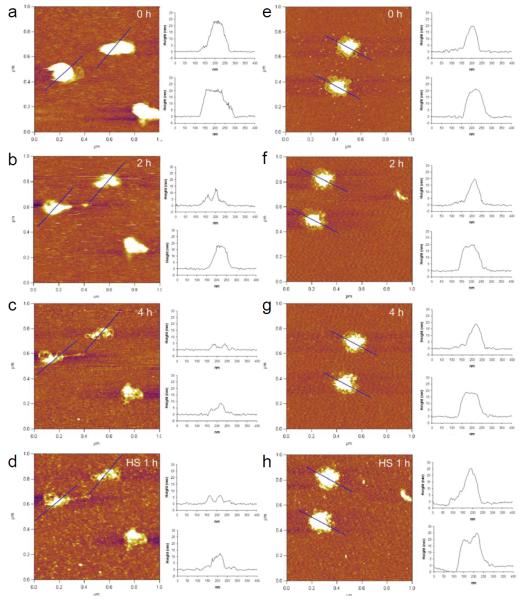
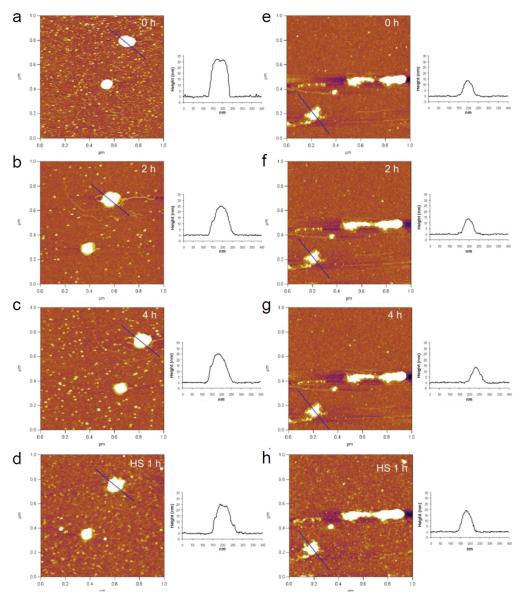
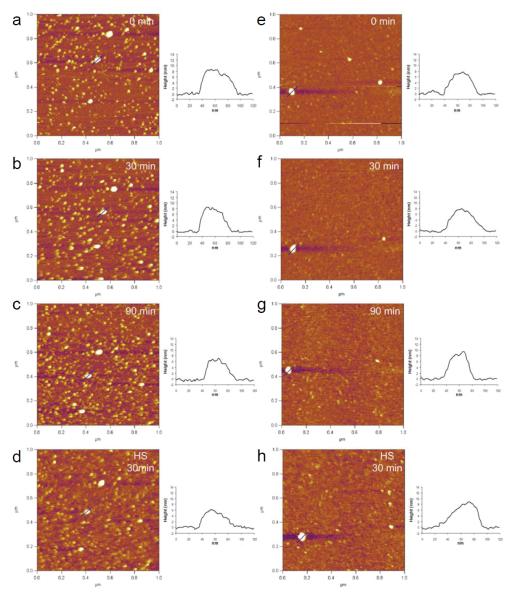
Structural changes of various DNA-complexing polyplexes prepared with acid-degradable ketalized and unmodified PEI under simulated mildly acidic endosomal (pH 5.0) and reducible cytosolic (presence of heparan sulfate) conditions were observed in differentially changing buffers. The positively charged (+20 - +30 mV) and fully hydrated DNA/PEI polyplexes were adsorbed onto negatively charged mica substrate and then imaged in buffers by in situ AFM in fluid-tapping mode (Figs. 3 and 4). As shown in Figs. 3 and 4, L-PEI was able to efficiently complex DNA regardless of its degradability, while only KB-PEI complexed DNA fairly-well in DI water. The majority of the well-complexed DNA-containing polyplexes were characterized as spheroids with dimensions of 20 - 25 nm in height and 80 - 100 nm in diameter, while relatively poorly complexed DNA by unmodified branched PEI (B-PEI) resulted in elongated morphology with a short height of approximately 15 nm. The diameter of the polyplexes was measured at half-height in order to minimize tip convolution in the lateral direction. Ketalization interfered with L-PEI’s DNA complexation (Fig. 3a and Fig. 4a), as indicated by the fact that height of DNA/L-PEI polyplexes was decreased from 32 nm to 20 nm with an elongated morphology. Interestingly, however, B-PEI’s DNA complexation capability was greatly enhanced by ketalization (i.e., as evidenced by the increased polyplexes’ height to length ratio demonstrating compact morphology) (Fig. 3e and Fig. 4e). In particular, DNA/KB-PEI polyplexes showed folded loops of DNA surrounding central cores (Fig. 3e), resembling the partially DNA-condensed polyplexes (Dunlap et al., 1997). Different ketalization effects on nucleic acid condensation efficiency were consistent with DLS results as shown in Fig.2.
Fig. 3.
In situ AFM images of DNA/KL-PEI polyplexes (a-d) and DNA/KB-PEI polyplexes (e-h). DNA/KL-PEI polyplexes (a-c) and DNA/KB-PEI polyplexes (e-g) were hydrolyzed in pH 5.0 acetate buffer containing 150 mM NaCl for different periods of time. After hydrolysis, the acidic acetate buffer was replaced with heparan sulfate (HS) solution containing 150 mM NaCl. DNA/KL-PEI polyplexes (d) and DNA/KB-PEI polyplexes (h) were incubated in the HS solution for 1 h. Cross-sectional profiles along the blue lines in the AFM images are represented. All images are 1 × 1 μm2 in scan size and 6 nm in z range.
Fig. 4.
In situ AFM images of DNA/L-PEI polyplexes (a-d) and DNA/B-PEI polyplexes (d-h). DNA/L-PEI polyplexes (a-c) and DNA/B-PEI polyplexes (e-g) were incubated in pH 5.0 acetate buffer containing 150 mM NaCl for different periods of time. After exchanging acetate buffer to HS-containing buffer (with 150 mM NaCl), DNA/L-PEI polyplexes (d) and DNA/B-PEI polyplexes (h) were incubated for 1 h. Cross-sectional profiles along the blue lines in the AFM images are represented. All images are 1 × 1 μm2 in scan size and 6 nm in z range.
AFM images of the same polyplexes under neutral and mildly acidic endosomal conditions were obtained by changing DI water to pH 5.0 acetate buffer containing 150 mM NaCl using a flow cell. Unmodified L-PEI and B-PEI polyplexes did not show significant morphological changes under the changed condition (Fig. 4), although it was expected that the acidic condition would strengthen DNA complexation due to elevated protonation. On the contrary, substantial structural changes were observed for DNA/KL-PEI (Figs. 3a-3c) polyplexes as a function of incubation time at pH 5.0. Clearly, the buried DNA loops in KL-PEI were exposed as acid-hydrolysis underwent, and almost bare DNA was observed after 4 h incubation at pH 5.0, because acid-hydrolyzed KL-PEI has only neutral hydroxyl side chains and is no longer capable of complexing DNA (Fig. 3c). This result indicates that, in order to be released as a complexed polyplex, DNA/KL-PEI should escape from the mildly acidic endosome within a short period of time (e.g., less than 2 h at 37 °C). Interestingly, no significant structural changes were observed for DNA/KB-PEI polyplexes under the identical acid-hydrolysis condition at pH 5.0 (Figs. 3e-3g). Because the majority (63%, Table 1) of the amines of KB-PEI is not ketalized, DNA remained complexed even after the remaining 37% of ketalized branches converted themselves to neutral hydroxyl arms under the endosomal conditions.
One of the unanswered intracellular processes in nonviral gene delivery is what happens to the polyplexes escaped from the endosome into the cytoplasm. DNA/polymer polyplexes released from the endosome will be under not only a neutral pH but also biochemically different conditions. For example, it has been conjectured that nucleic acid/cationic polymer complexes are disassembled by anionic biomacromolecules such as proteoglycans (e.g., heparan sulfate, HS), as demonstrated in vitro (Männistö et al., 2007). The DNA-complexing polyplexes were continuously imaged by changing the acidic acetate buffer to HS-containing buffer (pH 7.5). Unexpectedly, the presence of HS did not contribute to disassembly of DNA-complexing polyplexes. Rather, the overall sizes of the polyplexes slightly increased. It was noticed that fully exposed DNA loops did not increase size but the areas remained partially complexed (e.g., center of KL-PEI/DNA polyplexes) became larger (Figs. 3c and d; Figs. 3g and h). This indicates that HS simply adsorbed onto the remaining cationic polymer and slightly increased the size of polyplexes, rather than disassembling the polyplexes.
Dynamic Interactions of siRNA by Acid-degradable and Non-degradable PEI under Endosomal and Cytosolic Conditions
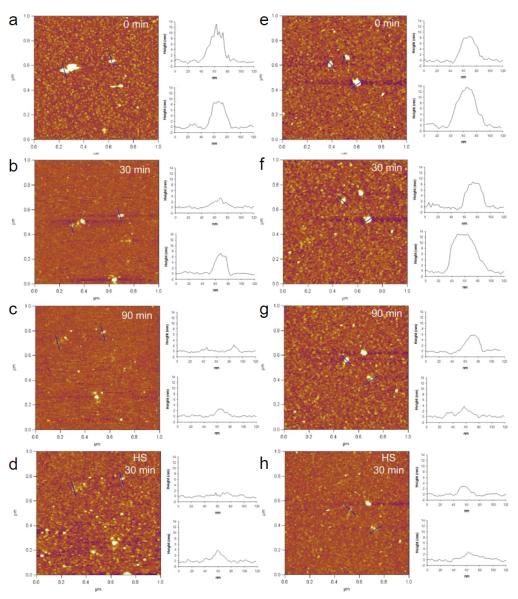
AFM captured time-course pH-dependent disassembly of siRNA from the polyplexes prepared with ketalized PEI (Fig. 5) and unmodified PEI (Fig. 6). The majority of siRNA/PEI polyplexes were spheroids with a diameter (measured at half-height) in the range of 40 to 60 nm with a height in the range of 8 to 13 nm. Dramatic morphological changes of both siRNA/KL-PEI and siRNA/KB-PEI polyplexes were observed shortly after being exposed to the endosomal pH (Fig. 5). The siRNA/KL-PEI polyplexes started to disassemble shortly after 30 min of acid-hydrolysis (Fig. 5b), and almost complete disassembly of the polyplexes was observed after 90 min (Fig. 5c), which was significantly faster than the disassembly of DNA/ketalized PEI polyplexes. The hydrolysis of siRNA/KB-PEI polyplexes was noticeably slower than that of siRNA/KL-PEI (Figs. 5e-5g). siRNA bound to the mica surface was not clearly observed because of substantially smaller size of siRNA (~ 23 bps) than plasmid DNA (~ 5 kbps). Complexing short nucleic acids (e.g., siRNA) is less efficient than complexing longer ones (e.g., plasmid DNA), which often requires chemical modification (Bolcato-Bellemin et al., 2007) or conjugation with larger molecules/carriers (Jeong et al., 2009). Relatively poor complexation seems to significantly contributed to rapid disassembly of the siRNA/ketalized PEI polyplexes (Figs. 2 and 5), compared with DNA/ketalized PEI polyplexes (Fig. 3). Regardless of loaded gene types (plasmid DNA or siRNA), more efficient disassembly from KL-PEI was observed than KB-PEI (Figs. 3 and 5). While KB-PEI was able to complex DNA even after 4 h of incubation at pH 5.0 (Fig. 3g), siRNA started to disassemble from KB-PEI after a 90 min of incubation under the same condition (Fig. 5g), due to short siRNA’s poor molecular interactions with the polymer. There was no noticeable siRNA disassembly from unmodified PEI for the entire incubation period at the endosomal pH (Fig. 6). Again, hydrolyzed KL-PEI lacking amine-bearing side chains rapidly released siRNA, while unketalized amine-bearing branches of KB-PEI were able to hold siRNA for a while, as explained for DNA/ketalized PEI polyplexes. Unlike DNA-complexing polyplexes, there was no size increase of siRNA-complexing polyplexes in the presence of HS regardless of structural difference (linear vs. branched) and acid-degradability.
Fig. 5.
In situ AFM images of siRNA/KL-PEI polyplexes (a-c) and siRNA/KB-PEI polyplexes (e-g) captured at different periods of time under acid-hydrolysis using pH 5.0 acetate buffer containing 150 mM NaCl. After hydrolysis, siRNA/KL-PEI polyplexes (d) and siRNA/KB-PEI polyplexes (h) were incubated in heparan sulfate (HS) solution containing 150 mM NaCl for 30 min. Cross-sectional profiles along the blue lines in the AFM images are represented. All images are 1 × 1 μm2 in scan size and 6 nm in z range.
Fig. 6.
In situ AFM images of siRNA/L-PEI polyplexes (a-d) and siRNA/B-PEI polyplexes (e-h). siRNA/L-PEI polyplexes (a-c) and siRNA/B-PEI polyplexes (e-g) were incubated in pH 5.0 acetate buffer containing 150 mM NaCl for different periods of time. After hydrolysis, siRNA/L-PEI polyplexes (d) and siRNA/B-PEI polyplexes (h) were incubated in heparan sulfate (HS) solution containing 150 mM NaCl for 30 min. Cross-sectional profiles along the blue lines in the AFM images are represented. All images are 1 × 1 μm2 in scan size and 6 nm in z range.
Regulated Transfection and Gene Silencing by the Dynamics of Polyplex Disassembly
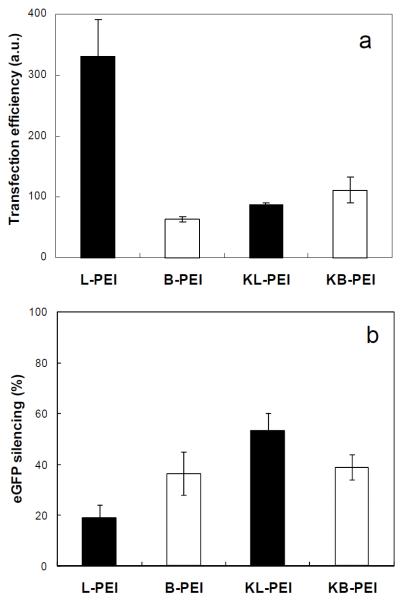
Various eGFP-encoding DNA-complexing polyplexes, which were used for AFM studies, transfected NIH 3T3 cells. As shown in Fig. 7a, unmodified L-PEI (N/P ratio of 40) showed the highest gene transfection efficiency. Importantly, ketalization significantly reduced the transfection capability of L-PEI, while DNA/KB-PEI polyplexes (N/P ratio of 120) showed slightly higher transfection efficiency than that of DNA/B-PEI (N/P ratio of 80). As shown in Figs. 3 and 4, only DNA/KL-PEI polyplexes (N/P ratio of 160) were efficiently disassembled under an endosomal pH, while the others remained almost unchanged. Therefore, it can be concluded that rapid disassembly of DNA from its carrier in the endosome inhibits transfection. This indicates that DNA should remain complexed when it is released into the cytoplasm for efficient diffusion to nucleus (Liu et al., 2003; Pollard et al., 1998) and transnuclear localization through nuclear pores (Elouahabi and Ruysschaert, 2005). Lower transfection by both KB-PEI and B-PEI than L-PEI can be explained by their poor DNA complexation efficiencies. Slightly increased gene expression by KB-PEI, compared with B-PEI, also indicates that DNA complexation is a key requirement for transfection. In other words, degradability of polymeric carriers (or efficient endosomal escape) alone does not guarantee their transfection capability when DNA is not efficiently complexed.
Fig. 7.
eGFP expression by NIH 3T3 cells that were transfected by various DNA/PEI polyplexes (a) and silenced gene expression by NIH 3T3/eGFP cells transfected by various siRNA/PEI polyplexes (b). Transfection efficiency (a) was determined by multiplying cell transfection rate by mean fluorescence intensity of the cells incubated with various polyplexes (counting total gene product quantity). Gene silencing efficiency (b) was quantified by comparing the mean fluorescence intensity (average expression level) of the cells incubated with various polyplexes, with that of the cells incubated without polyplexes.
Gene silencing efficiency of various anti-eGFP siRNA-complexing polyplexes showed that unmodified L-PEI polyplexes resulted in minimal RNA interference (Fig. 7b). Interestingly, however, gene silencing by siRNA/KL-PEI polyplexes was dramatically increased. On the other hand, both siRNA/B-PEI and siRNA/KB-PEI polyplexes showed a modest and consistent level of gene silencing. Release of siRNA might need to occur relatively quickly in order to achieve efficient RNA interference, considering the finding that slowly disassembling siRNA/KB-PEI polyplexes resulted in gene silencing similar to what non-disassembling siRNA/B-PEI polyplexes generated. Higher gene silencing by siRNA/KB-PEI polyplexes than siRNA/L-PEI polyplexes can be explained similarly for why DNA/KB-PEI polyplexes less efficiently transfected the cells than DNA/L-PEI. Lower gene silencing by siRNA/L-PEI polyplexes than siRNA/B-PEI polyplexes can be attributed to higher siRNA condensation without siRNA dissociation.
Therefore, from the DLS size analysis, AFM imaging, transfection, and gene silencing studies, optimal intracellular routes for siRNA delivery are 1 and 2 in Fig. 1. In comparison, DNA needs to be processed through the routes of 1, 3, and 6 in Fig. 1. Efficient disassembly of polyplexes in the cytoplasm by HS was not observed with nucleic acids/PEI polyplexes, and this process may have relatively insignificant effects on both transfection and RNA interference. It was not feasible to investigate transnuclear localization of free nucleic acids (route 5 in Fig. 1) in this brief study, although it is clear that this route should be prevented in order to achieve efficient RNA interference, as demonstrated previously (Shim and Kwon, 2009a).
DISCUSSION
Nonviral gene delivery processes include many extracellular and intracellular steps, such as adhesion of nucleic acid-complexing polyplexes on the cell surface, cellular internalization, escape from the endosomes (cytosolic release), and release of the nucleic acids into their intracellular target sites (e.g., the nucleus for DNA and the cytoplasm for siRNA). The significantly lower gene delivery efficiency by nonviral vectors than viral vectors is mainly attributed to poor intracellular processes of nonviral vectors whereas viral vectors have optimized these via evolution processes (Medina-Kauwe et al., 2005). Despites the importance of understanding intracellular trafficking of nonviral gene carriers, it has been a very daunting task to study interactions of nucleic acid/polymer in a cell. There are a number of molecules involved in the dynamic intracellular nonviral gene delivery processes, and it is not feasible to mimic them in vitro. As a result, current nonviral vectors are developed based on highly hypothetic theories for intracellular nonviral gene delivery pathways. For example, the eccentric theory for endosomal escape is the proton sponge effect that explains superiority of cationic polymers in nonviral gene delivery (Boussif et al., 1995). However, even this core theory has been questioned (Funhoff et al., 2004).
Ideal methodology to investigate molecular dynamics and interactions of nonviral vectors in a cell is tracking individual carriers, yet this is not technically feasible. For example, transmission electron microscopy (TEM) is not suitable to observe the dynamics of gene carriers on a live cell, due to required imaging conditions, and frequently it is difficult to distinguish individual gene carriers from intracellular molecules and organelles. Morphologies of nonviral gene carriers in dried TEM samples are expected to be very different from ones under aqueous conditions. The most commonly employed microscopic tool in investigating nonviral gene delivery processes is confocal laser scanning microscopy (CLSM). CLSM enables observing particles in a live cell. However, this technique requires fluorescent labeling of the particles and may obtain information on labeled molecules that could be significantly altered from unlabeled ones. In addition, the low resolution of the technique allows imaging of relatively large or aggregated particles, which would not explain intracellular trajectory of individual gene delivery carriers. Although AFM cannot differentiate nonviral gene carriers from other molecules, it has been essential tool in investigating polyplexes under biologically-inspired aqueous conditions at their fully hydrated state with high resolution. In comparison, dehydration during sample preparation for ambient AFM imaging results in shrinkage of the size (diameter and height) and morphological changes (Malkin et al., 2003) of biomolecules, polymers, and their complexes. For example, the typical dimensions of dehydrated and highly condensed DNA/PEI polyplexes were 30 - 60 nm in diameter (measured at half-height) and 5 - 10 nm in height (Itaka et al., 2004), which was different from the results of this study showing that the diameters of both highly condensed DNA/unmodified L-PEI and DNA/unmodified B-PEI polyplexes were around 100 and 70 nm, respectively (The heights of both polyplexes were 32 nm and 15 nm, respectively). Additionally, the artifacts caused by washing and drying of the samples for ex situ AFM imaging can be minimized by in situ imaging.
Our previous studies using CLSM demonstrated that siRNA complexed by KL-PEI was selectively localized and efficiently dissociated from KL-PEI in the cytoplasm, while the polyplexes of siRNA and L-PEI were colocalized not only in the cytoplasm but also in the nucleus without noticeable dissociation (Shim and Kwon, 2009b). We hypothesized that partially condensed siRNA in KL-PEI resulted in a large polyplex size, making the polyplexes localized in the cytoplasm rather than in the nucleus, followed by rapid dissociation for hybridization with a target mRNA. This study clearly supports the hypothesis. siRNA/KL-PEI polyplexes were efficiently disassembled under an endosomal condition (Fig. 5a-5d) and resulted in significantly enhanced gene silencing (Fig. 7b). For comparison, siRNA complexed with L-PEI and B-PEI, which did not disassemble siRNA under the same condition, showed significantly lower RNA interference (Fig. 6). KB-PEI, which disassembled siRNA relatively slower than KL-PEI was unable to silence the gene expression efficiently, which could not be measured even indirectly using CLSM or TEM. On the contrary, transfection requires delivery of DNA into the nucleus in a complexed form, which was also demonstrated in this study. DNA/KL-PEI polyplexes were disassembled quickly under an endosomal condition (Figs. 3a-3d) and resulted in substantially impaired gene expression (Fig. 7a). Transfection by DNA/KB-PEI polyplexes was enhanced mainly by improved complexation efficiency, compared with DNA/B-PEI polyplexes. For DNA/KB-PEI polyplexes, roles of acid-degradability were not clear and probably less crucial than complexation efficiency. KL-PEI and KB-PEI showed a remarkable difference in polyplex disassembly under the endosomal condition. KL-PEI presented a more rapid and efficient nucleic acid disassembly than KB-PEI. The primary amines in PEI are mainly responsible for attractive electrostatic interactions with nucleic acid (Reschel et al., 2002; Tang and Szoka, 1997; Wolfert et al., 1999), thus resulting in higher condensation efficiency than secondary and tertiary amines, due to higher protonation at a given pH as well as higher accessibility (Wolfert et al., 1999). KL-PEI completely lost its primary amines after acid-hydrolysis, while KB-PEI still had unketalized primary amines which can complex DNA. Therefore, KB-PEI was able to retain complexation of nucleic acids longer than KL-PEI in an acidic condition.
In order to explore how nucleic acid/polymer polyplexes disassemble after endosomal escape, heparan sulfate (HS) was used. In vitro studies indicated that anionic biomacromolecules such as proteoglycans (e.g., HS) are responsible for polyplex disassembly (Männistö et al., 2007). However, it is unclear whether there are sufficient proteoglycans in the cytoplasm. In addition, it is highly unlikely that relatively large proteoglycans effectively access to the nucleic acids buried in polymeric network, which was supported by this study. As shown in Fig. 3d, HS sulfate was not able to disassemble DNA/KL-PEI polyplexes but adsorbed on cationic polymers, slightly increasing the thickness of unhydrolyzed areas. This process might be even slower in a cell because this study was conducted in a slow flow of the HS-containing buffer but the cytoplasm will be relatively stationary. Likewise, polyplex disassembly would be expected to be much faster if polyplex and HS were well-mixed like most studies have been done to demonstrate nucleic acid release from a cationic polymer in the presence of proteoglycans (Liu and Reineke, 2006).
AFM analysis used in this brief study to image gene-carrying polyplexes in differentially changing buffers provided important information summarized in Fig. 1. However, it is also clear that further studies are necessary to obtain more conclusive and insightful results. For example, conditions more closely representing the endosomal and the cytoplasmic conditions, which are not currently available yet, need to be emulated. In addition, polyplex assembly and disassembly dynamics at 25 °C measured in this study may be substantially different from those at 37°. This can be easily addressed by conducting AFM studies on a temperature-controlled plate. Most importantly, AFM can image only surface-immobilized molecules; therefore, some fully dissociated nucleic acids could be washed away in flowing buffer or simply moved by the AFM tip. Additional microscopic techniques addressing this issue are needed to provide conclusive information on dynamics of nucleic acid/polymer polyplexes under biologically mimicking conditions.
ACKNOWLEDGMENT
This work was financially supported by the National Institute of Health (3R21DE19298-02S1), National Science Foundation (Electrochemistry and Surface Chemistry, 0748912), and Council on Research Computing and Libraries (CORCL) grant, University of California, Irvine.
REFERENCE
- Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Nucleic Acids Res. 2001;29:3882–3891. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcato-Bellemin A-L, Bonnet M-E, Creusat G, Erbacher P, Behr J-P. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci USA. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussif O, Lezoualćh F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr J-P. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim YTA, Lam JKW, Ma Y, Armes SP, Lewis AL, Roberts CJ, Stolnik S, Tendler SJB, Davies MC. Structural study of DNA condensation induced by novel phosphorylcholine-based copolymers for gene delivery and relevance to DNA protection. Langmuir. 2005;21:3591–3598. doi: 10.1021/la047480i. [DOI] [PubMed] [Google Scholar]
- Dunlap DD, Maggi A, Soria MR, Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elouahabi A, Ruysschaert J-M. Formation and intracellular trafficking of lipoplexes and polyplexes. Mol Ther. 2005;11:336–347. doi: 10.1016/j.ymthe.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Scheuring S, Müller SA, Engel A, Müller DJ. Imaging and manipulation of biological structures with the AFM. Micron. 2002;33:385–397. doi: 10.1016/s0968-4328(01)00026-9. [DOI] [PubMed] [Google Scholar]
- Funhoff AM, van Nostrum CF, Koning GA, Schuurmans-Nieuwenbroek NME, Crommelin DJA, Hennink WE. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules. 2004;5:32–39. doi: 10.1021/bm034041+. [DOI] [PubMed] [Google Scholar]
- Gadegaard N. Atomic force microscopy in biology: technology and techniques. Biotech Histochem. 2009;81:87–97. doi: 10.1080/10520290600783143. [DOI] [PubMed] [Google Scholar]
- Han S-O, Mahato RI, Sung YK, Kim SW. Development of biomaterials for gene therapy. Mol Ther. 2000;2:302–317. doi: 10.1006/mthe.2000.0142. [DOI] [PubMed] [Google Scholar]
- Hörber JKH, Miles MJ. Scanning probe evolution in biology. Science. 2003;302:1002–1005. doi: 10.1126/science.1067410. [DOI] [PubMed] [Google Scholar]
- Itaka K, Harada A, Yamasaki Y, Nakamura K, Kawaguchi H, Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J Gene Med. 2004;6:76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Mok H, Oh Y-K, Park TG. siRNA conjugate delivery systems. Bioconjugate Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- Kada G, Kienberger F, Hinterdorfer P. Atomic force microscopy in bionanotechnology. Nano Today. 2008;3:12–19. [Google Scholar]
- Kircheis R, Wightman L, Wagner E. Adv Drug Delivery Rev. 2001;53:341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- Kwon YJ, Yu H, Peng C-A. Enhanced retroviral transduction of 293 cells cultured on liquid-liquid interface. Biotechnol Bioeng. 2001;72:331–338. doi: 10.1002/1097-0290(20010205)72:3<331::aid-bit10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, Payne JM, Miller TJ, Brunovskis P, Fink TL, Muhammad O, Moen RC, Hanson RW, Cooper MJ. Nanoparticles of compacted DNA transfect postmitotic cells. J Biol Chem. 2003;278:32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery: stability of polyplexes and efficacy with cardiomyoblast cells. Bioconjugate Chem. 2006;17:101–108. doi: 10.1021/bc050275+. [DOI] [PubMed] [Google Scholar]
- Malkin AJ, McPherson A, Gershon PD. Structure of intracellular mature vaccinia virus visualized by in situ atomic force microscopy. J Virol. 2003;77:6332–6340. doi: 10.1128/JVI.77.11.6332-6340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männistö M, Reinisalo M, Ruponen M, Honkakoski P, Tammi M, Urtti A. Polyplex-mediated gene transfer and cell cycle: effect of carrier on cellular uptake and intracellular kinetics, and significance of lycosaminoglycans. J Gene Med. 2007;9:479–487. doi: 10.1002/jgm.1035. [DOI] [PubMed] [Google Scholar]
- Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr J-P, Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J Biol Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33:e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschel T, Koňák C, Oupicky D, Seymour LW, Ulbrich K. Physical properties and in vitro transfection efficiency of gene delivery vectors based on complexes of DNA with synthetic polycations. J Control Release. 2002;81:201–217. doi: 10.1016/s0168-3659(02)00045-7. [DOI] [PubMed] [Google Scholar]
- Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kwon YJ. Controlled Delivery of Plasmid DNA and siRNA to intracellular targets using ketalized polyethylenimine. Biomacromolecules. 2008;9:444–455. doi: 10.1021/bm7007313. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kwon YJ. Controlled cytoplasmic and nuclear localization of plasmid DNA and siRNA by differentially tailored polyethylenimine. J Control Release. 2009a;133:206–213. doi: 10.1016/j.jconrel.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kwon YJ. Acid-responsive linear polyethylenimine for efficient, specific, and biocompatible siRNA delivery. Bioconjugate Chem. 2009b;20:488–499. doi: 10.1021/bc800436v. [DOI] [PubMed] [Google Scholar]
- Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther. 1997;4:823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92:203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- Wolfert MA, Dash PR, Nazarova O, Oupicky D, Seymour LW, Smart S, Strohalm J, Ulbrich K. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjugate Chem. 1999;10:993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- You HX, Lowe CR. Progress in the application of scanning probe microscopy to biology. Curr Opin Biotechnol. 1996;7:78–84. doi: 10.1016/s0958-1669(96)80099-1. [DOI] [PubMed] [Google Scholar]