Abstract
Clinical applications of genetic therapies, including delivery of short, interfering RNAs (siRNAs) for RNA interference (RNAi), are limited due to the difficulty of delivering nucleic acids to specific cells of interest while at the same time minimizing toxicity and immunogenicity. The use of cationic polymers to deliver nucleic acid therapeutics has the potential to address these complex issues but is currently limited by low delivery efficiencies. While cell culture studies have shown that some polymers can be used to deliver siRNAs and achieve silencing, it is still not clear what physical or chemical properties are needed to ensure that the polymers form active polymer-siRNA complexes. In this study, we used multicolor fluorescence confocal microscopy to analyze the cellular uptake of siRNAs delivered by novel propargyl glycolide polymeric nanoparticles (NPs). Delivery by these vehicles was compared to delivery by linear polyethyleneimine (LPEI) and Lipofectamine 2000 (LF2K), which are both known as effective delivery vehicles for siRNAs. Our results showed that when LF2K and LPEI were used, large quantities of siRNA were delivered rapidly, presumably overwhelming the basal levels of mRNA to initiate silencing. In contrast, our novel polymeric NPs showed delivery of siRNAs but at concentrations that were initially too low to achieve silencing. Nonetheless, the exceptionally low cytotoxicity of our NPs, and the simplicity with which they can be modified, makes them good candidates for further study to optimize their delivery profiles and, in turn, achieve efficient silencing.
INTRODUCTION
Gene therapy has the potential to treat a wide variety of genetic and acquired diseases in which traditional molecular drugs have proven ineffective. The potential to design a highly specific nucleic acid therapy from the coding sequence of the disease protein is appealing over small molecule drugs which are more difficult to develop. These small molecules can have side effects through interactions with untargeted proteins and biomolecules. With the discovery and subsequent explosion of work in RNA interference (RNAi), the realm of nucleic acid therapies has expanded from the traditional focus of delivering genes to add functionality to cells, to include the delivery of a variety of nucleic acids for both upregulation and downregulation of specific proteins.
Clinical applications for nucleic acid delivery have been limited due to the lack of vehicles that survive the challenging in vivo environment. The vehicle must be able to protect the nucleic acid cargo as it is being transported, target only the cells of interest, efficiently deliver the cargo in its active form, and be non-toxic at the concentrations required for delivery. Delivery approaches are generally divided between viral and non-viral methods. Viral delivery methods, such as adenoviruses (Ghosh, 2006), retroviruses (Mori, 2005), and adeno-associated viruses (Peng, 2000), have an advantage of delivery efficiency but often result in safety concerns due to immunogenicity and resistance to repeated infection (Ghosh, 2006; Kaiser, 2007). Therefore, developing non-viral delivery methods for siRNAs has been and will continue to be a focus of significant research effort (recently reviewed in (Nel, 2009)).
Among non-viral approaches, the majority of delivery vehicles are based on cationic lipids (Hong, 1997; Cardoso, 2007) or cationic polymers (Pack, 2005). These cationic species allow electrostatic complex formation with anionic nucleic acids. A variety of commercial cationic lipid reagents, such as Lipofectamine 2000 (LF2K), are used for nucleic acid delivery to cultured cells. Unfortunately, these reagents are typically cytotoxic at concentrations only slightly higher than that required for effective delivery (Zhang, 2007), thus presenting a challenge for their use in vivo. Therefore, for eventual in vivo applications, the development of polymeric delivery vehicles has been an area of considerable study.
Polymeric delivery methods are most often used for the delivery of plasmid DNA (pDNA) (Farrell, 2007) and short interfering RNA (siRNA) (Whitehead, 2009), with antisense oligonucleotides (Chen, 2006) and aptamers (Que-Gewirth, 2007) also being studied. The challenges in delivering pDNA and siRNA are unique due to the differences between DNA and RNA and the dramatic difference in size. While pDNA has the ability to provide increased complex condensation (Spagnou, 2004) and extended transgene expression (Thierry, 1995), delivery is hampered by its large size and the need for the pDNA to reach the nucleus to be active (Gary, 2007). siRNAs are more susceptible to degradation by serum nucleases and require repeated delivery for sustained inhibition (Bartlett, 2006). However, they are considerably smaller than pDNA (~5 nm vs. > 30 nm), and therefore require smaller quantities of vehicle and need only to reach the cytoplasm to be active (Gary, 2007). Thus, delivery vehicles with the appropriate physical and chemical properties should be able to overcome the challenges presented by siRNA delivery.
Linear and branched polyethylenimine (LPEI, BPEI) have traditionally been the standard cationic polymers of choice (Mehrotra, 2009) even though they have been shown to be significantly toxic (Burke 2008). Modifications of PEI with the addition of poly(ethylene glycol) (PEG) (Mao, 2006), ethyl acrylate (Zintchenko, 2008), or cross-linking (Hu, 2009) have been able to increase delivery efficiency while decreasing toxicity. Other single polymer systems include poly (β-amino esters) (PBAE) (Lynn, 2000) or poly(amidoamine) (PAMAM) dendrimers (Tang, 1996). Combinations of polymers to create di- or tri-block polymers such as polyvinyl alcohol with poly(D,L-lactide-co-glycolide) (PVA-b-PLGA) (Nguyen, 2008) or poly(ethylene oxide) (PEO) with poly(ε-caprolactone) (PEO-b-PCL) (Xiong, 2009) are also promising methods of improving nucleic acid delivery.
Unfortunately, polymers synthesized with similar characteristics can have widely varying silencing efficiency, as seen in one study where only 46 of the 2350 PBAE derivatives demonstrated transfection efficacies better than that of PEI (Green, 2008). In most cases, the presence of amine groups is a common motif among all successful delivery vehicles, with secondary and tertiary amines showing the best nucleic acid binding and endosomal release (Green, 2008; Liu, 2005). Other studies have shown that the presence of disulfide bonds can also aid in the intracellular release of the complex (Breunig, 2008; Ou 2008; Rozema, 2007). Its reduction by cytoplasmic glutathione and thioredoxin can cause the breakdown of the polymer itself to release the siRNA from a non-covalent complex or sever the bond between the polymer and siRNA for a covalently-linked siRNA. However, no general rules exist for designing siRNA delivery vehicles.
It is believed that polymer-siRNA complexes enter cells through the endosomal pathway, eventually escaping into the cytoplasm, allowing for the release of the siRNA, and eventual silencing of a target gene. To date, siRNA studies have included microscopy images for delivery by LF2K (Berezhna, 2006) and at the point of silencing (Xiong, 2009). However, the delivery mechanism of polymeric siRNA complexes intracellularly over time, from the membrane, through endocytosis, to the entry of the siRNA into the cytosol, and silencing of the target mRNA remains unknown.
In our study, we used a poly(propargyl glycolide) (PPGL) polymeric backbone that allows for the addition of a variety of chemical functional groups through an alkyne-azide cycloaddition known as “click chemistry” (Jiang, 2008). For the polymers used in this study, we added combinations of primary amines, secondary amines, disulfide linkages, and cholesterol groups to create our delivery vehicles. Using confocal laser scanning microscopy, we imaged enhanced green fluorescent protein (EGFP) expressing cells at various time points after transfection of siRNAs by LF2K (a model lipid), LPEI (a model cationic polymer), and two of our PPGL nanoparticles (NPs). By using a fluorescently tagged siRNA and a 7-dimethylaminocoumarin acetic acid (DMACA) tagged particle, we are able to see cellular uptake of both the siRNA and polymer with respect to time. Our results showed that LF2K and LPEI bound more tightly to the siRNA and delivered siRNA to the target cells more quickly and in apparently larger complexes. We conclude that our NPs, which have similar chemical functionality to LPEI, do not result in silencing due to their inability to form the higher order complexes required to deliver a sufficiently high initial dose of siRNA to achieve measurable silencing.
MATERIALS AND METHODS
Chemicals
All synthesis reagents were purchased from Aldrich (St. Louis, MO) or Alfa Aesar (Ward Hill, MA), and were ACS reagent grade. Lipofectamine 2000 (LF2K) was purchased from Invitrogen (Carlsbad, CA). Linear poly(ethyleneimine) (LPEI, Mw = 25 kDa) was obtained from Polysciences, Inc (Warrington, PA). A working solution for LPEI was prepared in Milli-Q water at a concentration of 2.5 mg/mL. The solution was acidified using HCl to aid in solubility. Once dissolved, the solution was readjusted to pH 7.0 using NaOH. All polymer solutions were filtered through 0.22 μm Millex-GV PVDF sterile filters (Millipore) prior to being added to cells. The siRNA (Dharmacon; Lafayette, CO) sequence used was: siRNA guide strand - GAU GAA CUU CAG GGU CAG CUU; siRNA passenger strand - GCU GAC CCU GAA GUU CAU CUU. The dsDNA (IDT; Coraville, IA) was constructed to have identical hybridization structure to the siRNA (19 nucleotides hybridized with 2 nucleotide overhangs on each 3’ end). The dsDNA sequence used was: dsDNA strand 1 - CCA CTA CCT GAG CAC CCA GTT; dsDNA strand 2 - CAG GGT GCT CAG GTA GTG GTT. For microscopy, the 5’-end of the siRNA passenger strand was labeled with Dy547. For the binding studies, the dsDNA strand 1 5’-end was labeled with 6-carboxyfluorescein (6-FAM). Tris/Boric Acid/EDTA running buffer and nucleic acid sample loading buffer were purchased from Bio-Rad (Hercules, CA).
Poly(propargyl glycolide) polymer synthesis
Prior to use, chloroform was dried over phosphorous pentoxide, dichloromethane was dried over CaH2, and THF was dried over sodium and benzophenone. 4,4-N,N-dimethylaminopyridine (DMAP) was purified by sublimation at 35-40°C. 2- hydroxyethyldisulfide was distilled under vacuum. All glassware was oven-dried.
Synthesis of poly(propargyl glycolide) with disulfide initiator (PGSSPG)(Hou, 2009)
An oven dried Schlenk flask was charged with propargyl glycolide (PGL) and DMAP, the flask was evacuated for at least two hours, then bis-(2-hydroxyethyl)disulfide initiator solution (0.11M in CHCl3) [M]/[I] 10:1 was added, the monomer concentration was adjusted to 0.5 M by adding dry chloroform. The reaction mixture was degassed by 3 freeze-pump-thaw cycles, then backfilled with nitrogen gas. The flask was immersed in an oil bath at 60°C and was stirred for 12hrs. After polymerization, the solvent was removed and the white stringy solid was redissolved in THF and dialyzed against THF/Water 3:1 in a 1000 MWCO cellulose membrane. The polymer was then dried under vacuum. Nuclear Magnetic Resonance (NMR) was performed in 500 MHz or 300 MHz spectrometer. FTIR spectra were taken in a Mattson Galaxy series FTIR 3000. 1H NMR (CDCl3) δ5.47-5.35 (br, 1H) 5.26-5.22 (br,1H) 4.52-4.36 (m, 6H) 3.0-2.66 (br m, 2H) 2.12-2.03 (br, 1H)
Synthesis of poly(propargyl glycolide-b-ethylene oxide monomethyl ether) PEG8PG
An ampoule was loaded with propargyl glycolide and stir bar. The ampoule was made from 3/8 in. diameter glass tubing was connected via a cajon fitting to a T-shape vacuum adapter with a stopcock and an air free Teflon valve. The apparatus was connected to vacuum and evacuated through the Teflon valve, and then it was backfilled with nitrogen gas. Predetermined amounts of Tin II 2-ethylhexanoate catalyst (51mM) in toluene and PEG 350 initiator (100 mM) in toluene were transferred to the ampoule through the stopcock. The solution was stirred for 30 minutes. The solvent was removed via vacuum and the ampoule was flame-sealed. The polymerization was carried out for 30 minutes at 130°C. The polymerization was quenched in ice water, the ampoule was broken and the residue was dissolved in CH2Cl2 to a 10% solution. A small amount was withdrawn to evaluate conversion via 1H NMR. The rest was precipitated into ice cold methanol and the polymer was filtered and dried under vacuum. 1H NMR (CDCl3) δ: 5.43-5.33 (br, 1H) 3.70-3.66 (q, 2H) 3.64-3.61(dd, 2H) 3.54-3.51 (q, 2H) 3.36 (s, 3H) 3.0-2.82 (m, 2H) 2.14-2.06 (br s, 1H)
Standard Click Chemistry Procedure
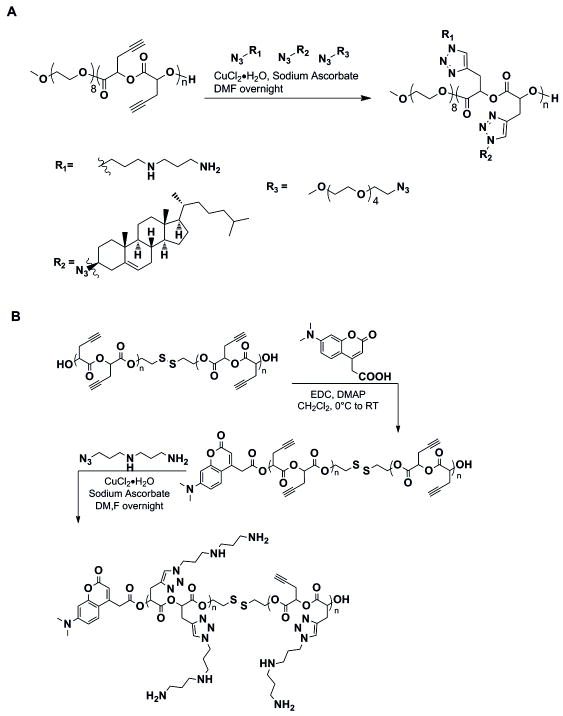
A schlenk flask was loaded with 40-60 mg of polymer, the desired mol % of azide ligand according to moles of alkyne in the polymer, and 24 mol% of sodium ascorbate. The mixture was dissolved in DMF and degassed by 3-4 freeze-pump-thaw cycles and backfilled with nitrogen gas. A 0.1 M solution of CuCl2·2H2O in DMF was added after degassing. The reaction mixture was stirred at room temperature overnight (Figure 1).
Figure 1. Click chemistry schematic.
A. The cholesterol particle was made using a long, continuous propargyl glycolide (n=45) backbone with PEG endgroups. The azide-alkene click reaction was used to add the primary amine, secondary amine, and cholesterol ligands. B. The DMACA tagged disulfide particle is synthesized by first adding the fluorescent end group onto two shorter propargyl glycolide backbones (n=5) linked by a disulfide bond. The amine ligand was added to 80% of the available alkynes.
Cholesteryl Iodide (R)
Modified procedure from Posner et al (1976) 1H NMR (CDCl3) 5.31 (t, 1H), 4.02 (m, 1H), 2.91 (t, 1H), 2.69-2.60 (dq, 1H) 2.32-1.03 (m, 24H), 1.02 (s, 3H), 0.97 (m, 2H), 0.89 (d, 3H), 0.84 (dd, 6H), 0.65 (s, 3H).
Cholesteryl Azide (S)
Cholesteryl iodide (2mmol, 1.03g) was combined with sodium azide (4mmol, 0.65g) and dissolved in 10 ml DMF. The reaction mixture was stirred at 80°C overnight. The solvent was removed under high vacuum. The solid was redissolved in diethyl ether and washed with water, sodium metabisulfite (Na2S2O3) and NaCl. The organic layer was dried over MgSO4. The solid was recrystallized from absolute ethanol (60 % yield) 1H NMR (CDCl3) δ: 5.38 (t, 1H), 3.86 (t, 1H), 2.54-2.45 (m, 1H), 2.21-2.13 (1H), 2.04-2.02 (2H), 1.86-1.02 (24H), 0.99 (s, 3H), 0.90 (d, 3H), 0.85 (dd, 6H), 0.66 (s, 3H)) 13C NMR (CDCl3) 138.08, 123.17, 58.28, 56.7, 56.1, 49.9, 42.3, 39.7, 39.5, 37.1, 36.2, 36.0, 35.8, 33.6, 31.82, 31.79, 28.2, 28.0, 26.1, 24.26, 23.8, 22.8, 22.5, 20.7, 18.9, 18.7, 11.8 FTIR 2080 cm-1 N3
Polymer Binding Gels
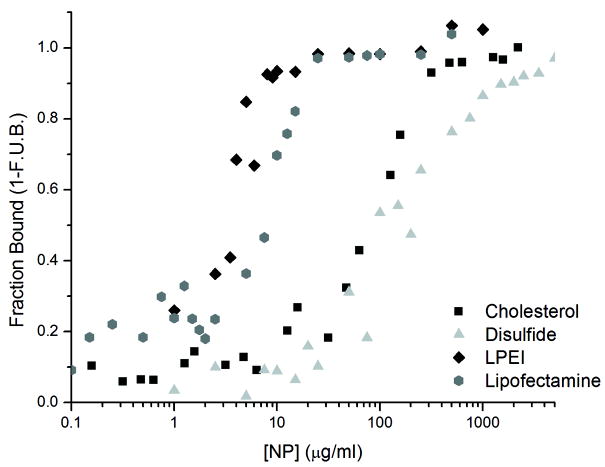
Solutions were prepared in Milli-Q water using 200nM dsDNA. To each sample, polymer was added and allowed to mix for 15 minutes. Bound complexes were resolved on 0.8% agarose gels. Signal intensities of free dsDNA were quantified from gel images and normalized to dsDNA only control lanes. Example: Fraction Bound = 1 − (intensity of unbound dsDNA from NP-containing lane/intensity of unbound dsDNA from lane without NP). A single plot of fraction bound versus NP concentration (in μg/ml) was constructed from all of the data from two independent experiments (Figure 2). The combined data were simultaneously fit by a modified form of the Hill equation (Hill, 1910) (equation 1) to obtain values for the binding constant (K) and a value to capture binding cooperativity (n) (Table 1). The K value reflects binding at 15min, which is not necessarily equivalent to the equilibrium dissociation constant since the time required to reach equilibrium varies with the NP composition and can approach up to ~24hrs for some NPs (data not shown). An n value greater than 1 indicates the possibility of cooperative binding.
Figure 2. Polymer/Nucleic Acid binding.
A graph of the fraction of dsDNA bound at varying polymer concentrations. The dsDNA concentration was maintained at 200nM, and the fraction bound to each particle was calculated as a mass balance from the amount of free dsDNA resolved on the gel.
Table 1. Hill Equation Parameters.
Fractional dsDNA binding was fit to a modified form of the Hill equation. The binding constant (K) reflects binding achieved at 15min. An exponential (n) greater than 1 is indicative of cooperative binding. The K values were compared using a two-tailed Student’s t-test for unequal variances and all four values were statistically significantly different at α=0.01.
| Hill Equation Parameters | LF2K | LPEI | Cholesterol NP | Disulfide NP |
|---|---|---|---|---|
| K (μg/mL) | 4.9 | 3.0 | 68 | 151 |
| error (μg/mL) | 0.7 | 0.4 | 8 | 14 |
| n (cooperativity) | 0.9 | 1.8 | 1.1 | 0.9 |
| error | 0.1 | 0.4 | 0.1 | 0.1 |
| (1) |
The K values were compared using a two-tailed Student’s t-test for unequal variances with α=0.01.
Cell Culture
Human lung carcinoma cells (H1299) constitutively expressing EGFP were generously provided by Dr. Jørgen Kjems, University of Aarhus, Denmark. Cell culture media was prepared with Dulbecco’s Modified Eagle’s Medium High Glucose (DMEM, #11965, Invitrogen) supplemented with 10% fetal bovine serum (FBS, GIBCO), 1% penicillin-streptomycin (GIBCO), and 1% Geneticin (GIBCO). Cells were maintained in an incubator at 37°C, 5% CO2, 100% relative humidity and subcultured approximately weekly by trypsinization. Leibovitz Medium (L-15, Sigma, St. Louis, MO) was used for cell imaging. All experiments were performed using cells with passages earlier than 20.
Transfection
Opti-Mem (GIBCO) was used for all transfections. H1299-EGFP cells were seeded in 4-well Nunc Lab-Tek chambered coverglasses (Fisher Scientific, Pittsburgh, PA) at a density of 37,500 cells/well in 0.5 mL of complete media without antibiotics. After 24 h, treatments were initiated in a staggered fashion so that all samples would be ready for processing and imaging at the same time. In other words, the 24 h time points were started first, with the 16 h points started 8 hours later, etc. 100 μL solutions of siRNA-NP were prepared in Opti-Mem and allowed to mix for 20 minutes prior to their addition to cells at final concentrations of 100 nM siRNA and 100 μg/mL NP. LPEI polyplexes and LF2K lipoplexes were prepared in a final volume of 600 μL at concentrations of 100 nM siRNA and either 10μg/mL LPEI or 2.3 μg/mL LF2K (1.4 μL of 1μg/μL stock solution). Cells were incubated in the transfection solutions for 24h, 16h, 8h, and 4h prior to imaging.
Confocal Microscopy
Immediately prior to imaging, the cell media was removed, the cells were rinsed, and 0.5mL of L-15 media were added. Confocal imaging was performed on an Olympus FluoView 1000 Inverted IX81 microscope, using a 40x oil objective. Images were taken sequentially, using a Kalman average of 2x. DMACA tagged particles were excited using a 405 nm diode laser and detected through a BA430-470nm emission filter. EGFP fluorescence was assayed using an excitation of 488 nm with a multi-line Argon laser and detected through a BA505-525nm emission filter. Dy547 fluorescence (siRNA) was excited at 543 nm by a HeNe laser and detected through a BA560-IFnm emission filter. The focal plane selected for each image was based on the highest intensity of green fluorescence. The line intensity graphs were constructed from the 4h, 160x magnification images. A line was drawn lengthwise across the cell so that it would intersect with the areas of highest intensity red fluorescence.
RESULTS
Polymer Binding Gels
It was first important to understand the association of our polymeric NPs with the siRNAs. To do this, we performed in vitro binding experiments using electrophoresis as the readout (Figure 2). For our binding experiments, a 21 base pair dsDNA of analogous structure to the GFP-targeting siRNA was used to help minimize cost. Both LPEI and LF2K showed markedly stronger binding to the dsDNA than either the Cholesterol NP or the disulfide NP (Figure 2 and Table 1). Only in the case of LPEI did the equation suggest possible binding cooperativity (n = 1.8). The strength of the binding interaction is important to the overall efficacy of the siRNA. Vehicles that form less-stable complexes will have to be used in greater excess concentration to ensure complete encapsulation of the siRNA cargo. Thus, based on our results, when performing cellular uptake experiments, we chose to use 10μg/mL LPEI and 100μg/mL polymer concentrations to maximize siRNA encapsulation while minimizing excess polymer concentration.
Cellular Uptake
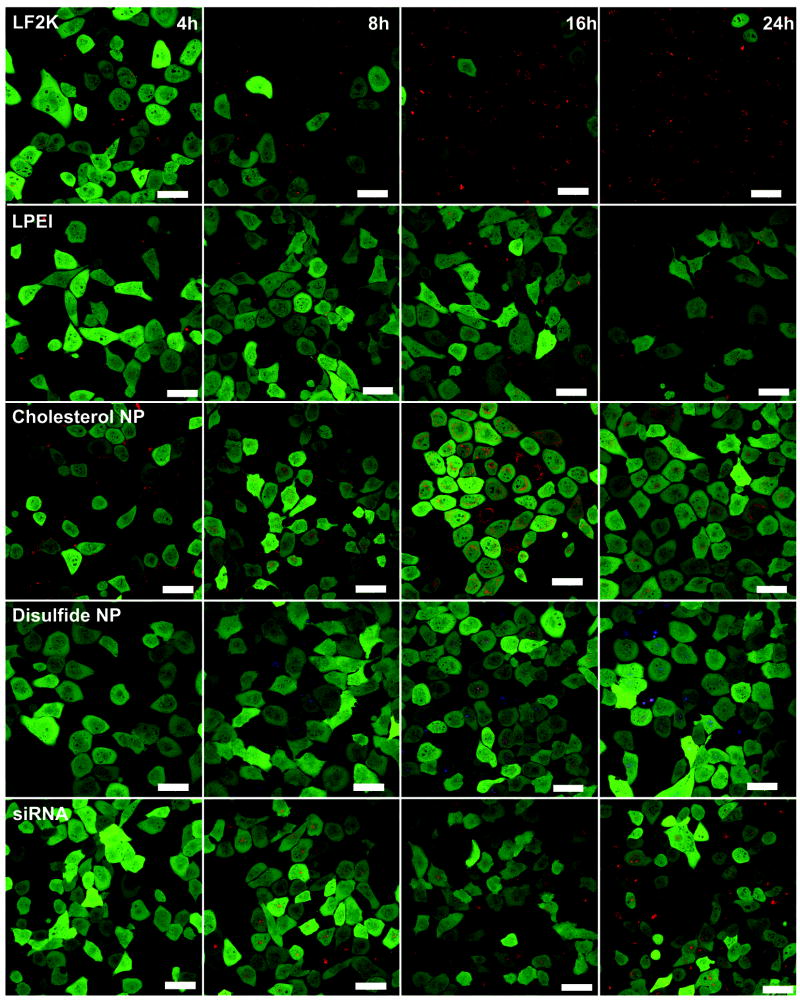
We monitored the delivery of polymer/siRNA complexes into GFP expressing H1299 cells over time using confocal microscopy (Figure 3). As early as 4h, both LF2K and LPEI begin to show intracellular accumulation. The disulfide NP and siRNA only cell populations do not show any intracellular siRNA signal, while the cholesterol NP shows minimal intracellular signal with significant extracellular accumulation around the cell membrane, perhaps due to increased aggregation and sedimentation of the hydrophobic cholesterol NP as compared to the other polymers. It should be noted that at this early time point, the siRNA signal from the LF2K- and LPEI-treated cells arises primarily from points of concentrated fluorescence (Figure 4), suggesting that these vehicles form structures that deliver multiple siRNAs simultaneously to a single cell.
Figure 3. Time series Microscopy pictures.
Confocal images of the uptake of various polymer/EGFPsiRNA complexes into GFP expressing H1299 cells. Pictures were taken at 4, 8, 16, and 24h time points. The green fluorescence represents the GFP expressing cells and the red fluorescence is of the Dy547-tagged siRNA. In the case of the disulfide particle, blue fluorescence indicates the presence of the particle. The scale bars are 50μm.
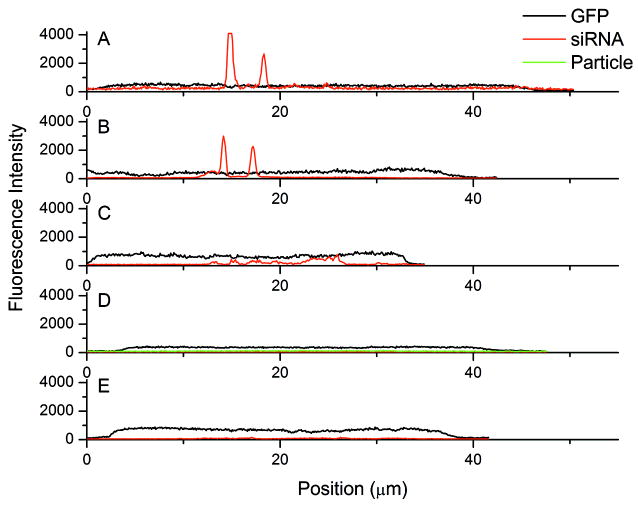
Figure 4. 4h line intensity graphs.
Fluorescence intensity graphs measured across individual cells at 4h after addition of LPEI (A), LF2K(B), Cholesterol NP (C), and Disulfide NP (D) complexes, as well as siRNA alone (E). For each image, the line was constructed by drawing across the longer diameter that intersected the areas of highest intensity red fluorescence. The GFP signal is indicative of the cell area. There was no visible siRNA uptake for the disulfide NPs or siRNA alone at the 4h time point.
By 8h, siRNA uptake can be seen for all 4 delivery vehicles and the siRNA only treatments (Figure 3). For LF2K and LPEI, the intracellular siRNA signal is still primarily concentrated in distinct intracellular locations. In contrast, the cholesterol and disulfide NPs show a more even siRNA distribution throughout the cell. This further suggests the LPEI and LF2K complexes that enter the cell are more densely packaged than those formed by the cholesterol and disulfide NPs. It is notable that the uptake and distribution pattern after delivery by the cholesterol and disulfide NPs more closely resembles that of the siRNA only treatment than those of the LF2K and LPEI.
After 16h, the LF2K-treated cells begin to show silencing of the GFP. Additionally, the siRNA has spread more uniformly throughout the cells. In the case of the LPEI treatment, changes in the cell morphology can be seen, indicative of the cytotoxicity of LPEI. As with the LF2K, the intracellular siRNA distribution has become more uniform throughout the cell. The cholesterol and disulfide particles, along with the siRNA only treatment, show widespread delivery of the siRNA throughout the cells but, in none of the cases, do the siRNAs significantly silence GFP. However, unlike the LPEI, our NPs show no effect on cell morphology, indicating far less cytotoxicity despite their higher concentration relative to LPEI (100 μg/mL vs. 10 μg/mL). Interestingly, the siRNAs delivered by the cholesterol NP appear to accumulate preferentially around the nuclei of the cells. This could be attributed to the hydrophobicity of the cholesterol causing it to associate with the nuclear membrane. For the disulfide particle, blue areas can also be seen inside the cells apart from the overlapping red siRNA signal. Due to the low binding affinity of this particle, it is likely that uncomplexed particles can also be taken up by the cell, potentially resulting in competition for uptake with complexed particles. Side view images and three dimensional rotational videos constructed from z-section images at this time point are available online (Figure S1 and Videos S1-S5, Supporting Information).
By 24h, it appears that two populations of cells have been generated by LF2K treatment, those cells that received siRNA and have had GFP nearly completely silenced and those cells that did not receive siRNA and have basal GFP expression. For the LPEI, a more graded silencing can be seen where more cells have intermediate fluorescence intensity. Additionally, the changes in morphology are more pronounced, with cells appearing more spindly and less regular in shape. For the cholesterol and disulfide NPs, despite the continuing accumulation of siRNA, there is no significant silencing. The same can be seen for the siRNA only images, where widespread uptake is seen without silencing. In some cells, the disulfide particle colocalizes with the siRNA, but this does not occur consistently across the cell population.
Intensity Analysis
To confirm the siRNA packaging found in LF2K and LPEI transfected cells are more highly concentrated as compared to the cholesterol NPs at the 4h time point, line intensity graphs were constructed across selected cells from the 160x magnification images (Figure 4). The lines were drawn lengthwise across regions of single cells where the siRNA-polymer complexes are present. In both the LF2K and LPEI graphs, there are highly localized regions of strong fluorescence from the siRNA. These fluorescence peaks are suggestive of the presence of multiple siRNAs at these points, likely still complexed with the vehicles. In contrast, the maximal siRNA signal when delivered using the cholesterol NPs, the only other polymeric delivery technique with any intracellular signal at this time point, is three- to six-fold lower than the signal from the LF2K and LPEI. The cholesterol complexes also show multiple siRNA peaks but at a much lower intensity. Because eventual silencing occurs for both the LF2K and LPEI, this suggests that the higher initial concentration of siRNA is necessary to initiate silencing.
DISCUSSION
Confocal microscopy is a valuable tool for analyzing the delivery and function of siRNAs to cultured cells at the resolution of a single cell. As we found, the uptake of siRNAs delivered by different vehicles can vary by rate, fraction of cells loaded, and the apparent structure of the complexes taken up. Moreover, we were able to identify, particularly for delivery using LF2K, that multiple cell populations could arise within a treatment group and that the distribution of the siRNAs within a cell changed markedly over time. Bulk measurements like western blotting or average fluorescence would have masked these important details of the delivery process. Additionally, we were able to visualize changes in cell morphology as a result of LPEI exposure, changes that did not occur upon exposure to our polymers. Combining this information improved our understanding of the siRNA delivery process and will greatly enhance our future ability to design more efficient vehicles for siRNA delivery based on our polymer system.
Many studies have reported on key compositional elements that are common among effective vehicles (Breunig, 2008; Green, 2008; Liu, 2005; Ou 2008; Rozema, 2007). Our binding data support that chemical composition is critical in forming complexes that can achieve silencing, with the more tightly bound complexes resulting in more active silencing (Figures 2 and 3). However, vehicles containing the key compositional features are not guaranteed to result in silencing when used for siRNA delivery (Green, 2008). Our data support that a fundamental aspect to achieving silencing may be in the physical structures of the vehicle-siRNA complexes that form, not just their chemical compositions.
These structures result from both the interaction of the vehicle with the siRNA but also from the aggregation of the complexes into higher order structures. In this way, the cooperativity of the interaction may also be a defining characteristic for the eventual function of the siRNAs. Elucidating the structure of the siRNA-vehicle complexes can be difficult. Modeling approaches could be used to predict the structures of polymer-siRNA complexes, in a similar manner to models used for predicting protein/peptide aggregation (Tjernberg, 1999). Several experimental techniques have proven useful for measuring the changes in the polymer complexes with time, concentration, and altered vehicle chemistry. These include dynamic light scattering (DLS) (Breunig, 2008; Ou, 2008), transmission electron microscopy (TEM) (Cohen, 2000), scanning electron microscopy (SEM) (Mehrotra, 2009), and atomic force microscopy (AFM) (Shifrina, 2009).
While siRNA delivery by our polymeric NPs did not result in silencing, we were encouraged that our polymers did not show any apparent toxicity, even at concentrations far in excess of that required for LPEI delivery. As we go forward in designing vehicles for siRNA delivery, we will place additional emphasis on understanding the structures formed from the vehicle-siRNA complexes. Moreover, we are investigating strategies for modifying our polymers to improve their binding with the siRNA cargo. Possible means to achieve this include covalent coupling of multiple siRNA to a single polymer and adding side-chains with greater positive charge density to improve the electrostatic attraction between the polymer and the siRNA. In all cases, confocal microscopy will serve as an essential tool for determining the siRNA uptake profiles for our various polymers.
Supplementary Material
Acknowledgments
We thank all the members of the Cellular and Biomolecular Laboratory (http://www.egr.msu.edu/cbl/) for their advice and support and Dr. Jørgen Kjems for providing us with the EGFP cells. Financial support for this work was provided in part by the MSU Foundation, National Institutes of Health (#GM079688, #RR024439), National Science Foundation (CBET 0941055), MUCI, and the Center for Systems Biology.
References
- Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nuc Acid Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhna SY, Supekova L, Supek F, Schultz PG, Deniz AA. siRNA in human cells selectively localizes to target RNA sites. PNAS. 2006;103:7682–7687. doi: 10.1073/pnas.0600148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig M, Hoza C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Burke RS, Pun SH. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconj Chem. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- Cardoso ALC, Simoes S, deAlmeida LP, Pelisek J, Culmsee C, Wagner E, Pedroso de Lima MC. siRNA delivery by a transferrin-associated lipid-based vector: a non-viral strategy to mediate gene silencing. J Gene Med. 2007;9:170–183. doi: 10.1002/jgm.1006. [DOI] [PubMed] [Google Scholar]
- Chen AM, Santhakumaran LM, Nair SK, Amenta PS, Thomas T, He H, Thomas TJ. Oligodeoxynucleotide nanostructure formation in the presence of polypropyleneimine dendrimers and their updake in breask cancer cells. Nanotechnology. 2006;17:5449–5460. [Google Scholar]
- Cohen H, Levy RJ, Gao J, Fishbein I, Kousaev V, Sosnowski S, Slomkowsk S, Golomb G. Sustained delivery and expression of DNA encapsulated in polymeric nanoparticles. Gene Ther. 2000;7:1896–1905. doi: 10.1038/sj.gt.3301318. [DOI] [PubMed] [Google Scholar]
- Farrell LL, Pepin J, Kucharski C, Lin X, Xu Z, Uludag H. A comparison of the effectiveness of cationic polymers poly-L-lysine (PLL) and polyethylenimine (PEI) for non-viral delivery of plasmid DNA to bone marrow stromal cells (BMSC) Eur J Pharm Biopharm. 2007;65:388–397. doi: 10.1016/j.ejpb.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Gary DJ, Puri N, Won Y-Y. Polymer-based siRNA delivery: Perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Gopinath P, Ramesh A. Adenoviral Vectors: A promising tool for gene therapy. App Biochem Biotech. 2006;133:9–29. doi: 10.1385/abab:133:1:9. [DOI] [PubMed] [Google Scholar]
- Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Phys. 1910;40:iv–vii. [Google Scholar]
- Hong K, Zheng W, Baker A, Papahadjopoulos D. Stabilization of cationic liposome-plasmid DNA complexes by polyamines and poly(ethylene glycol)-phospholipid conjugates for efficient in vivo gene delivery. FEBS Let. 1997;400:223–237. doi: 10.1016/s0014-5793(96)01397-x. [DOI] [PubMed] [Google Scholar]
- Hou X, Li Q, He Y, Jia L, Li Y, Zhu Y, Cao AJ. Visualization of spontaneous aggregates by diblock poly(styrene)-b-poly(L-lactide)/poly(D-lactide) pairs in solution with new fluorescent CdSe quantum dot labels. J Polym Sci Part B: Polym Phys. 2009;47:1393–1405. [Google Scholar]
- Hu P, Quick GK, Yeo Y. Gene delivery through the use of a hyaluronate-associated intracellulary degradable crosslinked polyethyleneimine. Biomat. 2009;30:5834–5843. doi: 10.1016/j.biomaterials.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Vogel EB, Smith MR, III, Baker GL. “Clickable” polyglycolides: Tunable synthons for thermoresponsive degradable polymers. Macromolecules. 2008;41:1937–1944. [Google Scholar]
- Kaiser J. Death prompts a review of gene therapy vector. Science. 2007;317:580. doi: 10.1126/science.317.5838.580. [DOI] [PubMed] [Google Scholar]
- Liu Y, Reineke TM. Hydroxyl Stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. JACS. 2005;127:3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- Lynn DM, Langer R. Degradable poly(β-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. JACS. 2000;122:10761–10768. [Google Scholar]
- Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physiochemical and biological properties of poly(ethylene imine)-graft-Poly(ethylene glycol) block copolymer/siRNA polyplexes. Bioconj Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, Lee I, Chan C. Multilayer mediated forward and patterned siRNA transfection using linear-PEI at extended N/P ratios. Acta Biomaterialia. 2009;5:1474–1488. doi: 10.1016/j.actbio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kiyono T, Imabayashi H, Takeda Y, Tsuchiya K, Miyoshi S, Makino H, Matsumoto K, Saito H, Ogawa S, Sakamoto M, Hata J-I, Umezawa A. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol Cell Biol. 2005;25:5183–5195. doi: 10.1128/MCB.25.12.5183-5195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel AE, Madler L, Velego D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nature Materials. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- Nguyen J, Steele TWJ, Merkel O, Reul R, Kissel T. Fast degrading polyesters as siRNA nano-carriers for pulmonary gene therapy. J Control Release. 2008;132:243–251. doi: 10.1016/j.jconrel.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou M, Wang X-L, Xu R, Chang C-W, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconj Chem. 2008;19:626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- Peng D, Qian C, Sun Y, Barajas MA, Prieto J. Transduction of hepatocellular carcinoma (HCC) using recombinant adeno-associated virus (rAAV): in vitro and in vivo effects of genotoxic agents. J Hepatol. 2000;32:975–985. doi: 10.1016/s0168-8278(00)80102-6. [DOI] [PubMed] [Google Scholar]
- Posner GH, Ting JS, Lentz CM. A mechanistic and synthetic study of organocopper substitution reactions with some homoallylic and cyclopropylcarbinyl substrates: Application to isoprenoid synthesis. Tetrahedron. 1976;32:2281–2287. [Google Scholar]
- Que-Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Blokhin AV, Hagstrom JE, Wolff JA. Dynamic polyconjugates for targeted in vivo delivery of siRNA to hepatocytes. PNAS. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifrina ZB, Kuchkina NV, Rutkevich PN, Vlasik TN, Sushko AD, Izumrudov VA. Water-soluble cationic aromatic dendrimers and their complexation with DNA. Macromolecules. 2009;42:9548–9560. [Google Scholar]
- Spagnou S, Miller AD, Keller M. Lipidic Carriers of siRNA: Differences in the formulation, cellular uptake and delivery with plasmid DNA. Biochem. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- Tang MX, Redemann CT, Szoka FC., Jr In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconj Chem. 1996;7:703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- Thierry AR, Lunardi-Iskandar Y, Bryant JL, Rabinovich P, Gallo RC, Mahan LC. Systemic gene therapy:biodistribution and long-term expression of a transgene in mice. PNAS. 1995;92:9742–9748. doi: 10.1073/pnas.92.21.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjernberg LO, Callaway DJE, Tjernberg A, Hahne S, Lilliehook C, Terenius L, Thyberg J, Nordstedt C. A molecular model of Alzheimer amyloid β-peptide fibril formation. J Biol Chem. 1999;274:12619–12625. doi: 10.1074/jbc.274.18.12619. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: Advances in siRNA delivery. Nat Rev. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X-B, Uludag H, Lavasanifar A. Biodegradable amphiphilic poly(ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nanocarriers for efficient siRNA delivery. Biomat. 2009;30:242–253. doi: 10.1016/j.biomaterials.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhao B, Jiang H, Wang B, Ma B. Cationic lipids and polymers mediated vectors for delivery of siRNA. J Control Release. 2007;123:1–10. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconj Chem. 2008;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.