Summary
Phosphorylation of proteins is a predominant reversible post-translational modification. It is central to a wide variety of physiological responses and signaling mechanisms. Recent advances have allowed the global scope of phosphorylation to be addressed by mass spectrometry using phosphoproteomic approaches. In this perspective we discuss four aspects of phosphoproteomics; namely insights and implications from recently published phosphoproteomic studies, and applications and limitations of current phosphoproteomic strategies. As about 50,000 known phosphorylation sites do not yet have any ascribed function, we present our perspectives on a major function of protein phosphorylation that may be of predictive value in hypothesis based investigations. Finally we discuss strategies to measure stoichiometry of phosphorylation in a proteome-wide manner which is not provided by current phosphoproteomic approaches.
Keywords: protein phosphorylation, phosphoproteomics, quantitative mass spectrometry, protein-protein interaction, stoichiometry of phosphorylation, kinases and phosphatases, cell signaling, cancer
Introduction
Phosphorylation of proteins is central to a wide variety of metabolic, hormonal, developmental, and stress responses [1]. It is an extensively employed mode of signal transduction, frequently involving cascades of phosphorylation events among kinases [2]. Importantly, protein phosphorylation has been a cornerstone in accelerating our understanding of health and disease [3,4]. Further, protein kinases have gained increasing focus as therapeutic targets for the treatment of cancer and chronic inflammatory diseases [5,6]. Accordingly, over the last decade and half, analysis of protein phosphorylation has been of significant interest to the practitioners of mass spectrometry [7]. Several approaches have been developed for the identification of phosphorylation sites [8–11]. These approaches take advantage of different types of chemistries for enrichment of phosphorylated peptides/proteins, and different technologies for ionization, fragmentation and mass-analysis of phosphorylated peptides. A general schematic for the phosphorylation analysis by mass spectrometry is provided in Figure 1. We point the reader to several excellent reviews on the details of methodological aspects [9,12–15].
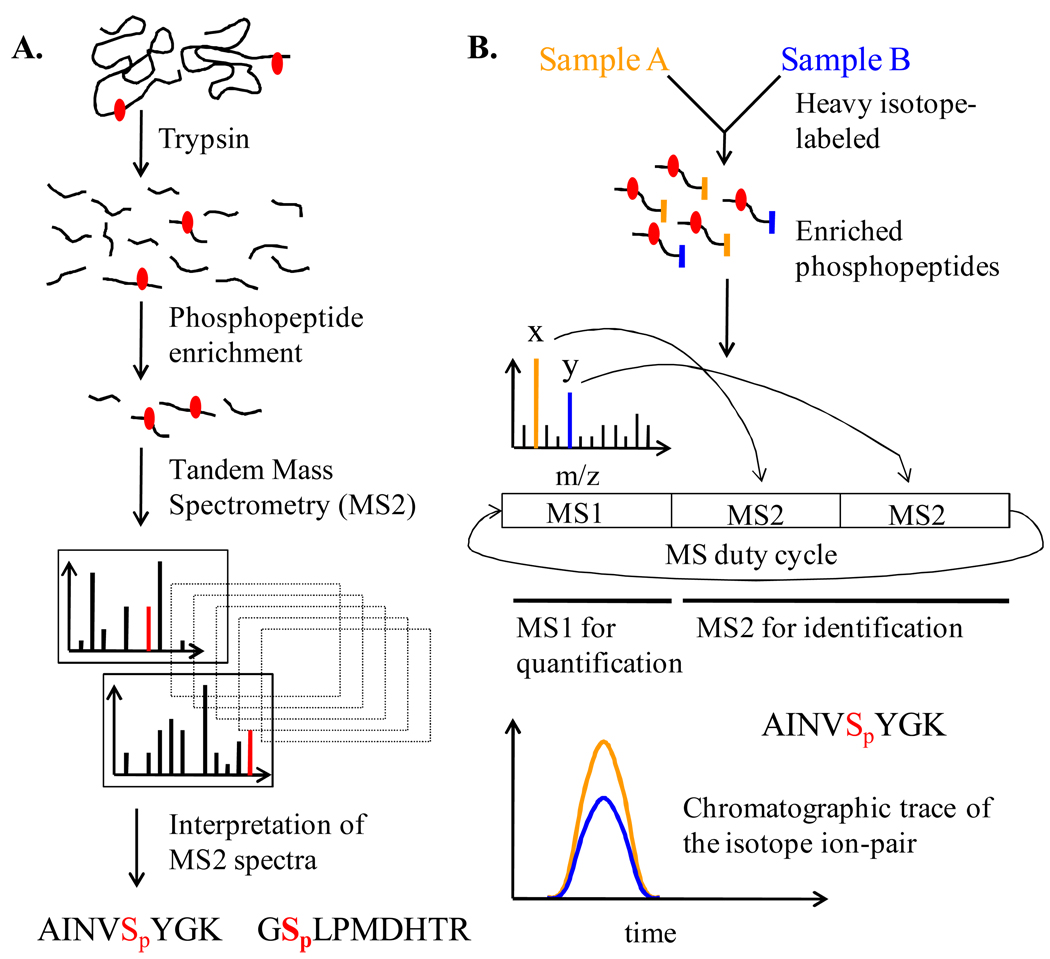
Figure 1. Generalized flowchart of important steps in the analysis of phosphorylation by mass spectrometry.
(A) Flowchart of important steps in the identification of phosphorylation sites by mass spectrometry. The red dot represents a phosphorylation site in the proteins/peptides. Interpretation of MS2 spectra involves both the identification of phosphopeptides and localization of the sites of phosphorylation. The red lines in the product ion (MS2) spectra represent site-determining ions. Note that variations and enhancements to this generalized procedure exist. (B) Schematic of a generalized approach to quantitative phosphoproteomics. Quantification of fold-change in phosphorylation relies on the application of isotope-dilution principle. One sample is heavy isotope labeled and mixed with the other sample for relative quantification. Natural and heavy isotope labels have been distinguished by using orange and blue colored codes. Data-dependent scheme wherein the two most intense ions (x and y that represent the isotope-ion pair) detected in the survey scan (MS1) are selected for triggering fragmentation and MS2 scans is shown. This duty cycle is continued over the time-course of chromatography. Ion trace of the isotope pair from the MS1 scan is used for calculating relative levels of the phosphopeptides and thereby fold-change in phosphorylation.
Phosphorylation of proteins is one of the most predominant reversible post-translational modifications in eukaryotes [1]. The human genome encodes 518 protein kinases, encompassing 1.7% of the genome [16]. Based on traditional double radiolabeling experiments, it was estimated that nearly a third of all encoded proteins are phosphorylated in mammalian cells [1,16]. The global scope of phosphorylation can now be addressed by mass spectrometry as a result of more recent advancements such as sequential enrichment strategies, assessment of false-discovery rate of identification, and automated assignment of site localization in phosphopeptides [17–21]. This has paved the way to a new sub-discipline called phosphoproteomics. In this article, we summarize general insights gained from the phosphoproteomic studies. We highlight applications of phosphoproteomics in making advances in cell biology and in settings of direct clinical relevance. We also offer our perspectives on the fundamental functions of protein phosphorylation. We present arguments and observations that point to modulation of protein-protein interaction (PPI) as a widespread function of serine/threonine phosphorylation. Finally, we point out that lack of information on stoichiometry of phosphorylation is a key limitation of the current phosphoproteomic approaches and present our thoughts on how to measure it in a proteome-wide manner.
Global trends observed in phosphoproteomic datasets
To date, large phosphoproteomic datasets with thousands of novel phosphorylation sites have been compiled for different model organisms and human cell lines [10,19,20,22–28]. Bioinformatic analyses of these phosphoproteomic datasets point to several global trends. We discuss the major global trends consistently observed in multiple datasets or by combining multiple datasets from different model organisms or biological sources. We propose that these trends are general, emergent features of most eukaryotic phosphoproteomes. We have been careful in generalizing the global trends observed in the different phosphoproteomic datasets as several factors influence the final compilation of identified phosphopeptides and phosphorylation sites.
Linear stretch of residues around the site of phosphorylation is a major determinant of kinase specificity towards protein substrates [29]. Consensus linear recognition motifs have been defined for many kinases and kinase sub-families [30]. Phosphoproteomic datasets have been classified based on these kinase-recognition motifs. Such classification has revealed that proline-directed, acidiphilic and basophilic phosphorylation sites are the predominant classes in eukaryotic cells [19,20,24,26–28]. Which among these is the most predominant is likely dependent on the particular biological state and thereby the activity status of kinases in the sample used for analysis. Similarly, further sub-classifications based on kinase recognition motifs do not show consistent trends across multiple datasets. The choice of different methodologies has also contributed to lack of consistent trends [10,22,26,28]. For example, enrichment of phosphopeptides using Titanium dioxide (TiO2) can be influenced by the presence of acidic and basic residues more than the other methods [25,31]. Further, immobilized metal affinity chromatography (IMAC) enriches multi-phosphorylated peptides better than TiO2 and phosphoramidate chemistry (PAC) [9,31]. In contrast, PAC and TiO2 enrich mono-phosphorylated peptides better than IMAC [9,31]. Similarly, evidence suggests that the use of endo-LysC and electron-transfer dissociation (ETD) will make more basophilic phosphorylation sites amenable for identification than the use of trypsin and collision activated dissociation (CAD) [10]. Thus different approaches for phosphoproteomic analysis are complementary and a compilation from multiple approaches is regarded more comprehensive [25,31–33].
Protein kinases themselves are the most over-represented class of phosphoproteins in multiple model organisms [10,22,24]. Similarly, more transcriptional regulators are targeted for phosphorylation than expected from their representation in the genomes of model organisms [10,22,24]. Both these observations are consistent with the classical signaling paradigm wherein phosphorylation cascades control the activation of multiple kinases culminating in the activation of transcription factors. In HeLa and Jurkat T-cell lines, more nuclear proteins tend to be targeted for phosphorylation than proteins localized in other sub-cellular organelles [19,24].
Both in yeast and in the Jurkat T-cell line, phosphoproteins participate in more PPIs overall and among themselves than expected from randomly chosen proteins irrespective of their phosphorylation status [10,24]. Network analyses of PPI datasets assembled from accumulated biological literature and large-scale yeast two-hybrid and affinity pull-down experiments were used to arrive at these conclusions. The observed correlation between phosphorylation and PPI has implications for signal transduction research and will be discussed further in a later section.
Several research articles and reviews on phosphoproteomics have implied that phosphorylation often occurs on signaling proteins of low abundance [9,34–36]. But phosphoproteomic datasets from yeast, drosophila, and the Jurkat T-cell line are not over-represented in proteins of lower abundance [10,22,24]. Instead, the profile of abundance distribution is similar for the phosphoproteome and the proteome. Interestingly, a recent bioinformatic analysis indicates that proteins of lower abundance in human cells tend to be phosphorylated at more sites than those of higher abundance [37]. However, this observation needs to be tested more rigorously in multiple model organisms.
The knowledge that linear stretch of residues around the site of phosphorylation is a major determinant of kinase specificity has structural implications [30]. This knowledge posits that phosphorylation tends to occur in unstructured regions of proteins. Accordingly, it has been observed that ends of proteins, ends of structured domains, and loops and hinges in protein structures, as well as large disordered stretches have more phosphorylation sites [20,38–40]. Also, serine/threonine (S/T) phosphorylation sites are located more on solvent-accessible surface residues [38,39]. A recent analysis also noted that phosphorylation sites tend to congregate around each other on the surface of multi-phosphorylated proteins [37,40].
Multiple studies have recently shown that S/T phosphorylation sites are conserved to a greater extent than expected for S/T residues irrespective of known phosphorylations [41–43]. These studies have used different sequence alignment approaches and statistical methods to investigate evolutionary conservation of phosphorylation sites. This evidence on conservation of phosphorylation sites further emphasizes the critical role of phosphorylation in physiology and also validates the efforts in the development of technologies for phosphoproteomic analyses.
Yachie et al. found that phosphoproteins in yeast and mammals followed a power-law distribution with respect to the number of phosphorylation sites [37]. A power-law distribution reflects that most proteins have few phosphorylation sites whereas a few proteins have disproportionately high number of phosphorylation sites. The observation of power-law distribution in the number of phosphorylation sites predicts that proteins with more phosphorylation sites tend to accumulate even more phosphorylation sites during evolution.
Availability of large-scale data has spawned and driven several sub-disciplines of bioinformatics in the past. On the evidence of investigations on the global trends discussed above, we anticipate that phosphoproteomic datasets will continue to feed bioinformatic analyses leading to new insights on the biology of protein phosphorylation. For example, even as this review was being compiled, important insights on the evolutionary aspects of phosphorylation based regulation have been obtained by novel bioinformatic analyses in the recent months [44–47]. Similarly, most of the global trends discussed above hold true mostly for S/T phosphorylation and focused bioinformatic analysis of tyrosine phosphorylation sites are yet to be published.
Insights and implications from quantitative phosphoproteomic studies
Quantitative mass spectrometric techniques involving heavy isotope labeling have been adapted to quantify changes in the abundance of phosphopeptides and thereby changes in the extent of phosphorylation on identified phosphopeptides (Figure 1B). This approach has been particularly fruitful in the study of cell signaling systems, identification of substrates for kinases, and comparative analysis of oncogenic signaling. We discuss insights from these quantitative phosphoproteomic studies.
Growth Factor signaling
Olsen et al. captured the temporal dynamics of the phosphoproteome in response to Epidermal Growth Factor (EGF) stimulation in HeLa cells [19]. Nearly a thousand phosphopeptides showed more than two-fold change in abundance over the five early time-points represented by different heavy isotope labels. This study revealed that the scope of stimulus-dependent changes in phosphorylation is much more than previously appreciated, both in terms of classes of proteins and number of proteins in each class. Apart from a large number of kinases, phosphatases, and transcription factors, the authors observed changes in phosphorylation on small GTPases, GTPase modulators, actin-binding proteins, RNA binding proteins and ubiquitin ligases. This study, along with others, also points to a prevalence of multisite phosphorylation during signaling [23,24].
TCR signaling
We have used quantitative phosphoproteomic strategies to identify novel TCR-responsive phosphorylation sites and gain insights on T Cell Receptor (TCR) signaling [24]. TCR signaling is central to maturation of T cells in the thymus and multiple aspects of the adaptive immune response. Using the Jurkat T cell line as a model system, we identified 10,665 unique phosphorylation sites, of which 696 showed TCR-responsive changes. Among these TCR-responsive sites 70 were tyrosine phosphorylation sites. Our major finding is that TCR-responsive phosphorylation extensively targets proteins involved in all of the salient T cell activation-associated cellular phenomena: patterning of certain cell-surface proteins, TCR endocytosis, formation of F-actin cup, activation of integrin by ‘inside-out’ mechanism, polarization of microtubules, alternative splicing, and cytokine transcription [24]. Although considerable understanding has come about on the regulation of cytokine transcription, little is known about how all the other associated phenomena are initiated and coordinated upon TCR stimulation. We conducted a detailed analysis of novel S/T TCR-responsive phosphorylation sites with the aim gaining potential insights on their role in TCR signaling. The results from a) conservation analysis, b) statistical analysis of network parameters, c) experimental results on TCR-responsive phosphorylation on tubulins, d) analysis of sequence stretches responsible for PPIs, and e) extrapolation from many functionally characterized phosphorylation sites enabled us to deduce that S/T phosphorylation modulates PPIs in a system-wide fashion to regulate diverse stimulus-dependent processes during TCR signaling [24].
Mitosis
Dephoure et al. used large-scale quantitative phosphoproteomic approaches to examine cell-cycle dependent phosphorylation changes in HeLa cells [23]. Cells arrested in G1 phase showed little change in phosphorylation, whereas cells arrested in the M phase showed phosphorylation changes in over a thousand proteins when compared to asynchronously growing cells. Majority of the phosphorylation sites showing up-regulation at M phase could be grouped into classes of motifs conforming to minimal substrate requirements of three major mitotic kinases, namely cyclin dependent kinase (Cdk1), Aurora Kinases, and Polo-like Kinase 1. Mitosis is accompanied by drastic changes to the cell [23]. Processes associated with protein synthesis such as transcription, splicing and translation are inhibited. In addition, the nucleolus dissolves and the nuclear envelope breaks down during the entry into mitosis. The chromosomes get condensed tremendously and get captured by microtubules to form the spindle structure. Finally the sister chromatids separate during anaphase, culminating in cytokinesis. Apart from known mitotic phosphorylations, the authors observed statistical over-representation of proteins involved in these processes to be phosphorylated extensively during mitosis. Thus many of the novel mitotic phosphorylations may have a functional and regulatory role in the mitotic phenomena.
The three systems discussed above have been extensively studied over the last two decades. On one hand, the available information on these signaling systems has allowed rigorous assessment of the capabilities of large-scale quantitative phosphoproteomic technologies. Simultaneously the quantitative data point to an extensive role for phosphorylation in regulating a wide variety of associated processes and classes of proteins that was previously unanticipated or under appreciated. Thus, these quantitative datasets allow for the formulation of concrete hypotheses on the true functional significance of the responsive phosphorylation sites. Based on these studies, it is reasonable for us to anticipate that quantitative phosphoproteomics will pave the way towards further characterization of many other signaling systems, including those of cytokines, hormones, morphogens, and those responsible for innate immunity.
Identification of kinase substrates
Several strategies and reagents exist to screen and identify substrates of protein kinases [16,30]. Advances in quantitative phosphoproteomics have provided hypothesis-free, proteome-wide screens for generating lists of putative in vivo substrates and their target sites of phosphorylation [16,30]. Matsuoka et al. aimed to identify putative substrates and target sites of ATM/ATR kinases that are activated upon DNA damage caused by ionizing radiation [48]. They used a panel of antibodies directed against many known substrate sites of ATM/ATR, as these kinases are known to target SQ/TQ motifs. The antibodies were used to enrich phosphopeptides conforming to this motif followed by quantitative mass spectrometry. More than 900 putative ATM/ATR substrate sites were identified in the study. The authors used siRNA to reduce the expression level of many of the novel candidates and provided evidence for their role in DNA damage response by multiple phenotypic assays. Most of the putative substrates identified in the study are known to play a role in processes that are modulated upon DNA damage such as DNA replication, recombination, and repair and cell-cycle checkpoint. Stokes et al. refined the substrate screening strategy further by using a phospho SQ/TQ motif-speicific antibody instead of a large panel of antibodies [49]. Smolka et al. used kinase-null strains to identify putative substrates of three check-point kinases Mec1, Tel1, and Rad53 in yeast upon DNA damage caused by a DNA alkylating agent [50].
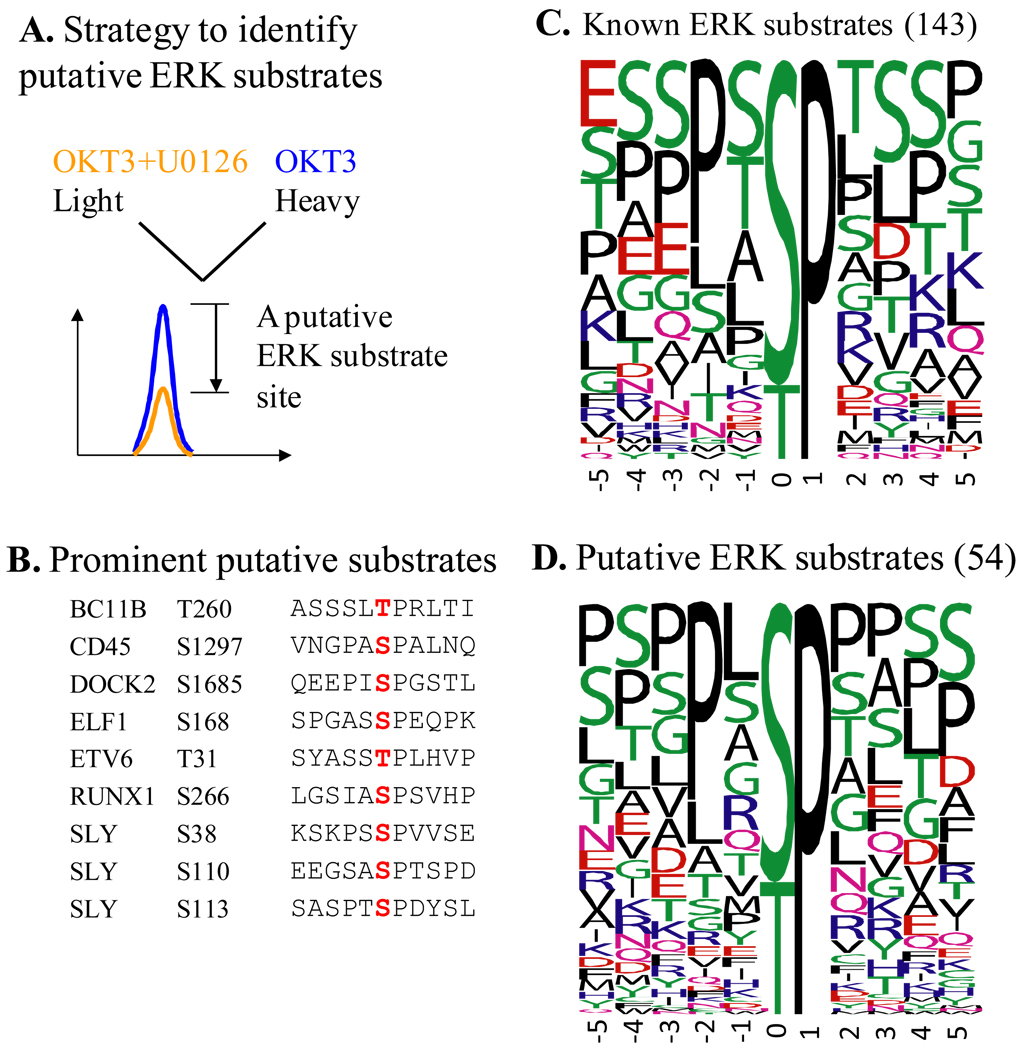
We have defined putative substrates of Erk kinases during the initial phase of TCR signaling by pharmacological means (Figure 2) [24]. U0126, a widely used inhibitor for Mek kinases was used to prevent activation of Erk and downstream kinases upon TCR stimulation. The frequency plot of the residues surrounding the phosphorylation sites with inhibitor-dependent response indicates that the candidates are likely to be bona fide substrates of Erk. Among the 52 novel putative Erk substrate target sites, eight are on proteins with known roles in TCR signaling. Among them we have validated T260 of Bcl11B to be a substrate of Erk using constitutively active and dominant negative forms of Mek [24].
Figure 2. Putative substrates of ERK during the initial phase of TCR signaling defined by quantitative phosphoproteomics.
(A) Stable Isotope Labeling of Cells in culture (SILAC) strategy was used to define TCR-responsive phosphorylation sites that are ERK-dependent, or in other words are ERK substrates. U0126 is a widely used, specific inhibitor of Mek kinases whose only known function is to activate ERK kinases. OKT3 is a functional antibody specific for CD3ε, which is used to crosslink CD3 coreceptors and initiate TCR signaling. Phosphopeptides that showed a >1.9 fold decrease in their abundance due to the inhibitor and harbored proline-directed phosphorylation sites were considered to contain substrate sites of ERK. (B) Putative substrates of ERK on seven proteins that are known to be involved in TCR signaling or are only found in lymphocytes. Residues marked in red are the actual substrate sites. (C) and (D) Frequency plots of residues surrounding known (C) and putative (D) substrate sites of ERK defined in the current study. The height of the residues represents the frequency with which they occur at the respective positions relative to the substrate site (designated 0 by convention) of phosphorylation. The color of the residues represents their physico-chemical properties. Note the preference for Pro and Leu in the −2 position in both the known and putative substrates of ERK. Also note the overall predominance of Ser, Thr, Pro, and Leu around the phosphorylation site. The frequency plots were generated with WebLogo (http://weblogo.berkeley.edu/). Among the 54 putative substrates from this study, Ser25 of stathmin and Ser266 of RUNX1 are known substrates of ERK. Known substrate sites were retrieved from the phosphoELM database. This figure is adapted from Mayya et al. (2009) [23].
Holt et al. used yeast strains expressing analog-sensitive kinases to identify 547 putative substrate sites of Cdk1 [47]. Conservation analysis of these phosphorylation sites revealed that the majority of putative Cdk1 substrate sites are located in rapidly evolving disordered or unstructured regions of proteins and that these have shifted positions as clusters in orthologs of the ascomycete lineage. This observation has important implications for the evolution of signaling by phosphorylation and resultant modulation of PPIs.
Although generating cell lines expressing analog-sensitive alleles is not seamless in mammals, Kevan Shokat’s lab has made available a handful of such reagents [51,52]. Analog-sensitive kinases function as wild-type counterparts, but are rapidly inhibitable by a specifically designed ATP analog [53]. These reagents provide highly specific means of inhibiting the desired kinase at any desired time-point for short periods. Phosphorylation changes observed with this experimental design are likely to be due to direct effects of the specific kinase, thus leading to the delineation of its substrate sites. But the approach may be biased towards substrate sites that are not under tight control of phosphatases [46].
Comparative analysis of oncogenic RTK signaling
Receptor Tyrosine Kinases (RTKs) function as oncogenes or ‘drivers’ both in solid tumors and hematologic cancers [4,54–57]. Therapeutic strategy of targeted inhibition of RTKs has been partially successful [6]. But, resistance towards tyrosine kinase inhibitors has been a major problem [56,58]. A better understanding of how the tumor cells uniquely rely on oncogenic signaling and how sensitivity or resistance towards RTK inhibitors develop in tumors is important for improvements in treating various malignancies. Here we discuss some of the recent quantitative phosphoproteomic studies that have contributed considerably towards this goal.
In order to better characterize phosphotyrosine signaling and identify putative RTK oncogenes in lung cancer, Rikova et al. conducted extensive profiling of phospho-tyrosine signaling in a large panel of non-small cell lung cancer (NSCLC) cell lines and tumors [59]. Peptides containing phosphotyrosine were enriched by immunoprecipitation and identified by mass spectrometry. The authors rank ordered RTKs based on the accumulated spectral count of all the constituent tyrosine phosphopeptides in each cell line or tumor sample. The rationale behind this approach was that the cumulative spectral count would serve as an index of the activity of RTKs and thus allow the identification of oncogenic RTKs. In known cases, oncogenic RTKs were found to be ranked high, validating their approach. Along with the known oncogenic kinases, ALK, ROS, PDGFRα, and DDR were most frequently found to be highly phosphorylated and active in the NSCLC cell lines and tumor samples. Of these, the authors showed that ROS and PDGFRα can act as ‘driver’ kinases in lung cancer. Another important observation was that NSCLC tumors expressed different combinations of active RTKs suggesting a need for individualized therapies.
Among NSCLC patients, only ~15% respond clinically to EGFR inhibitors and these responding patients frequently have tumors with activating mutations or genomic amplifications in EGFR. Guo et al. sought to understand the underlying mechanisms of sensitivity to EGFR inhibitors in tumors with high EGFR activity using NSCLC model cell lines [60]. The authors conducted quantitative phosphoproteomic analysis to identify tyrosine phosphorylations that decrease in response to EGFR inhibitors. In cell lines sensitive to EGFR inhibitors, the oncogenic EGFR activates multiple RTKs, including c-Met. Similar coactivation of other RTKs by the driver RTK has been observed in gastric cancer, glioblastoma, and other adenocarcinoma [58,60–62]. These studies highlight the need for targeting multiple RTKs in cancer therapy [58]. The above results also explain why some of the first generation tyrosine kinase inhibitors that were eventually recognized to inhibit more than one tyrosine kinase were remarkably effective in the clinic [58].
EGFR is frequently mutated in glioblastoma and 50% of tumors have a truncated, constitutively active form called EGFRvIII [61]. Huang et al. used a cell line model expressing varying amounts of EGFRvIII to understand the unique signaling properties of oncogenic form [61]. From quantitative phosphoproteomic analysis of tyrosine phosphorylation, the authors noted that only the mutated form but not the wild-type form of EGFR robustly activates PI3K. The authors also observed co-activation of c-Met that correlated with the level of expression of EGFRvIII. Their results from pharmacological experiments indicate that cotreatment strategies based on targeting of c-Met may be a viable approach to treat glioblastoma patients with EGFRvIII deletion mutations.
Expert Commentary
What is the major function of the large majority of S/T phosphorylation sites?
To date, more than 50,000 phosphorylation sites have been discovered in the human proteome with majority being S/T sites (Source: www.phosphosite.org). In a stark contract, an estimated 3000 of these phosphorylation sites have been functionally characterized to some extent. It is compelling to ask what functions do these thousands of newly discovered S/T phosphorylation sites serve. While one may argue that individual researchers focusing on particular proteins are best equipped to address this question rigorously in individual cases, we feel it is worthwhile to consider the above question in general. Often, a commonly observed mechanism serves as a basis for further fruitful hypothesis-based experimentation on more such instances in nature. For example, functional characterization in many signaling systems and cell types has been aided by the knowledge that most tyrosine phoshorylations recruit SH2 domain containing proteins in a sequence-restricted manner [63]. Are there similar rules or themes for signaling by S/T phosphorylations that may be of predictive value? We think that the major function is modulation of PPIs: inducing intra- or inter-molecular phosphorylation-dependent association or disruption. We present our arguments for this speculation below.
First, phosphoproteins participate in more PPIs than expected by chance alone [10,24]. Similarly, the case-by-case analysis as well as network analyses of proteins with TCR-responsive S/T phosphorylaitons point to system-wide modulation of PPIs during TCR signaling [24]. When tested experimentally, we found that all the known and novel S/T phosphorylation sites on tubulins abrogate their incorporation into microtubules. Based on the available crystal structure data, it appears that the S/T phosphorylation sites do not favor the three modes of PPIs during microtubule assembly. Further, we have observed that proteins belonging to large macromolecular complexes such as the spliceosome and the nuclear pore complex have more S/T phosphorylation sites than expected from an average number of about 3 phosphorylation sites per protein [24]. As extensive physical interactions are known to exist among proteins in macromolecular complexes, it is reasonable to speculate that S/T phosphorylations modulate these interactions. In fact, it has recently been proposed that extensive phosphorylations observed on nuclear pore proteins are responsible for the dissolution of the nuclear pore complex during mitosis [64]. Similarly, many proteins that are part of other macromolecular complexes such as the kinetochore, cohesin complex, APC, and the pre-replication complex are also phosphorylated during mitosis [23]. Finally, majority of S/T phosphorylation sites are found in disordered regions of proteins, which are also regions mediating majority of protein-protein interactions [40,46,47,65].
Accumulated knowledge on the role of S/T phosphorylation also points to modulation of PPIs as the predominant function. In our case-by-case analysis of TCR-responsive S/T phsophorylations, we found that majority of the functionally characterized sites were known to module PPIs [24]. Further, several conserved domains have been discovered that bind to S/T phosphorylation motifs to mediate PPIs [66]. While S/T phosphorylation has been documented to regulate protein half-life and intracellular localization in many cases, both are ultimately a consequence of modulation of PPIs [67]. Based on all the reasoning presented above, we propose that a major function of large majority of S/T phosphorylation sites is modulation of PPIs. One apparent difference between tyrosine versus S/T phosphorylation is that S/T phosphorylation seems to influence protein-protein association as well as disruption, while tyrosine phosphorylation has been recognized as promoting association in most cases via SH2 domains [24,63]. As S/T phosphorylation events are much more pervasive in cells, it is reasonable to speculate that protein complexes that are assembled due to phosphorylation-dependent associations (on S/T/Y residues) may also be disrupted by other site-specific S/T phosphorylation events. Thus, during a signal transduction event that initiates from the plasma membrane-associated kinases and permeates to the nucleus, a series of interlinked and dynamic phosphorylation-dependent assembly and disassembly of protein complexes may serve as an underlying mechanism of information transfer [24].
Quantifying the stoichiometry of phosphorylation in a proteome-wide manner
Stoichiometry of phosphorylation refers to the extent of phosphorylation or the fraction of protein that is phosphorylated at a given site. Traditionally, for most functional studies, a relative-change or fold-change in the stoichiometry of phosphorylation between different biological states is sufficient. While it may seem that measuring the change in stoichiometry is more crucial, knowledge on the actual stoichiometry is also valuable as elaborated below. Also, while biochemical methods based on radioisotope labeling are available [67–69], we present arguments for the need to quantify the stoichiometry of phosphorylation in a site-specific and proteome-wide manner.
It is becoming increasingly clear that intracellular signaling comprising of kinases and phosphatases occurs through networks of interactions and not just linear pathways [70]. Further, components of the classically defined linear pathways have been shown to participate in feedback, feed-forward regulations, and also bind to scaffolding proteins [71,72]. Mathematical and computational models have shown that these features of complexity allow signaling networks to not only transmit but also process, encode, and integrate extra- and intra-cellular signals [71,72]. However, in order to gain a comprehensive and quantitative understanding of the mechanisms, we need data-driven approaches to model-based analysis of intracellular signaling [73]. Herein the information on the stoichiometry of phosphorylation is probably the most important for data-driven modeling. Take, for example, the MAPK module comprising of three kinases that are activated in a cascade. The MAPK module has been explored extensively by modeling analyses and shown theoretically to possess versatile signal processing capabilities [74]. But yet, even fundamental systems-level properties such as amplification and ultrasensitivity, which is essential for the switch-like stimulus response, have not been characterized in cellulo due to the lack of data on stoichiometry of phosphorylation.
Multisite phosphorylation of proteins is a recurrent theme wherein the status of phosphorylation at one site controls that of another site [67,75,76]. Thus multisite phosphorylation enables intricate regulation of protein function and is an important feature of complexity of intracellular signaling. Rules for the regulation of multisite phosphorylation are currently interpreted from the measured fold-changes in phosphorylation on mutant forms of proteins [76]. Information on stoichiometry of phosphorylation allows direct and simultaneous comparison of multiple sites and thereby enables a more comprehensive and conclusive deciphering of rules for multisite phosphorylation.
It has recently been proposed that many of the known phosphorylations are non-functional or silent [77]. Unfortunately, non-functionality of specific phosphorylation sites cannot be proven definitively by experimentation, as one cannot exclude the possibility of the sites being functional under biological contexts other than those tested. For example, S454 of ATP:citrate lyase was generally considered to be a non-functional phosphorylation for 25 years, before a function was ascribed for it in 2000 [78]. Despite the difficulties in ascertaining non-functionality, three major arguments have been put forward to support the possible existence of vast number of non-functional phosphorylations [77]. One of them centers on the stoichiometry of phosphorylation: It can be argued that majority of the phosphorylations discovered by mass spectrometry have low stoichiometry due to extensive enrichment of phosphopeptides and improved sensitivity of mass spectrometers. Thus, even a phosphorylation with deleterious effect will effectively be non-functional if the stoichiometry of phosphorylation is low. Such non-functional phoshorylations can persist and accumulate due to lack of evolutionary pressure to eliminate them. While this argument is likely to be valid in isolation, it presupposes that functional phosphorylations have high stoichiometry of phosphorylation. But multiple counter-arguments can be made to suggest that the observed low stoichiometry phosphorylation sites are functional in vivo. a) Recent studies have demonstrated that even in isogenic populations there is considerable cell-to-cell variability in their states and responses [79,80]. Accordingly, one can expect the population of cells to show a wide distribution of individual stoichiometries, which when averaged during sample preparation can be low. b) Under many circumstances, as in neuronal and immunological synapses, phosphorylation changes in a small sub-cellular space dictate biological phenomena [24]. As large population of cells are disrupted and prepared for mass spectrometric analysis, measured stoichiometry of phosphorylation will be low. c) If a certain phosphorylation modulates enzymatic activity, then even a low stoichiometry of phosphorylation can have a significant biological consequence. In order to rigorously assess all the above arguments, we urgently need stoichiometry data on hundreds of phosphorylations with known functional consequences. Similarly, we need stoichiometry data on phosphorylation sites that are conserved to different degrees across phyla to establish clarity on issues relating function, evolution and stoichiometry of phosphorylation. Unfortunately, stoichiometry of phosphorylation is rarely quantified. To our knowledge, quantitative data on in cellulo stoichiometry of phosphorylation is available for ~10 sites and sites with ~5% phosphorylation have proven functional consequence [67,68,81].
Availability of proteome-wide abundance data in yeast has helped benchmark proteomic profiling technologies and acted as a guide for their incremental improvement [82,83]. Having proteome-wide stoichiometry data on phosphorylation in yeast will similarly help benchmark phosphoproteomic technologies and getter a better understanding of the chemical mechanisms behind these methods. This is especially crucial to establish as many options are available for protease treatment, phosphopeptide enrichment, and fragmentation of phosphopeptide ions [9].
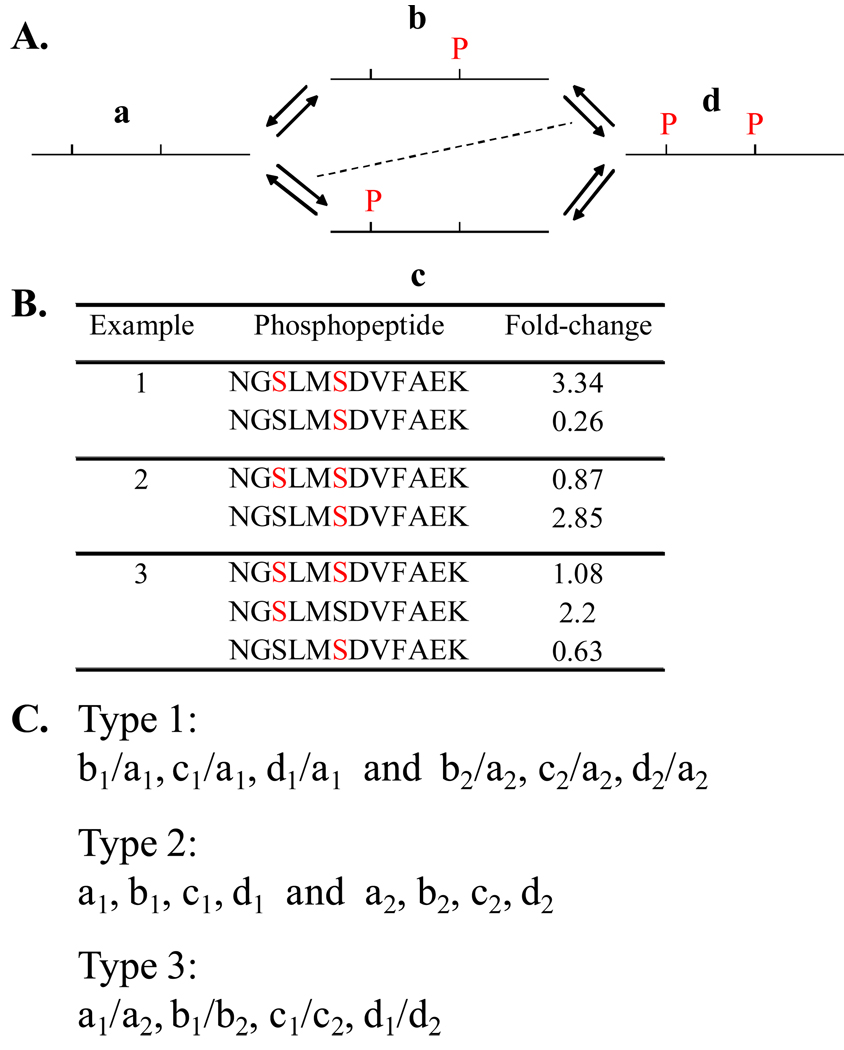
As described earlier, current phosphoproteomic methods rely on enriching phosphopeptides and disregarding non-phosphopeptide counterparts. Fold-change in the stoichiometry of phosphorylations on a proteome-wide scale is measured by enriching phosphopeptides and calculating their relative abundance based on incorporated heavy isotope labels (Figure 1B). Herein the phosphopeptides are identified from tandem mass spectra acquired in a data-dependent fashion. The corresponding parent ion traces are used for quantifying their relative abundance between different biological states. This approach works well if the phosphopeptide contains a single known phosphorylation site, as the fold-change in the phosphopeptide abundance equates to fold-change in the stoichiometry of phosphorylation. But the above approach presents limitations if there are multiple phosphorylations (Figure 3). In order to assign the fold-change in stoichiometry at each of the sites in a multiphosphorylated peptide one needs to have information on the relative abundance among each of its versions in different biological states, which is not provided by the current approaches (Figure 3C). Further, quantitative phosphoproteomic studies on cell signaling systems have revealed that the scope of phosphorylation is widespread [19,23,24]. But without the information on the actual stoichiometry of phosphorylation on a global scale, it is not possible to gain an understanding of how change in the activity status of a few protein species permeates into system-wide changes in phosphorylation. Thus, lack of information on stoichiometry of phosphorylation is a key limitation of the current phosphoproteomic approaches.
Figure 3. Issues in measuring stoichiometry of multisite phosphorylation.
(A) Reaction scheme for multisite phosphorylation in a peptide region. In case of multisite phosphorylation, phosphopeptides can exist in different versions accounting for different combinations. Here in the example, two neighboring phosphorylation sites give rise to four different versions (designated a, b, c, and d) of the phosphopeptide. It is important to note that a particular version can arise both by phosphorylation and dephosphorylation of other versions, which is represented by reverse arrow pairs. The dashed, diagonal line is shown to stress that reactions represented by diagonally opposite arrow pairs are equivalent by ‘mass action’ principles, if the phosphorylation sites are independent of each other. (B) Examples to explain limitations of current methods in ascertaining fold-change in multisite phosphorylation. In all three examples it is not possible to conclusively assign fold-changes to individual phosphorylation sites from the available data on fold-changes in phosphopeptide versions. Identical peptide backbone sequence is used in all three examples, which has two potential phosphorylation sites. Phosphorylated residue in each case is denoted in red. Real fold-change values from our datasets have been used, but with the surrogate peptide sequence for uniformity. First, it is important to note that information on non-phosphopeptide versions are rarely obtained by current methods as phosphopeptides are enriched. This is a limitation as explained further. In the first example, the doubly phosphorylated ‘d’ version increases in abundance accompanied by a concomitant decrease in the ‘b’ version. It can be said that this change is attributable to phosphorylation of the first site. But without the information on changes in the other two versions it is difficult to conclude so. It is not clear whether the status of phosphorylation on one site affects the status on the other site or if the inter conversions are independent of the phosphorylation status. If the sites are independent, version ‘c’ is also expected to show the same increase as version ‘d’ as per ‘mass-action’ principles. But without experimental data on all four versions one cannot be sure. Further, based on the specific rules for multisite phosphorylation other explanations for the observed changes can be given. Fold-changes in examples 2 and 3 are suggestive of specific rules for multisite phosphorylation. In example 2, if the sites were independent of each other, then the doubly phosphorylated ‘d’ version would have increased as much as the ‘b’ version. Simplistically, the data may be explained by phosphorylation of the second site on the non-phosphorylated ‘a’ version. But without complete information, other likely mechanisms combining phosphorylation and dephosphorylation among different versions may be proposed too. Thus the available information is not sufficient to either deduce the rules for multiphosphorylation or provide explanation for the observed fold-changes in the phosphopeptide versions. (C) Information needed for calculating fold-change in phosphorylation on two individual sites in a peptide. Any of the three different types of values (classified as type 1, 2, and 3) is sufficient. Variables represent the abundance of the four different versions (a, b, c, and d) of the phosphopeptide under two biological states (1 and 2). Thus, values from ‘type 1’ and ‘type 3’ are ratios, where as values from ‘type 2’ give the actual abundance of the phosphopeptide versions. More extensive data will be needed if there are more than two phosphorylation sites in a peptide. The type of information obtained is dependent on the experimental strategy as explained in Figure 4.
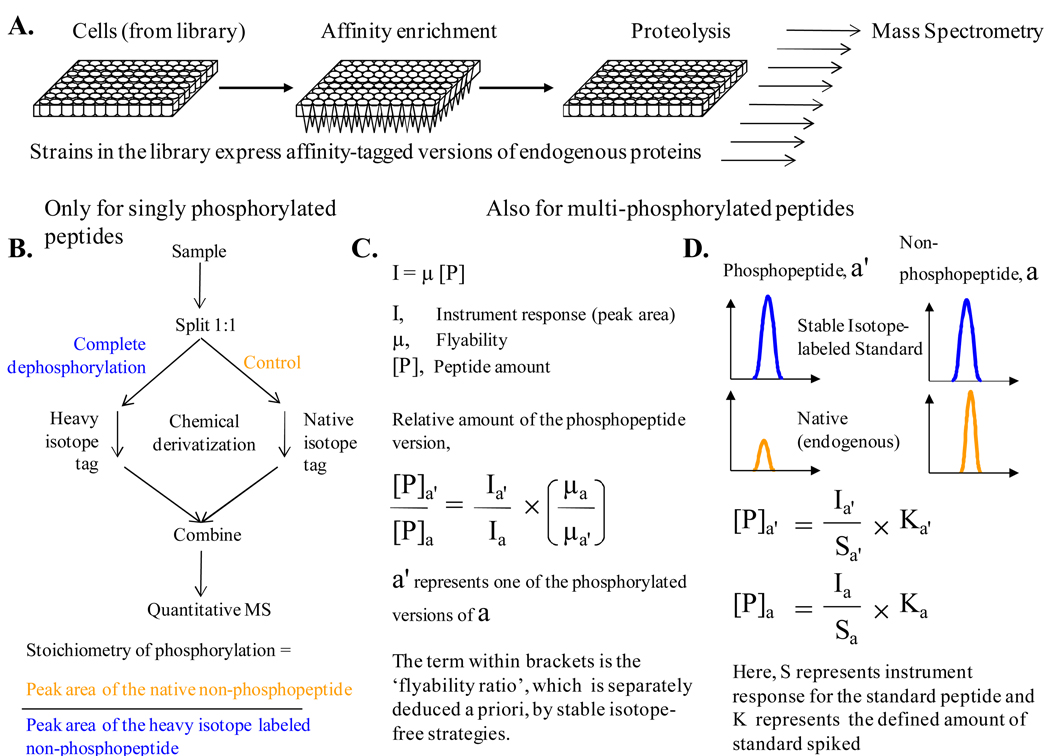
We propose to enrich a panel of endogenous proteins with affinity-tags from a library of cells in order to measure the stoichiometry of phosphorylation in a proteome-wide manner (Figure 4A). It must be mentioned that libraries have been developed for baker’s yeast and H1299 NSCLC cell line wherein affinity-tagged versions for over a thousand proteins are transcribed from their natural chromosomal locations [80,83]. These resources enable the application of high throughput strategies for simultaneous affinity-based enrichment of over a thousand endogenous proteins and subsequent proteolysis for mass spectrometric analysis [84]. It must be borne in mind that the activity of endogenous phosphatases must be minimized during affinity enrichment and proteolysis to avoid underestimation of stoichiometry of phosphorylation.
Figure 4. Strategies to measure stoichiometry of phosphorylation in a proteome-wide scale.
(A) Flow chart of important steps prior to mass spectrometric analysis for measuring stoichiometry of phosphorylation in a proteome-wide scale. The procedure relies on the availability of library of cells that express affinity-tagged versions of proteins from natural chromosomal locations. The initial steps can be performed simultaneously for the entire set in a 96-well format. But the subsequent steps for analysis by mass spectrometry have to be performed individually for each protein in the panel. This is represented by a group of arrows at the end of the schematic. (B) Flow chart for the measurement of stoichiometry of phosphorylation by means of complete dephosphorylation of one half of the sample. It is important to note that this method is applicable only if the peptide under consideration has only one known phosphorylation site. Isotope labeling is done during chemical derivatization of the peptides. (C) Formula for calculating the relative amount of a phosphopeptide version using previously determined ‘flyability ratio’. This approach provides information of ‘type 1’ as introduced in Figure 3C. ‘Flyability’ relates the instrument response to the actual peptide amount. ‘Flyability ratio’ is calculated from aliquots of the sample subjected to various gradations of controlled dephosphorylation or from serial dilutions of synthetic standard peptides. This method does not rely on isotope-dilution mass spectrometry (D) A known amount of Stable Isotope labeled Standard (SIS) peptide can be spiked into the sample before mass spectrometry to infer the amounts of phosphopeptide versions. This approach provides information of ‘type 2’ as introduced in Figure 3C. This method relies on the principle of isotope-dilution. Both the methods described in (C) and (D) are useful to calculate stoichiometry of multiple phosphorylation sites in a peptide.
There are three established MS-based approaches for measuring the stoichiometry of phosphorylation (Figure 4, B to D). In a simple and straightforward strategy, an identical sample that is labeled with a heavy isotope is subjected to complete dephosphorylation by alkaline phosphatase. The relative abundance of the unphosphorylated peptide gives the stoichiometry of phosphorylation (Figure 4B) [85]. Though straightforward, this approach has three major limitations. Firstly, it is valid only for peptides with a single known phosphorylation site. Secondly, the efficiency of proteolytic action at the desired residues is known to be influenced by neighboring phosphorylated residues, which may lead to inaccurate measurement of stoichiometry. Thirdly, the results for low stoichiometry phosphorylation are going to be inaccurate due to the variability in the measurement of ion currents. Irrespective of the type of instrumentation and mass-analysis used we anticipate that the values will be unreliable for stoichiometry values that are less than 10%. Nonetheless, we feel this approach is useful in finding out how many of the phosphorylation sites are of low stoichiometry.
Steen et al. have developed an isotope-free approach to quantify the stoichiometry that can be extended to multiphosphorylated peptides (Figure 4C) [86]. The crux of the approach lies in calculating the ‘flyability’ ratio for each of the phosphopeptide versions with respect to that of their unphosphorylated counterpart. The ‘flyability’ ratio can be calculated from subjecting aliquots of a sample to controlled dephosphorylation or by using the corresponding set of synthetic peptide standards. The authors note that ‘flyability’ ratios calculated from the cost-effective approach of controlled dephosphorylation are reliable only if the actual stoichiometry of phosphorylation in the original sample is more than 10%. For the alternate approach, peptides can now be synthesized in a high throughput fashion at nominal rates of 25$ per peptide [67]. An important note of caution on the isotope-free method is that it has not yet been used to measure in vivo stoichiometry of phosphorylation on endogenous proteins. Finally, use of known amount of Stable Isotope labeled Standard (SIS) peptides to quantify the abundance of different versions of the phosphopeptide is probably the most superior and accurate method available to measure stoichiometry of phosphorylation (Figure 4D) [67,68,87]. But the cost of procuring thousands of SIS peptides that have been purified and quantified accurately is a constraint in employing this strategy.
Five-year view
As mentioned previously, over 50,000 phosphorylation sites have been catalogued in human cells, of which about 3000 may have been functionally characterized. Although pinpointing the role of phosphorylation can be very hard, we hope there will be a concerted effort to functionally characterize the rest of them. As discussed before, this is crucial to advance our understanding on the issues relating function, evolution, and stoichiometry of phosphorylation.
In the previous sections, we have highlighted that the lack of information on stoichiometry of phosphorylation is a key limitation of the current phosphoproteomic approaches. But there are many other limitations currently and further advancements are desired in the coming years. One aspect wherein a lot of improvement is still desired is the amount of sample needed for quantitative phosphoproteomic experiments. Currently, around 100 million cells are required if one aims to monitor thousands of phosphorylation sites. Thus, most of the newly discovered phosphorylation sites have come from a few transformed human cell lines [17–19,23,24,33]. On the contrary, we are aware of very few reports on the large scale phosphoproteomic analysis of clinical samples or primary cells and tissues [88], which are likely to deliver important insights. We also anticipate that such studies will identify thousands of novel phosphorylation sights, as the repertoire of cellular states is going to be much richer in clinical samples and primary cells/tissues. With these studies, we may have a better estimate on the total size of the mammalian phosphoproteomes.
As mentioned before, multisite phosphorylation is a recurrent theme in signal transduction [67,68,75,76]. Multisite phoshorylation can be employed for combinatorial control of protein function, wherein specific combinations of phosphorylations code for distinct outcomes, as in histone modifications [67,76]. The current shotgun approach to phosphoproteomics precludes the analysis of phosphorylation sites on entire proteins. ‘Top-down’ or ‘middle-down’ schemes are necessary for the discovery of these distinctly phosphorylated subsets of proteins [89].
Most of the quantitative phosphoproteomic studies published to date have compared a two or three biological states. We hope that the future studies will seek better temporal resolution to gain an understanding of how change in the activity status of a few protein species permeates into system-wide changes in phosphorylation. Another related aspect is the measurement of phosphorylation turn over rates in a proteome-wide fashion. Computational modeling studies have established that intracellular information is encoded not only in states, but also in rates [71,72]. Thus, efforts to measure phosphorylation turn over rates are paramount to further our understanding of signaling mechanisms.
Quantitative phosphoproteomics has provided very effective tools to identify putative substrates of kinases. Currently the list of putative substrates is being generated from a single end-point comparison. However, there is enough evidence to suggest that many kinases have different sets of substrates depending on the biological context [51,52]. We anticipate that analog sensitive alleles will be of immense utility in unraveling such context-dependent activity of kinases over time-course [53]. We also hope that quantitative phosphoproteomics will be applied towards cataloguing putative substrates of phosphatases, which are increasingly being implicated in disease pathology [90–92]. Further, the principles of substrate recognition by protein phosphatases is increasingly becoming clear [90,92], which allows for more rational design of quantitative phosphoproteomic screens for phosphatase substrates.
Particular sets of kinases and phosphatases either sensitize or render cancer cells resistant to different types of insults [93,94]. Quantitative phosphoproteomics will be useful in understanding how such effects of kinases and phosphatases are manifested in cancer cells [56,93]. Protein kinases are given foremost importance as targets for therapies against cancers [4–6,58]. Many agents that inhibit the function of protein kinases are in clinical practice and many more are in clinical trials [4–6,58]. We anticipate that quantitative phosphoproteomics will help in providing a good understanding of their mode of action and off-target effects, as well as differences among their variants [56]. Similarly, quantitative phosphoproteomics may be applicable to reveal fundamental mechanisms of resistance towards kinase inhibitors that is frequently observed [56,58]. Most importantly, we anticipate that quantitative phosphoproteomics strategies will drive the identification of phosphorylation-dependent dynamically-interacting protein networks such as the lymphoma / leukemia / hematopoiesis protein network that we have deduced [24]. We propose that identification of disease-specific signaling protein networks will serve as a blueprint for targeting rational drug design and to specifically inhibit multiple key signaling nodes [54,93].
Key issues
Recent advancements in phosphoproteomic methodologies are leading to the discovery of unprecedented number of novel phosphorylation sites. But only a small fraction of all known phosphorylation sites have been functionally characterized to date.
Global trends in phosphoproteomic datasets have provided insights into the biology of phosphorylation.
Quantitative phosphoproteomic studies on well characterized cell signaling systems have revealed an extensive role for phosphorylation in regulating a wide variety of processes initiated by the signaling.
Quantitative phosphoproteomic strategies are very useful for cataloguing putative substrates of kinases and phosphatases.
Quantitative phosphoproteomics has clinical relevance, especially with regard to cancer.
Current approaches for quantitative phosphoproteomics do not provide information on the stoichiometry of phosphorylation and there is a need for measuring it in a proteome-wide manner.
Acknowledgement
Authors’ work discussed here was supported by NIH Grants RO1 HL 67569 and PO1 HL 70694 to DKH. The authors also acknowledge critical inputs from the anonymous reviewers.
Contributor Information
Viveka Mayya, Center for Vascular Biology, Department of Cell Biology, University of Connecticut Health Center, 263 Farmington Ave., Farmington, CT 06030, Tel: 860-679-2775, Fax: 860-679-1201, viveka.mayya@gmail.com.
David K. Han, Center for Vascular Biology, Department of Cell Biology, University of Connecticut Health Center, 263 Farmington Ave., Farmington, CT 06030, Tel: 860-679-2444, Fax: 860-679-1201, han@nso.uchc.edu.
References
- 1.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4(5):E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T, Scott JD. Protein phosphorylation in signaling--50 years and counting. Trends Biochem Sci. 2005;30(6):286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268(19):5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 4.Hunter T. Tyrosine phosphorylation in cell signaling and disease. Keio J Med. 2002;51(2):61–71. doi: 10.2302/kjm.51.61. [DOI] [PubMed] [Google Scholar]
- 5. Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1(4):309–315. doi: 10.1038/nrd773.• Reviews major breakthroughs that eventually lead to protein kinases being regarded as major drug targets.
- 6.Bozulic L, Morin P, Jr, Hunter T, Hemmings BA. Meeting report: targeting the kinome--20 years of tyrosine kinase inhibitor research in Basel. Sci STKE. 2007;2007(374):pe8. doi: 10.1126/stke.3742007pe8. [DOI] [PubMed] [Google Scholar]
- 7.Loyet KM, Stults JT, Arnott D. Mass spectrometric contributions to the practice of phosphorylation site mapping through 2003: a literature review. Mol Cell Proteomics. 2005;4(3):235–245. doi: 10.1074/mcp.R400011-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol. 2001;5(5):591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 9. Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9(6):1451–1468. doi: 10.1002/pmic.200800454.•• Excellent and comprehensive review on the methodological and analytical aspects of phosphoproteomics.
- 10. Chi A, Huttenhower C, Geer LY, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2193–2198. doi: 10.1073/pnas.0607084104.• Introduction of a new peptide fragmentation scheme for the identification of phosphopeptides
- 11.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76(13):3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 12.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1(4):1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 13.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4(10):798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 15.Garcia BA, Shabanowitz J, Hunt DF. Analysis of protein phosphorylation by mass spectrometry. Methods. 2005;35(3):256–264. doi: 10.1016/j.ymeth.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 16. Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat Methods. 2005;2(1):17–25. doi: 10.1038/nmeth731.• Reviews methods for deciphering protein kinase-substrate relationships.
- 17. Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101.•• The introduction of robust methodological breakthrough combining dual enrichment strategies employing strong cation exchange (SCX) chromatography followed by immobilized metal affinity chromatography (IMAC) to make large-scale phosphoproteomics a reality.
- 18. Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240.• Introduced a probablistic approach thereby obviating the need for manual interpretation of phosphorylation site localization.
- 19. Olsen JV, Blagoev B, Gnad F, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026.•• First large-scale quantitative phosphoproteomic study to be reported.
- 20.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J Proteome Res. 2008;7(7):3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodenmiller B, Malmstrom J, Gerrits B, et al. PhosphoPep--a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dephoure N, Zhou C, Villen J, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mayya V, Lundgren DH, Hwang SI, et al. Quantitative Phosphoproteomic Analysis of T Cell Receptor Signaling Reveals System-Wide Modulation of Protein-Protein Interactions. Sci Signal. 2009;2(84) doi: 10.1126/scisignal.2000007.•• Based on a large-scale quantitative phosphoproteomic analysis of TCR signaling, this study proposes system-wide modulation of protein-protein interactions by serine/threonine phosphorylation.
- 25.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7(3):1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 26.Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 2008;7(4):1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemeer S, Pinkse MW, Mohammed S, et al. Online automated in vivo zebrafish phosphoproteomics: from large-scale analysis down to a single embryo. J Proteome Res. 2008;7(4):1555–1564. doi: 10.1021/pr700667w. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Gerber SA, Rudner AD, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6(3):1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 29.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 30.Manning BD, Cantley LC. Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE. 2002;2002(162):PE49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 31. Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4(3):231–237. doi: 10.1038/nmeth1005.• Demonstrated that different enrichment methods allow for identification of reproducible but distinct sets pools of phosphopeptides.
- 32.Gauci S, Helbig AO, Slijper M, et al. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81(11):4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 33.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delom F, Chevet E. Phosphoprotein analysis: from proteins to proteomes. Proteome Sci. 2006;4:15. doi: 10.1186/1477-5956-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goshe MB, Veenstra TD, Panisko EA, et al. Phosphoprotein isotope-coded affinity tags: application to the enrichment and identification of low-abundance phosphoproteins. Anal Chem. 2002;74(3):607–616. doi: 10.1021/ac015528g. [DOI] [PubMed] [Google Scholar]
- 36.Mann M, Ong SE, Gronborg M, et al. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20(6):261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 37.Yachie N, Saito R, Sugahara J, Tomita M, Ishihama Y. In silico analysis of phosphoproteome data suggests a rich-get-richer process of phosphosite accumulation over evolution. Mol Cell Proteomics. 2009;8(5):1061–1071. doi: 10.1074/mcp.M800466-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gnad F, Ren S, Cox J, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8(11):R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez JL, Hegemann B, Hutchins JR, Peters JM, Durbin R. A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database. Genome Biol. 2007;8(5):R90. doi: 10.1186/gb-2007-8-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins MO, Yu L, Campuzano I, Grant SG, Choudhary JS. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol Cell Proteomics. 2008;7(7):1331–1348. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Klemke RL. PhosphoBlast, a computational tool for comparing phosphoprotein signatures among large datasets. Mol Cell Proteomics. 2008;7(1):145–162. doi: 10.1074/mcp.M700207-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Boekhorst J, van Breukelen B, Heck AJ, Snel B. Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol. 2008;9(10):R144. doi: 10.1186/gb-2008-9-10-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malik R, Nigg EA, Korner R. Comparative conservation analysis of the human mitotic phosphoproteome. Bioinformatics. 2008;24(12):1426–1432. doi: 10.1093/bioinformatics/btn197. [DOI] [PubMed] [Google Scholar]
- 44.Beltrao P, Trinidad JC, Fiedler D, et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7(6):e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan CS, Bodenmiller B, Pasculescu A, et al. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal. 2009;2(81):ra39. doi: 10.1126/scisignal.2000316.• Introduced new approaches to consider protein phosphorylation in the evolutionary context.
- 46.Collins MO. Cell biology. Evolving cell signals. Science. 2009;325(5948):1635–1636. doi: 10.1126/science.1180331. [DOI] [PubMed] [Google Scholar]
- 47.Holt LJ, Tuch BB, Villen J, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325(5948):1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321.•• Reports an extensive phosphoproteomic screen to identify substrates of DNA damage response kinases.
- 49.Stokes MP, Rush J, Macneill J, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104(50):19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104(25):10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burkard ME, Randall CL, Larochelle S, et al. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci U S A. 2007;104(11):4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levin SE, Zhang C, Kadlecek TA, Shokat KM, Weiss A. Inhibition of ZAP-70 kinase activity via an analog-sensitive allele blocks T cell receptor and CD28 superagonist signaling. J Biol Chem. 2008;283(22):15419–15430. doi: 10.1074/jbc.M709000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3(12):739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pawson T, Warner N. Oncogenic re-wiring of cellular signaling pathways. Oncogene. 2007;26(9):1268–1275. doi: 10.1038/sj.onc.1210255. [DOI] [PubMed] [Google Scholar]
- 55.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 56.Huang PH, White FM. Phosphoproteomics: unraveling the signaling web. Mol Cell. 2008;31(6):777–781. doi: 10.1016/j.molcel.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torkamani A, Verkhivker G, Schork NJ. Cancer driver mutations in protein kinase genes. Cancer Lett. 2009;281(2):117–127. doi: 10.1016/j.canlet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21(2):140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025.• Reports a semi-quantitative phosphoproteomic strategy to identify aberrantly activated kinases in lung cancer.
- 60.Guo A, Villen J, Kornhauser J, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105(2):692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A. 2007;104(31):12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 63.Songyang Z, Cantley LC. ZIP codes for delivering SH2 domains. Cell. 2004;116(2 Suppl):S41–S43. doi: 10.1016/s0092-8674(04)00041-8. 42 p following S48. [DOI] [PubMed] [Google Scholar]
- 64.Glavy JS, Krutchinsky AN, Cristea IM, et al. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc Natl Acad Sci U S A. 2007;104(10):3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunker AK, Oldfield CJ, Meng J, et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9 Suppl 2:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7(7):473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 67.Mayya V, D KH. Proteomic applications of protein quantification by isotope-dilution mass spectrometry. Expert Rev Proteomics. 2006;3(6):597–610. doi: 10.1586/14789450.3.6.597. [DOI] [PubMed] [Google Scholar]
- 68.Mayya V, Rezual K, Wu L, Fong MB, Han DK. Absolute quantification of multisite phosphorylation by selective reaction monitoring mass spectrometry: determination of inhibitory phosphorylation status of cyclin-dependent kinases. Mol Cell Proteomics. 2006;5(6):1146–1157. doi: 10.1074/mcp.T500029-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Sefton BM. Measurement of stoichiometry of protein phosphorylation by biosynthetic labeling. Methods Enzymol. 1991;201:245–251. doi: 10.1016/0076-6879(91)01022-t. [DOI] [PubMed] [Google Scholar]
- 70.Jordan JD, Landau EM, Iyengar R. Signaling networks: the origins of cellular multitasking. Cell. 2000;103(2):193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhalla US, Iyengar R. Functional modules in biological signalling networks. Novartis Found Symp. 2001;239:4–13. doi: 10.1002/0470846674.ch2. discussion 13–15, 45–51. [DOI] [PubMed] [Google Scholar]
- 72.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7(3):165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8(11):1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 74.Sauro HM, Kholodenko BN. Quantitative analysis of signaling networks. Prog Biophys Mol Biol. 2004;86(1):5–43. doi: 10.1016/j.pbiomolbio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 75.Holmberg CI, Tran SE, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27(12):619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 76.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24(10):1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 77. Lienhard GE. Non-functional phosphorylations? Trends Biochem Sci. 2008;33(8):351–352. doi: 10.1016/j.tibs.2008.05.004.•• Argues that a major portion of phosphorylation sites recently discovered by mass spectrometry are non-functional.
- 78.Potapova IA, El-Maghrabi MR, Doronin SV, Benjamin WB. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry. 2000;39(5):1169–1179. doi: 10.1021/bi992159y. [DOI] [PubMed] [Google Scholar]
- 79.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459(7245):428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cohen AA, Geva-Zatorsky N, Eden E, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165.• Reports construction of a library of lung cancer cells with endogenously tagged proteins.
- 81.Richardson CJ, Broenstrup M, Fingar DC, et al. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr Biol. 2004;14(17):1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 82.de Godoy LM, Olsen JV, de Souza GA, et al. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7(6):R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghaemmaghami S, Huh WK, Bower K, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 84.Dephoure N, Howson RW, Blethrow JD, Shokat KM, O'Shea EK. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc Natl Acad Sci U S A. 2005;102(50):17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, Jin QK, Carr SA, Annan RS. N-Terminal peptide labeling strategy for incorporation of isotopic tags: a method for the determination of site-specific absolute phosphorylation stoichiometry. Rapid Commun Mass Spectrom. 2002;16(24):2325–2332. doi: 10.1002/rcm.864. [DOI] [PubMed] [Google Scholar]
- 86. Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc Natl Acad Sci U S A. 2005;102(11):3948–3953. doi: 10.1073/pnas.0409536102.• Introduces a stable isotope-free strategy to measure stoichiometry of phosphorylation that is also applicable to multiphosphorylated peptides.
- 87.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100(12):6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci U S A. 2006;103(18):7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A. 2007;104(7):2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33(5):537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304(5674):1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 92.Alonso A, Sasin J, Bottini N, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 93.Manning BD. Challenges and opportunities in defining the essential cancer kinome. Sci Signal. 2009;2(63):pe15. doi: 10.1126/scisignal.263pe15. [DOI] [PubMed] [Google Scholar]
- 94.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7(6):591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]