Abstract
Cameroon is a country in West Central Africa in which all four groups of HIV-1 (M, N, O, and P), some circulating recombinant forms (CRFs) and unique recombinant forms (URFs) are prevalent. The CRF22 was initially identified through a novel URF strain, 01CM53122, and later defined from two additional sequences; however, the genomic properties of CRF22 have never been demonstrated in detail. In this study, we describe the characterization of five CRF22_01A1 strains, 02CMLT72, 01CM1867LE, 01CM001BBY, 02CM3097MN, and 02CM1917LE, identified in Cameroon without apparent epidemiological links. A typical CRF22_01A1 strain contains five fragments that can be assigned to the CRF01_AE and subsubtype A1 radiations. Forty-eight percent of the genome is classified as CRF01_AE, spanning the entire region of the gag gene, part of the pol gene, and accessory genes as well as the beginning and the end of the env gene and nef gene. Fifty-two percent of the genome is subsubtype A1 including regions mostly in the pol, vif, and env genes. The five CRF22_01A1 viruses formed a deep branch outside the groups of CRF01_AE and displayed similar mosaic structure but were moderately different from the original strain of CRF22_01A1, 01CM53122. Further analysis of the 01CM53122 genome showed that this virus represents a diverse set of mosaic genomes from CRF22_01A1, including a 446-nt segment of 01CM53122 in the env region, but unlike other CRF22 strains, clustered with CRF01_AE rather than the A1 sequence, suggesting that the 01CM53122 strain is a recombinant of CRF22_01A1 and CRF01_AE.
Introduction
Human immunodeficiency virus (HIV) is among the most genetically variable human retroviruses and is characterized by high rates of mutation, viral replication, and recombination.1–3 One of the most significant biological characteristics of HIV-1 is its broad genetic diversity, which may be attributed to the low fidelity of its reverse transcriptase, high turnover rate of replication, and genetic recombination between different HIV-1 viruses during replication. HIV-1 comprises four groups: M, N, O, and the newly described group P.4–6 HIV-1 group M is predominant in HIV-1 infections worldwide and can be further divided into nine subtypes (A–D, F–H, J, K), five subsubtypes (A1–A3, F1, and F2), and numerous recombinant forms. To date, 45 circulating recombinant forms (CRFs) of HIV-1 have been categorized in the Los Alamos HIV database.7,8 Some of them are called second-generation recombinant viruses (SGRs) since they contain the genetic material of at least one CRF. In addition, numerous unique recombinant forms (URFs) of HIV-1 have been reported.8,9
The factors that contribute to the emergence of HIV-1 variants and their epidemics and evolution over time are not well known. Molecular surveillance has revealed significant heterogeneity in geographic distribution of HIV-1 variants worldwide and continuous change of HIV-1 variants over the years. CRF01_AE virus has been documented at low frequencies in several Central African countries, such as Central African Republic, Cameroon, and the Democratic Republic of Congo,10–12 but at a high prevalence in Southeast Asia.13–15 Cameroon is a country in West Central Africa in which HIV-1 infection is endemic, and the natural reservoirs of HIV-1 group M, N, O, and P have been identified. The genetic diversity of HIV-1 is very broad in Cameroon where all group M clades and several CRFs (in particular CRF01_AE, CRF02_AG, CRF06_cpx, CRF09_cpx, CRF11_cpx, CRF13_cpx, CRF22_01A1, CRF36_cpx, and CRF37_cpx) have been identified.7,16–22 The cocirculation of different subtypes and CRFs results in the continuing emergence of new intersubtype recombinants. The current epidemic of HIV-1 in Cameroon is dominated by the circulating recombinant CRF02_AG. However, numerous HIV-1 variants have been emerging due to ongoing genetic mutation and recombination.23–27
CRF22_01A1 was initially defined as a recombinant of CRF01_AE and subsubtype A1 by Carr et al. in 2001 based on a URF, 01CM53122. Its sequences have been deposited in GenBank in two fragments: AY037284 and AY037285.28 The genetic mosaic structure of CRF22_01A1 listed in the Los Alamos HIV Databases was based on a full-length sequence of 01CM001BBY virus (accession no. AY371159). However, the detailed mosaic structure of CRF22_01A1 has not yet been described. Our previous study identified an HIV-1 virus 02CMLT72, which shared high homology with CRF22_01A1 based on partial sequence data.18 Since then, we have obtained three near full-length sequences of CRF22_01A1 viruses, including 02CMLT72, 02CM3097MN, and 02CM1917LE, and identified one HIV-1 virus (02CM1867LE) sequence in the GenBank database. These four near full-length sequences together with the CRF22_01A1 reference strain 01CM001BBY allow us to characterize the structure of this recombinant form of HIV-1 with an improved degree of accuracy. Here we describe the genomic mosaic structure of CRF22_01A1, which is moderately different from the original strain 01CM53122, indicating that 01CM53122 was a recombinant of CRF22_01A1 and CRF01_AE.
Materials and Methods
Samples
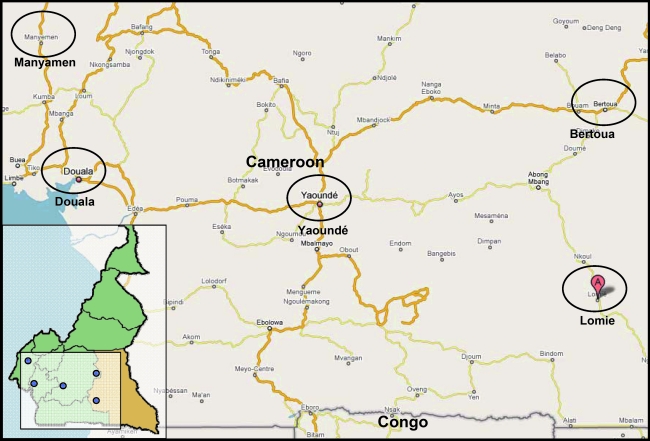
The blood samples from HIV-1-seropositive individuals were collected at Douala (02CMLT72), Lomie (02CM1917LE), and Manyamen (02CM3097MN) in Cameroon in 2001 and 2002, respectively (Fig. 1). These patients were not epidemiologically linked.
FIG. 1.
Map of the Southeast and West Province of Cameroon. Approximate locations from which the specimens were obtained are named and indicated in circles. Douala, 02CMLT72; Lomie, 02CM1867LE and 02CM1917LE; Manyamen, 02CM3097MN; Yaoundé, 01CM001BBY; Bertoua, CM53122. (Color image can be found at www.liebertonline.com).
RNA extraction, PCR, and sequencing
Viral RNA was extracted from plasma samples using the QIAamp Viral RNA Mini kit (Qiagen Inc., Valencia, CA). Reverse transcription was performed using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. A nested polymerase chain reaction (PCR) was performed to amplify around 1.7–3.0 kb of four overlapping segments, which span the entire HIV-1 genome.21 Purified PCR products were sequenced directly by primer walking using the ABI Prism BigDye Terminator Cycle Sequencing kit and ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences were assembled using the Vector NTI software, version 10.3.0 (Invitrogen, Carlsbad, CA), and analyzed as described below. The primers used have been described previously.21
Phylogenetic analysis
Phylogenetic analyses were performed in partial genome segments and full-length genomes using the MEGA 4.1 software package. Pairwise evolutionary distances were generated using the Kimura's two-parameter method; major gaps in the alignment were masked out prior to analysis and phylogenetic trees were constructed by neighbor-joining.29,30 Nucleotide sequences 02CMLT72, 02CM3097MN, 02CM1917LE, 02CM1867LE, 01CM001BBY, and 01CM53122 were aligned by the Clustal W program30 using the complete genomes of 42 subtypes as reference sequences. All reference subsubtypes and CRFs were obtained from the Los Alamos HIV Sequence Database and initially used to construct the trees.7,26,30,31 Some references have been omitted during the analysis for clarity and all positions with alignment gaps were removed. The alignment data are available on request.
Recombination analysis
Similarity and bootstrap plot analysis were performed using the SimPlot 3.5.1 software of the Phylip package to analyze the recombinant structure of the genome (http://sray.med.som.jhmi.edu/RaySoft/Simplot/).32 The reliability of plot topologies was assessed by bootstrapping with 1000 replicates, 1000-bp window, a step size of 50 bp, with a bootstrap support of >70% being required to define a phylogenetic cluster.33 Similarity plot analysis was also performed using a Recombinant Identification Program (RIP 3.0)34 and bootscan plot was performed using jpHMM-HIV software.35 The genetic distances of each segment of the novel strain were derived using SUDI program. RIP 3.0, jpHMM, and SUDI software are available at the Los Alamos site (www.hiv.lanl.gov/content/sequence/HIV/HIVTools).
Results
Characterization of specimen 02CMLT72
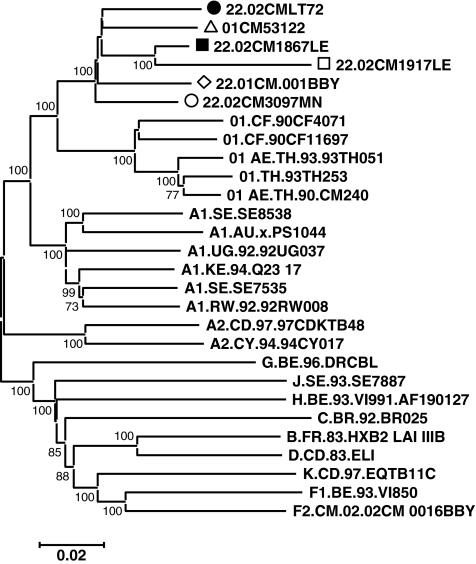
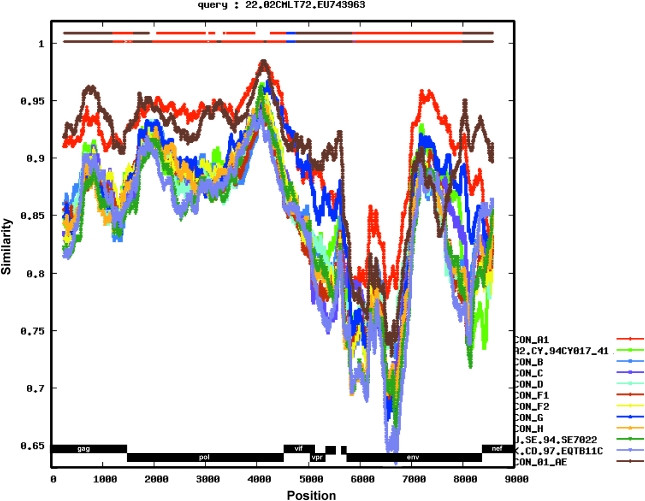
In our previous study, 135 HIV-1-positive plasma samples were collected from blood centers in Douala and Yaoundé, in Cameroon. There was a high proportion (94%) of samples identified as HIV-1 CRFs or URFs including recombinants of CRF02_AG with subtype D, F2, or CRF11_cpx and CRF19_cpx in this population.18 One of the viruses, 02CMLT72, was identified as a URF of CRF22gag-Aenv based on the partial sequencing data of HIV-1 gag and env. To further determine the subtype of this virus, we have sequenced the whole genome and performed detailed phylogenetic analysis of the sequence data. In total, 8807 nucleotides (nt) of the 02CMLT72 genome were sequenced and the blast search data showed several HIV-1 viruses isolated from Cameroon with a high score, including 02CM1867LE, a URF identified in Lomie in 200236; 01CM001BBY, a CRF22_01A1 strain isolated from the Yaoundé blood bank36; and 01CM53122, originally identified from Bertoua as a URF with genetic materials of CRF01_AE and subsubtype A1.28 Our study also revealed that two viral sequences, 02CM3097MN, isolated in Manyamen in the Southwest Province in Cameroon, and 02CM1917LE, isolated in Lomie, closely clustered with the 02CMLT72 virus and shared a similar phylogenetic structure. Therefore we analyzed the sequences of the six viruses mentioned above, and performed phylogenetic analysis with all known HIV-1 group M subtype, subsubtype, and CRF reference sequences, which are available at the Los Alamos HIV sequence database (www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) using the CLUSTAL W program.30 The phylogenetic trees generated by neighbor-joining methods showed that the six viruses clustered together to form a unique branch of CRF22_01A1 with high bootstrap value (100%) to CRF01_AE (Fig. 2).
FIG. 2.
Phylogenetic analysis of the newly derived near full-length sequence 02CMLT72 and their relatedness. All reference subsubtypes and available CRFs were initially used to construct the tree; some references have been omitted for clarity. Reference strains are identified by subtype or CRF number classification followed by sequence ID. The scale bar represents 2% genetic distance. Sequences from Cameroon specimens are indicated as •, 02CMLT72; ▪, 02CM1867LE; ○, 02CM3097MN; ▵, 01CM53122; ⋄, 01CM001BBY; and □, 02CM1917LE throughout the article. Bootstrap values above 70% are shown; numbers at the nodes of the tree represent maximum parsimony bootstrap values.
Determination of the mosaic genomic structure of CRF22_01A1
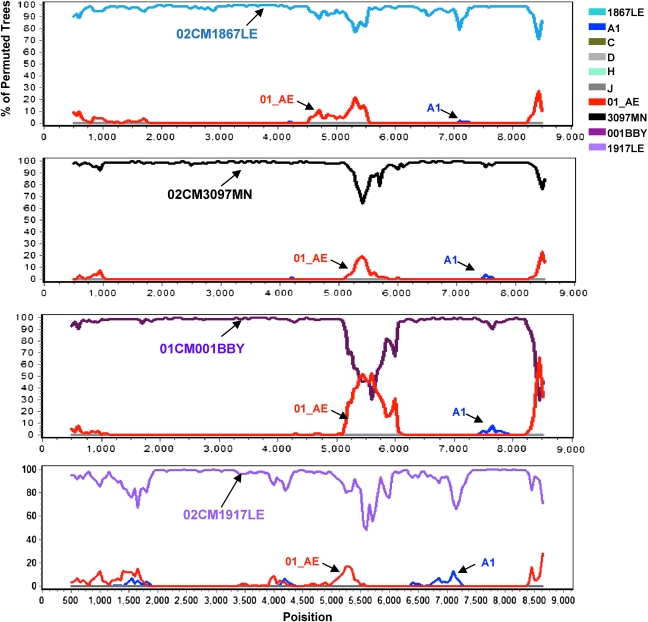
The sequences of HIV-1 CRF22_01A1 were further analyzed by bootscanning using the SimPlot software package. Each sequence was plotted against all known pure subtypes of HIV-1 group M and CRF01_AE. Four sequences, 02CM1867LE, 02CM3097MN, 01CM001BBY, and 02CM1917LE, clustered phylogenetically with 02CMLT72 throughout the entire genome when queried against the 02CMLT72 sequence (Fig. 3). These results indicated that the five viruses shared the same mosaic genome, but there were minor differences in the plot patterns from the 01CM53122 virus. A separate bootscanning analysis of 01CM53122 genome is described in the section below (Fig. 9A).
FIG. 3.
Bootscan analysis of 02CMLT72 queried against 02CM1867LE, 02CM3097MN, 01CM001BBY, and 02CM1917LE. Bootscan analysis was performed using SimPlot 3.5.1 software configured with 1000 bootstrap replicates, 1000 bp window, and a step size of 50 bp. The x-axis shows all aligned nucleotides of the sequence analyzed and the y-axis shows the bootstrap value. (Color image can be found at www.liebertonline.com).
FIG. 9.
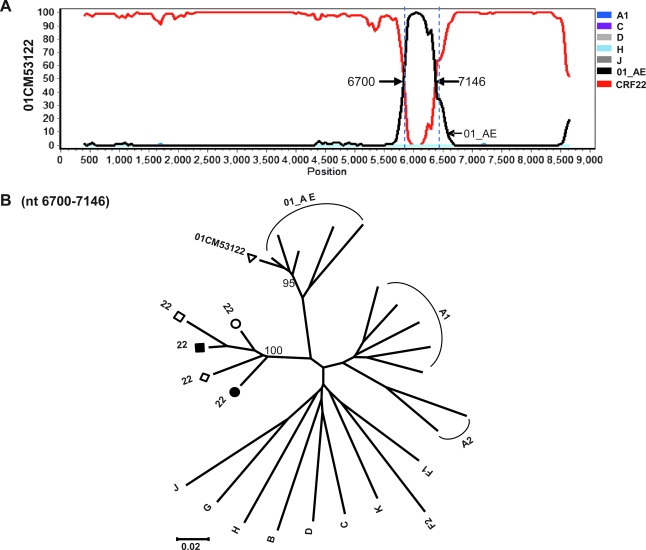
Reanalysis of the 01CM53122 genome. Bootscan plots were performed as described in Fig. 3; breakpoints are indicated. (A) Bootscanning analysis of 01CM53122 queried against CRF22_01A1 reference strains including 02CMLT72, 02CM1867LE, 02CM3097MN, 01CM001BBY, and 02CM1917LE. (B) The segment was analyzed by neighbor-joining representative trees and is illustrated for the region nt 6700–7146 (HXB2 numbering) in 01CM53122. (Color image can be found at www.liebertonline.com).
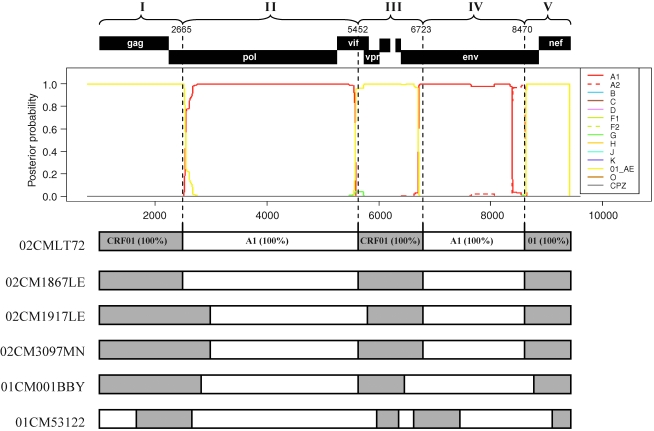
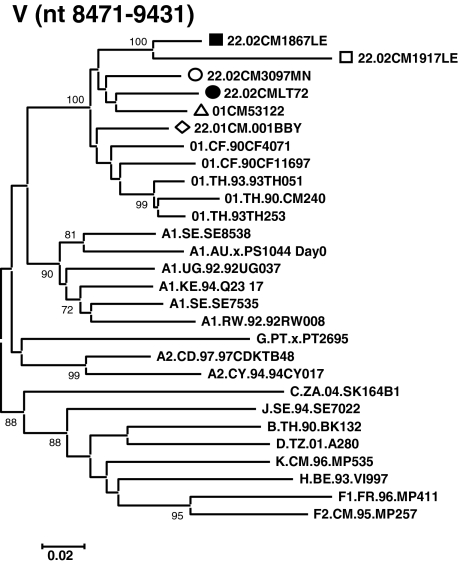
To define the subtype identities of the various recombinant segments, the 02CMLT72 strain was used as a representative of the new cluster of CRF22_01A1 and queried against all sets of the default HIV-1 subtype and CRF01_AE reference sequences in the RIP 3.0 online software to identify the recombination. As shown in Fig. 4, the 02CMLT72 genome was mainly broken into five segments. Because the 02CMLT72 genome was found to be composed of CRF01_AE and subsubtype A1 parental sequences, we also used online jpHMM-HIV software to predict the phylogenetic recombination breakpoints for the 02CMLT72 virus. This prediction method is based on a precalculated multiple alignment of the major HIV-1 subtypes including CRF01_AE references, and is more accurate than the competing methods used for phylogenetic breakpoint detection.35 The jpHMM software revealed four recombinant breakpoints at the position of nt 2665, 5452, 6723, and 8470 (Fig. 5, numbered according to the nucleotide sequence of HXB2, GenBank accession number K03455). In parallel, the similar recombinant breakpoints identified in 02CMLT72 were also found in the viruses of 02CM1867LE, 02CM3097MN, 01CM001BBY, and 02CM1917LE (Fig. 5). These results confirmed that the five CRF22_01A1 viruses share a similar mosaic backbone, which consists of five segments. The neighbor-joining trees were further built for each segment (Fig. 6). The segments I, III, and V spanning the gag, the beginning of pol, vif, vpr, the beginning and the end of env, and nef genes clustered within CRF01_AE radiation with 100% bootstrap value; the segment IV consisting of the majority of the env gene clustered with the lineage of subsubtype A1 radiation with a high bootstrap value of 97% (Table 1).
FIG. 4.
Identification of recombinant of 02CMLT72. Similarity plots were performed using RIP 3.0 software configured with Window Size, 500; Significance threshold, 0.9; Gap Option, 1; and Multistate characters, yes. The standard HIV-1 subtype alignment was selected as the default background and consisted of consensus sequences for subtypes A1, A2, B, C, D, F1, F2, G, H, and CRF01. Genomic regions are indicated at the bottom of the plot. (Color image can be found at www.liebertonline.com).
FIG. 5.
Prediction of genomic recombination in CRF22_01A1. Genomic regions are indicated at the top of the plot with the segment numbers and breakpoints (HBX2 numbering). In the center, the genomic recombinants of cluster HIV-1 subtypes are plotted for the 02CMLT72 genome based on jpHMM output. At the bottom, each of the resulting six genomes was analyzed by jpHMM separately; recombinant structures of 02CMLT72, 02CM1867LE, 02CM1917LE, 02CM3097MN, 01CM001BBY, and 01CM53122 were determined and listed. Open bar, subsubtype A1; gray, CRF01_AE. (Color image can be found at www.liebertonline.com).
FIG. 6.
Phylogenetic analysis of CRF22_01A1 regional segments. Regions of the sequence alignment were extracted according to the indicated breakpoints. Each segment was analyzed separately by the phylogenetic neighbor-joining method with the bootstrap value above 70%. Representative trees are illustrated for regions I through V. Loci of genomic segments is based on the HXB2 numbering engine. The genetic distance corresponding to the lengths of the branches is shown by the bottom line.
Table 1.
Phylogenetic Prediction of CRF22_01A1 Recombinant Segments
| Segment no. | HIV-1 gene | Location in HXB2 | Nearest strain | Bootstrap value (%) | SUDI distance (closest strain) |
|---|---|---|---|---|---|
| I | gag, pol | 791–2665 | 01_AE | 100 | Intrasubtype (01_AE) |
| II | pol, vif | 2666–5452 | A1a | 100 | Intersubsubtype (A1) |
| III | vif, vpr, env | 5453–6723 | 01_AE | 100 | Intrasubtype (01_AE) |
| IV | env | 6724–8470 | A1 | 97 | Intrasubtype (A1) |
| V | env, nef | 8471–9431 | 01_AE | 100 | Intrasubtype (01_AE) |
Relatedness to specific lineage in the subtype radiation was determined by Mage 4, RIP, and jpHMM software.
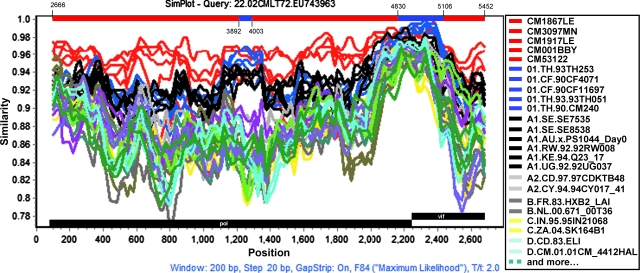
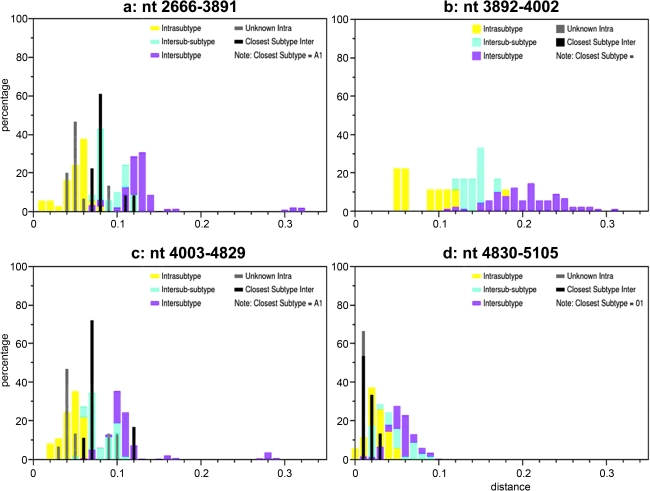
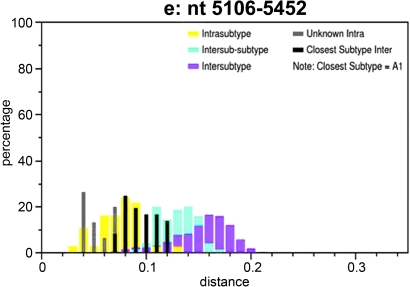
Interestingly, phylogenetic analysis of segment II (nt 2666–5452), which covers the majority of the pol and vif genes, showed that all CRF22_01A1 strains fell into the radiation of subsubtype A1 (bootstrap value, 100%), which include branches of CRF01_AE and CRF22_01A1 (Fig. 6). Within the lineage subsubtype A1 radiation, both branches clustered in the nearest node with subsubtype A1, but the topologies of neighbor-joining trees could not be supported by significant bootstrap values (bootstrap values 67%, data not shown, Fig. 6). Therefore, additional similarity plots analyses of segment II were performed by utilizing SimPlot software and including CRF22_01A1 strains, CRF01_AE, and subsubtype A1 sequences as references against the query sequence of 02CMLT72. As shown in Fig. 7, the segment II was composed of five blocks (a, nt 2666–3891; b, nt 3892–4002; c, nt 4003–4829; d, nt 4830–5105; and e, nt 5106–5452). Although the majority of 02CMLT72 segment II clustered with CRF22_01A1 in the block a, c, and e with high bootstrap values, two small blocks, b and d, clustered with CRF01_AE but not CRF22_01A1. This result suggested that segment II is a recombinant of the subsubtype A1 and CRF01_AE sequence, and may be a new HIV-1 CRF. This finding was also supported by a pairwise distance analysis with the Subtyping Distance Tool (SUDI) (http://www.hiv.lanl.gov/content/sequence/SUDI/sudi.html). The SUDI distance analysis showed that the entire segment II sequence was defined as intersubsubtype A1 (data not shown), blocks a, c, and e were defined as intersubsubtype A1, and block d (276 nt) was determined to be CRF01_AE. Block b (111 nt) could not be assigned to any known subtypes or CRFs (Fig. 8). Blast search results indicated that block d had the highest similarity (99%) with the representative sequences of Thai204 and CM240, which are old CRF01_AE viruses isolated in Thailand. These combined results indicated that 85% of the segment II sequence is subsubtype A1 and 11% of the sequence is closely associated with CRF01_AE.
FIG. 7.
Subtype identities of 02CMLT72 segment II (nt 2666–5452). Similarity plots were created using SimPlot 3.5.1 software configured with Window size, 200 nt; Step, 20 nt; GapStrip, On; F84 (maximum likelihood). Breakpoints are indicated at the top of the plot. There are five blocks (block a, nt 2666–3891; b, nt 3892–4002; c, nt 4003–4829; d, nt 4830–5105; and e, nt 5106–5452). Reference sequences of the subsubtypes A1, A2, B, C, D, F1, F2, G, H, J, and K, as well as of CRF22_01A1 and CRF01_AE are listed on the right side; red, CRF22_01A1; blue, CRF01_AE; black, subtype A1. (Color image can be found at www.liebertonline.com).
FIG. 8.
SUDI analysis of genome segment II. The input alignment includes all query sequences from each block in segment II of CRF22_01A1 strains, custom selected reference sequences of the group M subsubtypes A1, A2, B, C, D, F1, F2, G, H, J, and K, as well as of CRF01_AE (01). The subsubtype comparisons considered in the analyses were (A1 × A2), (B × D), (F1 × F2), (K × F1), (K × F2), and (01 × A1), (01 × A2) where applicable. A group O sequence, 98CMU2901, was used as an outlier. (Color image can be found at www.liebertonline.com).
Reexamination of the unique 01CM53122 strain
The designation of CRF22_01A1 was initially proposed based on the partial sequence of 01CM53122, and subsequently on the full-length sequence of 01CM001BBY. The near full-length sequence of 01CM53122 was obtained by joining two fragments: AY037284 (gag-vpr region, nt 790–5984 relative to HXB2) and AY037285 (vpu-nef region, nt 5999–9012). There was a 14-nt gap between these two fragments. The phylogenetic recombination breakpoints of 01CM53122 virus predicted by jpHMM-HIV software were slightly different from those for CRF22_01A1, and thus resulted in an alternative mosaic structure (Fig. 5). To further investigate the genomic recombination difference between CRF22_01A1 and 01CM53122 virus, bootscanning analysis was performed by using 01CM53122 as a query sequence and 02CMLT72, 02CM1867LE, 02CM3097MN, 01CM001BBY, and 02CM1917LE as reference strains. These results showed that the CRF22_01A1 sequences matched well with 01CM53122 throughout the whole genome except for a 446-nt fragment (5% of the whole genome), which spans the region from codon 159 to 308 of gp120 (nt 6700–7146, Fig. 9A). The neighbor-joining tree showed that the 446-nt fragment of 01CM53122 clustered with CRF01_AE radiation with a bootstrap value of 95% (Fig. 9B). Blast searches indicated that it had the highest similarity (91%) with sequences of TH235 and CM240, which are old CRF01_AE strains isolated in Thailand in 1990. It is clear that 01CM53122 has a slightly different mosaic structure from CRF22_01A1, and may be a new URF of HIV-1 with the recombinant genome sequences of CRF22_01A1 and CRF01_AE.
Discussion
To our best knowledge, this is the first report describing the precise mosaic structure of CRF22_01A1. In this study, we characterized five near full-length sequences of HIV-1 isolates 02CMLT72, 02CM1867LE, 02CM3097MN, 01CM001BBY, and 02CM1917LE that are identified in five apparently unlinked individuals from different areas in Cameroon (Fig. 1). They exhibited similar genomic mosaic backbone and were designated as CRF22_01A1 references. A typical CRF22_01A1 mosaic structure was constructed based on two isolates, 02CMLT72 and 02CM1867LE (Fig. 10). The 01CM53122 virus was originally assigned as CRF22_01A1 and phylogenetic analysis showed that the genome closely clustered within the CRF22_01A1 branch (Fig. 2), however, further comparison of these five CRF22_01A1 strains with the 01CM53122 virus demonstrated a moderate difference in mosaic structure. The 01CM53122 virus was reclassified as a URF with 95% of CRF22_01A1 and 5% of CRF01_AE rather than subsubtype A1 sequence in the env region (Fig. 9A). We concluded that the 01CM53122 strain is a recombinant of CRF22_01A1 and CRF01_AE, suggesting the existence of a new recombinant of CRF22_01A1 with other subtype/CRFs in Cameroon.
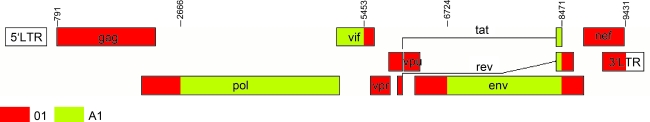
FIG. 10.
The mosaic structure of the CRF22_01A1. Breakpoints were determined from two sequences, 02CMLT72 and 02CM1867LE, using the jpHMM program. The schematic structure was created using the Recombinant HIV-1 Drawing Tool. Both programs are available on the Los Alamos HIV sequence Database website, http://www.hiv.lanl.gov/content/sequence/HIV/HIVTools.html. (Color image can be found at www.liebertonline.com).
CRF22_01A1 is composed of five genomic segments that can be assigned to the subsubtype A1 and CRF01_AE lineages, respectively. Within the subtype A lineages, three CRF22_01A1 segments (I, III, and V) are positioned close to the internal node of CRF01_AE viruses indicating a close relationship between CRF22_01A1 and CRF01_AE. CRF22_01A1 may display some structural features similar to a CRF01_AE sequence. The segment IV is within subtype A radiation, which does not include CRF01_AE. The segment II (nt 2666–5452) is the only one that all three clusters of subsubtype A1, CRF01_AE, and CRF22_01A1 diverge from the same point on the main trunk; their nearest node was found among some of the early subtype A lineages. Further phylogenetic analysis and SUDI distance analysis indicated that segment II is a recombinant of A1 and CRF01_AE, suggesting that different recombination events were involved in the evolution of HIV-1. CRF01_AE was previously reported to be a recombinant of subsubtype A1 and E and consists of at least 10 segments of A1 and E backbone.7,37,38 CRF01_AE is designated as subsubtype A1 in the region of nt 790–5096 and subtype E in the region of nt 5097–5320, respectively.7
The presence of CRF01_AE in segment II suggests that CRF22_01A1 may contain a subtype E sequence in this region. However, it will be difficult to determine the recombinant nature of segment II in CRF22_01A1 due to the lack of pure subtype E sequences.39 Interestingly, CRF22_01A1 showed two of the same recombinant breakpoints as CRF01_AE in the accessory gene (nt 5452) and env gene (nt 8470) regions. A possible explanation for this is that CRF22_01A1 may share ancestry with CRF01_AE. If so, similar to CRF01_AE, the CRF22_01A1 strain may have existed and circulated in Cameroon or other areas for a long time.37,38 CRF22_01A1 could represent recombination between contemporary strains derived from lineages CRF01_AE or subsubtype A1. Subtype A of HIV-1 has spread in humans for a long time and is one of the parent strains involved in many viral recombinants such as CRF01_AE and CRF02_AG.26,37–40 In this study, we present evidence that CRF22_01A1 was also formed by the recombination of subtype A and CRF01_AE. Five CRF22_01A1 viruses exhibited similar mosaic structure and formed a new CRF, although some minor sequence variations were observed within the different viruses. The genetic diversity of CRF22_01A1 viruses may be due to the duration of infections in the patients,41–43 but further studies are needed to examine this issue.
Identification of CRF22_01A1 from different patients who have no epidemiological links indicates that CRF22_01A1 is apparently another CRF of HIV-1 that is spreading in humans, and may behave like CRF01_AE and CRF02_AG by forming a new recombinant variant of HIV-1 with other subtypes/CRFs capable of geographic spread in Cameroon and perhaps other parts of the world. We have previously reported a novel CRF36_cpx in Cameroon that was assigned as a recombinant of CRF01_AE, CRF02_AG, subtype A and G radiations, however, the pol and env genes of CRF36_cpx were found to cluster with CRF22_01A1.21 As described above, the 01CM53122 strain is another example of the recombination of CRF22_01A1 with CRF01_AE. Brennan et al. reported that 22.4% of HIV-1 URFs contain the genome of CRF22_01A1 in blood donor samples collected between 1996 and 2004 in Cameroon. CRF22_01A1 is the second most frequent CRF found among HIV-1 URFs identified during a 9-year period of HIV surveillance in Cameroon.24 The reported CRF22_01A1 recombinants include CRF22gag/CRF02pol/CRF02env,24 CRF22gag/-/CRF11env, CRF22gag/-/CRF02env, and CRF22gag/-/Aenv.18 These results suggest that CRF22_01A1 may be a new HIV-1 variant that could potentially have emerged due to a founder effect that resulted in the formation of new HIV-1 recombinant variants in Cameroon.
Recent reports indicate that HIV-1 genetic diversity was relatively stable in Cameroon during the past decade and that some dominant CRFs found in Cameroon are not currently prevalent in the global HIV-1 epidemic.24 However, increasing global travel may contribute to the spread of HIV-1 infection worldwide44 as evidenced from the detection of CRF22_01A1 from HIV-1-infected patients in the United States45 and Saudi Arabia.46 According to a recent report from the Centers for Disease Control and Prevention (CDC), between 2003 and 2006, 5.1% of 3130 HIV-infected individuals in the 11 states in the United States were diagnosed as having HIV-1 non-B infections.47
The prevalence of non-B subtypes in the United States has increased,45 and the majority of non-B subtypes (80.8%) came from recent immigrants from Africa.48 It has been noted previously that strains emerging as dominant variants in Cameroon and West Central Africa have later spread to other regions of the world and emerged as new infections in other geographically distinct countries. It is thus necessary to continuously monitor the evolution of strains in this region as they can be predictive of the phylogenetic nature of future dominating strains in the HIV pandemic. Highly divergent HIV strains could affect HIV pathogenesis, ease of spread in a population, susceptibility to antiretroviral treatment, or vaccine development strategies. Studies of HIV-1 genetic diversity could have a potential impact on the diagnosis of HIV infection and could provide useful reference reagents for standardization of assays. For these reasons it is important to study the evolution of CRF22_01A1 in these regions and characterize its biological characteristics as it may potentially become a major HIV strain, similar to CRF02_AG, globally in the future.
Sequence Data
The nucleotide sequences of 02CMLT72, 02CM3097MN, and 02CM1917LE strains used in this study were submitted to GenBank with the following accession numbers; EU743963, GQ229529, and GQ229530, respectively.
Acknowledgments
The authors wish to acknowledge The National Heart, Lung, and Blood Institute for funding part of this work through an IAA–National Heart, Lung and Blood InterAgency Agreement BY1-HB-5026-01. This work was also supported in part by the Global Viral Forecasting Initiative. We wish to acknowledge Drs. Jinhai Wang, Krishnakumar Devadas, and Hira Nakhasi for review of the manuscript. The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Robertson DL. Hahn BH. Sharp PM. Recombination in AIDS viruses. J Mol Evol. 1995;40(3):249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 2.Ho DD. Neumann AU. Perelson AS. Chen W. Leonard JM. Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein J. Mathematics vs. evolution: Mathematical evolutionary theory. Science. 1989;246(4932):941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- 4.Keele BF. Van Heuverswyn F. Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Heuverswyn F. Li Y. Neel C, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444(7116):164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 6.Plantier JC. Leoz M. Dickerson JE, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15(8):871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 7.The Circulating Recombinant Forms (CRFs) Los Alamos National Laboratory (online) http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html. 2009. http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html (Last modified: Monday February 2, 12:40, 2009).
- 8.Robertson DL. Anderson JP. Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 9.McCutchan FE. Understanding the genetic diversity of HIV-1. AIDS. 2000;14(Suppl 3):S31–44. [PubMed] [Google Scholar]
- 10.Muller-Trutwin MC. Chaix ML. Letourneur F, et al. Increase of HIV-1 subtype A in Central African Republic. J Acquir Immune Defic Syndr. 1999;21(2):164–171. [PubMed] [Google Scholar]
- 11.Nkengasong JN. Janssens W. Heyndrickx L, et al. Genotypic subtypes of HIV-1 in Cameroon. AIDS. 1994;8(10):1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Vidal N. Peeters M. Mulanga-Kabeya C, et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74(22):10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menu E. Reynes JM. Muller-Trutwin MC, et al. Predominance of CCR5-dependent HIV-1 subtype E isolates in Cambodia. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20(5):481–487. doi: 10.1097/00042560-199904150-00011. [DOI] [PubMed] [Google Scholar]
- 14.Motomura K. Kusagawa S. Kato K, et al. Emergence of new forms of human immunodeficiency virus type 1 intersubtype recombinants in central Myanmar. AIDS Res Hum Retroviruses. 2000;16(17):1831–1843. doi: 10.1089/08892220050195793. [DOI] [PubMed] [Google Scholar]
- 15.Piyasirisilp S. McCutchan FE. Carr JK, et al. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J Virol. 2000;74(23):11286–11295. doi: 10.1128/jvi.74.23.11286-11295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burda ST. Konings FA. Williams CA. Anyangwe C. Nyambi PN. HIV-1 CRF09_cpx circulates in the North West Province of Cameroon where CRF02_AG infections predominate and recombinant strains are common. AIDS Res Hum Retroviruses. 2004;20(12):1358–1363. doi: 10.1089/aid.2004.20.1358. [DOI] [PubMed] [Google Scholar]
- 17.Konings FA. Haman GR. Xue Y, et al. Genetic analysis of HIV-1 strains in rural eastern Cameroon indicates the evolution of second-generation recombinants to circulating recombinant forms. J Acquir Immune Defic Syndr. 2006;42(3):331–341. doi: 10.1097/01.qai.0000219784.81163.2e. [DOI] [PubMed] [Google Scholar]
- 18.Machuca A. Tang S. Hu J, et al. Increased genetic diversity and intersubtype recombinants of HIV-1 in blood donors from urban Cameroon. J Acquir Immune Defic Syndr. 2007;45(3):361–363. doi: 10.1097/QAI.0b013e318053754c. [DOI] [PubMed] [Google Scholar]
- 19.Ndongmo CB. Pieniazek D. Holberg-Petersen M, et al. HIV genetic diversity in Cameroon: Possible public health importance. AIDS Res Hum Retroviruses. 2006;22(8):812–816. doi: 10.1089/aid.2006.22.812. [DOI] [PubMed] [Google Scholar]
- 20.Powell RL. Zhao J. Konings FA, et al. Circulating recombinant form (CRF) 37_cpx: An old strain in Cameroon composed of diverse, genetically distant lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23(7):923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- 21.Powell RL. Zhao J. Konings FA, et al. Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23(8):1008–1019. doi: 10.1089/aid.2006.0289. [DOI] [PubMed] [Google Scholar]
- 22.Luk KC. Holzmayer V. Ndembi N, et al. Near full-length genome characterization of an HIV type 1 CRF25_cpx strain from Cameroon. AIDS Res Hum Retroviruses. 2008;24(10):1309–1314. doi: 10.1089/aid.2008.0124. [DOI] [PubMed] [Google Scholar]
- 23.Laurent C. Bourgeois A. Faye MA, et al. No difference in clinical progression between patients infected with the predominant human immunodeficiency virus type 1 circulating recombinant form (CRF) 02_AG strain and patients not infected with CRF02_AG, in Western and West-Central Africa: A four-year prospective multicenter study. J Infect Dis. 2002;186(4):486–492. doi: 10.1086/341833. [DOI] [PubMed] [Google Scholar]
- 24.Brennan C. Bodelle P. Coffey R, et al. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr. 2008;49(4):432–439. doi: 10.1097/QAI.0b013e31818a6561. [DOI] [PubMed] [Google Scholar]
- 25.Andersson S. Norrgren H. Dias F. Biberfeld G. Albert J. Molecular characterization of human immunodeficiency virus (HIV)-1 and −2 in individuals from Guinea-Bissau with single or dual infections:Predominance of a distinct HIV-1 subtype A/G recombinant in West Africa. Virology. 1999;262(2):312–320. doi: 10.1006/viro.1999.9867. [DOI] [PubMed] [Google Scholar]
- 26.Carr JK. Laukkanen T. Salminen MO, et al. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS. 1999;13(14):1819–1826. doi: 10.1097/00002030-199910010-00003. [DOI] [PubMed] [Google Scholar]
- 27.Nyambi P. Zekeng L. Kenfack H, et al. HIV infection in rural villages of Cameroon. J Acquir Immune Defic Syndr. 2002;31(5):506–513. doi: 10.1097/00126334-200212150-00008. [DOI] [PubMed] [Google Scholar]
- 28.Carr JK. Torimiro JN. Wolfe ND, et al. The AG recombinant IbNG and novel strains of group M HIV-1 are common in Cameroon. Virology. 2001;286(1):168–181. doi: 10.1006/viro.2001.0976. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S. Nei M. Dudley J. Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 31.Thomas L. Bette K. Marcus D. Charles C. Brian F. HIV-1 subtype, circulating recombinant form (CRF) reference sequences. Los Alamos National Laboratory (online) http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/LEITNER2005/leitner.html. 2005. http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/LEITNER2005/leitner.html
- 32.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hillis DM. Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 2008. 1993;42(2):182–192. [Google Scholar]
- 34.Siepel AC. Halpern AL. Macken C. Korber BT. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11(11):1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 35.Schultz AK. Zhang M. Bulla I, et al. jpHMM: Improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009;37(Web Server issue):W647–651. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kijak GH. Sanders-Buell E. Wolfe ND, et al. Development and application of a high-throughput HIV type 1 genotyping assay to identify CRF02_AG in West/West Central Africa. AIDS Res Hum Retroviruses. 2004;20(5):521–530. doi: 10.1089/088922204323087778. [DOI] [PubMed] [Google Scholar]
- 37.Carr JK. Salminen MO. Koch C, et al. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70(9):5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao F. Robertson DL. Morrison SG, et al. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70(10):7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson JP. Rodrigo AG. Learn GH, et al. Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J Virol. 2000;74(22):10752–10765. doi: 10.1128/jvi.74.22.10752-10765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carr JK. Salminen MO. Albert J, et al. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247(1):22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 41.Nelson KE. Costello C. Suriyanon V. Sennun S. Duerr A. Survival of blood donors and their spouses with HIV-1 subtype E (CRF01 A_E) infection in northern Thailand, 1992–2007. AIDS. 2007;21(Suppl 6):S47–54. doi: 10.1097/01.aids.0000299410.37152.17. [DOI] [PubMed] [Google Scholar]
- 42.Zhu T. Korber BT. Nahmias AJ. Hooper E. Sharp PM. Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391(6667):594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 43.Carr JK. Avila M. Gomez Carrillo M, et al. Diverse BF recombinants have spread widely since the introduction of HIV-1 into South America. AIDS. 2001;15(15):F41–47. doi: 10.1097/00002030-200110190-00002. [DOI] [PubMed] [Google Scholar]
- 44.Coovadia HM. Hadingham J. HIV/AIDS: Global trends, global funds and delivery bottlenecks. Global Health. 2005;1:13. doi: 10.1186/1744-8603-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyne MT. Holzmayer V. John Hackett J. Hillyardl DR. Analysis of HIV subtype prevalence and geographic distribution in a large U.S. population. 16th Conference on Retroviruses and Opportunistic Infections; Montreal, Canada. Feb 8–11;2009 ; Poster #292. [Google Scholar]
- 46.Alrajhi AA. Halim MA. Al-Abdely HM. Mode of transmission of HIV-1 in Saudi Arabia. AIDS. 2004;18(10):1478–1480. doi: 10.1097/01.aids.0000131344.91536.64. [DOI] [PubMed] [Google Scholar]
- 47.Wheeler W. Mahle K. Bodnar U, et al. Antiretroviral drug-resistance mutations and subtypes in drug-naïve persons newly diagnosed with HIV-1 infection, United States, March 2003 to October 2006. 14th Conference on Retroviruses and Opportunistic Infections; Los Angeles, CA. Feb 25–28;2007 ; Poster #648. [Google Scholar]
- 48.Carr J. Flynn C. Maheshwari V. Blattner W. Zhao R. Detection of HIV-1 non-B subtypes in the city of Baltimore and state of Maryland. 16th Conference on Retroviruses and Opportunistic Infections, Montreal; Canada. Feb 8–11;2009 ; Poster #291. [Google Scholar]