Abstract
Background and purpose:
Increased circulating levels of L-α-lysophosphatidylinositol (LPI) are associated with cancer and LPI is a potent, ligand for the G-protein-coupled receptor GPR55. Here we have assessed the modulation of breast cancer cell migration, orientation and polarization by LPI and GPR55.
Experimental approach:
Quantitative RT-PCR was used to measure GPR55 expression in breast cancer cell lines. Cell migration and invasion were measured using a Boyden chamber chemotaxis assay and Cultrex® invasion assay, respectively. Cell polarization and orientation in response to the microenvironment were measured using slides containing nanometric grooves.
Key results:
GPR55 expression was detected in the highly metastatic MDA-MB-231 breast cancer cell line. In these cells, LPI stimulated binding of [35S]GTPγS to cell membranes (pEC50 6.47 ± 0.45) and significantly enhanced cell chemotaxis towards serum. MCF-7 cells expressed low levels of GPR55 and did not migrate or invade towards serum factors. When GPR55 was over-expressed in MCF-7 cells, serum induced a robust migratory and invasive response, which was further enhanced by LPI and prevented by siRNA to GPR55. The physical microenvironment has been identified as a key factor in determining breast tumour cell metastatic fate. LPI endowed MDA-MB-231 cells with the capacity to detect shallow (40 nm deep) grooved slides and induced marked cancer cell polarization on both flat and grooved surfaces.
Conclusions and implications:
LPI and GPR55 play a role in the modulation of migration, orientation and polarization of breast cancer cells in response to the tumour microenvironment.
This article is part of a themed issue on Cannabinoids. To view the editorial for this themed issue visit http://dx.doi.org/10.1111/j.1476-5381.2010.00831.x
Keywords: GPR55, GPCR, breast cancer, CBD, LPI
Introduction
GPR55 is an orphan G-protein-coupled receptor (GPCR) (nomenclature follows Alexander et al., 2009), which is thought to be expressed in a variety of cells, including cells in the bone marrow, endothelial cells, neurons in dorsal root ganglia and microglia. While the pharmacology of GPR55 remains controversial, the weight of evidence indicates that GPR55 is activated by certain cannabinoid ligands and by the lysophospholipid, L-α-lysophosphatidylinositol (LPI), as first discovered by Oka et al. (2007) (see Oka et al., 2007; Lauckner et al., 2008; Ross, 2009). Cannabidiol (CBD), the main non-psychoactive component of Cannabis sativa, appears to be an antagonist of this receptor (Ryberg et al., 2007; Whyte et al., 2009). Increased circulating levels of LPI, as well as lysophosphatidic acid (LPA), have been found in patients with ovarian cancer, suggesting that these lysophospholipids may be potential biomarkers for diagnosis and prognosis (Xiao et al., 2000; Sutphen et al., 2004). However, in contrast to LPA, whose molecular targets and pro-tumour effects have been well described (Stähle et al., 2003), little is known about the role of LPI in cancer.
GPR55 couples to G12 proteins (there is evidence for involvement of both Gα12 and Gα13) to activate RhoA, Cdc42 and Rac1 (Ryberg et al., 2007), possibly in an agonist- and tissue-dependent manner (see Ross, 2009). Indeed, LPI activates RhoA in cells expressing recombinant human GPR55 and in cultured human and mouse osteoclasts, which express native GPR55 (Henstridge et al., 2009; Whyte et al., 2009). A number of studies have highlighted the role of G12 signalling in promoting cancer cell migration and metastasis (Kelly et al., 2007; Juneja and Casey, 2009). RhoA activation is involved in regulation of the actin cytoskeleton, cell polarity, microtubule dynamics, transcription factor activity and cell growth (Kelly et al., 2007).
Thus, the cellular signalling pathways associated with GPR55 and LPI suggest potential roles in the modulation of tumour cell migration and polarity. Material surface topography has a major influence on the cytoskeleton, cell morphology and cell migration. In particular, micro- and nano-metre scale grooved surfaces, fabricated using electron beam lithography techniques, stimulate cell migration along grooves, cell elongation (polarization) parallel to grooves and cell alignment (orientation) with grooves (Kaiser et al., 2006). The depth and width of grooves both influence cell polarization and orientation, with RhoA signalling controlling the orientation of cells (Rajnicek et al., 2008). Measuring elongation of cancer cells is an accepted measure of polarity, with a more elongated phenotype associated with increased aggressiveness (Dalby et al., 2008). As part of this study we investigated the ability of cancer cell lines to detect nanometre scale grooves by measuring the orientation index (OI) and elongation of cells seeded onto grooves, and the modulation of these effects by LPI.
The aim of this study was to investigate the effects of LPI on breast cancer cell migration, orientation and polarization. In this manuscript we present the first evidence that LPI increases breast cancer cell chemotaxis and induces cell polarization and orientation on grooved substrata with features similar in scale to those present in the environment through which tumour cells metastasize. Furthermore, we demonstrate that GPR55, a receptor for LPI, is present in breast cancer cells that have a metastatic phenotype, and that over-expression of this receptor in MCF-7 cells that have low endogenous expression levels of GPR55 induces a migratory phenotype.
Methods
GPR55 plasmid and siRNA
A pcDNA3.1 vector containing human GPR55 cDNA tagged with a triple hemagglutinin epitope (HA) at the N-terminus (HA-GPR55) preceded by an optimized artificial signal sequence from the human growth hormone (hGH; residues 1–33) to ensure efficient surface expression (Henstridge et al., 2009) was used. For knock-down of GPR55 expression, a set of four siRNA duplexes against GPR55 was used (ON-TARGETplus siRNA from Dharmacon, cat. no. LQ-005581-00). Negative control siRNA (Ambion) was used to control for effects of transfection.
Tissue culture and transient cell transfections
Human breast carcinoma MDA-MB-231 cells were purchased from ATCC (Middlesex, UK) and maintained in Leibovitz L-15 medium (Sigma) supplemented with 10% foetal bovine serum (FBS) and 2 mM L-glutamine, and were incubated at 37°C in a CO2-free environment. Human breast carcinoma MCF-7 cells (ATCC) were maintained in DMEM (Sigma) supplemented with 10% FBS and 2 mM L-glutamine, and were incubated at 37°C with 5% CO2. MCF-7 cells were transfected with HA-GPR55 or empty vector plasmid, with or without GPR55 or negative control siRNA, by electroporation in an Amaxa Nucleofector II using solution V and program P-020, according to the recommended protocols for this cell line. Cells were incubated for 24 h post-transfection, before proceeding with further assays.
Immunofluorescence staining, flow cytometry and confocal microscopy
Immunostaining was performed on fixed and permeabilized cells using the BD Cytofix/Cytoperm Kit (BD Biosciences, Oxford, UK), according to the manufacturer's instructions. HA-GPR55 was detected with a mouse monoclonal anti-HA antibody (Covance) followed by an Alexa Fluor 488-conjugated goat-anti-mouse secondary antibody (Molecular Probes). Immunostained cells were analysed on a FACSCalibur or LSRII flow cytometer (BD Biosciences), or cytospun onto slides, counterstained with the nuclear dye Sytox Orange, and analysed on a ZEISS LSM510 META laser-scanning confocal microscope system using AIM software for image acquisition and analysis.
Quantitative RT-PCR
Cells were grown to 80% confluence in 75 cm2 tissue culture flasks, and RNA was extracted using TRIzol reagent (Invitrogen, Paisley, UK). RNA purification was performed using a Qiagen RNeasy minikit following the manufacturer's instructions (VWR, Vienna, Austria). The RNA concentration of purified samples was determined using a NanoDrop® Spectrophotometer. cDNA was synthesized from 2 µg total RNA using random hexamer primers and the SuperScript II Reverse Transcriptase kit (Invitrogen), according to the manufacturer's instructions. TaqMan® primers for GPR55 were from AppliedBiosystems (Warrington, UK). Lightcycler® 480 probe master mix was purchased from Roche (West Sussex, UK). Quantitative PCR (qPCR) reactions were performed in triplicate on a Roche LightCycler® 480 qPCR system. Expression levels were normalized to GAPDH and quantified using standard curves generated from a cell line expressing recombinant GPR55. GPR55 expression was further confirmed by traditional RT-PCR using a second set of primers designed with Primer3 software and supplied by Sigma Genosys (Paisley, UK). Forward: 5′-TCTCCCTCCCATTCAAGATG, reverse: 5′-AAGGAGGAAGCCAAACACCT; amplicon length 353 bp. For both primer sets, amplicon lengths of PCR products were validated by agarose gel electrophoresis.
[35S]GTPγS in MDA-MB-231 cells
Agonist-stimulated [35S]GTPγS-binding to G-proteins was performed as previously described (Anavi-Goffer et al., 2007). Briefly, cells were serum starved for 48 h then harvested and membranes reconstituted in TME buffer (50 mM Tris-HCl, 3 mM MgCl2, 1 mM EGTA, pH 7.4). Binding was initiated by the addition of 10 µg of membranes into glass tubes containing 0.5 nM [35S]GTPγS, and 30 µM GDP in GTPγS binding buffer (50 mM Tris-HCl, 100 mM NaCl, MgCl2, 0.2 mM EGTA, 0.1% fatty acid free bovine serum albumin, pH 7.4). Membranes were incubated for 40 min at 30°C. Non-specific binding was assessed in the presence of 50 µM unlabelled GTPγS. Drugs were diluted into a final concentration of 0.11% dimethyl sulphoxide (DMSO) (v/v).
Boyden chamber cell chemotaxis assay
Cells were grown to confluence in a 75 cm2 tissue culture flask and incubated in serum-free medium overnight. The lower wells of the Boyden chamber were loaded with 27 µL of medium containing the chemoattractant (10% FBS), and/or vehicle [0.01–0.02% DMSO (v/v)] or test compound. A polyvinylpyrrolidone-free polycarbonate filter with pores 8 µm in diameter was placed on the lower wells, and then upper wells were loaded with 45 µL of cell suspension at a density of 1 × 106 cells·mL−1. For modulation assays, cells were pre-incubated for 30 min with test compounds or vehicle prior to loading into the upper wells of the Boyden chamber, and lower wells contained corresponding concentrations of test compound or vehicle, plus 10% FBS (chemoattractant). The Boyden chamber was incubated for 2 or 4 h at 37°C, with or without CO2 depending on cell type. Following incubation, the filter was removed from the chamber and placed non-migrated side down in a Petri dish containing 70% ethanol for 7 min, then distilled H2O for a further 3 min. The non-migrated side was then drawn over a wiper blade several times to remove any non-migrated cells. The filter was left to air dry, and migrated cells were fixed and stained with a Diff-Quik stain set. Finally, the filters were mounted onto microscope slides using xylene and p-xylene-bis-pyridinium bromide. Cells in four fields from each well were counted under an inverted microscope at 20× magnification. Migration of cells towards FBS and test compounds was calculated as a percentage of the relative vehicle control. For modulation assays, migration was calculated relative to FBS plus vehicle.
Cell invasion assay
Cultrex® 24-well BME invasion assays (AmsBio, UK), were used according to the manufacturer's instructions. Briefly, cells were cultured to 80% confluence, serum starved overnight, then harvested and re-suspended to a density of 5 × 105 cells·mL−1 in serum-free medium. One hundred microlitres of cell suspension were added to the top chamber, while 500 µL of media, with or without FBS and/or test compound, were added to the lower well. The chambers were incubated at 37°C in a humidified environment for 24 h. The following day, cells invading the lower wells were detected by calcein-AM staining followed by fluorescence measurement on a Synergy™ microplate reader at 485 nm excitation and 520 nm emission.
Measuring viable cell numbers
Cells were seeded in 96-well plates at a density of 2 × 104 cells·well−1. Twenty-four h later cells were serum-starved overnight then treated with compounds of interest at various concentrations in replicates of 6 for 20 h or as stated. Ten microlitres of AlamarBlue® (Invitrogen, Paisley, UK) were then added to each well and cells were incubated for a further 4 h. The number of viable cells was determined by measuring fluorescence at 530 nm excitation and 590 nm emission using a Synergy™ HT microplate-reader.
Measuring cell polarity and orientation on grooved substratum
Grooved substrata with evenly spaced ridges and grooves, with approximately right angle cross section, were prepared by the Department of Electronics and Electrical Engineering, Glasgow University, as previously described (Rajnicek et al., 1997). MDA-MB-231 cells were grown to 80% confluence then serum-starved overnight. Cells were harvested and seeded onto grooved substrata in serum-free medium containing 1 µM LPI or vehicle [0.01% DMSO (v/v)]. Following incubation for 4 h at 37°C, phase-contrast images of live cells on grooves of width 4 µm and depth of either 520 nm or 40 nm were captured using a Zeiss Axiovert 200 inverted microscope. The length-to-width ratio was determined in Axiovision 4.7 software using the length tool, by measuring the length and widths of individual cells. Analysis of cell alignment (OI) was performed on the same images of cells which were isolated from their neighbouring cells. The angle of the longest cell diameter was calculated relative to the groove direction, as previously described (Rajnicek et al., 2008).
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5 software (GraphPad Software Inc., San Diego, CA, USA). Data are expressed as the mean ± SEM from at least three individual experiments. Analysis was by one-way anova and Newman Keuls multiple comparison tests, unless otherwise stated. A P-value of <0.05 was considered significant.
Materials
L-α-lysophosphatidylinositol was purchased from Sigma-Aldrich (Dorset, UK). CBD was purchased from Cayman Chemicals (Tyne and Wear, UK). All other reagents were from Sigma-Aldrich (Dorset, UK), unless otherwise stated. Stock solutions of test compounds were prepared in DMSO. In all assay incubations, test ligands were applied at 0.01–0.02% DMSO (v/v), unless specifically stated.
Results
GPR55 is expressed in human breast cancer cell lines
Quantitative RT-PCR analysis revealed that GPR55 was expressed in the highly metastatic MDA-MB-231 cell line, while the low-metastatic MCF-7 cell line expressed around 30-fold lower levels (Figure 1A). Expression of GPR55 was further confirmed by traditional RT-PCR analysis using a different primer set, and amplicon length was validated using gel electrophoresis (not shown).
Figure 1.
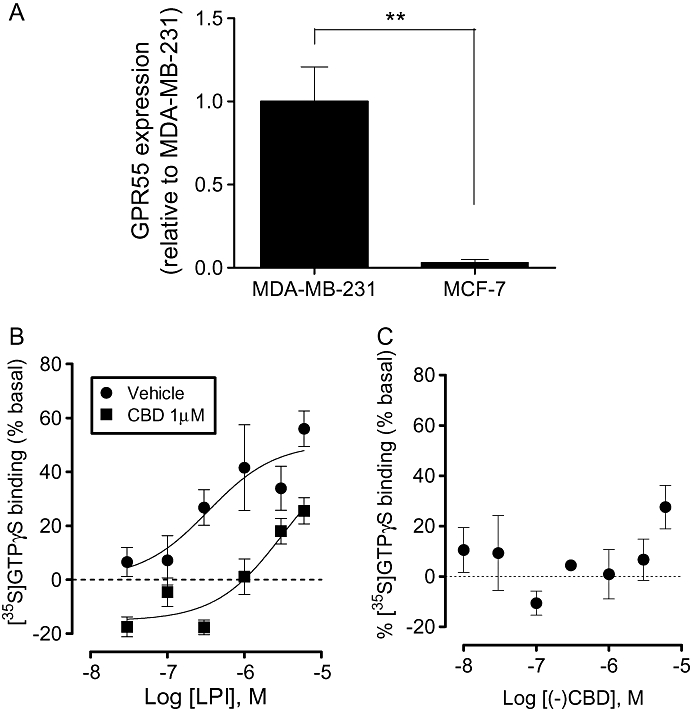
GPR55 expression in breast cancer cell lines. (A) Histogram showing expression levels of GPR55 in MDA-MB-231 and MCF-7 cell lines. Data are expressed as the mean ± SEM (n= 3). **P < 0.01, analysed by unpaired t-test. (B) LPI stimulated [35S]GTPγS binding to cell membranes from MDA-MB-231 cells with an Emax of 51 ± 8 % (pEC50 6.47 ± 0.45); an effect that was attenuated in the presence of CBD (pEC50 5.5 ± 0.38) (data are mean ± SEM, n= 6). (C) CBD alone had no effect on basal [35S]GTPγS binding to cell membranes from MDA-MB-231 cells (1 nM–1 µM); at 10 µM the compound produced a stimulation of 27.5 ± 9%. CBD, cannabidiol; LPI, L-α-lysophosphatidylinositol.
Stimulation of [35S]GTPγS binding by LPI in MDA-MB-231 cells
LPI produced a concentration-related stimulation of [35S]GTPγS binding in membranes prepared from MDA-MB-231 cells, with Emax and pEC50 values of 51 ± 11% and 6.47 ± 0.45 (340 nM), respectively (Figure 1B). This effect is indicative of activation of a GPCR. The stimulation was significantly attenuated in the presence of the putative GPR55 receptor antagonist, CBD (1 µM).
MDA-MB-231 cell chemotaxis is enhanced by LPI
In a Boyden chamber transwell chemotaxis assay, the endogenous GPR55 agonist LPI (1 µM) did not act as a chemotactic factor for MDA-MB-231 cells in the absence of FBS within a 4 h incubation period (Figure 2A). However, LPI modulated chemotaxis of MDA-MB-231 cells towards 10% FBS. Pre-incubation of cells with the GPR55 agonist LPI, at a concentration of 1 µM, significantly enhanced FBS-induced migration (P < 0.05; Figure 2B). Following co-incubation of LPI with the putative GPR55 receptor antagonist, CBD (both 1 µM), migration towards FBS was decreased (P < 0.001; Figure 2C). Pretreatment with CBD alone, however, also caused a significant decrease in FBS-induced migration (P < 0.001; Figure 2D). LPI at 1 µM had no effect on MDA-MB-231 cell invasion as measured using the Cultrex® BME cell invasion assay (data not shown). To investigate whether the effects of LPI on cell migration could have been affected by alterations in viable cell number, we used a fluorimetric AlamarBlue® viability assay. LPI had no effect on the number of viable MDA-MB-231 cells after 4 h of incubation at concentrations of 0.1 nM to 1 µM (data not shown).
Figure 2.
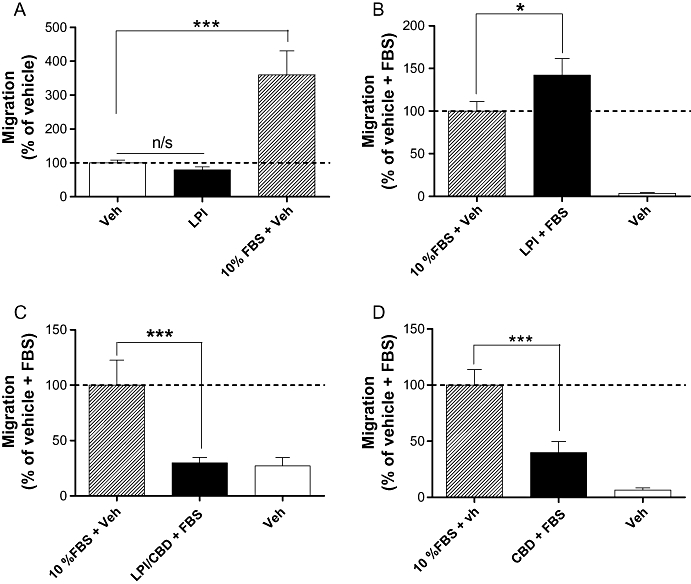
Histograms showing the migration of MDA-MB-231 cells in a Boyden chamber transwell assay. (A) Cells migrating towards LPI or FBS placed in lower wells of the chamber; (B) cells that have been pre-incubated with LPI (1 µM) migrating towards FBS placed in lower chamber; (C) cells that have pre-incubated with LPI and CBD migrating towards FBS placed in the lower chamber; (D) cells that have been pre-incubated with CBD (1 µM) migrating towards FBS placed in the lower chamber. The data represent mean ± SEM (n= 3–4). *P < 0.05; ***P < 0.001; n/s, not significant; one-way anova followed by Newman–Keuls multiple comparison tests. CBD, cannabidiol; FBS, foetal bovine serum; LPI, L-α-lysophosphatidylinositol.
Several attempts were made to knock down native GPR55 expression in MDA-MB-231 cells with siRNA against GPR55, but these were unsuccessful (qRT-PCR, data not shown). However, the same siRNA successfully knocked down artificially expressed GPR55 in these cells (data not shown). Furthermore, in MCF-7 cells, successful siRNA knock-down ofartificially over-expressed GPR55 was also achieved (see next section).
Over-expression of GPR55 induces a migratory and invasive phenotype in MCF-7 cells
MCF-7 cells were found to express relatively low levels of endogenous GPR55 compared with MDA-MB-231 cells. Over-expression of GPR55 in MCF-7 cells was found to alter the migratory phenotype of these cells. MCF-7 cells were transiently transfected with pcDNA3.1 plasmid encoding human HA-GPR55, and efficient expression of HA-GPR55 on the cell surface was confirmed by immunostaining with an antibody against the HA-tag, using confocal microscopy (Figure 3A). Transfection efficiency was 50–60%, as determined by flow cytometry, and co-transfection with siRNA against GPR55, but not negative control siRNA, efficiently knocked down HA-GPR55 protein expression (Figure 3B). This was further confirmed by Western blotting, demonstrating an approximately 90% knock-down by co-transfection with GPR55 siRNA (not shown). In a Boyden chamber transwell chemotaxis assay, vector-transfected MCF-7 cells (Figure 3C) did not migrate towards FBS and were unaffected by LPI. However, MCF-7 cells over-expressing recombinant GPR55 migrated towards FBS (Figure 3C). Furthermore, LPI enhanced FBS-induced migration in these cells (Figure 3C), in a similar manner to that observed in MDA-MB-231 cells (Figure 3C). LPI-enhanced migration of MCF-7 cells transfected with HA-GPR55 was completely prevented by co-transfection with siRNA to GPR55 (Figure 3C). MCF-7 cells over-expressing GPR55 also showed a significant increase in invasion towards FBS in a Cultrex® BME cell invasion assay (Figure 3D).
Figure 3.
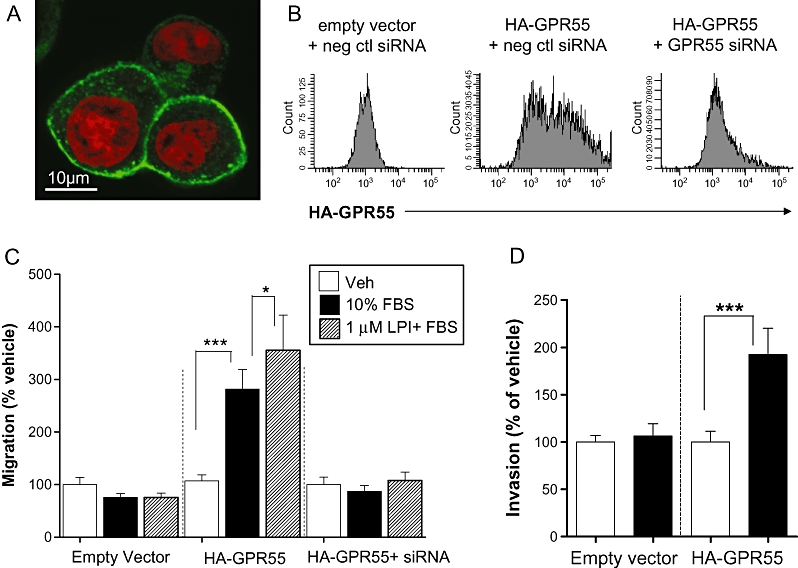
Histograms showing the migration of MCF-7 cells over-expressing GPR55 in a Boyden chamber transwell assay and Cultrex® 24-well BME invasion assay. (A) Detection of HA-GPR55 expression in MCF-7 cells by confocal microscopy using an anti-HA antibody. Green: HA-GPR55; Red: Nuclei. (B) Detection of HA-GPR55 over-expression in MCF-7 cells, and knock down using siRNA to GPR55, by flow cytometry. (C) MCF-7 cells over-expressing GPR55 migrated towards 10% FBS, whereas empty vector transfected cells did not. In GPR55 over-expressing cells, FBS-induced migration was significantly enhanced by pre-incubation of cells with LPI (1 µM), and was completely abolished using siRNA to GPR55. (D) MCF-7 cells over-expressing GPR55 invaded towards 10% FBS, whereas empty vector transfected cells did not. The data represent mean ± SEM (n= 3). *P < 0.05; ***P < 0.001, one-way anova followed by Newman–Keuls multiple comparison tests. FBS, foetal bovine serum; LPI, L-α-lysophosphatidylinositol.
LPI stimulates elongation and orientation of MDA-MB-231 cells on grooved substrata
Briefly, MDA-MB-231 cells were plated onto nanogrooved quartz slides in the presence or absence of LPI (1 µM). After 4 h the OI and length-to-width ratio (elongation) were calculated for individual cells (see Figure 4), as previously described (Rajnicek et al., 2008). MDA-MB-231 cells on nanogrooves 4 µm wide and 520 nm deep elongated (polarized) in response to groove detection when incubated with vehicle [0.01% (v/v) DMSO] compared with cells on flat surfaces (Figure 5A). Polarization was enhanced further in the presence of the GPR55 agonist LPI (1 µM), which caused a robust elongation on both grooved and flat surfaces. Shallower (40 nm deep) nanogrooves failed to polarize MDA-MB-231 cells significantly. However, as was observed on deeper grooves, LPI stimulated cell elongation on both flat and grooved substrata (Figure 5B). Under control conditions, almost all MDA-MB-231 cells detected and aligned with the 520 nm deep grooves, with an average OI of almost -1 (fully aligned) (Figure 5C). The OI values for vehicle-treated cells and those for LPI-treated cells were not different from control values. When cultured on 40 nm deep grooves, however, vehicle (DMSO)-treated cells were unable to detect (align with) the grooves; the OI was not significantly different from that on flat surfaces (P > 0.05; n= 3) (Figure 5D). In the presence of LPI, however, the OI was significantly different from vehicle-treated cells (P < 0.05; n= 3) and significantly different from the flat surface (P < 0.01; n= 3). Cells exposed to LPI also exhibited many more cell protrusions at the periphery than vehicle-treated cells (see representative images in Figure 5E). These may represent polarized lamellipodia.
Figure 4.
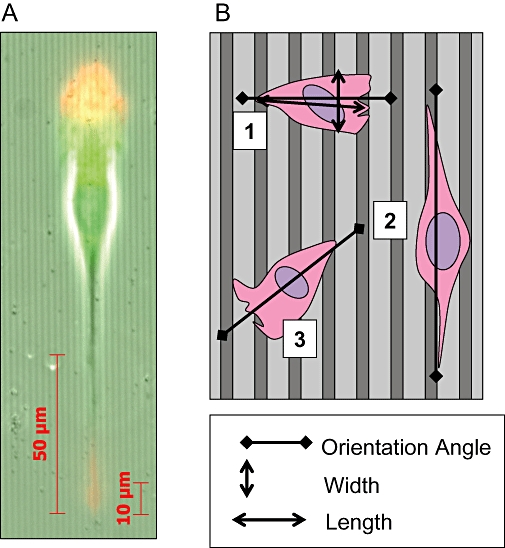
(A) Image showing a polarized MDA-MB-231 cell on grooved substratum, red: actin, green: tubulin. (B) Diagram representing the analysis of the effects of grooved substrata on cancer cells. Cell 1 is orientated at 90° relative to groove direction and is unaligned with an orientation index (OI) of +1. Cell 2 is orientated at 0° relative to groove direction and is fully aligned with an OI of −1. Cell 3 is orientated at 45° relative to groove direction with an OI of 0. The cell elongation (polarization) is represented as the length/width ratio.
Figure 5.
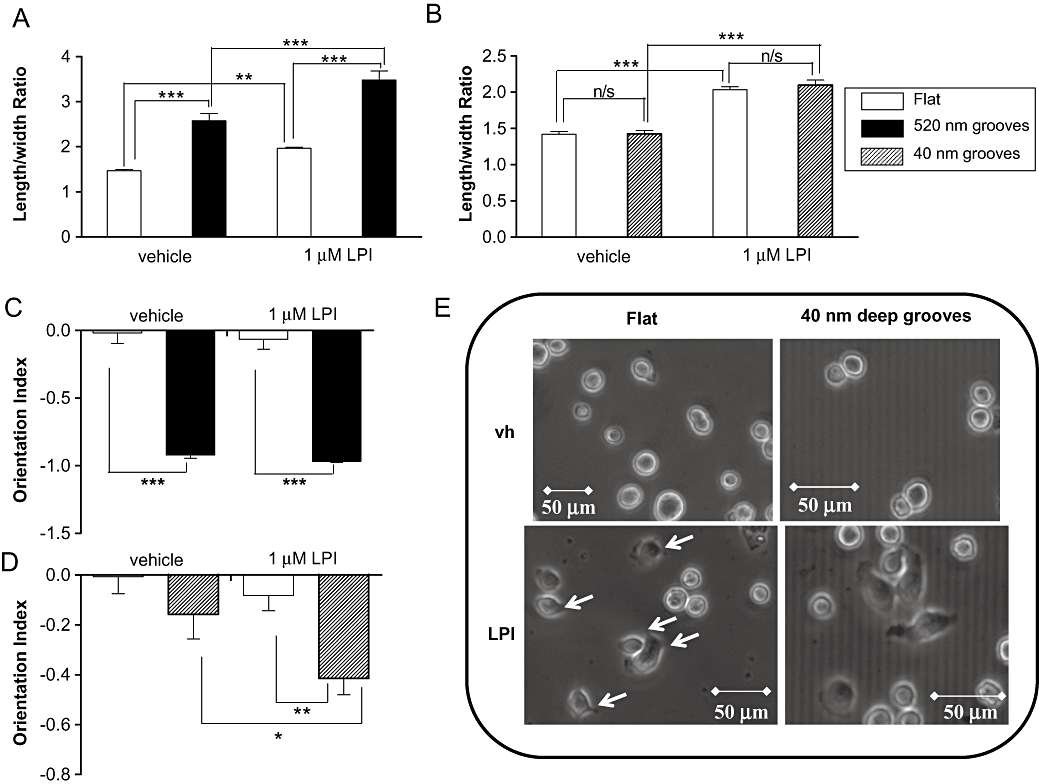
Histograms showing that MDA-MB-231 cells (A) become polarized (elongated) in response to 520 nm deep grooves, with a significant enhancement of polarity in response to LPI on both the flat and grooved surface, (B) do not elongate in response to 40 nm deep grooves, but elongate in the presence of LPI, (C) almost fully align in response to 520 nm deep nanogrooved slides, and (D) do not align in response to 40 nm deep grooves when vehicle-treated, but align in response to these grooves following incubation with LPI. Data are the mean ± SEM, n= 58–97 cells (from three separate experiments). (E) Representative phase contrast images of MDA-MB-231 cells cultured on nanogrooved slides showing vehicle-treated and LPI-treated MDA-MB-231 cells on flat surface, and on 4 µm wide, 40 nm deep grooves. *P < 0.05; **P < 0.01; ***P < 0.001, one-way anova followed by Newman–Keuls multiple comparison tests. LPI, L-α-lysophosphatidylinositol.
Over-expression of GPR55 increases orientation of MCF-7 cells
MCF-7 cells artificially over-expressing GPR55, or vector transfected control cells, were seeded on 520 nm-deep grooved slides. MCF-7 cells over-expressing GPR55 detected and aligned with the 1 µm wide grooves with an average OI significantly different from vector transfected control cells (P < 0.05; n= 39–76 cells) (Figure 6A). In contrast, while both GPR55- and vector-transfected control cells elongated in response to grooved slides, no significant differences were observed between these groups (P > 0.05; n= 19–54 cells) (Figure 6B). Representative images of GPR55- and vector-transfected control cells can be seen in Figure 6C.
Figure 6.
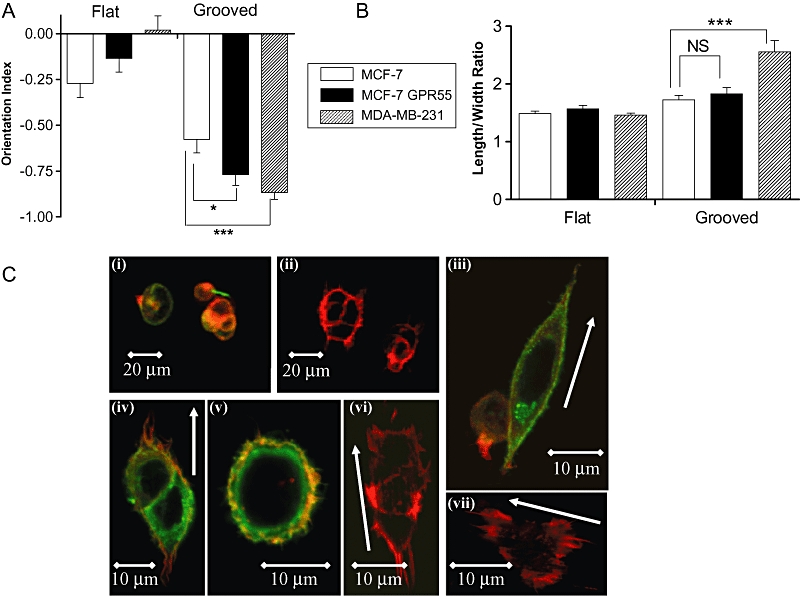
Histograms showing that (A) MCF-7 cells expressing GPR55 display a significantly greater degree of alignment to grooved substrata (1 µm wide, 520 nm deep) after 4 h than empty vector (EV) transfected cells, which display significantly less alignment than MDA-MB-231 cells. Data are the mean ± SEM, n= 39–76 cells. *P < 0.05; ***P < 0.001. (B) Polarization of MCF-7 cells expressing GPR55 is not significantly different from empty vector transfected cells; MDA-MB-231 cells display significantly greater elongation as compared with MCF-7 cells. ***P < 0.001, analysed by Students's unpaired t-test. (C) Representative images of MCF-7 cells artificially expressing GPR55, or EV transfected control cells, stained for HA-GPR55 (green) and actin (red). (i) HA-GPR55-expressing cells on flat surface, wide-view; (ii) vector-transfected cells on flat surface, wide-view; (iii and iv) HA-GPR55-expressing cells aligned along grooves; (v) HA-GPR55-expressing cells on flat surface; (vi and vii) vector-transfected cells on grooves. White arrows indicate groove direction.
Discussion
Cancer cell metastasis underlies the vast majority of cancer-related deaths. The immediate extracellular chemical and physical milieu (microenvironment) of a tumour cell is a key factor in determining metastatic fate (Robinson et al., 2009). In the context of anti-cancer drug development, it is therefore imperative to understand the signals that stimulate tumour cells metastasis. To our knowledge, this is the first report showing that the GPCR GPR55 is expressed in breast cancer cells. Furthermore, we found that the highly metastatic MDA-MB-231 cell line expressed 30-fold higher levels of GPR55 than the low-metastatic MCF-7 cell line, suggesting a possible link between elevated GPR55 expression levels and the metastatic potential of breast carcinomas. In addition, we demonstrate that over-expression of recombinant GPR55 in MCF-7 cells endowed these cells with the ability to migrate in response to serum factors and enhanced their ability to detect nanometric grooved substratum. This suggests that signals from GPR55 could control metastasis, in response to cues present in the microenvironment.
Oka et al. recently identified LPI as an endogenous ligand for GPR55 (Oka et al., 2007). Increased circulating levels of LPI, as well as LPA, are associated with late-stage or recurrent ovarian cancer, highlighting a potential role for this lysophospholipid in cancer progression (Xiao et al., 2000; Sutphen et al., 2004; Chen and Xu, 2001). However, in contrast to LPA, whose molecular targets and pro-tumour effects are well described (Stähle et al., 2003), little is known about the role of LPI or GPR55 in cancer cells. In line with a role for a GPCR in the actions of LPI, we demonstrated that LPI stimulated [35S]GTPγS binding to membranes prepared from MDA-MB-231 cells. Notably, the EC50 value of 340 nM for LPI is consistent with the potency of this compound as a GPR55 agonist (Henstridge et al., 2009; Oka et al., 2009). Furthermore, the stimulation of [35S]GTPγS binding to MDA-MB-231 cell membranes by LPI was attenuated by CBD, which has been reported to antagonize GPR55 (see Ross, 2009; Whyte et al., 2009). It is notable that, while CBD had no effect alone on basal levels of [35S]GTPγS binding, in combination with LPI there appeared to be a decrease in the basal binding, a finding that may be indicative of an allosteric interaction.
Here we demonstrated for the first time that LPI (1 µM) significantly enhanced migration of MDA-MB-231 cells. Clearly, future studies will be required to determine the potency of LPI as a stimulator of tumour cell migration. The stimulatory effect of 1 µM LPI was absent in the presence of CBD. However, this may not necessarily be due to direct antagonism, because CBD alone also significantly inhibited migration of MDA-MB-231 cells. When MCF-7 cells, which express low native levels of GPR55, were transiently transfected with GPR55, LPI significantly enhanced migration. This effect was abolished by siRNA against GPR55 and was not observed in cells transfected with empty vector. These findings demonstrate that GPR55 over-expression induced a pro-migratory phenotype in breast cancer cells, and suggested that the GPR55 ligand LPI may mediate its pro-migratory effects on breast cancer cells via GPR55 receptor activation. Furthermore, elevated GPR55 expression could be linked to the metastatic potential of tumour cells; therefore this receptor may mediate the oncogenic effects of LPI. A recent report demonstrated that LPI induces migration of prostate cancer cells via activation of TRPV2 (Monet et al., 2009). Here we report that LPI, by itself, did not act as a chemotactic factor for breast cancer cells (Figure 2A). To further implicate GPR55 as the receptor underlying the effects of LPI, numerous attempts were made to knock down native GPR55 in MDA-MB-231 cells; however, we were unable to detect any significant knock-down at the mRNA level. Interestingly, using the same pools of siRNAs, we were able to knock down recombinant (artificial) GPR55 expression in both MDA-MB-231 and MCF-7 cells.
GPR55 couples to Gα12/13 and activates the Rho family of small GTPases (see Ryberg et al., 2007; Ross, 2009). LPI has been demonstrated to activate RhoA in a cell line over-expressing recombinant GPR55 (Henstridge et al., 2009). Furthermore, LPI activates RhoA in both human and mouse osteoclasts; an effect that was absent in osteoclasts derived from GPR55-/- mice (Whyte et al., 2009). Tumour cell migration requires coordinated reorganization of the actin cytoskeleton, a process that requires the Rho family GTPases, Rho, Rac and Cdc42. Kelly et al. (2007) reported that the expression of Gα12 was significantly up-regulated in the early stages of breast cancer and that metastasis of breast cancer cells in vivo was attenuated by inhibition of Gα12 signalling.
Acquisition of a migrating ‘mesenchymal’ or elongated phenotype allows the cells to migrate during oncogenesis (Etienne-Manneville, 2008). Activation of Gα12 signalling promotes tumorigenesis and progression of nasopharyngeal carcinoma by modulating actin cytoskeleton reorganization and expression of epithelial-to-mesenchymal transition-related genes (Liu et al., 2009). Tumour cells adopt ‘elongated’ or ‘rounded’ forms of movement in response to their microenvironment; this is orchestrated by Rho and Rac (Croft and Olson, 2008). Mesenchymal-type movement is characterized by an elongated cellular morphology (Sanz-Moreno et al., 2008). Cancer cell elongation has been used as a measure of cancer cell polarity (Dalby et al., 2002; Ahmed et al., 2008) and MDA-MB-231 cells exhibit an elongated morphology associated with rapid, directed migration in response to mechanical contact guidance cues (Irimia and Toner, 2009). Here we demonstrated that on flat surfaces, LPI caused MDA-MB-231 cells to adopt an elongated phenotype. In this study we have used nanogrooved surfaces to provide an improved representation of the physiological microenvironment that promotes contact guidance of cancer cells. MDA-MB-231 cells cultured on 4 µm wide, 520 nm deep nanogrooved surfaces elongated to a greater degree than on flat surfaces, suggesting that these cells are highly responsive to environmental cues. On the 520 nm grooved surfaces, LPI caused a marked increase in cancer cell elongation. On shallow, 40 nm deep grooves, which more closely mimic the protrusions and pits of the basement membrane (Dalby, 2007), MDA-MB-231 cells did not elongate in response to grooves. However, in the presence of LPI these cells significantly elongated. These data suggest that LPI promotes a mesenchymal phenotype in these cells and indicate that modulation of cell polarity may underlie the pro-migratory effects of LPI in the chemotaxis assay. After 4 h incubation, MDA-MB-231 cells fully aligned with the 520 nm deep grooves. While vehicle-treated cells cultured on 40 nm deep grooves did not respond to the grooved substratum, those treated with LPI were significantly aligned. The increased cancer cell detection of nanogrooves in the presence of LPI, which activates GPR55, is in line with the signalling pathways which underlie cell orientation. Cells treated with LPI appear to develop filopodia, which are known to be involved in the cell sensing of nanogrooved surfaces (Dalby, 2007). Subsequent cell alignment involves actin cytoskeleton reorganization mediated by Rho activation but which is independent of signals downstream of Rac or Cdc42 activation (Rajnicek et al., 2008). In line with a role for GPR55 in cell orientation, MCF-7 cells over-expressing GPR55 displayed significantly greater alignment with grooved substratum as compared with vector controls.
In summary, we show here, for the first time, that the bioactive lipid LPI enhanced breast cancer cell chemotaxis and increased cellular polarization and orientation on nanogrooved substratum. LPI is a potent agonist at GPR55 and we demonstrated that artificial over-expression of GPR55 induced a pro-migratory phenotype in breast cancer cells, which was further enhanced by LPI. Moreover, artificial over-expression of GPR55 enhanced cancer cell orientation in response to grooved substratum. Our findings suggest that effects on the actin cytoskeleton may contribute to the documented pro-tumour effects of LPI and that GPR55 may play an important role in the modulation of the metastasis of breast cancer cells. In this study we were unable to knock down endogenous expression of GPR55 in cancer cells, and future experiments will be necessary to directly implicate GPR55 as the receptor mediating the effects of LPI in these cells.
Acknowledgments
L.F. was supported by a grant from Schering-Plough. R.A.R. is funded by NIH grants DA-03672, DA-09789.
Glossary
Abbreviations:
- CBD
cannabidiol
- FBS
foetal bovine serum
- GPCR
G-protein-coupled receptor
- GPR55
G-protein-coupled receptor 55
- LPA
lysophosphatidic acid
- LPI
L-α-lysophosphatidylinositol
- RT-PCR
reverse transcription-polymerase chain reaction
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- Ahmed T, Shea K, Masters JR, Jones GE, Wells CM. A PAK4-LIMK1 pathway drives prostate cancer cell migration downstream of HGF. Cell Signal. 2008;20:1320–1328. doi: 10.1016/j.cellsig.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Mathie A, Peters JA. Guide to receptors and channels (GRAC), 4th edn. Br J Pharmacol. 2009;158:S1–S254. doi: 10.1111/j.1476-5381.2009.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anavi-Goffer S, Fleischer D, Hurst DP, Lynch DL, Barnett-Norris, Shi S, et al. Helix 8 Leu in the CB1 cannabinoid receptor contributes to selective signal transduction mechanisms. J Biol Chem. 2007;282:25100–25113. doi: 10.1074/jbc.M703388200. [DOI] [PubMed] [Google Scholar]
- Chen YL, Xu Y. Determination of lysophosphatidic acids by capillary electrophoresis with indirect ultraviolet detection. J Chromatogr B Biomed Sci Appl. 2001;753:355–363. doi: 10.1016/s0378-4347(00)00582-x. [DOI] [PubMed] [Google Scholar]
- Croft DR, Olson MF. Regulating the conversion between rounded and elongated modes of cancer cell movement. Cancer Cell. 2008;14:349–351. doi: 10.1016/j.ccr.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Dalby MJ. Cellular response to low adhesion nanotopgraphies. Int J Nanomedicine. 2007;2:373–381. [PMC free article] [PubMed] [Google Scholar]
- Dalby MJ, Reihle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Nucleus alignment and cell signalling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2002;284:272–280. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- Dalby MJ, Hart A, Yarwood SJ. The effect of the RACK1 signalling protein on the regulation of cell adhesion and cell contact guidance on nanometric grooves. Biomaterials. 2008;29:282–289. doi: 10.1016/j.biomaterials.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- Henstridge CM, Balenga N, Ford LA, Ross RA, Irving AJ. The GPR55 ligand L-α-lysophosphatidylinositol promotes RhoA-dependant Ca2+ signalling and NFAT activation. FASEB. 2009;23:183–193. doi: 10.1096/fj.08-108670. [DOI] [PubMed] [Google Scholar]
- Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja J, Casey PJ. Role of G12 proteins in oncogenesis and metastasis. Br J Pharmacol. 2009;158:32–40. doi: 10.1111/j.1476-5381.2009.00180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser JP, Reinmann A, Bruinink A. The effect of topographic characteristics on cell migration velocity. Biomaterials. 2006;30:5230–5241. doi: 10.1016/j.biomaterials.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kelly P, Casey PJ, Meigs TE. Biological functions of the G12 subfamily of heterotrimeric G proteins: growth, migration, and metastasis. Biochemistry. 2007;46:6677–6687. doi: 10.1021/bi700235f. [DOI] [PubMed] [Google Scholar]
- Lauckner JE, Jensen JB, Chen HY, Lu HC, Hille B, Mackie K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc Natl Acad Sci USA. 2008;105:2699–2704. doi: 10.1073/pnas.0711278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SC, Jen YM, Jiang SM, Chang JL, Hsjung CS, Wang CH, et al. Gα12-mediated pathway promotes invasiveness of nasopharyngeal carcinoma by modulating actin cytoskeleton reorganisation. Cancer Res. 2009;69:6122–6130. doi: 10.1158/0008-5472.CAN-08-3435. [DOI] [PubMed] [Google Scholar]
- Monet M, Gkika D, Lehen'kyi V, Pourtier A, Vanden AF, Bidaux G, et al. Lysophospholipids stimulate prostate cancer cell migration via TRPV2 channel activation. Biochim Biophys Acta. 2009;1793:528–539. doi: 10.1016/j.bbamcr.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Oka S, Nakajima K, Yamashita S, Sugiura T. Identification of GPR55 as a lysophosphatidyinositol receptor. Biochem Biophys Res Commun. 2007;362:928–934. doi: 10.1016/j.bbrc.2007.08.078. [DOI] [PubMed] [Google Scholar]
- Oka S, Toshida T, Maruyama K, Nakajima K, Yamashita A, Sugiura T. 2-Arachidonoyl-sn-glycero-3 phosphoinositol: a possible natural ligand for GPR55. J Biochem. 2009;145:13–20. doi: 10.1093/jb/mvn136. [DOI] [PubMed] [Google Scholar]
- Rajnicek AM, Britland S, McCaig CD. Contact guidance of CNS neurites on grooved quartz: influence of groove dimensions, neuronal age and cell type. J Cell Sci. 1997;110:2905–2913. doi: 10.1242/jcs.110.23.2905. [DOI] [PubMed] [Google Scholar]
- Rajnicek AM, Foubister LE, McCaig CD. Alignment of corneal and lens epithelial cells by co-operative effects of substratum topography and DC electric fields. Biomaterials. 2008;29:2082–2095. doi: 10.1016/j.biomaterials.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Robinson BD, Sica GL, Liu YF, Rohan TE, Gertler FB, Condeelis JS, et al. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res. 2009;15:2433–2441. doi: 10.1158/1078-0432.CCR-08-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RA. The enigmatic pharmacology of GPR55. Trends Pharmacol Sci. 2009;30:156–163. doi: 10.1016/j.tips.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Ryberg E, Larsson N, Sjogren S, Hjorth S, Hermansson NO, Leonova J, et al. The orphan receptor GPR55 is a novel cannabinoid receptor. Br J Pharmacol. 2007;152:1092–1101. doi: 10.1038/sj.bjp.0707460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Stähle M, Veit C, Bachfischer U, Schierling K, Skripczynski B, Hall A, et al. Mechanisms in LPA-induced tumor cell migration: critical role for phoshorylated ERK. J Cell Sci. 2003;116:3835–3846. doi: 10.1242/jcs.00679. [DOI] [PubMed] [Google Scholar]
- Sutphen R, Wilbanks GD, Fiorica J, Grendys EC, Jr, LaPolla JP, Arango H, et al. Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1185–1191. [PubMed] [Google Scholar]
- Whyte LS, Ryberg E, Sims NA, Ridge SA, Mackie K, Greasley PJ, et al. The putative cannabinoid receptor GPR55 affects osteoclast function in vitro and bone mass in vivo. Proc Natl Acad Sci U S A. 2009;106:16511–16516. doi: 10.1073/pnas.0902743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Chen Y, Kennedy AW, Belinson J, Xu Y. Evaluation of plasma lysophospholipids for diagnostic significance using electrospray ionization mass spectrometry (ESI-MS) analyses. Ann NY Acad Sci. 2000;905:242–259. doi: 10.1111/j.1749-6632.2000.tb06554.x. [DOI] [PubMed] [Google Scholar]


