Abstract
The majority of fast inhibitory synaptic transmission in the mammalian nervous system is mediated by GABAA receptors (GABAARs). Here we report a novel interaction between the protein Maf1 and GABAAR β-subunit intracellular domains. We find Maf1 to be highly expressed in brain and enriched in the hippocampus and cortex. In heterologous cells and neurons we show Maf1 co-localises with GABAARs in intracellular compartments and at the cell surface. In neurons, Maf1 is found localised in the cytoplasm in dendrites, partially overlapping with GABAARs and inhibitory synapses and in addition is enriched in the neuronal nucleus. We also report that Maf1 interacts with a novel coiled-coil domain containing protein that we have called Macoco (for Maf1 interacting coiled-coil protein). Like Maf1, Macoco can also be found localised to inhibitory synapses and directly interacts with GABAARs. Expressing Macoco in neurons increases surface GABAAR levels. Our results suggest that Maf1 and Macoco are novel GABAAR interacting proteins important for regulating GABAAR surface expression and GABAAR signalling in the brain.
Introduction
GABAARs are chloride-selective ligand gated ion channels that mediate the majority of fast inhibitory synaptic transmission in adult mammalian central nervous system (CNS). They are heteropentameric oligomers assembled from seven subunit classes (α1–6, β1–3, γ1–3, δ, ε, π and θ). It is generally assumed that most benzodiazepine-sensitive GABAARs in the brain are assembled from at least 2α, 2β and 1 γ2 subunit (Arancibia-Carcamo and Kittler, 2009). Function of GABAARs is critically dependent on their correct assembly, transport and targeting to inhibitory synapses and the number of surface and synaptic GABAARs is an important determinant of inhibitory synapse strength (Arancibia-Carcamo and Kittler, 2009; Kittler and Moss, 2003). Surface receptor number can in part be regulated by trafficking and surface stability of receptors but the molecular machinery that regulates the forward transport and stability of receptors at the plasma membrane remains unclear. GABAAR subunit intracellular domains (ICDs) play a key role in regulating these processes as they are sites for post-translational modifications (including phosphorylation and palmitoylation), and protein interactions that are important for regulating receptor function, intracellular transport and localisation (Arancibia-Carcamo and Kittler, 2009).
There has been considerable interest in identifying GABAAR associated proteins important for regulating receptor distribution and signalling. To better understand GABAAR function and regulation we screened for GABAAR associated protein complexes using a yeast two-hybrid (Y2H) screen (Kittler, 2006). Here we report the characterisation of the proteins Maf1 and Macoco in the brain, which we identify as novel GABAAR β-subunit interacting proteins. We show that Maf1 is expressed in neuronal dendrites and at the cell surface but interestingly, is also found enriched in the nucleus. We also identify a novel protein complex between Maf1 and a new protein Macoco, which we find contains multiple coiled-coil domains. We demonstrate that Maf1 and Macoco are expressed in the nervous system with high levels of expression in the hippocampus and cortex and that altering the function of the Maf1/Macoco complex in neurons by expressing Macoco affects surface GABAAR number. Our results suggest that the Maf1/Macoco complex may be important for regulating GABAAR surface trafficking and stability.
Methods
Yeast two-hybrid screening
The major intracellular domain of the rat GABAAR β3 subunit was amplified by PCR and subcloned into the yeast expression vector pPC97 (Brakeman et al., 1997). The construct was transformed into yeast strain Y190 and transformants were selected on leucine-deficient (Leu) medium and co-transformed with a rat hippocampal cDNA library subcloned into pPC86 vector. Positive clones were selected on –Leu/-His/-Trp media containing 25 mM 3-aminotriazole and assayed for β-galactosidase activity. Plasmids from positive clones were then co-transformed with either the pPC97- α1, pPC97-α2, pPC97-β1 or pPC97-β3 (encoding the intracellular domain of the specified subunit) or empty pPC97 into yeast to confirm the specificity of the interaction. For the Y2H screen with Maf1, full length Maf1 was subcloned from pPC86 into pPC97 and used to screen the same hippocampal cDNA library following the same procedure as described above.
cDNA cloning
The Maf1YFP construct was generated by PCR cloning full length rMaf1 into the pEYFP-N1 (Clonetech) vector using HindIII/BamHI restriction sites. rMaf1 was also cloned by PCR into pRK5-myc using BamHI/HindIII restriction sites. A subsequent Y2H screen was performed using the full length Maf1 sequence as bait. This identified a sequence encoding a partial amino acid fragment (Macoco-Maf1 Binding Fragment). After a BLAST search, a full length sequence was assembled by overlapping BLAST searches of human EST clones. The predicted ORF was too large to be found in a single EST, so PCR of a human brain cDNA pool (Stratagene) was performed to amplify the full length human Macoco sequence. Full length Macoco and Macoco-MBF were N-terminally myc tagged by subcloning into pRK5-myc using BamHI/EcoR1 restriction sites. GST-Macoco was produced by subcloning full length Macoco into the BamHI/XhoI sites of pGEX-4T3 and transformed into E. coli BL21 cells. The GST-Macoco fusion protein was then produced as previously described (Kittler et al., 2005).
Antibodies
The following primary antibodies were used: mouse monoclonal to GFP (Roche) (WB 1:1000), rabbit anti-VIAAT (a kind gift from B. Gasnier, (Dumoulin et al., 1999) (IF 1:1000)), mouse anti-MAP2 (Sigma)(IF 1:500), guinea pig anti-γ2 (serum, (Benke et al., 1994; Kittler et al., 2001)(IF 1:100)), rabbit anti-Myc (Santacruz) (IP 1μg), mouse monoclonal to β3 (culture supernatant (WB 1:10) and affinity purified (IF 1:100), Neuromab), goat anti-LaminB (Santacruz) (IF 1:500), mouse monoclonal anti-gephyrin (Connex GmbH) (IF 1:100), rabbit polyclonal anti-Macoco to an epitope in a central region of the protein (anti-ccdc46; Atlas Antibodies) (WB 1:500), mouse monoclonal anti-Myc was obtained from 9E10 hybridoma cells (WB and IF, supernatant 1:100), mouse monoclonal anti-GAD was obtained from GAD6 hybridoma cells (IF 1:100), mouse monoclonal anti-SV2 was obtained from SV2 hybridoma cells (IF 1:100). Rabbit anti-β3 was raised against MBP-β3-(345–408) and purified on a GST-β3-(345–408) column (WB 1:100). For generation of the Maf1 antibodies, the cDNA encoding amino acids 1–260 of rat Maf1 was subcloned from the yeast pPC86 vector into the SaII/NotI sites of pGEX4T2 (Pharmacia) and transformed into E. coli BL21 cells. The fusion protein was produced and purified as described previously (Kittler et al., 2005). The purified fusion proteins were then used to inoculate rabbits (Cocalico Biologicals) and the resulting sera was affinity purified against GST-Maf1. For generation of the anti-Macoco N-terminal region antibodies, amino acids 1–111 of Macoco were sublcloned into the EcoR1/HindIII sites of pMAL-c2X (New England BioLabs) and transformed into E. coli BL21 cells. The fusion protein was produced and purified by immobilisation on amylose resin (New England BioLabs) and used to inoculate rabbits (as above). Macoco sera was affinity purified on a GST-Macoco column.
Cell culture
Cultures of cortical and hippocampal neurons were prepared from E18 Sprague-Dawley rats as described previously (Brandon et al., 1999). Cortical neurons were transfected by nucleofection before plating as previously described (Kittler et al., 2004; MacAskill et al., 2009b). Hippocampal neurons were transfected by calcium phosphate precipitation (MacAskill et al., 2009a). COS-7 cells were maintained in DMEM (GIBCO), supplemented with 10% heat inactivated foetal bovine serum and penicillin-streptomycin and transfected by electroporation (Kittler et al., 2004).
GST fusion protein pull-down assays from rat brain homogenate
Pull-downs from brain were performed as previously described (Smith et al., 2008). Briefly, adult rat brain was homogenised in pull-down buffer (50 mM HEPES pH 7.5, 0.5 % triton X-100, 150 mM NaCl, 1 mM EDTA, 1mM PMSF with antipain, pepstatin and leupeptin at 10 μg/ml) and solubilised for 2 hours. Solubilised material was ultracentrifuged and the supernatant (solubilised protein) was exposed to 10–40 μg GST fusion protein attached to glutathione agarose beads for 1 hour at 4°C. Beads were then washed 4 times with pull-down buffer to remove any non-specific binding and analysed by SDS-PAGE and western blotting.
Pull-down or immunoprecipitation experiments with transfected cell-lines
Transfected COS-7 cells were solubilised in 0.5 ml pull-down buffer (as above) for 1 hour and then centrifuged. The supernatant was incubated with 10–40 μg fusion protein or 2 μg of antibody for 2 hours. Protein A beads were added to the immunoprecipitation experiments for a subsequent 1hr incubation. Beads were washed 4 times with pull-down buffer and analysed by SDS-PAGE and western blotting.
In vitro translation of 35S-Methionine labelled protein and GST pull down
Macoco and Maf1 were in vitro translated using TNT SP6 Quick coupled Transcription/Translation system (Promega) and labelled with [35S]-Methionine (Amersham Biosciences) following the manufacturer’s protocol (Smith et al., 2008). 5 μl of labelled protein was incubated with 10μg GST fusion protein in pull-down buffer (as above) for 2 hours at 4°C. The beads were washed 3 times in pull-down buffer and resolved by SDS-PAGE and stained with coomassie blue. Radioactivity was detected with a phosphor storage screen and phosphorimager.
Surface biotinylation of cortical neurons
Biotinylation of cortical neuron cultures was carried out as previously described (Kittler et al., 2008). Briefly, cortical neurons were incubated on ice with biotin solution (Sulpho-NHS-biotin (PIERCE) at 0.5 mg/ml in PBS containing Ca2+/Mg2+) and quenched with quench buffer (PBS Ca2+/Mg2+ containing 1 mg/ml BSA). The cells were solubilised for 1 hour in RIPA buffer (50 mM Tris pH 7.5, 1 mM EDTA, 2 mM EGTA, 150 mM NaCl, 1 % NP40, 0.5 % DOC, 0.1 % SDS, 1 mM PMSF with antipain, pepstatin and leupeptin 10 μg/ml) and the lysates were then centrifuged to pellet cell debris. 15 % of the supernatant was taken to use as a total protein sample and the remainder was incubated for 2 hours with 50 μl Ultralink immobilised NeutrAvidin (PIERCE) 50 % slurry at 4°C to precipitate biotin labeled membrane proteins. Beads were then washed three times in RIPA buffer and analysed by SDS-PAGE and western blotting. Biotinylated surface GABAARs were identified by using anti-β3 primary antibody and detection of enhanced chemilluminescence from HRP-coupled anti-rabbit secondary antibodies followed by detection with an ImageQuant LAS4000 mini imaging system and analysis with ImageQuant software (GE Healthcare).
Immunocytochemistry
Coverslips for surface or whole cell staining were fixed with PFA (4% paraformaldehyde/4% sucrose/PBS pH7) for 4 or 10 minutes respectively before being blocked with block solution (PBS, 10 % horse serum, 0.5 % BSA) for ten minutes. Coverslips for whole cell staining were incubated in block solution containing 0.2 % Triton X-100. Antibody dilutions were carried out in block solution and washes with PBS. One hour incubations with primary antibodies (described above) were followed with one hour incubations with secondary antibodies: Alexa 633 conjugated anti-rabbit, Alexa 594 conjugated anti-guinea pig, anti-goat and anti-mouse, Alex 488 conjugated anti-rabbit and anti-mouse (Molecular Probes 1:1000). After extensive washing, coverslips were mounted on microscope slides using ProLong Gold antifade reagent (Invitrogen) and sealed with nail varnish. Images were aquired using a Zeiss LSM 510 META confocal microscope and digitally captured using LSM software. Quantification of colocalisation was performed as previously described (Twelvetrees et al., 2010). Briefly, 25μm sections of dendrite were straightened using the ImageJ plugin Straighten (Eva Kocsis, http://rsbweb.nih.gov/ij/plugins/straighten.html) which allowed the plane of the dendrite to be rotated by 180°. The integrated colocalisation of GABAARs with Maf1 or Macoco was then determined using Metamorph software and then the GABAAR plane was rotated by 180° to give a random distribution of GABAAR and the integrated colocalisation was determined again (using Metamorph).
In situ hybridisation
DIG-labelled riboprobes representing the coding sequence of mouse Maf1 (Accession number NM_026859) and Macoco (NM_029606) were hybridised to cryosections as previously described (Isaacs et al., 2003). Briefly, whole mouse brains were frozen in OCT (Merck, Darmstadt, Germany) on dry ice, and 14 μm parasagittal cryosections were cut and mounted on positively charged slides. Digoxigenin-labeled riboprobe synthesis and hybridization were performed essentially as described previously (Wilkinson, 1992).
Immunohistochemistry
Male C57BL/6 mice, 8–12 weeks old, were deeply anesthetized and intracardially perfused with saline solution followed by 4% paraformaldehyde, post-fixed overnight and cryoprotected in 30% sucrose. Free-floating sections were cut at 40 μm using a freezing microtome and stored in cryoprotectant at −20°C until processing. Immunostaining was performed as previously described (Kuramoto et al., 2007; Liu et al., 2008). Sections were mounted onto slides, air-dried, dehydrated through graded alcohols followed by xylenes and then mounted for viewing. Sections were visualized with an Olympus BX51 microscope (Olympus Optical).
Results
A novel GABAAR β-subunit associated protein Maf1 and its expression in brain
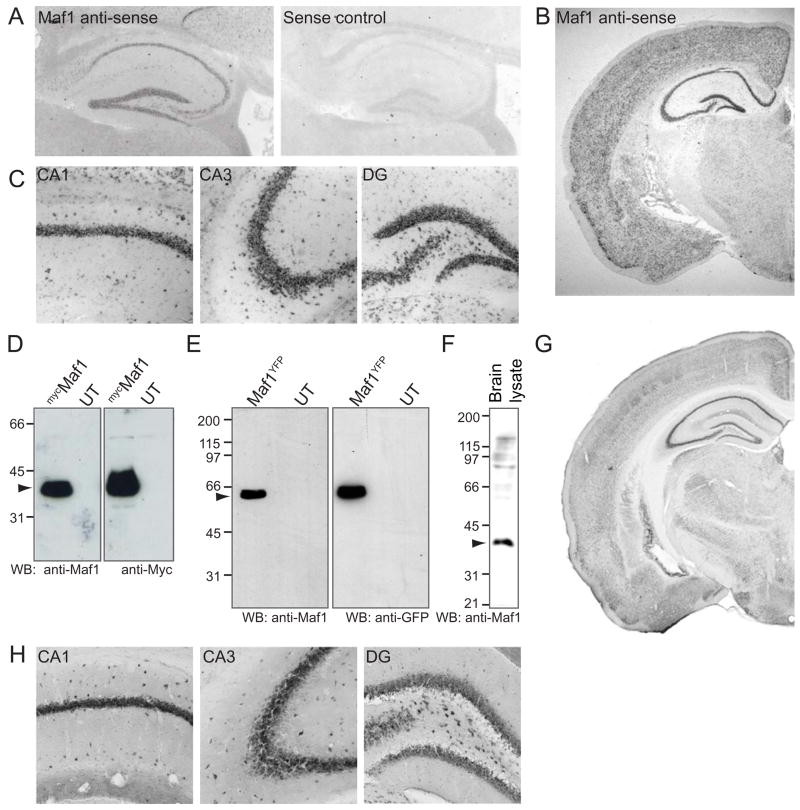
To identify proteins that regulate the activity and function of GABAARs, we carried out a yeast two-hybrid (Y2H) screen of a rat hippocampal library with a bait encoding the major intracellular domain of the GABAAR β3 subunit (amino acids 302–425). This screen identified an interacting clone encoding the sequence of a rat protein of 260 amino acids (Supplementary Fig. 1), blast analysis of which revealed that it was very similar to a previously reported human Maf1 sequence (Johnson et al., 2007). Y2H analysis revealed that Maf1 interacted strongly with the GABAAR β3–subunit and to a lesser extent with the GABAAR β1-subunit, but not GABAAR α1 or α2 subunits (data not shown). In situ hybridisation analysis of mouse brain revealed significant mRNA localisation of Maf1 in brain, with high levels of expression in hippocampus (Fig. 1A) and cortex (Fig. 1B). In the hippocampus, Maf1 mRNA was localised in CA1, CA3 and dentate gyrus (Fig. 1C). Northern blot analysis using full length rMaf1 cDNA confirmed the expression of Maf1 RNA in adult brain (data not shown).
Figure 1. Maf1 is highly expressed in the hippocampus and cortex.
(A) In situ hybridisation of mouse parasagittal cryosections with Maf1 anti-sense or sense control DIG-labelled riboprobes showing Maf1 RNA localisation in the hippocampus. (B,C) In situ hybridisation of whole brain showing Maf1 localisation in the cortex. Close-up images of hippocampus showing Maf1 localisation in CA1, CA3 and dentate gyrus. (D–H) Characterisation of Maf1 antibody. (D) Lysates of COS cells expressing mycMaf1 were probed with anti-Maf1 and anti-Myc antibodies demonstrating the specificity of the Maf1 antibody. (E) Lysates of COS cells expressing Maf1YFP were probed with anti-Maf1 and anti-GFP antibodies demonstrating the specificity of the Maf1 antibody. (F) 50 μg brain lysate probed with anti-Maf1 identifies a band of approximately 35 kDa. (G,H) Immunohistochemistry of mouse brain sections with anti-Maf1, showing a high level of Maf1 expression in the hippocampus, specifically in CA1, CA3 and the dentate gyrus (H).
We tagged rat Maf1 at its amino terminus with the myc epitope tag (mycMaf1) or at its carboxyl terminus with yellow fluorescent protein (YFP; Maf1YFP). As no Maf1 antibodies were available, to further study the Maf1 protein we generated antibodies to full length rat Maf1. These antibodies could also specifically recognise myc (Fig. 1D) or YFP tagged Maf1 from COS7 cell lysates by western blotting (Fig. 1E). Western blotting of brain lysate demonstrated that the anti-Maf1 antibodies also recognised a band of approximately 35 kDa, the expected molecular weight of Maf1 (Fig. 1F). In addition, we showed by immunofluorescence of mycMaf1 transfected cells, almost complete overlap between fluorescent signals for Maf1 as detected by anti-myc antibodies and anti-Maf1 antibodies (Supplementary Fig. 2A). Thus our Maf1 antibody appears to be highly specific for Maf1 protein. We used the Maf1 antibody to examine the expression of Maf1 in mouse brain sections using immunohistochemistry. Maf1 was found to be expressed in most brain regions with high levels of expression in the hippocampus and cortex (Fig. 1G), similar to the distribution of Maf1 revealed by in situ hybridisation (Fig. 1A, B). In the hippocampus, Maf1 showed high levels of expression in CA1, CA3 and dentate gyrus (Fig. 1H). Thus, Maf1 appears to be highly expressed in the adult nervous system.
Maf1 interacts directly with GABAAR β-subunit intracellular domains
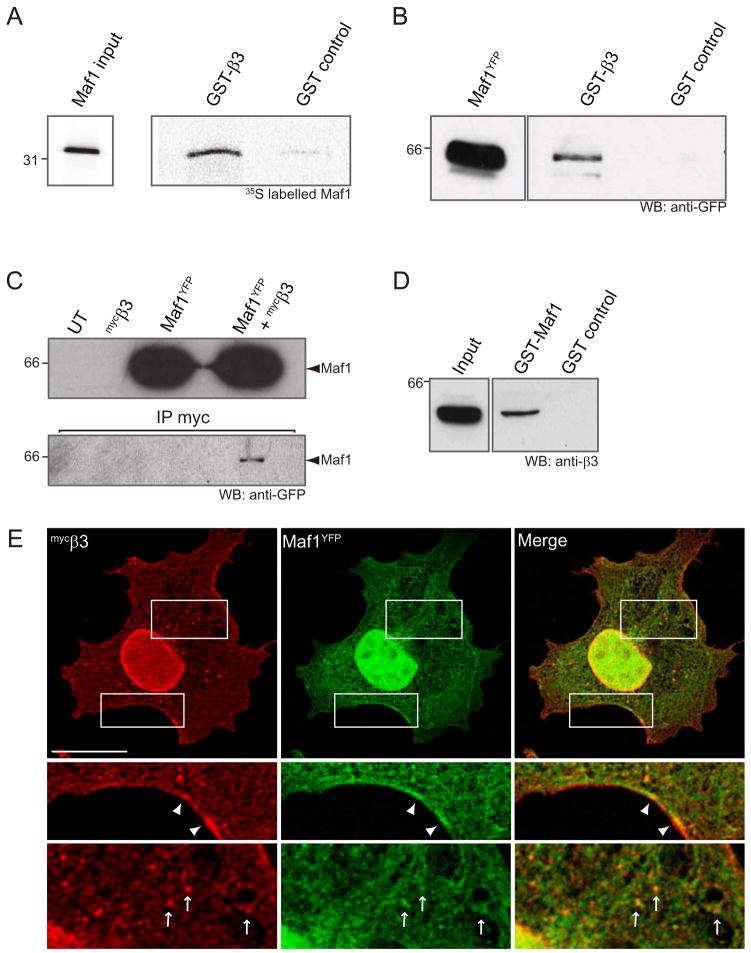
To confirm the direct interaction of Maf1 with GABAAR β-subunit intracellular domains observed using the Y2H system we used glutathione-S-transferase (GST) affinity chromatography with a GST fusion protein of the GABAAR intracellular domain and investigated its interaction with in vitro translated 35S labelled full length Maf1. Using this approach, we observed a direct interaction of Maf1 with the GABAAR β3-subunit intracellular domain (Fig. 2A), supporting our Y2H results.
Figure 2. Maf1 interacts directly with the intracellular domain of the β3 subunit of GABAARs.
(A) Pull down of [35S]-methionine labelled Maf1 with GST-β3 or GST alone. (B) Pull down from COS cell lysates expressing Maf1YFP with GST-β3 or GST alone. (C) Co-immunoprecipitation of myc tagged GABAAR β3-subunits (mycβ3) and Maf1YFP complexes with anti-myc antibodies from COS cell lysates. (D) Pull-down of GABAARs from brain lysate with GST-Maf1 or GST alone. (E) Immunofluorescence of Maf1 and co-transfected GABAAR β3 subunits co-expressed in COS cells demonstrates co-localisation at the plasma membrane (arrowheads) and in intracellular compartments (arrows) (scale bar = 20μm).
To further investigate the interaction of GABAAR β3-subunits with Maf1 in a more cellular context, we exposed GST fusion protein of the GABAAR β3 subunit ICD or GST alone to lysates of COS cells expressing Maf1YFP. Proteins bound to GST-β3 were then resolved by SDS-PAGE followed by western blotting with anti-GFP antibodies. In agreement with the in vitro translation pull down assay, the GST-β3 subunit fusion protein interacted with Maf1YFP (Fig. 2B). Given that receptor β-subunits are essential components for the functional expression of GABAARs these results suggest that Maf1 binding may be a common property for most GABAAR subtypes. We also tested the interaction of Maf1 with GABAARs via co-immunoprecipitation of GABAAR subunits and Maf1 expressed in COS cells. We exploited the ability of GABAAR β3 subunits to assemble into pentameric cell surface homomeric ion channels, a property that depends on residues within the N terminus of this subunit (Taylor et al., 1999; Wooltorton et al., 1997). Maf1YFP and mycβ3 cDNAs were transiently transfected into COS cells. Antibodies against myc were used to immunoprecipitate GABAAR β3 subunits and bound precipitated complexes were resolved by SDS-PAGE and analysed by western blotting with antibodies against GFP. Using this approach a small amount of Maf1 was found to co-immunoprecipitate with GABAAR β3-subunits from cell lysates (Fig. 2C). We also carried out pull down assays to investigate the interaction of Maf1 with GABAARs in brain. As our antibodies to Maf1 were raised to GST-Maf1 we were unable to carry out pull down experiments with the intracellular domain of the GABAAR β3 subunit and probe for Maf1 (due to cross reactivity with GST). Instead, we performed pull down assays from brain lysates with GST-Maf1 and probed with antibodies to the GABAAR β3 subunit (Fig. 2D). This confirmed that GABAAR β3 containing receptors could be readily pulled down by GST-Maf1 from brain, further supporting the interaction of these two proteins in native systems. We were unable to co-immunoprecipitate Maf1 and GABAARs in vivo, a similar problem as encountered with a number of other GABAA and GABAB receptor associated proteins including Grif-1 (Beck et al., 2002), 14-3-3 (Couve et al., 2001), GODZ (Keller et al., 2004) and gephyrin (Tretter et al., 2008). This may be due to this being a low affinity, transient interaction in neurons.
To further study the interaction of Maf1 with GABAARs we co-expressed Maf1YFP and myc tagged GABAAR β3 subunits, taking advantage of their ability to form monomeric receptors that can access the cell surface when expressed in cells lines (Arancibia-Carcamo et al., 2009; Taylor et al., 1999) and investigated the distribution of these proteins. When expressed in COS cells, Maf1 exhibited significant cytoplasmic distribution in addition to a substantial enrichment in the nucleus (Fig. 2E). Nuclear localisation of Maf1 was confirmed by immunofluorescent co-localisation with endogenous lamin B to mark the nucleus (Supplementary Fig 2B). When co-expressed with GABAAR β3 subunits, Maf1 exhibited co-localisation with these receptors in puncta in the cell cytoplasm and could also be detected at the plasma membrane where it co-localised with surface β3-subunits (Fig. 2E).
Subcellular distribution of Maf1 in neurons
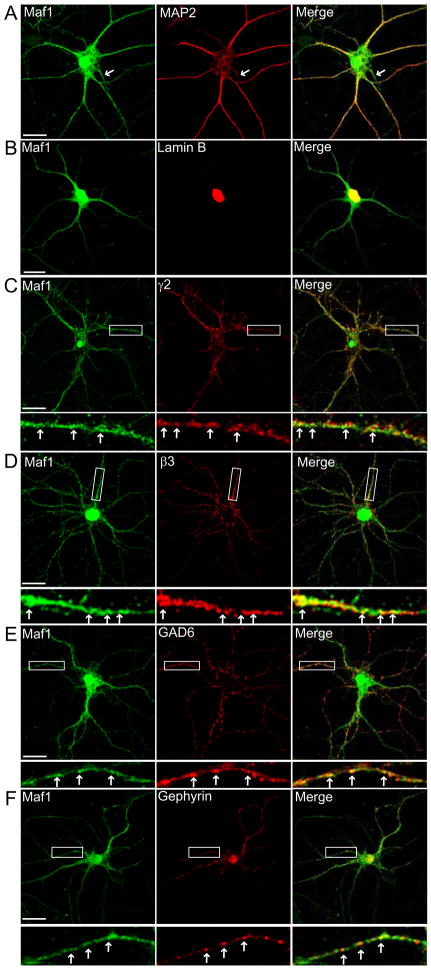
We investigated the subcellular distribution of endogenous Maf1 in cultured hippocampal neurons by immunofluorescence and confocal microscopy. In order to determine in which neuronal compartments Maf1 is localised, we carried out double labelling experiments with our antibody to Maf1 and an antibody to the neuronal marker MAP2 which is localised to the somatodendritic region of hippocampal neurons in culture (Craig and Banker, 1994). Maf1 was detected in MAP2-positive cell soma and processes (Fig. 3A). Maf1 in the cell soma appears to be localised both to the cytosol and, in addition, is also strongly localised to the nucleus, similar to that observed in COS cells. Nuclear Maf1 localisation in neurons was confirmed by co-localisation with the nuclear localised protein lamin B (Fig. 3B). In addition, Maf1 was also clearly localised along MAP2 positive processes where it had a clustered distribution along dendrites (Fig. 3A). Maf1 could also be detected in a MAP2-negative axonal process (Fig. 3A, see arrows). These results suggest that Maf1 can be found in both somatodendritic and axonal compartments of neurons in addition to being localised to the neuronal nucleus. A similar distribution of Maf1 was also observed for Maf1YFP transfected hippocampal neurons (Supplementary Fig. 2C). Maf1YFP is clearly distributed out along neuronal processes in addition to being enriched in the neuronal somata and nucleus.
Figure 3. Maf1 subcellular localisation in hippocampal neurons.
Maf1 is localised to the dendrites, axons and nuclei of hippocampal neurons as demonstrated by co-staining with antibodies to MAP2 (arrows= MAP2 negative process) (A), and Lamin B (B). (C,D) Maf1 co-localises with both the γ2 and β3 GABAAR subunits in hippocampal neurons. (E,F) Maf1 is found in clusters apposed to GAD6 labelled terminals and co-localised with gephyrin, a major component of the inhibitory postsynapse (scale bar = 20μm, arrows = co-localised puncta).
The subcellular distribution of Maf1 with respect to inhibitory synaptic domains and GABAARs in cultured hippocampal neurons was investigated using antibodies against the GABAAR β3 and γ2 subunits. The GABAAR β3 subunits or γ2 subunits appeared as membrane clusters in addition to abundant intracellular aggregates (Fig. 3C,D) and a proportion of Maf1 staining in neuronal processes was found to significantly co-localise with GABAAR staining (Fig. 3C,D; arrows), compared to a randomised control (Supplementary Fig. 4A). We could also demonstrate co-localisation of Maf1 with other markers of inhibitory synapses including glutamic acid decarboxylase (GAD), a marker of inhibitory presynaptic terminals and the synaptic scaffold protein gephyrin, a marker of inhibitory post-synaptic domains (Fig. 3E, F).
Association of Maf1 with a novel protein Macoco
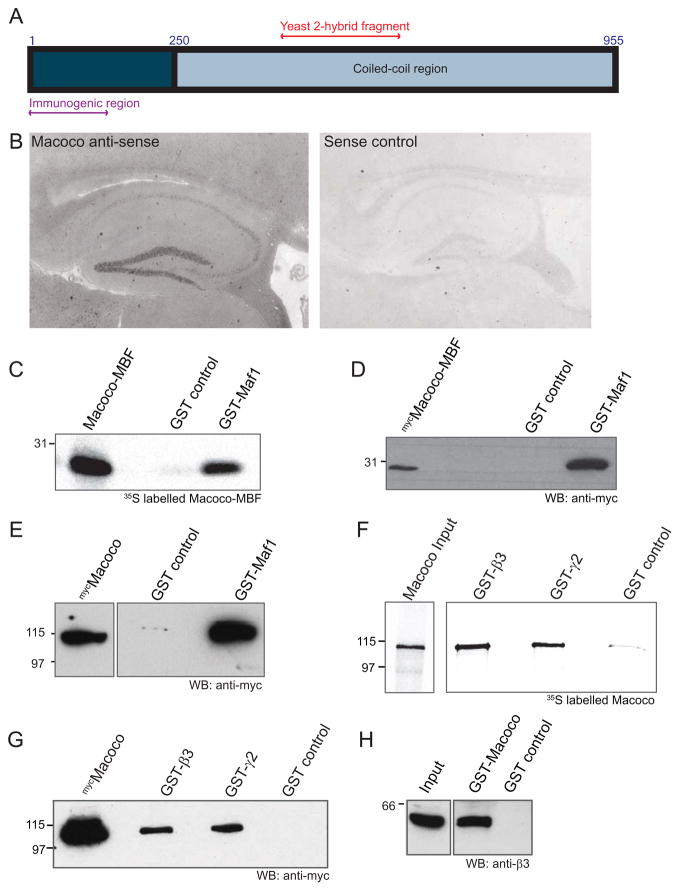
We reasoned that identifying proteins that interact with Maf1 may help to further understand the function of Maf1 in the brain and with respect to the GABAAR. This strategy has been proven to be effective in the identification of several proteins important for regulating the construction and plasticity of synapses, and for example led to the identification of GKAPs and proSAP/SHANKs as downstream interactors of PSD-95 and NMDA receptors (Boeckers et al., 2002; Sheng and Kim, 2000) and collybistin as an interactor of gephyrin (Kins et al., 2000). To identify proteins that interact with Maf1, we carried out a Y2H screen of a rat hippocampal library, with a bait encoding the full length sequence of rat Maf1 and identified an interacting clone that encoded a partial amino acid sequence. Database searches revealed a match to a highly conserved large predicted open reading frame protein containing multiple coiled-coil regions. We cloned the full length human version of this protein (highly homologous to rat and mouse sequences of the protein) by PCR from a human brain 1st strand cDNA pool and named this protein Macoco (for Maf1 associated coiled-coil protein; for alignment of the full length human protein sequence with the rat partial clone see supplementary Fig. 3). Human Macoco encodes for a protein of 955 amino acids, highly enriched in coiled-coil domains and with a predicted molecular weight of approximately 115 kDa (Fig. 4A). We confirmed the expression of Macoco in mouse brain by in situ hybridisation analysis which revealed localisation of Macoco mRNA in neurons, including in hippocampus (Fig. 4B), confirming that Maf1 and Macoco can both be found expressed in the same neuronal regions and cell types.
Figure 4. Maf1 associates with a novel 113 kDa protein Macoco.
(A) Schematic diagram of human Macoco showing its large coiled-coil domain, the region used to produce the anti-Macoco antibody and the region from the Y2H screen. (B) In situ hybridisation of mouse parasagittal cryosections with Macoco anti-sense or sense control DIG-labelled riboprobes shows the localisation of Macoco in the hippocampus. (C) The Macoco fragment pulled out of the Y2H screen with Maf1 (Macoco-Maf1 Binding Fragment) was in vitro translated, labelled with [35S]-methionine and subjected to a pull-down assay with GST-Maf1 or GST alone. (D,E) Pull-down from lysates of COS cells expressing mycMacoco-MBF or full length mycMacoco with GST-Maf1 or GST alone. (F) Pull-down of [35S]-methionine labelled full length Macoco with GST-β3, GST-γ2 or GST alone. (G) Pull down from COS cell lysates expressing full length mycMacoco with GST-β3, GST-γ2 or GST alone. (H) Pull-down of GABAARs from brain lysate with GST-Macoco or GST alone.
To confirm the direct interaction of Maf1 and Macoco, we first used GST affinity chromatography with GST-Maf1 and the in vitro translated 35S labelled Macoco fragment pulled out from our Y2H screen with Maf1 (termed Macoco-Maf1 Binding Fragment; Macoco-MBF), followed by SDS-PAGE and phosphorimaging. 35S-labelled Macoco-MBF (which expresses as an approximately 30 kD fragment) could be readily pulled-down by GST-Maf1 in this assay (Fig. 4C). Similar pull-down experiments from COS7 cell lysates expressing a myc tagged version of the Macoco-MBF further supported the interaction of the Macoco-MBF with Maf1 in a cellular system (Fig. 4D). Finally we performed pull-down experiments with GST-Maf1 and a myc tagged version of full length Macoco (mycMacoco) and similarly found that full length Macoco readily interacted with GST-Maf1 in the pull-down assay (Fig. 4E).
We also tested the binding of Macoco to the GABAAR. GST-fusion proteins of the intracellular domains of GABAAR β3 and γ2 subunits (GST-β3 and GST-γ2), or GST alone as a control, were exposed to 35S methionine labelled Macoco. The pull-down assays revealed a direct interaction of Macoco with GST-β3 and GST-γ2 subunit intracellular domains (Fig 4F). The interaction of Macoco with GABAARs was further confirmed with pull-down assays with GST-β3 and GST-γ2 from lysates of COS cells expressing mycMacoco (Fig 4G). We then carried out pull-down assays from brain lysates with GST-Macoco and probed with antibodies to the GABAAR β3 subunit (Fig. 4H) which confirmed that GABAAR β3 subunit containing receptors could be readily pulled-down by GST-Macoco from brain, further supporting the interaction of these two proteins in an endogenous system. Similar to our experiments with Maf1, we were unable to co-immunoprecipitate Macoco with GABAARs from brain lysates (as for several other GABAAR associated proteins) further supporting the possibility that the GABAAR Maf1/Macoco complex is either a transient or low affinity interaction.
Co-localisation of Macoco with Maf1 and GABAARs in cell lines
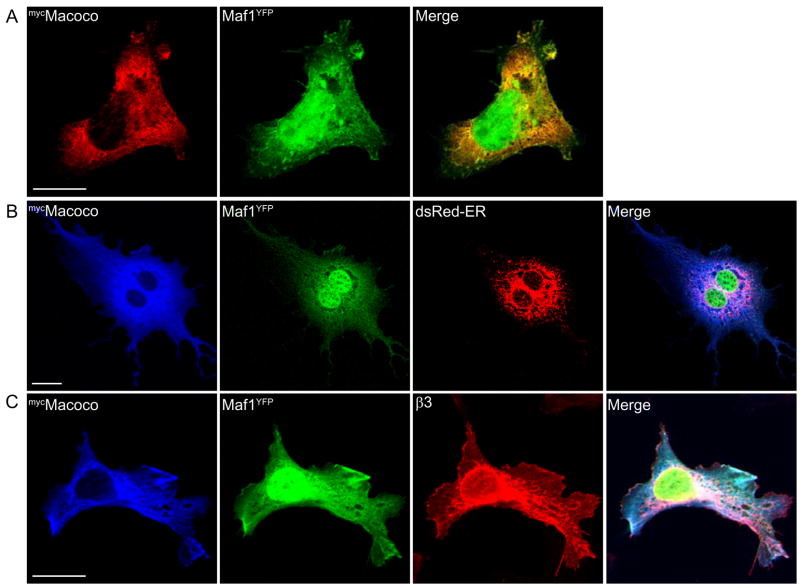
To further investigate the interaction of Maf1 and Macoco, we co-expressed mycMacoco with Maf1YFP in cells lines and investigated the distribution of these proteins. When mycMacoco was co-expressed with Maf1YFP, significant co-localisation of the two proteins could be observed in the cell cytoplasm. In contrast to Maf1 however, we did not observe localisation of Macoco in the cell nucleus (Fig. 5A). A substantial amount of Maf1YFP and mycMacoco were found to be localised to a perinuclear compartment that substantially overlapped with the endoplasmic reticulum, as determined by triple co-localisation with co-transfected DsRed-ER (Fig. 5B). Triple expression of Maf1YFP, mycMacoco and GABAAR β3 subunits revealed substantial overlap of the three proteins in a perinuclear compartment as well as at the plasma membrane (Fig. 5C).
Figure 5. Co-localisation of Macoco with Maf1 and GABAARs in cell lines.
Immunofluorescent staining of COS cells co-expressing mycMacoco and Maf1YFP(A), mycMacoco, Maf1YFP and dsRed-ER (B) and mycMacoco, Maf1YFP and the β3 subunit of GABAARs (C) (scale bar = 20 μm).
Characterisation of Macoco antibodies and localization of Macoco in neurons
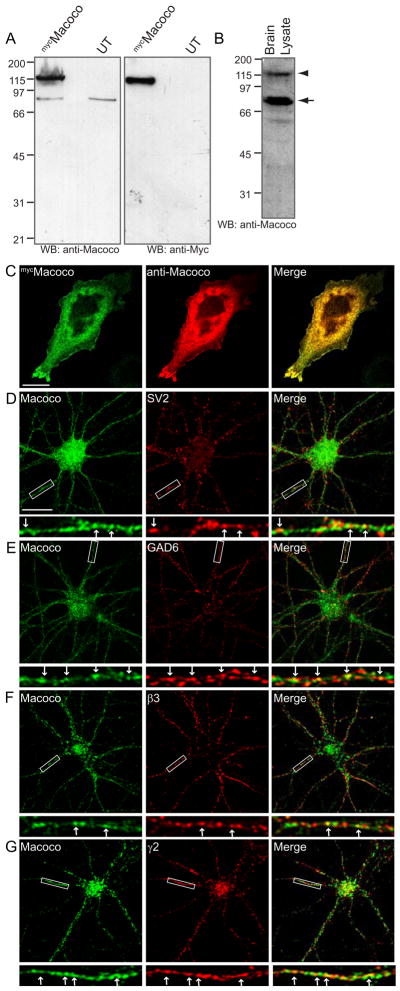
To further characterise the Macoco protein expression in neurons, we generated anti-Macoco antibodies and examined the localisation of Macoco proteins in cultured hippocampal neurons using immunofluorescence and confocal microscopy. An antibody was raised to an N-terminal region in Macoco (Fig 4A and supplementary Fig. 3) and the specificity of this antibody was initially tested in cell lines. Western blots of mycMacoco transfected cell lysates readily detected Macoco using either anti-myc antibodies or the anti-Macoco antibody (Fig. 6A). The anti-macoco antibody also recognised a similar sized band at 115 kDa in rat brain (Fig. 6B); arrowhead. In western blots of brain lysate the antibody also recognised an additional band at approximately 75 kDa (arrow) which likely represents a Macoco splice variant or degradation product (Fig. 6B). In agreement with this, a second commercially available anti-Macoco antibody raised to a central epitope in Macoco (Supplementary Fig. 3), and which also readily detected mycMacoco from transfected COS cell lysates (Supplementary Fig. 5B) also recognised bands at 115 and 75 kDa in western blots of brain lysate (Supplementary Fig. 5A).
Figure 6. Macoco antibody characterisation and localisation of Macoco in hippocampal neurons.
(A–B) Anti-Macoco antibody characterisation. (A) Lysates of COS cells expressing mycMacoco were probed with anti-Macoco or anti-myc antibodies. (B) 50μg brain lysate probed with anti-Macoco identifies bands of approximately 115 (arrowhead) and 75 kDa (arrow). (C) mycMacoco transfected COS cells stained with anti-myc and anti-Macoco (scale bar = 20 μm). (D–G) Localisation of Macoco in hippocampal neurons. Macoco co-localises with the synaptic marker SV2 (D) and the inhibitory synaptic marker GAD6 (E). (F,G) Macoco co-localises with the β3 and γ2 GABAAR subunits (scale bar = 20 μm, arrowheads = co-localised puncta).
Our N-terminal anti-Macoco antibody also readily recognised Macoco expressed in COS cells as confirmed in double immunofluorescence co-localisation experiments with antibodies to Macoco and to the myc epitope (Fig 6C). Using our anti-Macoco antibody to stain hippocampal neurons that had been maintained in culture for 2–3 weeks, the distribution of endogenous Macoco was investigated and found to be similar to that of hippocampal neurons transfected with mycMacoco (Supplementary Fig. 2E). Neurons double labelled with anti-Macoco and antibodies to the synaptic marker SV2 (Fig. 6D) demonstrated that Macoco could be found localised to synapses in neurons. Similar double staining experiments were performed to examine the localisation of Macoco specifically at inhibitory synapses using antibodies to inhibitory presynaptic terminals (GAD6). Importantly, co-localisation between Macoco and GAD6, seen as yellow staining in the merged panels, could be detected in dendrites (Fig. 6E). Additionally, a proportion of Macoco was found to be clustered and significantly co-localised with both the β3 and γ2 subunits of GABAARs (Fig. 6F,G and Supplementary Fig. 4B).
The Maf1/Macoco complex regulates the number of surface GABAARs
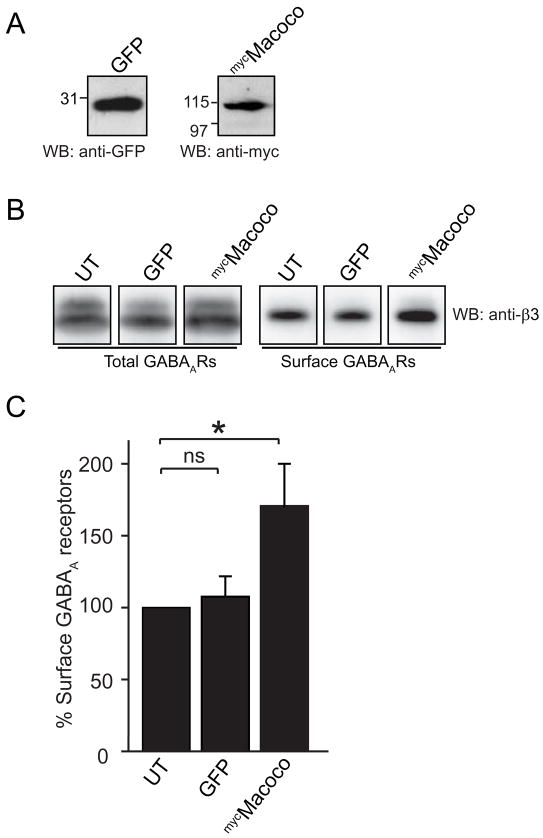
To further examine the role of the Maf1/Macoco protein complex in regulating the function of the GABAAR we exogenously expressed Macoco in neurons to investigate the consequences for GABAAR trafficking of increased Macoco protein levels in cultured cortical neurons. We have previously shown that nucleofection can achieve high transfection efficiency of cultured cortical neurons with exogenous cDNA constructs (Kittler et al., 2004). We hypothesised that increasing expression of Macoco would enhance endogenous Macoco function. A similar expression strategy has been effectively used to study the functional consequences of other GABAAR associated proteins such as GABARAP (Boileau et al., 2005; Chen et al., 2005; Leil et al., 2004), Plic1 (Saliba et al., 2008) and HAP1 (Kittler et al., 2004). Cultured cortical neurons were nucleofected with full length mycMacoco or GFP as a control, and we were able to routinely detect expression of both constructs by western blotting lysates of the nucleofected neurons (Fig. 7A). We then examined the fate of surface GABAARs by using a surface biotinylation assay to compare surface GABAAR expression levels in neurons transfected with Macoco or control GFP compared to untransfected neurons. We detected a significant 71.0 ± 30 % (P < 0.05, n=5) increase of surface GABAARs in cells transfected with mycMacoco compared with untransfected neurons (Figure 7B,C) whereas expression levels of GABAARs in GFP transfected neurons were unaffected. We were unable to investigate the consequences of increasing the expression levels of Maf1 using the same system as Maf1 nucleofection caused cytotoxicity in our cultures. Our results suggest that the Maf1/Macoco complex may play an important role in controlling surface levels of GABAARs.
Figure 7. Over expression of Macoco causes changes in surface GABAAR number.
(A) GFP and mycMacoco expression in lysates of nucleofected neurons. (B,C) Over-expression of mycMacoco increases surface expression of GABAARs by 71.0 ± 30 % (P < 0.05, n=5) in neurons as demonstrated by surface biotinylation and western blotting with anti-GABAAR-β3 antibodies. There is no significant difference between surface expression of GABAARs in neurons transfected with GFP compared with untransfected neurons.
Discussion
GABAARs are essential mediators of neuronal inhibition and therefore it is of fundamental importance to understand the cellular mechanisms used by neurons to control GABAAR surface number and activity. To identify proteins important for regulating GABAAR trafficking and signalling we searched for novel receptor binding partners and identified an interaction between the intracellular domain of the GABAAR β3 subunits and the proteins Maf1 and Macoco. Maf1 was distributed in dendrites where it could be found to co-localise with GABAARs at inhibitory synapses. Maf1 was also found to be highly expressed in neuronal nuclei. We further demonstrated that Maf1 interacts with a novel protein called Macoco, rich in coiled-coil domains and which also interacted with GABAAR β-subunits and partly localised to inhibitory synapses. Moreover, increasing expression of Macoco in neurons increased surface GABAAR levels. Thus the Maf1/Macoco complex appears to play a role in regulating surface GABAAR levels and trafficking.
Maf1 mRNA was found to be expressed at high levels in the hippocampus, which was confirmed by immunohistochemistry with a Maf1 specific antibody. In terms of subcellular distribution, Maf1 was found expressed in the somatic and dendritic cytoplasm. Maf1 exhibited extensive localisation in dendrites and both endogenous Maf1 (detected with a Maf1 specific antibody) and Maf1YFP expressed in neurons could be detected in the dendritic cytoplasm of mature neurons, hundreds of microns away from the soma. Macoco was similarly found to be expressed in the hippocampus and cortex with substantial dendritic localisation. Both Maf1 and Macoco exhibited only a partial overlap with inhibitory synapses and GABAARs. The partial overlap of Maf1 and Macoco with GABAARs at inhibitory synapses may be due to other roles for these proteins in the cell unrelated to their regulation of GABAARs and is also likely to be due to the fact that they may be important for the trafficking of the receptors, rather than their anchoring at synaptic sites. Similar results have been found for the co-localisation of GABAARs with several other GABAAR associated trafficking proteins reported to alter the number of surface GABAARs including GABARAP, Plic, BIG2 and GODZ (Bedford et al., 2001; Charych et al., 2004; Keller et al., 2004; Kittler et al., 2001). In addition to being localised to dendrites, Maf1 was also highly enriched in the neuronal nucleus as confirmed by co-staining with a nuclear marker. While we show that Maf1 interacts with Macoco we found no evidence for Macoco localisation to the cell nucleus, either in cell lines or neurons, suggesting that the Maf1/Macoco complex in dendrites important for regulating surface GABAAR levels may have a different role to the nuclear Maf1.
To study the functional role of the Maf1/Macoco complex we expressed exogenous Macoco in neurons which increased surface GABAAR levels (a similar approach with Maf1 was not possible due to cytotoxicity). This expression strategy has been successfully used in previous studies to better understand the role of other GABAAR associated proteins including GABARAP, Plic1 and HAP1 in regulating GABAAR function (Boileau et al., 2005; Chen et al., 2005; Kittler et al., 2004; Leil et al., 2004; Saliba et al., 2008). Interestingly, Macoco was found to interact with both GABAARs and Maf1, similarly to other GABAAR associated proteins. For example, while the large proteins PRIP1/2, NSF and gephyrin were all identified to interact with the small molecular weight protein GABARAP, they were also found to interact directly with GABAAR subunit intracellular domains (Goto et al., 2005; Kneussel et al., 2000; Terunuma et al., 2004; Tretter et al., 2008). Similarly, while Macoco was initially identified as an interacting protein for Maf1, it can itself interact directly with the GABAAR β and γ2 subunits.
The mechanisms that underlie the enhanced surface expression of GABAARs conferred by expressing Macoco remain to be fully determined. The Macoco protein is very rich in coiled-coil domains, which are often found in scaffold and trafficking proteins. Indeed, coiled-coil tethering proteins and multisubunit tethers (e.g. Golgins) have emerged as key regulators of membrane traffic and organellar architecture (Kummel and Heinemann, 2008; Short et al., 2005; Sztul and Lupashin, 2006). Thus Macoco, via its multiple coiled-coils, may be a novel ‘tethering factor’ that plays a role linking GABAARs to components of the trafficking pathway, which is in agreement with the co-localisation of these two proteins in intracellular compartments and co-localisation with markers of the ER. Macoco may also play a role via its coiled-coil domains in stabilising surface GABAARs, since it could also be found to co-localise with surface GABAARs.
We were unable to immunoprecipitate GABAARs and Maf1/Macoco complexes from brain homogenates. Similar difficulties were reported for the interaction between GABAARs and several other GABAAR and GABABR interacting proteins (Beck et al., 2002; Couve et al., 2001; Keller et al., 2004; Tretter et al., 2008). The reason behind this is unknown, but possible explanations include that the interaction with Maf1/Macoco may be transient or highly regulated in neurons. This would not be the case in transfected cells, where the overexpression of the receptor could overcome the instability of the interaction. The association is observed much more readily in pull down assays where the immobilised domain of GABAAR β3 subunit is present in high concentrations and the levels of Maf1 and Macoco in transfected cell extracts or brain extracts are also high.
In contrast to Macoco, mammalian Maf1 has no obvious domain structure apart from a nuclear localisation signal (Pluta et al., 2001) and previous studies in non-neuronal cells have shown that Maf1 associates with nuclear protein complexes that participate in transcriptional repression (Upadhya et al., 2002). This is in agreement with our findings that Maf1 is partly localised to the nucleus in neurons. Maf1 may have evolved more than one role in postmitotic cells such as neurons. Examples of such a dual function include the protein gephyrin which has a ubiquitous function in molybdenum co-factor synthesis in addition to having a synaptic scaffolding role in the brain (Feng et al., 1998), and polo like kinases which are key regulators of the cell cycle, but in post-mitotic neurons are also regulators of homeostatic plasticity (Seeburg and Sheng, 2008). In its complex with Macoco (which was not found in the nucleus) Maf1 may be directly linked to GABAAR trafficking processes independently from its role in the nucleus as has recently been observed for other proteins initially implicated in transcriptional and translational regulation. For example EIF3e, a component of the translation initiation complex, is also important for activity-dependent endocytic trafficking of calcium channels (Green et al., 2007). Similarly mDpy30, a component of several histone methyltransferase complexes important for gene regulation and chromatin organisation, and which resides in both the trans Golgi network (TGN) in addition to the nucleus, has recently been found to have an additional role in endosome to TGN transport (Xu et al., 2009).
Maf1’s role with respect to the GABAAR trafficking may be functionally distinct and separate from its signalling role in transcriptional regulation or alternatively, Maf1’s nuclear function (e.g. as a regulator of transcription) could be linked to its ability to interact with GABAARs and Macoco. In addition to its central role as a fast inhibitory neurotransmitter, GABA also has a number of trophic roles in the developing and adult nervous system, influencing multiple processes including growth cone dynamics, dendritic branching and synapse maturation, in addition to neuronal proliferation, migration, differentiation and death (Ben-Ari, 2002; Ben-Ari et al., 2007; Owens and Kriegstein, 2002). While many of GABA’s trophic roles are mediated by GABAAR activation, the underlying mechanisms remain unclear. Both depolarising GABAAR activity leading to elevations in intracellular calcium and hyperpolarising (calcium independent) actions of GABAARs are implicated in trophic effects of GABA. For example in stem cells, activation of GABAARs results in cellular hyperpolarisation, activation of S-phase kinase and accumulation of stem cells in S-phase, resulting in a rapid decrease in cell proliferation (Andang et al., 2008). Thus an intriguing possibility is that Maf1 may additionally be implicated in GABAAR signalling, for example by shuttling between inhibitory synaptic contacts in dendrites and the nucleus providing a mechanism for GABAAR dependent transcriptional repression. Interestingly, shuttling between excitatory synaptic scaffolds and the nucleus has recently been demonstrated for the protein AIDA-1d which can undergo calcium-independent translocation from synaptic PSD-95 complexes to the nucleus where it increases protein synthesis by increasing nucleolar numbers (Jordan et al., 2007). We speculate that Maf1 could be important for regulating GABAAR signalling which could be implicated in some of the trophic roles of GABA. An additional possibility is that Maf1 signalling to the nucleus is linked to its role in the Maf1/Macoco complex for regulating surface GABAAR levels, perhaps by acting as a feedback mechanism between GABAAR trafficking pathways and GABAAR signalling. Maf1 could control the translation, or protein stability of GABAARs either directly or via transcriptional regulation of GABAAR trafficking proteins. Further work will be needed to investigate these possibilities.
To conclude, we have identified and characterised two novel GABAAR associated proteins, Maf1 and the coiled-coil domain containing protein Macoco. We show they are both highly expressed in several brain regions including hippocampus and cortex. The subcellular localisation of Maf1 and Macoco in dendrites and at synapses and Macoco’s ability to regulate surface GABAAR levels suggest the Maf1/Macoco complex plays a role in the transport to or stabilisation of GABAARs at the cell surface domains. Maf1/Macoco complex may therefore have an important role in the production and maintenance of GABAergic synapses. Moreover we also demonstrate that Maf1 is enriched in the neuronal nucleus and thus may have additional roles in the nervous system linked to its role in nuclear signalling.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andang M, Hjerling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, Pozas E, Bryja V, Halliez S, Nishimaru H, Wilbertz J, Arenas E, Koltzenburg M, Charnay P, El Manira A, Ibanez CF, Ernfors P. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- Arancibia-Carcamo IL, Kittler JT. Regulation of GABA(A) receptor membrane trafficking and synaptic localization. Pharmacol Ther. 2009;123:17–31. doi: 10.1016/j.pharmthera.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Arancibia-Carcamo IL, Yuen EY, Muir J, Lumb MJ, Michels G, Saliba RS, Smart TG, Yan Z, Kittler JT, Moss SJ. Ubiquitin-dependent lysosomal targeting of GABA(A) receptors regulates neuronal inhibition. Proc Natl Acad Sci U S A. 2009;106:17552–17557. doi: 10.1073/pnas.0905502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Brickley K, Wilkinson HL, Sharma S, Smith M, Chazot PL, Pollard S, Stephenson FA. Identification, molecular cloning, and characterization of a novel GABAA receptor-associated protein, GRIF-1. J Biol Chem. 2002;277:30079–30090. doi: 10.1074/jbc.M200438200. [DOI] [PubMed] [Google Scholar]
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci. 2001;4:908–916. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y. Excitatory actions of gaba during development: the nature of the nurture. Nat Rev Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Gaiarsa JL, Tyzio R, Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Benke D, Fritschy JM, Trzeciak A, Bannwarth W, Mohler H. Distribution, prevalence, and drug binding profile of gamma-aminobutyric acid type A receptor subtypes differing in the beta-subunit variant. J Biol Chem. 1994;269:27100–27107. [PubMed] [Google Scholar]
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- Boileau AJ, Pearce RA, Czajkowski C. Tandem subunits effectively constrain GABAA receptor stoichiometry and recapitulate receptor kinetics but are insensitive to GABAA receptor-associated protein. J Neurosci. 2005;25:11219–11230. doi: 10.1523/JNEUROSCI.3751-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Uren JM, Kittler JT, Wang H, Olsen R, Parker PJ, Moss SJ. Subunit-specific association of protein kinase C and the receptor for activated C kinase with GABA type A receptors. J Neurosci. 1999;19:9228–9234. doi: 10.1523/JNEUROSCI.19-21-09228.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charych EI, Yu W, Miralles CP, Serwanski DR, Li X, Rubio M, De Blas AL. The brefeldin A-inhibited GDP/GTP exchange factor 2, a protein involved in vesicular trafficking, interacts with the beta subunits of the GABA receptors. J Neurochem. 2004;90:173–189. doi: 10.1111/j.1471-4159.2004.02481.x. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Chang CS, Leil TA, Olcese R, Olsen RW. GABAA receptor-associated protein regulates GABAA receptor cell-surface number in Xenopus laevis oocytes. Mol Pharmacol. 2005;68:152–159. doi: 10.1124/mol.104.009878. [DOI] [PubMed] [Google Scholar]
- Couve A, Kittler JT, Uren JM, Calver AR, Pangalos MN, Walsh FS, Moss SJ. Association of GABA(B) receptors and members of the 14-3-3 family of signaling proteins. Mol Cell Neurosci. 2001;17:317–328. doi: 10.1006/mcne.2000.0938. [DOI] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Dumoulin A, Rostaing P, Bedet C, Levi S, Isambert MF, Henry JP, Triller A, Gasnier B. Presence of the vesicular inhibitory amino acid transporter in GABAergic and glycinergic synaptic terminal boutons. J Cell Sci. 1999;112 ( Pt 6):811–823. doi: 10.1242/jcs.112.6.811. [DOI] [PubMed] [Google Scholar]
- Feng G, Tintrup H, Kirsch J, Nichol MC, Kuhse J, Betz H, Sanes JR. Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science. 1998;282:1321–1324. doi: 10.1126/science.282.5392.1321. [DOI] [PubMed] [Google Scholar]
- Goto H, Terunuma M, Kanematsu T, Misumi Y, Moss SJ, Hirata M. Direct interaction of N-ethylmaleimide-sensitive factor with GABA(A) receptor beta subunits. Mol Cell Neurosci. 2005;30:197–206. doi: 10.1016/j.mcn.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Green EM, Barrett CF, Bultynck G, Shamah SM, Dolmetsch RE. The tumor suppressor eIF3e mediates calcium-dependent internalization of the L-type calcium channel CaV1.2. Neuron. 2007;55:615–632. doi: 10.1016/j.neuron.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs AM, Oliver PL, Jones EL, Jeans A, Potter A, Hovik BH, Nolan PM, Vizor L, Glenister P, Simon AK, Gray IC, Spurr NK, Brown SD, Hunter AJ, Davies KE. A mutation in Af4 is predicted to cause cerebellar ataxia and cataracts in the robotic mouse. J Neurosci. 2003;23:1631–1637. doi: 10.1523/JNEUROSCI.23-05-01631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–379. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kins S, Betz H, Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci. 2000;3:22–29. doi: 10.1038/71096. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Chen G, Honing S, Bogdanov Y, McAinsh K, Arancibia-Carcamo IL, Jovanovic JN, Pangalos MN, Haucke V, Yan Z, Moss SJ. Phospho-dependent binding of the clathrin AP2 adaptor complex to GABAA receptors regulates the efficacy of inhibitory synaptic transmission. Proc Natl Acad Sci U S A. 2005;102:14871–14876. doi: 10.1073/pnas.0506653102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler JT, Chen G, Kukhtina V, Vahedi-Faridi A, Gu Z, Tretter V, Smith KR, McAinsh K, Arancibia-Carcamo IL, Saenger W, Haucke V, Yan Z, Moss SJ. Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit. Proc Natl Acad Sci U S A. 2008;105:3616–3621. doi: 10.1073/pnas.0707920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler JT, Moss SJ. Modulation of GABAA receptor activity by phosphorylation and receptor trafficking: implications for the efficacy of synaptic inhibition. Curr Opin Neurobiol. 2003;13:341–347. doi: 10.1016/s0959-4388(03)00064-3. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Oliver P. Genomic and post-genomic tools for studying synapse biology. In: Kittler JT, Moss SJ, editors. The dynamic synapse: molecular methods in ionotropic receptor biology. CRC/Taylor & Francis; Boca Raton, FL: 2006. pp. 205–239. [Google Scholar]
- Kittler JT, Rostaing P, Schiavo G, Fritschy JM, Olsen R, Triller A, Moss SJ. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol Cell Neurosci. 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Thomas P, Tretter V, Bogdanov YD, Haucke V, Smart TG, Moss SJ. Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type A receptor membrane trafficking. Proc Natl Acad Sci U S A. 2004;101:12736–12741. doi: 10.1073/pnas.0401860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M, Haverkamp S, Fuhrmann JC, Wang H, Wassle H, Olsen RW, Betz H. The gamma-aminobutyric acid type A receptor (GABAAR)-associated protein GABARAP interacts with gephyrin but is not involved in receptor anchoring at the synapse. Proc Natl Acad Sci U S A. 2000;97:8594–8599. doi: 10.1073/pnas.97.15.8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummel D, Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- Kuramoto N, Wilkins ME, Fairfax BP, Revilla-Sanchez R, Terunuma M, Tamaki K, Iemata M, Warren N, Couve A, Calver A, Horvath Z, Freeman K, Carling D, Huang L, Gonzales C, Cooper E, Smart TG, Pangalos MN, Moss SJ. Phospho-dependent functional modulation of GABA(B) receptors by the metabolic sensor AMP-dependent protein kinase. Neuron. 2007;53:233–247. doi: 10.1016/j.neuron.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leil TA, Chen ZW, Chang CS, Olsen RW. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J Neurosci. 2004;24:11429–11438. doi: 10.1523/JNEUROSCI.3355-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Day M, Muniz LC, Bitran D, Arias R, Revilla-Sanchez R, Grauer S, Zhang G, Kelley C, Pulito V, Sung A, Mervis RF, Navarra R, Hirst WD, Reinhart PH, Marquis KL, Moss SJ, Pangalos MN, Brandon NJ. Activation of estrogen receptor-beta regulates hippocampal synaptic plasticity and improves memory. Nat Neurosci. 2008;11:334–343. doi: 10.1038/nn2057. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Brickley K, Stephenson FA, Kittler JT. GTPase dependent recruitment of Grif-1 by Miro1 regulates mitochondrial trafficking in hippocampal neurons. Mol Cell Neurosci. 2009a;40:301–312. doi: 10.1016/j.mcn.2008.10.016. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009b;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DF, Kriegstein AR. Is there more to GABA than synaptic inhibition? Nat Rev Neurosci. 2002;3:715–727. doi: 10.1038/nrn919. [DOI] [PubMed] [Google Scholar]
- Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5031–5040. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Pangalos M, Moss SJ. The ubiquitin-like protein Plic-1 enhances the membrane insertion of GABAA receptors by increasing their stability within the endoplasmic reticulum. J Biol Chem. 2008;283:18538–18544. doi: 10.1074/jbc.M802077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg DP, Sheng M. Activity-induced Polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J Neurosci. 2008;28:6583–6591. doi: 10.1523/JNEUROSCI.1853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113 ( Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Short B, Haas A, Barr FA. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim Biophys Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Smith KR, McAinsh K, Chen G, Arancibia-Carcamo IL, Haucke V, Yan Z, Moss SJ, Kittler JT. Regulation of inhibitory synaptic transmission by a conserved atypical interaction of GABA(A) receptor beta- and gamma-subunits with the clathrin AP2 adaptor. Neuropharmacology. 2008;55:844–850. doi: 10.1016/j.neuropharm.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am J Physiol Cell Physiol. 2006;290:C11–26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- Taylor PM, Thomas P, Gorrie GH, Connolly CN, Smart TG, Moss SJ. Identification of amino acid residues within GABA(A) receptor beta subunits that mediate both homomeric and heteromeric receptor expression. J Neurosci. 1999;19:6360–6371. doi: 10.1523/JNEUROSCI.19-15-06360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terunuma M, Jang IS, Ha SH, Kittler JT, Kanematsu T, Jovanovic JN, Nakayama KI, Akaike N, Ryu SH, Moss SJ, Hirata M. GABAA receptor phospho-dependent modulation is regulated by phospholipase C-related inactive protein type 1, a novel protein phosphatase 1 anchoring protein. J Neurosci. 2004;24:7074–7084. doi: 10.1523/JNEUROSCI.1323-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter V, Jacob TC, Mukherjee J, Fritschy JM, Pangalos MN, Moss SJ. The clustering of GABA(A) receptor subtypes at inhibitory synapses is facilitated via the direct binding of receptor alpha 2 subunits to gephyrin. J Neurosci. 2008;28:1356–1365. doi: 10.1523/JNEUROSCI.5050-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, Kittler JT. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Wilkinson JW. In situ Hybridization: A Practical Approach. IRL Press; Oxford: 1992. Whole-mount in situ hybridization of vertebrate embryos; pp. 75–83. [Google Scholar]
- Wooltorton JR, Moss SJ, Smart TG. Pharmacological and physiological characterization of murine homomeric beta3 GABA(A) receptors. Eur J Neurosci. 1997;9:2225–2235. doi: 10.1111/j.1460-9568.1997.tb01641.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Gong Q, Xia B, Groves B, Zimmermann M, Mugler C, Mu D, Matsumoto B, Seaman M, Ma D. A role of histone H3 lysine 4 methyltransferase components in endosomal trafficking. J Cell Biol. 2009;186:343–353.h. doi: 10.1083/jcb.200902146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.