Abstract
GDE1 is a mammalian glycerophosphodiesterase (GDE) implicated by in vitro studies in the regulation of glycerophopho-inositol (GroPIns) and possibly other glycerophospho (GroP) metabolites. Here, we show using untargeted metabolomics that GroPIns is profoundly (> 20-fold) elevated in brain tissue from GDE1(-/-) mice. Furthermore, two additional GroP-metabolites not previously identified in eukaryotic cells, glycerophospho-serine (GroPSer) and glycerophospho-glycerate (GroPGate), were also highly elevated in GDE1(-/-) brains. Enzyme assays with synthetic GroP-metabolites confirmed that GroPSer and GroPGate are direct substrates of GDE1. Interestingly, our metabolomic profiles also revealed that serine (both L-and D-) levels were significantly reduced in brains of GDE1 (-/-) mice. These findings designate GroPSer as a previously unappreciated reservoir for free serine in the nervous system and suggest that GDE1, through recycling serine from GroPSer, may impact D-serine-dependent neural signaling processes in vivo.
The glycerophosphodiester phosphodiesterases (GDEs) are an ancient and ubiquitous family of enzymes with numerous members found in all three domains of life. These enzymes are metal-dependent phosphodiesterases that cleave the phosphodiester bond of various glycerophosphodiester (GroP) metabolites, such as the deacylated phospholipids glycerophospho-choline (GroPCho) and glycerophospho-inositol (GroPIns), to release glycerol-3-phosphate (G3P) and the corresponding head-group alcohol. Several prokaryotic GDEs have been characterized and shown to be necessary for catabolism of environmental GroP-metabolites, suggesting a role for these enzymes in phospholipid remodelling and nutrient scavenging (Larson, et al., 1983; Tommassen, et al., 1991). Transporters for GroP-metabolites have also been identified in microbes and mammals, indicating that dedicated systems exist for GroP-metabolite uptake and catabolism (Fisher, et al., 2005; Mariggio, et al., 2006; Patton-Vogt and Henry, 1998).
Following the characterization of microbial GDEs, a group of mammalian GDEs was identified that contains seven members (GDE1-7) with diverse patterns of expression (Yanaka, 2007). In contrast to microbial GDEs, which are predominantly periplasmic or cytoplasmic, six of the seven mammalian GDEs are membrane-bound and possess multiple membrane-spanning domains, suggesting that their functions may have diverged significantly from their prokaryotic ancestors. The physiological roles ascribed to mammalian GDEs are diverse. GDE2 has been shown to regulate motor neuron differentiation by a mechanism that depends on the integrity of its catalytic residues (Rao and Sockanathan, 2005; Yan, et al., 2009), and a separate report implicated GDE2 in the regulation of GroPCho levels in the kidney (Gallazzini, et al., 2008). GDE3 has been found to promote osteoblast differentiation upon transient overexpression (Yanaka, et al., 2003). Recently, GroPIns was identified as a substrate for GDE3, and, interestingly, GDE3 displays phospholipase C-type activity (releasing inositol-phosphate and glycerol) rather than the D-type phospholipase activity exhibited by other characterized GDEs (Corda, et al., 2009). Finally, GDE1 has been shown to interact with regulators of G-protein signaling (Zheng, et al., 2000), and its catalytic activity can be modulated by stimulation of G-protein coupled receptors (Zheng, et al., 2003). GroPIns is a substrate for GDE1 in vitro (phospholipase D-type hydrolysis), and several other GroP-metabolites, including GroP-serine (GroPSer) and GroP-inositol-4,5-bisphosphate, can competitively inhibit the action of GDE1 on GroPIns (Zheng, et al., 2003), suggesting that these GroP compounds might also be direct GDE1 substrates. Finally, we discovered that GDE1, but not other mammalian GDEs, also accepts GroP-N-acyl ethanolamines (GP-NAEs) as substrates, implicating this enzyme in the biosynthesis of NAE transmitters, such as the endocannabinoid anandamide (Simon and Cravatt, 2008). Consistent with this premise, we have found that mice harbouring a targeted disruption of the GDE1 gene display a moderate impairment in acute NAE biosynthesis (Simon and Cravatt, 2010). The extent to which other GroP-metabolites are altered in GDE1(-/-) mice, however, has not yet been examined.
Here, we present a global analysis of GDE1(-/-) mice using a mass spectrometry (MS)-based metabolomics platform that measures aqueous-soluble, polar analytes, including GroP-metabolites and their derivatives. We confirmed that GroPIns is a physiological substrate of GDE1 that is profoundly elevated in the central nervous system of mice lacking this enzyme. Additionally, we discovered other GDE1-regulated metabolites, including GroPSer and GroPGate, that, to our knowledge, have not yet been described as natural products in eukaryotes. Finally, our broad metabolomic profiles also revealed lower serine levels in brains of GDE1(-/-) mice, suggesting that GroPSer serves as a previously unappreciated reservoir for free serine in the nervous system.
Results
Analysis of GDE1(-/-) mice by untargeted metabolomics
The ability to measure and study GroPIns and its derivatives has been facilitated by the availability of 3H-inositol, which can be used to metabolically label cultured cells to track the fate of inositol-containing metabolites (Emilsson and Sundler, 1984; Falasca, et al., 1997; Falasca, et al., 1996). While this technique is powerful, it is not readily applicable to organismal studies, and is dependent on the availability and facile metabolic incorporation of radioactive standards. Consequently, comparatively little is known about GroPIns metabolism in vivo or the existence and enzymatic regulation of other GroP-metabolites due to a dearth of synthetic and analytical chemistry techniques for their characterization and measurement. MS-based metabolomics has emerged as a powerful strategy to globally investigate biochemical pathways in living systems and has succeeded in assigning novel substrates and products to both characterized and uncharacterized enzymes (Chiang, et al., 2006; Dang, et al., 2009; Saghatelian, et al., 2004; Tagore, et al., 2009; Tang, et al., 2009; Vinayavekhin and Saghatelian, 2009). Such MS-based metabolomics experiments can be performed in two different ways: targeted or untargeted. Targeted analyses focus on a defined set of known metabolites that are typically measured by selected ion monitoring or multiple reaction monitoring with internal standards for absolute or relative quantitation (Jackson, et al., 2008; McNally, et al., 2007; McNally, et al., 2006; Rabinowitz and Kimball, 2007). In contrast, untargeted metabolomics surveys a broad mass range for relative changes in both known and unknown metabolites.
Untargeted profiling involves a comparative liquid chromatography (LC)-MS analysis of two sets of samples, e.g. cells/tissues from wild-type and enzyme-knockout disrupted systems, to identify differences in the intensity of mass ion peaks that share mass-to-charge ratios and retention times. We and others have routinely applied untargeted metabolomics to analyze organically extracted fractions of cells and tissues, which provide preferential access to hydrophobic, non-polar metabolites (a process often referred to as “lipidomics”) (Garrett, et al., 2009; Guan, et al., 2007; Ivanova, et al., 2009; Saghatelian, et al., 2004). However, many of the putative substrates for GDE1 are hydrophilic, polar metabolites that would likely remain in the aqueous fractions of cell/tissue extracts and thus go unsampled in conventional lipidomic experiments. We therefore set out to establish a method to analyze the aqueous-soluble metabolome that combines normal phase LC-MS conditions as pioneered by Rabinowitz and co-workers for their targeted metabolomic studies (Bajad, et al., 2006) with an untargeted profiling protocol suitable for the discovery of both known and novel metabolites (Saghatelian, et al., 2004).
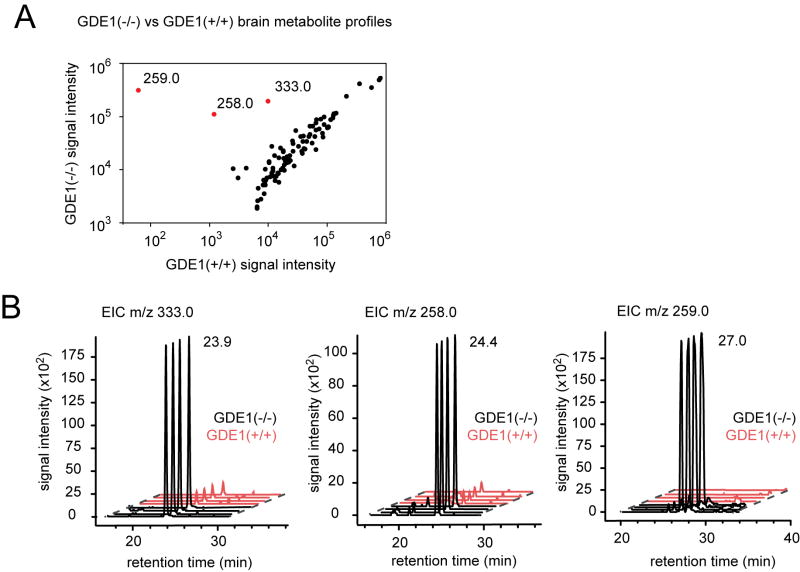
We identified optimal extraction and chromatography methods by testing a diverse collection of aqueous-soluble metabolites, including nucleosides, amino acids, and synthetic GroP-metabolites under normal phase chromatography conditions (Table S1). GroP-metabolites were obtained from their respective phospholipids by alkaline hydrolysis and subsequent purification (see Experimental Procedures). We next applied our optimized extraction and LC-MS conditions to perform an untargeted comparative analysis of water-soluble metabolites from brain tissue of GDE1(+/+) and (-/-) mice. Initial full-scan LC-MS surveys in negative mode analyzed an m/z range from 100 to 1,200, and the XCMS software (Benton, et al., 2008; Smith, et al., 2006) was used for comparative data analysis. Three metabolites with different m/z values (258.0, 259.0 and 333.0) and LC retention times (24.4, 27.0, and 23.9 min, respectively) were found to be dramatically elevated (> 20-fold) in GDE1(-/-) brains (Figure 1A and 1B). Metabolomes from spinal cord, showed similar overall profiles to brain (Table S2).
Figure 1.
GDE1(-/-) brains contain markedly elevated levels of multiple GroP-metabolites. (A) Mass ion peak intensity measurements from GDE1(+/+) versus (-/-) brains. Data are derived from untargeted metabolite profiling experiments performed in the negative ionization mode. Red spots represent m/z ions that are significantly elevated in GDE1(-/-) brains (in each case, fold-change > 10) and their m/z values are displayed. Isotopologues, adducts, and ions produced by in-source fragmentation were manually removed for clarity. (B) Extracted ion chromatograms (EICs) from individual LC-MS runs of GDE1(+/+) and (-/-) brain metabolomes. The retention times for the relevant peaks are shown in minutes. See also Figure S1, and Tables S1 and S3.
Structural characterization of GDE1-regulated brain metabolites
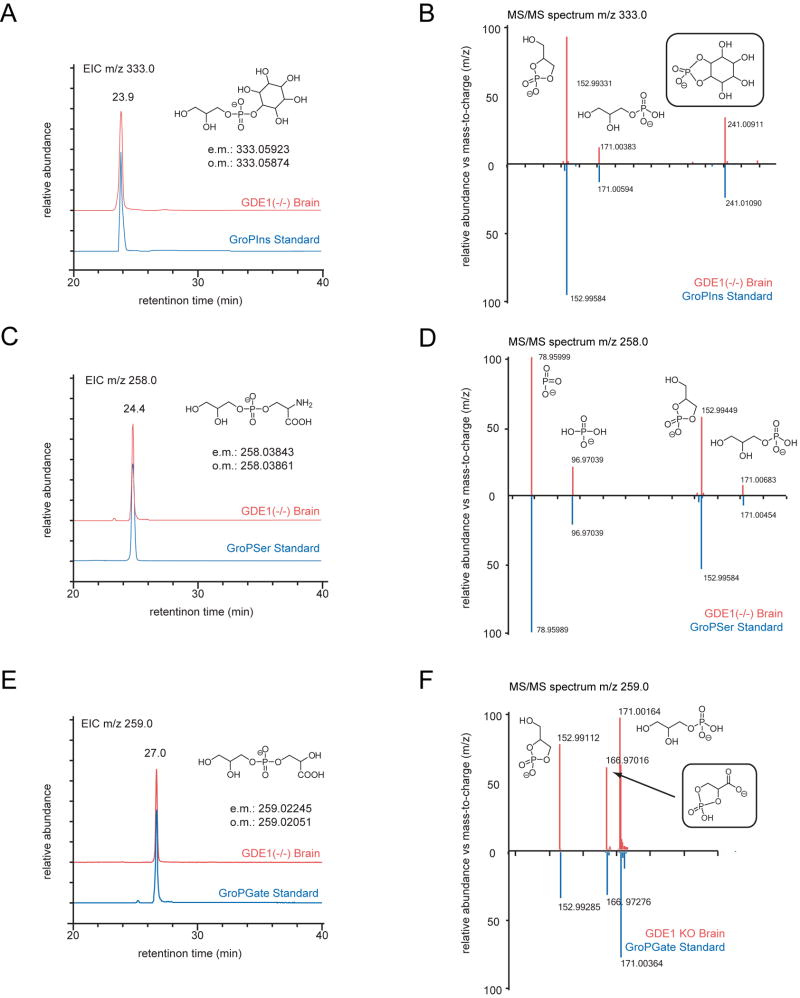
Previous studies by the Farquhar group showed that GDE1 is a GroPIns phosphodiesterase in vitro (Zheng, et al., 2003). As such, we presumed that the m/z 333.0 metabolite was likely GroPIns, a premise supported by high-resolution mass spectrometry (HRMS) measurements, which assigned an exact mass value within 0.1 ppm of the expected value for GroPIns (Table S4). Chemical synthesis of GroPIns and comparison of its LC retention-time and MS/MS fragmentation pattern to the natural GDE1-regulated metabolite confirmed its identity as GroPIns (Figure 2A, B and Figure S1A).
Figure 2.
Structural characterization of GDE1-regulated brain metabolites. (A, C, E) Chromatographic behaviour on normal-phase LC of synthetic GroPIns, GroPSer, and GroPGate compared to the natural metabolites elevated in brains of GDE1(-/-) mice. Insets show observed and calculated masses (o.m. and c.m., respectively) for natural metabolites, as measured by quadrupole time-of-flight (QTOF) mass spectrometry (for higher-resolution FT-ICR mass-measurements see Table S4). (B, D, F) MS/MS spectra showing daughter ions that correspond to the glycerophosphate backbone (m/z 78.95, 96.97, 152.99, 171.00). Endogenous daughter ions matched their synthetic counterparts within 20 ppm which is typical for MS/MS ions obtained via QTOF mass spectrometry. The existence of the inositol (B) and glycerate (C) head-groups was confirmed by diagnostic peaks representing inositol- and dehydrated glycerate-phosphate, respectively. MS/MS profiles of synthetic (blue) and natural (red) metabolites are shown. See also Figure S2 and Tables S2 and S4.
In the aforementioned Farquhar study (Zheng, et al., 2003), competition experiments revealed that several GroP-metabolites, including GroPSer, could inhibit GroPIns hydrolysis by GDE1. These data suggested that GroPSer might also serve as a GDE1 substrate, although, to our knowledge, GroPSer had not previously been identified as a natural metabolite in eukaryotes. The monoisotopic mass of the GroPSer anion ([M-H]-) is 258.04, providing a candidate identity for the m/z 258.0 metabolite that was dramatically elevated in GDE1(-/-) brains. Preparation of GroPSer via alkaline hydrolysis of phosphatidylserine (PtdS) yielded a molecule with an identical LC-elution profile, high-resolution mass (within 0.3 ppm), and MS/MS fragmentation pattern as the natural m/z 258 metabolite, thus confirming its identity as GroPSer (Figure 2C, D, Figure S1B, and Table S4).
Structural assignment of the m/z 259 metabolite proved more challenging, as this mass did not match those of any known or predicted GDE1 substrates. We obtained HRMS data for this metabolite and found that the anion ([M-H]-) had an exact mass of 259.022, which suggested a molecular formula of C6H13NO8P-. Searches of publicly available metabolite databases revealed that this is the chemical formula for inositol-monophosphates, and we thus chose inositol-1-phosphate as an initial candidate structure. However, fragmentation studies performed on the endogenous m/z 259 metabolite revealed fragment ions with m/z values of 152.993 and 171.004, which are characteristic of a glycerol-phosphate backbone (Figure 2F), and, indeed, comparison with synthetic inositol-monophosphate standards revealed that these species did not co-elute with the m/z 259 metabolite (Figure S1D). Working with the knowledge that the m/z 259 metabolite likely contains a glycerol-phosphate backbone, we surmised that the alcohol head-group could represent glyceric acid (glycerate), which would engender a small-molecule glycerophosphoglycerate (GroPGate) with an exact mass value that matches the natural metabolite within 0.2 ppm (Table S4).
Key to the ultimate structural assignment was the chemical synthesis of GroPGate as described in Experimental Procedures. Briefly, starting from GroPSer, the α-amino group of serine was converted into an alcohol by diazotation in the presence of sodium nitrite to afford the carboxylic acid after hydrolysis (Lencina, et al., 2008). The fragmentation of the synthetic GroPGate standard revealed an m/z 166.970 ion corresponding to the exact mass of the dehydrated phosphate adduct of the glycerate head-group (Figure 2F). An identical fragmentation pattern was obtained for the endogenous m/z 258 brain metabolite. Additionally the synthetic and natural species displayed identical LC-elution profiles (Figure 2E and Figure S1C). Together, these data indicate that GroPGate is a novel natural product of the mouse brain and a principal endogenous substrate for GDE1. It should be noted, however, that we cannot formally exclude the possibility that the m/z 259 metabolite represents GroPGate conjugated via the 2′- rather than 3′ position on the glycerate head-group, as it is not clear whether these two GroPGate variants would show different retention times under our chromatographic conditions. We suggest based on potential biosynthetic pathways (see Discussion) that the 3′-conjugated GroPGate is a more likely structure.
Finally, it is important to note that not all detectable GroP-metabolites were elevated in GDE1(-/-) mice. Peaks corresponding to GroP-ethanolamine (GroPEtn) and GroP-choline (GroPCho) were identified in our metabolomic data by comparison to synthetic standards (Figure S2, Table S4) and displayed similar levels in GDE1(+/+) and (-/-) brains (Table 1). These findings indicate that GroPEtn and GroPCho are not endogenous substrates for GDE1, which is consistent with a previous study showing that neither GroPEtn nor GroPCho competitively inhibits GDE1 activity in vitro (Zheng, et al., 2003).
Table 1.
Endogenous levels of GroP metabolites and serine in nmols/g wet brain tissue as measured by LC-MS. Data represent the average value of six independent experiments per group ± standard error.
| Metabolite | GDE1 (-/-) Brain | GDE1 (+/+) Brain | Ratio (-/-)/(+/+) | P value |
|---|---|---|---|---|
| GroPIns | 647±14 | 26.5±0.1 | 25 | 0.02 |
| GroPSer | 933±67 | 8.3±0.2 | 112 | 0.03 |
| GroPGate | 1426±10 | ≤5a | ≥285a | 0.01 |
| GroPEtn | 3415±360 | 3919±381 | 0.87 | |
| GroPCho | 10306±1605 | 10240±1182 | 1.01 | |
| Serine | 990±22 | 1626±101 | 0.61 | 0.03 |
GroPGate signals from GDE1 (+/+) tissue were below the detection limit and are listed as ≤5 nmol/g tissue leading to a minimal (-/-)/(+/+) ratio of ≥ 285.
Evaluation of GroP-metabolites as GDE1 substrates
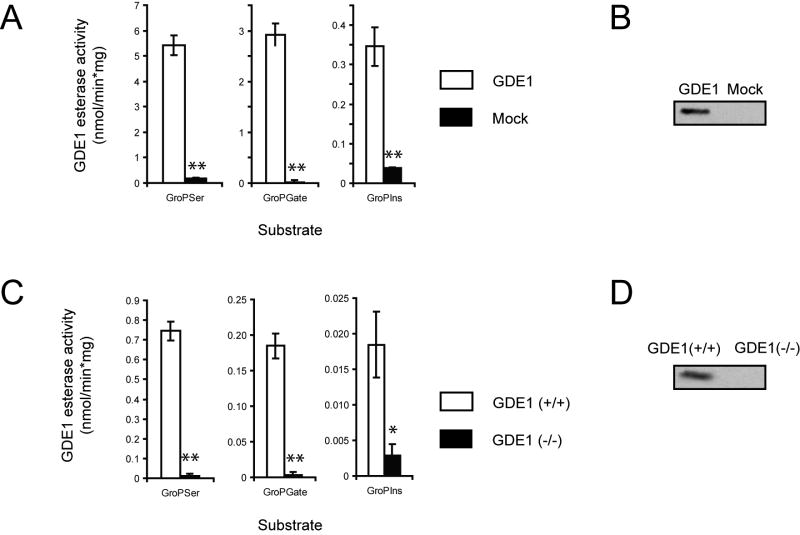
We next asked whether GroPSer and GroPGate, like GroPIns, are direct substrates for GDE1. In support of this premise, the membrane proteome from GDE1-transfected cells showed significantly greater hydrolytic activity towards all three GroP substrates compared to the membrane proteome from mock-transfected control cells (Figure 3A and B). Substrate assays were also performed with brain membrane proteomes from GDE1(+/+) and (-/-) mice, and much greater activity was observed in GDE1(+/+) samples for all three GroP-substrates (Figure 3C and D). GDE1 (either recombinant or natural) was found to hydrolyze GroP-metabolites with relative efficiencies of GroPSer > GroPGate > GroPIns. Together, these studies indicate that GroPSer and GroPGate, like GroPIns, are direct substrates for GDE1.
Figure 3.
GroPSer, GroPIns, and GroPGate are direct substrates of GDE1. Membrane preparations from GDE1-transfected COS-7 cells (A) or mouse brain samples (C) were tested for activity against synthetic GroP-metabolites with different head-groups – serine (GroPSer), glycerate (GroPGate), or inositol (GroPIns). Enzyme activity was determined by quantification of the release of free head group (serine, glycerate, and inositol from GroPSer, GroPGate, and GroPIns, respectively) by LC-MS in recombinant GDE1 assays. For mouse brain samples, enzyme activity against GroPSer, GroPGate was measured in the same way, and, for GroPIns, was determined by measuring reductions in consumed substrate (due to the presence of high background isobaric mass signals for inositol in brain samples.) All assays were performed with n=3, **, p < 0.01; * p < 0.05, student's t-test.
Serine levels are reduced in brain tissue from GDE1(-/-) mice
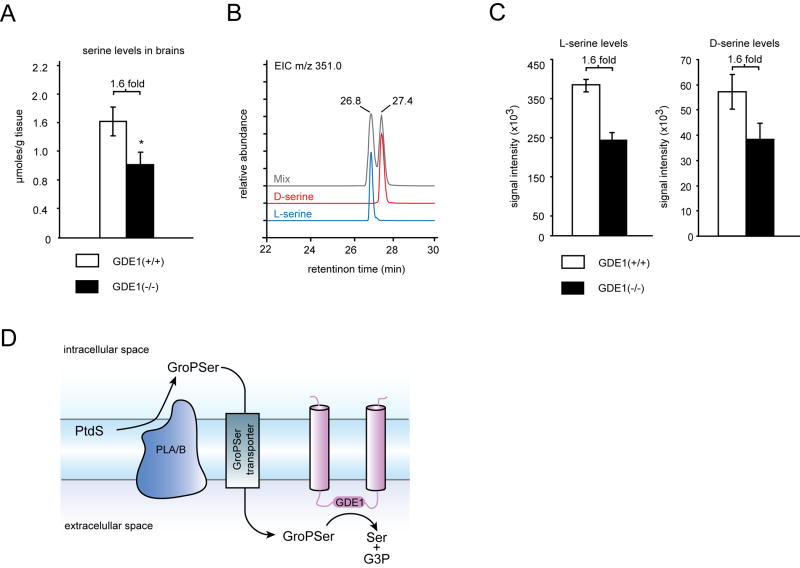
Our metabolomic profiles had the potential to reveal additional biochemical changes beyond the dramatic elevations in GroP-metabolites caused by GDE1 disruption. A survey of known polar metabolites, including amino acids and nucleosides, revealed that most were unaltered in GDE1(-/-) brains (Supplemental Table 3). A 1.6-fold reduction was, however, observed for the amino acid serine in GDE1(-/-) brains. Subsequent targeted analysis using selected ion monitoring and a deuterated (d3)-serine standard confirmed the significance of this 1.6-fold decrease in serine in GDE1(-/-) brains (Figure 4A). Furthermore, the addition of d3-serine as an internal standard permitted precise quantitation of absolute serine concentrations, which were found to be approximately 1.6 μmols/g brain tissue in GDE1(+/+) mice, consistent with previous measurements (Klivenyi, et al., 2005; Nagata, et al., 1994). To evaluate the possibility that GroPSer serves as a metabolic precursor to serine in the brain, we compared the magnitude of the GroPSer elevation with the reduction in serine observed in GDE1(-/-) mice. The synthesis of isotopically labeled GroP-metabolites was impractical so we compared the ionization of endogenous GroP-metabolites to standard curves generated with synthetic, unlabeled GroPs (using d3-serine as a normalizing internal standard) to give estimates of endogenous GroP levels in GDE1(+/+) and (-/-) brains. We found that the absolute levels of GroPSer, GroPGate, and GroPIns were 933, 1426, and 647 nmols/g in GDE1(-/-) brain and 8, ≤ 5, and 26 nmols/g in GDE1(+/+) brain (Table 1). Thus, in GDE1(-/-) brain, the absolute elevation in GroPSer levels (∼900 nmols/g increase) is roughly “mass-balanced” by the reduction in corresponding serine levels (∼650 nmols/g decrease), lending support to a model where ∼40% of free serine is generated from GDE1-catalyzed hydrolysis of GroPSer.
Figure 4.
GDE1 regulates free serine levels in mouse brain. (A) Levels of serine are reduced by 1.6-fold in GDE1(-/-) brains; Data represent the average of six independent experiments per group ± standard error. *,p < 0.05. (B) Derivitization with BocPheOSu enables resolution of L- and D-serine via reverse-phase chromatography. (C) BocPheOSu derivitization of brain metabolomes reveals that both L- and D- serine levels are reduced by a similar magnitude in GDE1(-/-) brains. *, p < 0.05 for both enantiomers. (D) Working model for the role of GDE1 in serine metabolism. Phosphatidylserine (PtdS) which is exclusively located on the inner leaflet of the plasma membrane is de-acylated by A- or B-type phospholipases (PLA/B) whereupon the resulting GroPSer is released into the cytosol and is transported across the plasma membrane by a putative GroPSer transporter. Subsequently, extra-cellular GroPSer is exposed to the catalytic domain of GDE1 resulting in the release of serine and glycerol-3-phosphate. In the absence of GDE1, GroPSer accumulates, resulting in corresponding reductions in serine. See also Figure S3.
Serine exists in both L- and D- enantiomers in brain, and the latter metabolite serves as a neurotransmitter that activates NDMA receptors (Mothet, et al., 2000; Snyder and Kim, 2000). We therefore asked whether L- and D-serine were similarly altered in GDE1(-/-) brains. To answer this question, we derivatized both enantiomers of serine with the chiral coupling partner N-(N-α-t-Boc-phenylalanyloxy)succinimide (BocPheOSu) to generate diastereomers that could be separated by HPLC (Figure 4B). Derivatization of brain extracts with BocPheOSu confirmed that both L- and D-serine are decreased by 1.6 fold in GDE1(-/-) brains. Thus, GDE1(-/-) mice have constitutively lower brain levels of the neuroactive substance D-serine.
Discussion
GroP-metabolites are thought to be formed by the action of A/B-type phospholipases on membrane phospholipids, which generate free fatty acids and water-soluble glycerol phosphodiesters. It has been shown in yeast that deacylation of phosphatidylcholine (PtdC) by a B-type phospholipase (Nte1p, the orthologue of mammalian neuropathy target esterase) yields GroPCho, which is catabolized by Gde1p (YPL110c) in a pathway that is important for PtdC recycling (Dowd, et al., 2001; Patton-Vogt, 2007). Less is known about the existence of other GroP-metabolites or how they are enzymatically formed and degraded in mammalian cells. This is an important problem not only from the perspective of basic metabolism, but also signaling, since GroPIns and its phosphorylated derivatives are dynamically regulated by various cell stimuli and have been shown to impact processes such as cancer invasion and cytoskeletal remodeling (reviewed in Corda, et al., 2009). The development of robust methods to measure GroP-metabolites in complex biological systems is critical to any effort aimed at characterizing mammalian enzymes involved in GroP metabolism.
In this study, we established an LC-MS-based metabolomics platform capable of detecting a wide range of aqueous-soluble, polar analytes, including monosaccharides, nucleosides, amino acids, and GroP-metabolites. We used this platform to characterize the polar metabolome of brains from mice lacking the glycerophosphodiesterase GDE1. Previous studies have shown that GDE1 acts as a GroPIns phosphodiesterase in vitro (Zheng, et al., 2003), and, accordingly, we observed dramatic (>20-fold) elevations in brain GroPIns from GDE1(-/-) mice. These findings confirm that GroPIns is a physiological substrate for GDE1 in vivo.
Because our metabolomic profiling was performed in an untargeted manner, we also observed several other changes in the polar metabolome. Most striking were two additional metabolites that displayed large-magnitude elevations in GDE1(-/-) brains and showed evidence of belonging to the GroP class of metabolites by MS/MS fragmentation. Chemical synthesis and comparison with standards revealed that these two substances were GroP-metabolites with serine and glycerate head-groups (GroPSer and GroPGate, respectively). To our knowledge, neither GroPSer nor GroPGate have been previously reported as natural products in eukaryotes. This may be rationalized, at least in part, by the low levels of GroPSer and GroPGate found in GDE1(+/+) mice (< 10 nmol/g brain tissue). These results thus underscore one of the attributes of performing metabolomics on enzyme-disrupted systems, which has the potential to discover new metabolites that are too rare to detect in wild-type models. In contrast, other GroP-metabolites, including GroPEtn and GroPCho were not substantially altered in GDE1(-/-) mice. These results indicate that GDE1 plays a broad, but not exclusive role in regulating the “GroP-metabolome”, which appears to be much larger and more chemically diverse than previously appreciated.
Beyond the GroP-metabolome, we also observed a ∼40% reduction in free serine in GDE1(-/-) brains. Quantitation of the absolute magnitudes of serine reduction and GroPSer elevation indicate that these changes are mass-counterbalanced, suggesting that GroPSer serves as a hitherto unappreciated metabolic precursor to serine in the mammalian brain. The biosynthesis and physiological role of serine in the nervous system is a research topic of much interest. Serine is classified as a non-essential amino acid because most eukaryotic cells contain the enzymes necessary to biosynthesize it from 3-phosphoglycerate. In contrast, it was recently discovered that one of these enzymes, 3-phosphoglycerate dehydrogenase (3PGDH), is down-regulated in neurons compared to astroglia, where it is highly expressed (Yamasaki, et al., 2001). Cerebellar Purkinje neurons, in particular, lack detectable 3PGDH and therefore cannot synthesize serine de novo (Furuya, et al., 2000; Yamasaki, et al., 2001). These findings have led to a model wherein serine is classified as a neurotrophic factor that is likely supplied by proximal glia in vivo. Our data point to an important role for GDE1 in maintaining the homeostatic balance of serine in the nervous system. Intriguingly, GDE1 appears to be highly expressed in Purkinje neurons (Figure S3), suggesting that this enzyme may provide certain neuronal populations with the ability to scavenge serine from the interstitial space by catabolizing GroPSer.
The biosynthesis of GroPSer itself is likely the result of A/B-type phospholipase activity on membrane phospholipids. As phosphatidylserine is localized exclusively on the inner-leaflet of the plasma membrane (Martin, et al., 1995), the resulting GroPSer is presumably released into the cytosol whereupon it must be transported outside the cell by transporters (Mariggio, et al., 2006; Patton-Vogt and Henry, 1998) prior to exposure to GDE1, whose catalytic domain faces the luminal/extracellular side of the membrane (Figure 4D) (Zheng, et al., 2003). The resulting free serine generated by GDE1 may be taken up by neurons and utilized directly, or taken up by glia for conversion to D-serine by serine racemase (Mothet, et al., 2000; Wolosker, et al., 1999; Yamamoto, et al., 2003).
Serine is also of special relevance to the nervous system because its D-enantiomer is an endogenous co-agonist of brain NMDA receptors (Mothet, et al., 2000; Snyder and Kim, 2000). Interestingly, Ca2+-mediated release of D-serine from astrocytes is required for NMDAR-mediated long-term potentiation at nearby excitatory synapses (Henneberger, et al., 2010). We found that both L- and D-serine are reduced by approximately 40% in the brains of GDE1(-/-) mice. While these mice are overtly normal, it may be interesting to look for more specific differences in nervous system function and behaviour, as serine racemase(-/-) mice, which lack D-serine, show elevated anxiety (Basu et al 2009) and are resistant to various forms of neurotoxicity (Inoue et al 2010). Additionally, several neurological disorders are caused by extreme serine deficiency in humans owing in part to loss of D-serine (de Koning and Klomp, 2004).
The biosynthetic origin(s) of GroPGate are more perplexing. Whereas the deacylation of membrane phospholipids to form GroP-metabolites is a well-documented phenomenon (Emilsson and Sundler, 1984; Murray and McMaster, 2005; Simon and Cravatt, 2006), the existence of a suitable phospholipid precursor for GroPGate via deacylation (e.g., phosphatidylglycerate) has not been reported. As such, GroPGate may be produced through another mechanism, possibly metabolic conversion from GroPSer.
In conclusion, we have shown using an untargeted metabolomics approach that GDE1 regulates several GroP-metabolites in the mammalian brain, including GroPIns, GroPSer, and GroPGate. All three metabolites were subsequently verified to be direct substrates of GDE1 in vitro. Because the catalytic domain of GDE1 faces the extracellular side of the membrane (Zheng, et al., 2003), our findings indicate that these metabolites likely transit through the interstitial space in the brain, raising the intriguing possibility that they may serve as signaling molecules in their own right, a possibility consistent with some of the proposed functions of GroPIns and its phosphorylated derivatives (Corda, et al., 2009). Further investigation of the potential metabolic and signaling functions of GroP-metabolites will benefit from the availability of animal models, such as GDE1(-/-) mice, that have substantial alterations in the GroP-metabolome.
Significance
Glycerophosphodiesterases (GDEs) catalyze the hydrolysis of the phosphodiester bond in glycerophospho (GroP)-metabolites. Numerous GDE enzymes have been characterized in prokaryotes, but it is only recently that we have begun to appreciate the roles played by these enzymes in mammals. Here we have characterized the polar metabolome of mice bearing a targeted disruption in the GDE1 gene. This represents the first unbiased attempt to identify the physiological substrates of a GDE enzyme in a eukaryotic organism. By applying an untargeted LC-MS-based metabolomic profiling technique, we found that GroP-inositol (GroPIns), a known in vitro substrate for GDE1, is dramatically elevated in the central nervous system of GDE1(-/-) mice, as were two other metabolites. Analytical and synthetic chemistry methods were used to elucidate the structures of these metabolites as GroP-serine (GroPSer) and GroP-glycerate (GroPGate), neither of which, to our knowledge, have been previously described as natural products in eukaryotes. All three GroP metabolites were confirmed to be direct GDE1 substrates. Our untargeted metabolomic profiles also revealed a 40% reduction in free serine levels in the brain of GDE1(-/-) mice. Interestingly, this reduction was similar in magnitude to the corresponding elevation in GroPSer in these animals, indicating that GroPSer may serve as a metabolic reservoir for serine in the central nervous system. Subsequent derivitization studies demonstrated that this 40% reduction was observed for both L-and D-serine, a significant result given that the latter amino acid is a neurotransmitter involved in long-term potentiation and neuroprotection. Taken together, these findings indicate that the mammalian “GroP-metabolome” is quite diverse in structure and function and designate GDE1 as one of its principal enzymatic regulators in vivo.
Experimental Procedures
Metabolome preparation
For the isolation of water-soluble metabolites a methanol-water extraction protocol was used, essentially as previously described (Rabinowitz and Kimball, 2007). Briefly, male GDE1(+/+) and (-/-) mice (8-12 weeks old) were sacrificed and tissue was isolated and flash frozen in liquid nitrogen. Frozen brains were weighed and placed into a dounce tissue homogenizer with 1.5 mL ice-cold 80:20 CH3OH:water and homogenized with 10 strokes of the pestle. Each sample was sonicated and centrifuged at 10,000 × g to separate insoluble material. The resulting pellet was re-extracted with 500 μL of 80:20 CH3OH:water and centrifuged once again. The combined supernatant was dried down with a SpeedVac at 37 °C, resuspended in 50 μL of water, and stored at -80°C or injected directly into the mass spectrometer.
LC-MS
Tissue metabolome samples were analysed using an Agilent 1100 LC-MSD SL instrument with ionization achieved via electrospray ionization (ESI). The scan range was set to 100-1,200 Da with a source voltage of 3.0 kV. The fragmentor voltage was set to 100 V, the drying gas flow rate was 10 L/min, and the nebulizer pressure was 35 psi. Normal-phase chromatography was performed with a Luna-5 μm NH2 column (50 × 4.60 mm, Phenomenex, Torrance, CA). Mobile phases were as follows, Buffer A: MeCN, Buffer B: 95:5 H2O:CH3CN with 50mM NH4OAc and 0.2% NH4OAc with a resulting pH of 9.3. A 30 min gradient was applied: 0 to 100% buffer B followed by 10 min of isocratic flow with 100% buffer B. The flow rate was 0.5 mL/min. For each run the injection volume was 10 μL.
The obtained data were exported as common data format (.CDF) files. Differentially abundant metabolites between sample pairs were identified with the XCMS analyte profiling software, which aligns and quantifies the relative signal intensities of mass peaks from multiple LC-MS traces (Smith, et al., 2006). Significant peak changes between samples were confirmed by manual quantification by calculating the area under the peak from raw chromatograms. For some experiments an internal d3-serine standard (85 nmol) was added to the extraction solution. Absolute GroP levels were estimated by comparison to external calibration curves with synthetic GroP-metabolites and d3-serine.
Tandem MS and high mass-accuracy measurements
Initial mass-measurements and MS/MS experiments were performed using an Agilent 6520 Accurate Mass QTOF instrument coupled to an Agilent 1100 LC system. Samples were chromatographed as described above and analyzed in negative ionization mode. The source temperature was set to 350 °C with a cone gas flow of 11 l/hr, a desolvation gas temperature of 300 °C, and a nebulization pressure of 45 psig. The capillary voltage was set at -4 kV, the fragmentor voltage to 100 V and the skimmer voltage to 64 V. The collision energy was set at 35 V. Data were collected using a mass range of 100-1,200 Da with an acquisition time of 1.0 sec per spectrum. For MS/MS data GroPIns (333.0), GroPSer (258.0) and GroPGate (259.0) ions were targeted for fragmentation.
High mass-accuracy measurements (Supplemental Table 4) were obtained via Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry using an Apex II 7.0T FT-ICR mass spectrometer (Bruker Daltonics, Billerica, MA) coupled to an Agilent 1100 LC. This system is equipped with a custom electrospray source with two nebulizers for dual spray ionization enabling operation in “lock-mass” mode via the constant infusion of three compounds (omeprazole at 344.1074 m/z, hymecromone sulphate at 254.9969, and indoxyl sulphate at 212.0023) for internal calibration. A 1 megaword file size was used resulting in an acquisition time of 0.9 seconds per spectrum. Using these parameters, 247,000 resolving power was achieved at 259 m/z.
Preparation and purification of GroPIns, GroPSer, and GroPEtn
To obtain GroPIns, GroPSer, and GroPEtn, commercially available phosphatidylinositol, phosphatidylserine, and phosphatidylethanolamine were hydrolyzed according to the following procedure. In a 4 mL glass vial provided with a magnetic stirring bar 25 mg phospholipid was dissolved in 3 mL 2:1 CHCl3:CH3OH. 1mL 2 N NaOH was added and the mixture was stirred at room temperature. The progress of the reaction was checked by TLC (developed with 80:20 CH2Cl2:CH3OH) and, after complete hydrolysis was observed (usually after 1-2 hours), the reaction was neutralized by the addition of 1 mL 2 N HCl. To separate the desired GroP products from the liberated free fatty acids the reaction mixture was extracted twice with 2 mL of water. The combined aqueous phases were washed with 2 mL CHCl3, frozen in liquid nitrogen and lyophilized over night.
To separate the crude product from small molecules by-products and inorganic salts gel filtration chromatography was performed by dissolving the lyophilate in a minimum amount of water and passing it over a column packed with Sephadex® LH-20 material (Sigma Aldrich) using 80:20 MeOH:water as mobile phase to obtain the desired products.
GroPSer: 1H-NMR (300 MHz, D2O): δ 3.6-3.8 (m, 2H); 3.87 (m, 1H); 3.95 (m, 3H); 4.0-4.2 (m, 2H). ESI-MS calculated for C6H13NO8P-: 258.03843, found 258.03861. GroPEtn: 1H-NMR (300 MHz, D2O): δ 3.25 (t, 2H, J = 5.4 Hz), 3.5-3.7 (m, 2H), 3.8-4.0 (m, 5H). ESI-MS calculated for C5H13NO6P-: 214.04860, found 214.04702.
Chemical synthesis of bis((2,2-dimethyl-1,3,-dioxolan-4-yl)methyl) phenyl phosphate
To a solution of phenyl dichlorophosphate (600 μL, 4.0 mmol) in 20 mL dichloromethane and DIEA (4 mL) at 0°C, solketal (930 μL, 7.5 mmol) was added dropwise and the reaction was stirred at r. t. for 3 hrs. Phosphate buffer (pH 7) was added to a reaction, and the reaction was stirred for additional 1 hr to hydrolyze extra phenyl dichlorophosphate. After an addition of dichloromethane, organic layer was collected, and further washed with phosphate buffer (pH 7) three times to give bis((2,2-dimethyl-1,3,-dioxolan-4-yl)methyl) phenyl phosphate (935 mg, 62%). 1H-NMR (300 MHz, CDCl3): δ 1.36 (s, 6H), 1.45 (s, 3H), 1.47 (s, 3H), 3.87 (m, 2H), 4.10 (m, 2H), 4.2-4.3 (m, 4H), 4.40 (m, 2H), 7.2-7.3 (m, 2H), 7.38 (m, 3H). ESI-MS (ESI+): m/z 403 (M + H)+.
Chemical Synthesis of GroPGate
To a solution of glycerophosphoserine (GroPSer) (26 mg, 100 μmol) in 0.5 N HCl (2 mL) was added NaNO2 (15 mg, 220 μmol) at 4 °C with stirring. After 12 hrs, 100μL NaOH were added and the reaction mixture was flash frozen with liquid nitrogen and lyophilized over night. The resulting white solid was dissolved in a minimum amount of water and purified using Sephadex® LH-20 gel filtration material as described above affording GroPGate as a white solid. 1H-NMR (300 MHz, D2O): δ 3.7-3.9 (m, 2H); 3.98 (m, 1H); 4.05 (m, 2H); 4.20 (m, 1H); 4.33 (m, 2H). ESI-MS calculated for C6H12O9P-: 259.02244, found 259.02241.
Analysis of D- and L-serine levels in brain tissue
To effect the transformation of the serine enantiomers into diastereomers by derivatization with N-(N-α-t-Boc-phenylalanyloxy)succinimide (BocPheOSu) D-serine, L-serine or a mixture thereof (104 μg, 1 μmol) was dissolved in 500 μL PBS and BocPheOSu (10 mg, 25 μmol) was added. After stirring at room temperature for 3 h the reaction mixture was directly analyzed by LC-MS on an Agilent Eclipse XDB-C18 column (5 μM, 4.6 mm×150 mm). Mobile phase A consisted of 95:5 water and methanol, with 0.1% of 28% ammonium hydroxide, and mobile phase B consisted of 60:55:5 2-propanol, CH3OH, and water, with 0.1% of 28% ammonium hydroxide. The gradient started at 5% B and then linearly increased to 45% B over 40 min followed by 100% B for 10 min at a flow rate of 0.5 mL/min. In this way the retention times of BocPheOSu-derivatized D- and L-serine were determined. For serine level analysis in brains, tissue samples were prepared and metabolites were extracted as described above. The resuspended metabolome (10 μL of the 50 μL resuspension) was derivatized with BocPheOSu following the derivatized metabolite mixture was analyzed by reverse-phase LC-MS as described above.
Enzyme assays
Enzyme assays were performed with LC-MS by observing the formation of serine and glycerate in the cases of GroPSer and GroPGate, and by observing formation of inositol (for recombinant GDE1 assays) or reduction of substrate (for mouse brain samples) in the case of GroPIns. For recombinant GDE1 assays an N-terminal Myc-fusion of GDE1 was transiently overexpressed in COS-7 cells and membrane fractions prepared as described previously (Simon and Cravatt, 2008). The membrane samples were resuspended in assay buffer (50 mM Tris, 2 mM MgCl2, pH 8.0). Assays were carried out in 100 μL total reaction volume. For each reaction 200 μM of substrate was incubated with a final concentration of 0.1 mg/mL cell-lysate or 0.5 mg/mL mouse brain membrane preparation respectively. Reactions were incubated at 37°C for 3 hours (in the case of GroPSer and GroPGate) or 9 hours (in the case of GroPIns) before quenching with 1.0 mL CH3OH. Subsequently, 25 nmoles of D3-serine were added to each reaction as an internal standard. Samples were centrifuged at 1,400 × g for 3 min, and dried down under vacuum. For normal phase LC-MS analysis samples were resuspended in 30 μL water and injected into an Agilent 1100 LC-MSD SL instrument applying the following gradient: 15-90 % buffer B over 15 min followed by isocratic flow with 100 % B for 5 min at a flow rate of 0.5 mL/min. For each run the injection volume was 30 μL. Each assay was conducted with an n = 3.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Benton HP, Wong DM, Trauger SA, Siuzdak G. XCMS2: processing tandem mass spectrometry data for metabolite identification and structural characterization. Anal Chem. 2008;80:6382–6389. doi: 10.1021/ac800795f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Corda D, Kudo T, Zizza P, Iurisci C, Kawai E, Kato N, Yanaka N, Mariggio S. The developmentally regulated osteoblast phosphodiesterase GDE3 is glycerophosphoinositol-specific and modulates cell growth. J Biol Chem. 2009;284:24848–24856. doi: 10.1074/jbc.M109.035444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D, Zizza P, Varone A, Filippi BM, Mariggio S. The glycerophosphoinositols: cellular metabolism and biological functions. Cell Mol Life Sci. 2009;66:3449–3467. doi: 10.1007/s00018-009-0113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning TJ, Klomp LW. Serine-deficiency syndromes. Curr Opin Neurol. 2004;17:197–204. doi: 10.1097/00019052-200404000-00019. [DOI] [PubMed] [Google Scholar]
- Dowd SR, Bier ME, Patton-Vogt JL. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- Emilsson A, Sundler R. Differential activation of phosphatidylinositol deacylation and a pathway via diphosphoinositide in macrophages responding to zymosan and ionophore A23187. J Biol Chem. 1984;259:3111–3116. [PubMed] [Google Scholar]
- Falasca M, Carvelli A, Iurisci C, Qiu RG, Symons MH, Corda D. Fast receptor-induced formation of glycerophosphoinositol-4-phosphate, a putative novel intracellular messenger in the Ras pathway. Mol Biol Cell. 1997;8:443–453. doi: 10.1091/mbc.8.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca M, Marino M, Carvelli A, Iurisci C, Leoni S, Corda D. Changes in the levels of glycerophosphoinositols during differentiation of hepatic and neuronal cells. Eur J Biochem. 1996;241:386–392. doi: 10.1111/j.1432-1033.1996.00386.x. [DOI] [PubMed] [Google Scholar]
- Fisher E, Almaguer C, Holic R, Griac P, Patton-Vogt J. Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J Biol Chem. 2005;280:36110–36117. doi: 10.1074/jbc.M507051200. [DOI] [PubMed] [Google Scholar]
- Furuya S, Tabata T, Mitoma J, Yamada K, Yamasaki M, Makino A, Yamamoto T, Watanabe M, Kano M, Hirabayashi Y. L-serine and glycine serve as major astroglia-derived trophic factors for cerebellar Purkinje neurons. Proc Natl Acad Sci U S A. 2000;97:11528–11533. doi: 10.1073/pnas.200364497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallazzini M, Ferraris JD, Burg MB. GDPD5 is a glycerophosphocholine phosphodiesterase that osmotically regulates the osmoprotective organic osmolyte GPC. Proc Natl Acad Sci U S A. 2008;105:11026–11031. doi: 10.1073/pnas.0805496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TA, Raetz CR, Richardson T, Kordestani R, Son JD, Rose RL. Identification of phosphatidylserylglutamate: a novel minor lipid in Escherichia coli. J Lipid Res. 2009;50:1589–1599. doi: 10.1194/jlr.M800549-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Li S, Smith DC, Shaw WA, Raetz CR. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry (Mosc) 2007;46:14500–14513. doi: 10.1021/bi701907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger C, Papouin T, Oliet SH, Rusakov DA. Long-term potentiation depends on release of D-serine from astrocytes. Nature. 2010;463:232–236. doi: 10.1038/nature08673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova PT, Milne SB, Myers DS, Brown HA. Lipidomics: a mass spectrometry based systems level analysis of cellular lipids. Curr Opin Chem Biol. 2009;13:526–531. doi: 10.1016/j.cbpa.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AU, Werner SR, Talaty N, Song Y, Campbell K, Cooks RG, Morgan JA. Targeted metabolomic analysis of Escherichia coli by desorption electrospray ionization and extractive electrospray ionization mass spectrometry. Anal Biochem. 2008;375:272–281. doi: 10.1016/j.ab.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Klivenyi P, Kekesi KA, Hartai Z, Juhasz G, Vecsei L. Effects of mitochondrial toxins on the brain amino acid concentrations. Neurochem Res. 2005;30:1421–1427. doi: 10.1007/s11064-005-8512-x. [DOI] [PubMed] [Google Scholar]
- Larson TJ, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem. 1983;258:5428–5432. [PubMed] [Google Scholar]
- Lencina CL, Dassonville-Klimpt A, Sonnet P. New efficient enantioselective synthesis of 2-oxopiperazines: a practical access to chiral 3-substituted 2-oxopiperazines. Tetrahedron-Asymmetry. 2008;19:1689–1697. [Google Scholar]
- Mariggio S, Iurisci C, Sebastia J, Patton-Vogt J, Corda D. Molecular characterization of a glycerophosphoinositol transporter in mammalian cells. FEBS Lett. 2006;580:6789–6796. doi: 10.1016/j.febslet.2006.11.039. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LaFace DM, Green DR. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally DJ, Aubry AJ, Hui JP, Khieu NH, Whitfield D, Ewing CP, Guerry P, Brisson JR, Logan SM, Soo EC. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J Biol Chem. 2007;282:14463–14475. doi: 10.1074/jbc.M611027200. [DOI] [PubMed] [Google Scholar]
- McNally DJ, Hui JP, Aubry AJ, Mui KK, Guerry P, Brisson JR, Logan SM, Soo EC. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J Biol Chem. 2006;281:18489–18498. doi: 10.1074/jbc.M603777200. [DOI] [PubMed] [Google Scholar]
- Mothet JP, Parent AT, Wolosker H, Brady RO, Jr, Linden DJ, Ferris CD, Rogawski MA, Snyder SH. D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc Natl Acad Sci U S A. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JP, McMaster CR. Nte1p-mediated deacylation of phosphatidylcholine functionally interacts with Sec14p. J Biol Chem. 2005;280:8544–8552. doi: 10.1074/jbc.M413999200. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Horiike K, Maeda T. Distribution of free D-serine in vertebrate brains. Brain Res. 1994;634:291–295. doi: 10.1016/0006-8993(94)91932-1. [DOI] [PubMed] [Google Scholar]
- Patton-Vogt J. Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim Biophys Acta. 2007;1771:337–342. doi: 10.1016/j.bbalip.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Patton-Vogt JL, Henry SA. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics. 1998;149:1707–1715. doi: 10.1093/genetics/149.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Kimball E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal Chem. 2007;79:6167–6173. doi: 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- Rao M, Sockanathan S. Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science. 2005;309:2212–2215. doi: 10.1126/science.1117156. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry (Mosc) 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Anandamide biosynthesis catalyzed by the phosphodiesterase GDE1 and detection of glycerophospho-N-acyl ethanolamine precursors in mouse brain. J Biol Chem. 2008;283:9341–9349. doi: 10.1074/jbc.M707807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Characterization of mice lacking candidate N-acyl ethanolamine biosynthetic enzymes provides evidence for multiple pathways that contribute to endocannabinoid production in vivo. Mol Biosyst. 2010 doi: 10.1039/c000237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J Biol Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Snyder SH, Kim PM. D-amino acids as putative neurotransmitters: focus on D-serine. Neurochem Res. 2000;25:553–560. doi: 10.1023/a:1007586314648. [DOI] [PubMed] [Google Scholar]
- Tagore DM, Nolte WM, Neveu JM, Rangel R, Guzman-Rojas L, Pasqualini R, Arap W, Lane WS, Saghatelian A. Peptidase substrates via global peptide profiling. Nat Chem Biol. 2009;5:23–25. doi: 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Martin MV, Guengerich FP. Elucidation of functions of human cytochrome P450 enzymes: identification of endogenous substrates in tissue extracts using metabolomic and isotopic labeling approaches. Anal Chem. 2009;81:3071–3078. doi: 10.1021/ac900021a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommassen J, Eiglmeier K, Cole ST, Overduin P, Larson TJ, Boos W. Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol Gen Genet. 1991;226:321–327. doi: 10.1007/BF00273621. [DOI] [PubMed] [Google Scholar]
- Vinayavekhin N, Saghatelian A. Regulation of alkyl-dihydrothiazole-carboxylates (ATCs) by iron and the pyochelin gene cluster in Pseudomonas aeruginosa. ACS Chem Biol. 2009;4:617–623. doi: 10.1021/cb900075n. [DOI] [PubMed] [Google Scholar]
- Wolosker H, Blackshaw S, Snyder SH. Serine racemase: a glial enzyme synthesizing D-serine to regulate glutamate-N-methyl-D-aspartate neurotransmission. Proc Natl Acad Sci U S A. 1999;96:13409–13414. doi: 10.1073/pnas.96.23.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nishizaki I, Furuya S, Hirabayashi Y, Takahashi K, Okuyama S, Yamamoto H. Characterization of rapid and high-affinity uptake of L-serine in neurons and astrocytes in primary culture. FEBS Lett. 2003;548:69–73. doi: 10.1016/s0014-5793(03)00742-7. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Yamada K, Furuya S, Mitoma J, Hirabayashi Y, Watanabe M. 3-Phosphoglycerate dehydrogenase, a key enzyme for l-serine biosynthesis, is preferentially expressed in the radial glia/astrocyte lineage and olfactory ensheathing glia in the mouse brain. J Neurosci. 2001;21:7691–7704. doi: 10.1523/JNEUROSCI.21-19-07691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Sabharwal P, Rao M, Sockanathan S. The antioxidant enzyme Prdx1 controls neuronal differentiation by thiol-redox-dependent activation of GDE2. Cell. 2009;138:1209–1221. doi: 10.1016/j.cell.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaka N. Mammalian glycerophosphodiester phosphodiesterases. Biosci Biotechnol Biochem. 2007;71:1811–1818. doi: 10.1271/bbb.70062. [DOI] [PubMed] [Google Scholar]
- Yanaka N, Imai Y, Kawai E, Akatsuka H, Wakimoto K, Nogusa Y, Kato N, Chiba H, Kotani E, Omori K, et al. Novel membrane protein containing glycerophosphodiester phosphodiesterase motif is transiently expressed during osteoblast differentiation. J Biol Chem. 2003;278:43595–43602. doi: 10.1074/jbc.M302867200. [DOI] [PubMed] [Google Scholar]
- Zheng B, Berrie CP, Corda D, Farquhar MG. GDE1/MIR16 is a glycerophosphoinositol phosphodiesterase regulated by stimulation of G protein-coupled receptors. Proc Natl Acad Sci U S A. 2003;100:1745–1750. doi: 10.1073/pnas.0337605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chen D, Farquhar MG. MIR16, a putative membrane glycerophosphodiester phosphodiesterase, interacts with RGS16. Proc Natl Acad Sci U S A. 2000;97:3999–4004. doi: 10.1073/pnas.97.8.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.