Abstract
Adult regenerative myogenesis is vital for restoring normal tissue structure after muscle injury. Muscle regeneration is dependent on progenitor satellite cells, which proliferate in response to injury, and their progeny differentiate and undergo cell–cell fusion to form regenerating myofibers. Myogenic progenitor cells must be precisely regulated and positioned for proper cell fusion to occur. Chemokines are secreted proteins that share both leukocyte chemoattractant and cytokine-like behavior and affect the physiology of a number of cell types. We investigated the steady-state mRNA levels of 84 chemokines, chemokine receptors and signaling molecules, to obtain a comprehensive view of chemokine expression by muscle cells during myogenesis in vitro. A large number of chemokines and chemokine receptors were expressed by primary mouse muscle cells, especially during times of extensive cell–cell fusion. Furthermore, muscle cells exhibited different migratory behavior throughout myogenesis in vitro. One receptor–ligand pair, CXCR4–SDF-1α (CXCL12), regulated migration of both proliferating and terminally differentiated muscle cells, and was necessary for proper fusion of muscle cells. Given the large number of chemokines and chemokine receptors directly expressed by muscle cells, these proteins might have a greater role in myogenesis than previously appreciated.
Keywords: Fusion, Myoblast, Myocyte, CXCR4, SDF-1α, Regeneration
Introduction
Skeletal muscle degeneration can occur as a result of disease or injury; however, this tissue has an extensive ability to regenerate. Adult regenerative myogenesis is dependent on progenitor cells called satellite cells. Satellite cells are normally quiescent, but proliferate in response to injury, and their progeny myoblasts differentiate into fusion-competent myocytes, which fuse with one another or with existing myofibers to restore normal tissue architecture. In vitro studies demonstrate that migration is a key process during myogenesis. Migration is crucial to achieve cell–cell adhesion, which is necessary for differentiation (Kang et al., 2004), as well as formation and growth of myotubes in vitro (Bae et al., 2008; Jansen and Pavlath, 2006; Mylona et al., 2006; O'Connor et al., 2007). Identification of molecules that regulate cell migration might reveal potential molecular targets for improving muscle regeneration and the efficiency of cell-transplantation therapies (Galvez et al., 2006; Hill et al., 2006; Palumbo et al., 2004).
A number of extracellular molecules are known to regulate muscle cell migration in vitro. Secreted factors such as hepatocyte growth factor, fibroblast growth factor, platelet-derived growth factor and IL-4 have key roles during myogenesis (Bischoff, 1997; Corti et al., 2001; Lee et al., 1999; Robertson et al., 1993; Horsley et al., 2003; Lafreniere et al., 2006). In addition, extracellular matrix (ECM) proteins and ECM-associated molecules, such as laminin, fibronectin, CD44, decorin and N-cadherin, as well as matrix metalloproteinases, are crucial for regulating cell migration during myogenesis (Echtermeyer et al., 1996; Lluri and Jaworski, 2005; Lluri et al., 2008; Mylona et al., 2006; Ocalan et al., 1988; Olguin et al., 2003; Yao et al., 1996). Overall, a complex interplay among many types of proteins is required for proper migration of muscle cells.
Chemokines are secreted proteins, approximately 8–10 kDa in size, with 20–70% homology in amino acid sequences, that share both leukocyte chemoattractant and cytokine-like behavior (Baggiolini et al., 1995; Luster, 1998). Chemokines are important for the migration of muscle precursor cells during embryonic myogenesis (Vasyutina et al., 2005; Yusuf et al., 2006) and for macrophage infiltration into damaged muscle tissue (McLennan, 1996; Robertson et al., 1993). Furthermore, chemokines and their receptors are expressed by diseased or regenerating muscle tissue (Hirata et al., 2003; Porter et al., 2003; Sachidanandan et al., 2002; Warren et al., 2005; Warren et al., 2004; Civatte et al., 2005; Demoule et al., 2009). Finally, chemokines are known to regulate migration of several cell types postnatally, such as immune cells, sperm and metastasizing cancer cells (Kim, 2004; Kim, 2005; Stebler et al., 2004; Bleul et al., 1996; Isobe et al., 2002; Miyazaki et al., 2006; Vandercappellen et al., 2008; Muciaccia et al., 2005a; Muciaccia et al., 2005b). However, no studies have comprehensively examined the expression of these molecules specifically by muscle cells at different phases of myogenesis.
Our studies indicate that a large number of chemokines and chemokine receptors are expressed by primary mouse muscle cells in vitro, especially during times of extensive cell–cell fusion. Furthermore, muscle cells exhibited different migratory behavior throughout myogenesis in vitro. One receptor–ligand pair, CXCR4–SDF-1α (CXCL12), regulated the migration of both proliferating and terminally differentiated muscle cells, and was necessary for proper fusion of muscle cells.
Results
Many chemokines and their receptors are expressed during myogenesis
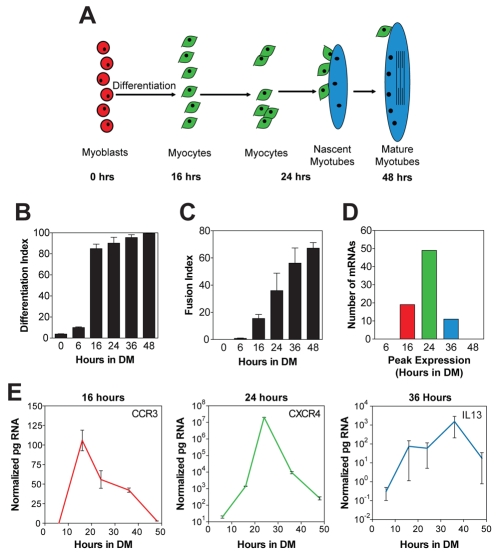
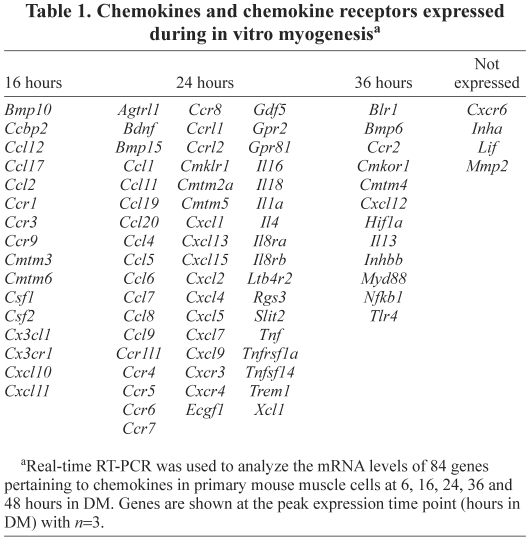
To determine which chemokine receptors and ligands are expressed by muscle cells at different time points during myogenesis, pure cultures of primary mouse muscle cells were used because they follow a predictable time-course of myogenesis. Upon removal of serum, myoblasts differentiate into myocytes that fuse to form nascent myotubes, which are small and contain few nuclei. Subsequently, myocytes fuse with nascent myotubes creating mature myotubes, which are large and contain many nuclei (Fig. 1A). In our culture conditions, by 16 hours in differentiation medium (DM), the majority of cells were terminally differentiated myocytes as indicated by the high percentage of embryonic myosin-heavy-chain-positive (eMyHC+) cells (Fig. 1B). After 24 hours in DM, ~40% of myocytes were fused with each other to form nascent myotubes. By 48 hours, ~70% of myocytes were fused, creating mature myotubes (Fig. 1C). A real-time RT-PCR array was used to investigate the mRNA steady-state levels of 84 chemokines, chemokine receptors and signaling molecules, to obtain a comprehensive view of chemokine expression during myogenesis. Approximately 80 of these mRNAs were detected during myogenesis, indicating that many chemokine receptors and ligands are expressed directly by muscle cells in vitro. The steady-state levels of these mRNAs varied drastically; a small subset of genes had extremely high steady-state levels, ~10,000- to 1-million-fold higher than other genes (supplementary material Table S1). Furthermore, no genes were constitutively expressed at a stable level throughout myogenesis; instead the mRNA levels of all genes increased after differentiation. Very few mRNAs were present after 6 or 48 hours in DM; rather, most mRNA steady-state levels were highest between 16 and 36 hours in DM (Table 1; Fig. 1D,E), which were time points of extensive differentiation and fusion of myocytes.
Fig. 1.
Chemokines and their receptors are expressed during in vitro myogenesis. (A) During myotube formation, the majority of myoblasts (red) terminally differentiate into myocytes (green) which migrate, adhere and fuse with one another to form small nascent myotubes with few nuclei (blue). Subsequently, nascent myotubes fuse with myocytes to form large mature myotubes with many nuclei (blue). (B) Primary mouse muscle cells were immunostained for eMyHC at different times in DM and the percentage of nuclei within eMyHC+ cells (differentiation index) was determined. By 16 hours in DM, most nuclei were in eMyHC+ cells. (C) The fusion index, or percentage of nuclei in myotubes, increased with time, and by 48 hours the majority of nuclei were within myotubes. (D) A real-time RT-PCR array was used to analyze the time-course of expression in vitro for 84 genes pertaining to chemokines. Positive results were obtained for 80 genes. Three patterns of expression were observed with mRNA steady state levels peaking at 16, 24 or 36 hours in DM, times of extensive differentiation and fusion. The number of genes with peak expression levels at each time point is shown. (E) Time course of expression for three representative genes peaking at 16 (CCR3), 24 (CXCR4) or 36 hours (IL13) in DM. Data are means ± s.e.m., n=3.
Table 1.
Chemokines and chemokine receptors expressed during in vitro myogenesisa
Many chemokine receptors and ligands known to be expressed by skeletal muscle cells or tissue were shown in this assay to be expressed directly by muscle cells (Bischoff, 1997; Chazaud et al., 2003; Chong et al., 2007; Civatte et al., 2005; De Rossi et al., 2000; Hirata et al., 2003; Odemis et al., 2007; Peterson and Pizza, 2009; Porter et al., 2003; Ratajczak et al., 2003; Sachidanandan et al., 2002; Summan et al., 2003; Warren et al., 2005; Warren et al., 2004). For example, IL4, an important pro-myogenic factor expressed during myogenesis in vitro and in vivo (Horsley et al., 2003; Lafreniere et al., 2006), was identified by this chemokine array (Table 1). However, a few chemokine receptors and ligands not previously known to be expressed by skeletal muscle cells or tissue were also identified, including angiotensin receptor-like 1 (AGTRL1, Aplnr, apelin receptor), bone morphogenic protein 10 (BMP10), CXCL13, and its receptor CXCR5 (Burkitt's lymphoma receptor 1, BLR1). The large number of chemokine receptor–ligand pairs expressed directly by muscle cells suggests a complex spatial and temporal control of migration during myogenesis.
The migratory behavior of muscle cells changes during myogenesis
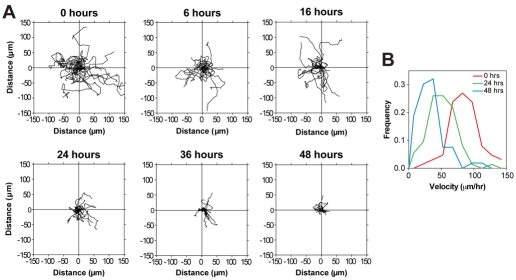
To conduct an in-depth analysis of the migratory behavior of muscle cells during myogenesis, time-lapse microscopy was performed for 3 hours at different time points (Fig. 2A). Myocytes displayed distinct differences in migration compared with myoblasts. At 0 hours, myoblasts migrated far from their point of origin, whereas over the course of myogenesis, myocytes stayed progressively closer to their point of origin (Fig. 2A). The proportion of slow-moving cells also increased during myogenesis (Fig. 2B), causing a concomitant decrease in mean velocity from 56 μm/hour at 0 hours to 22 μm/hour at 48 hours in DM. The diminished velocity of myocytes at 48 hours was not due to a loss in cell motility or viability because the addition of fresh serum-free DM increased cell migration (data not shown). The enhancement of cell migration by fresh DM might be due to elimination of inhibitory factors secreted by cells into the medium during myogenesis. Thus, muscle cells are migratory throughout myogenesis; as most investigations have focused on myoblast migration, the majority of receptor–ligand pairs that regulate myocyte migration are unknown.
Fig. 2.
Changes in migratory behavior with muscle cell differentiation. (A) Migratory paths of mononucleated primary mouse muscle cells at 0, 6, 16, 24, 36 and 48 hours in DM. Tracks were taken from 3 hours of time-lapse microscopy with pictures every 5 minutes. Representative graphs are shown from one of three independent isolates with 20 cells each. (B) Frequency distribution of cell velocity at different times in myogenesis. A total of 60 cells were analyzed. Data are n=3.
Myoblasts and myocytes migrate to distinct factors
To determine whether myocytes migrate in response to canonical myoblast chemoattractants, cell migration was analyzed in Boyden chambers using hepatocyte growth factor (HGF) and platelet-derived growth factor (PDGF), potent myoblast chemoattractants (Bischoff, 1997; Corti et al., 2001). We enriched for myocytes by culturing cells in DM for 24 hours at low density to prevent myotube formation yielding 96% of nuclei in eMyHC+ cells, and only 7% of nuclei in myotubes. Both HGF and PDGF greatly enhancing the migration of myoblasts; however, neither factor stimulated myocyte migration (Fig. 3), suggesting that intrinsic differences exist between the two cell types, such as differential expression of chemoattractant receptors. However, myocytes exhibited a 65-fold increase in migration to conditioned media (CM), which contains the factors secreted by muscle cells during differentiation and fusion, compared with control medium (Fig. 3). Migration to CM suggests that migratory factors, such as chemokines, are secreted during myogenesis and control migration during the process of cell fusion to form myotubes. Together, these data suggest that factors which regulate myoblast migration might not regulate myocyte migration during myogenesis in vitro.
Fig. 3.
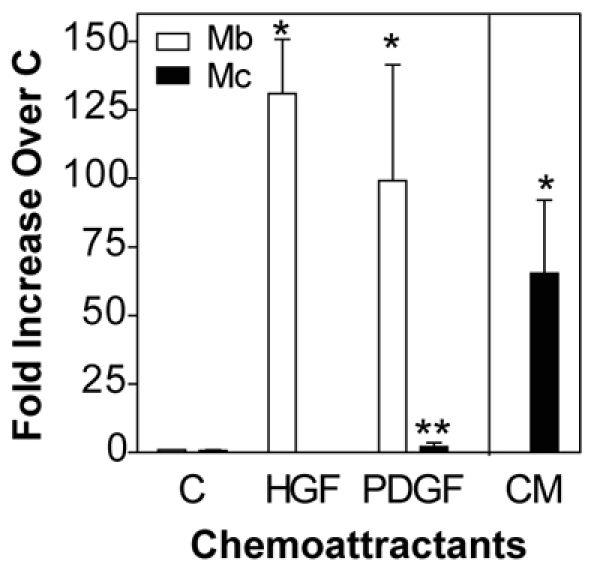
Myocytes do not migrate to canonical myoblast migratory factors. Primary mouse myoblasts (Mb) and myocytes (Mc, 24 hours in DM) were allowed to migrate in Boyden chambers to control medium (C) or medium containing 100 ng/ml HGF or PDGF for 5 hours. Myocyte migration to conditioned medium (CM) from cultures in DM for 24 hours was also tested. Data are mean ± s.e.m., n=3–5 (*P<0.05 compared with control; **P<0.05 compared with myoblasts).
Myocytes exist during muscle regeneration
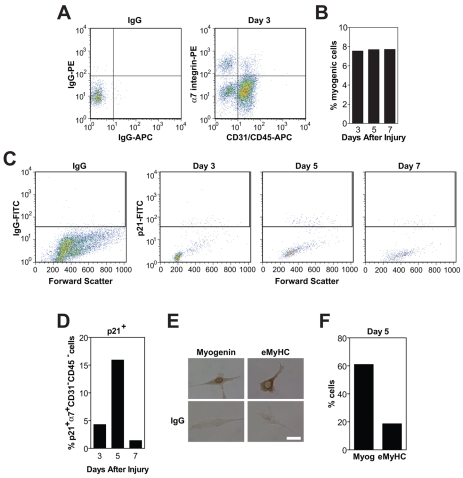
We next quantified the percentage of myocytes during adult regenerative myogenesis in vivo. Regenerative myogenesis is an asynchronous process that requires both spatial and temporal coordination. Upon injury, satellite cells proliferate and then terminally differentiate to become fusion-competent myocytes, which express differentiation-specific proteins such as myogenin, p21 and eMyHC and then fuse with each other and with myofibers to restore normal tissue architecture. Mononucleated cells were isolated from injured mouse muscles and analyzed by flow cytometry (Fig. 4A). Muscle cells were defined as α7-integrin-positive cells, which were also negative for endothelial and hematopoietic lineage markers (CD31 and CD45) (Blanco-Bose et al., 2001; Kafadar et al., 2009). As muscle cells are quiescent before injury (Schultz et al., 1978) and in days immediately following injury, the majority of mononucleated cells in muscle tissue are immune cells (Allbrook, 1981; McLennan, 1996; Tidball, 2005); day 3 was the earliest time point analyzed. At later time points, myogenic cells are fusing into newly regenerating myofibers (Allbrook, 1981), therefore day 7 was the latest time point assayed. The relative percentage of mononucleated muscle cells did not change during these time points of regeneration (Fig. 4B). To determine whether differentiated α7-integrin+ CD31− CD45− muscle cells exist during regeneration, cells were also immunostained for p21, which marks terminally differentiated cells (Andres and Walsh, 1996). The peak percentage of terminally differentiated p21+ myogenic cells was observed at day 5 after injury (Fig. 4C,D).
Fig. 4.
Myocytes exist during muscle regeneration. (A) Mononucleated cells were isolated from gastrocnemius muscles at days 3, 5 and 7 after injury and immunostained with antibodies against CD31 (APC), CD45 (APC) and α7 integrin (PE): CD31+ CD45+, to identify endothelial and immune cells and α7-integrin+ CD31− CD45− for myogenic cells. Myogenic cells constituted ~8% of the total mononucleated cells at day 3. Isotype controls were used to determine proper gating (left panel). (B) The percentage of myogenic cells remained stable during muscle regeneration. (C) Mononucleated cells were isolated from gastrocnemius muscles at indicated days after injury and immunostained with antibodies against CD31 (APC), CD45 (APC), α7 integrin (PE) and p21 (FITC) to identify terminally differentiated muscle cells. Myogenic α7-integrin+ CD31− CD45− cells were analyzed for p21. Isotype control was used to determine proper gating (left panel). (D) The percentage of p21+ myogenic cells was highest at day 5 after injury. (E) Mononucleated α7-integrin+ CD31− CD45− myogenic cells isolated from gastrocnemius muscles 5 days after injury were plated in vitro and immunostained for differentiation markers, myogenin (top left) and eMyHC (top right) or appropriate IgG controls (bottom). Scale bar: 10 μm. (F) The percentage of myogenin+ and eMyHC+ cells in E; ~60% of cells were myogenin+ a marker for earlier stages of differentiation, and ~20% were eMyHC+, a marker for later stages of differentiation. Data are from a pool of ten mice.
We used several markers to determine the progression of muscle cells through the continuum of differentiation. As muscle cells progress through differentiation, first myogenin is expressed, then p21 and finally MyHC (Andres and Walsh, 1996). Therefore, cells at later stages of differentiation are myogenin+ p21+ eMyHC+ and these cells are not likely to accumulate because they should be fusing to form newly regenerated myofibers. To determine the percentage of muscle cells at early and late stages of differentiation, myogenic cells were isolated from gastrocnemius muscles at day 5 after injury by FACS, and immunostained for myogenin and eMyHC in vitro (Fig. 4E). Approximately 60% of myogenic cells were myogenin+ and 18% were eMyHC+ (Fig. 4F). Therefore, regenerating muscle tissue at day 5 is a mixture of myogenic cells at various stages of differentiation. As the expression of chemokine receptor–ligand pairs increased after differentiation of muscle cells in vitro, these factors are likely to be involved in the regulation of differentiating myogenic cells in vivo.
CXCR4 and SDF-1α are expressed during myogenesis in vitro and in vivo
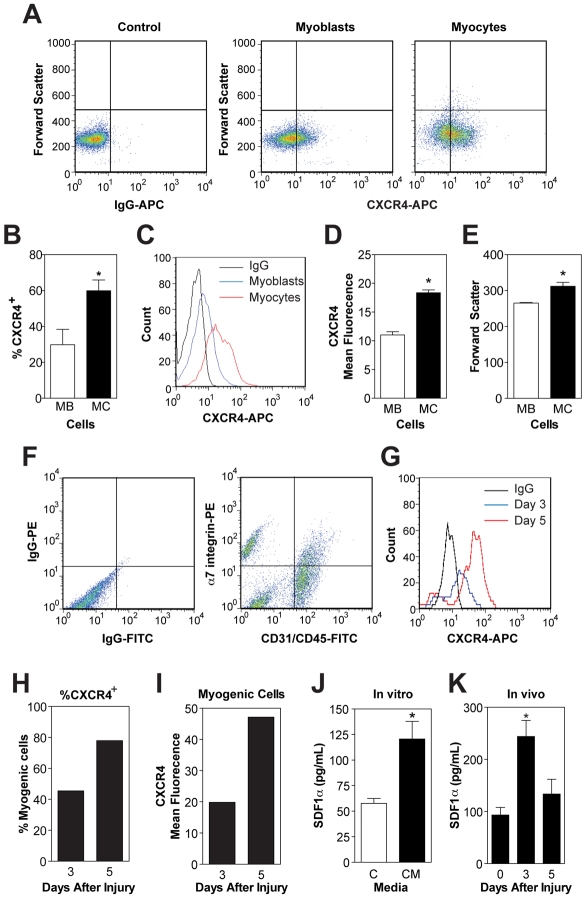
We examined the role of the most highly expressed chemokine receptor CXCR4 and its ligand, CXCL12 or SDF-1α, in more detail. The receptor CXCR4 and ligand SDF-1α were of specific interest because several studies have shown expression of these proteins by muscle cells or tissue, but conflicting reports exist regarding their role during myogenesis (Bae et al., 2008; Chong et al., 2007; Melchionna et al., 2010; Odemis et al., 2007; Odemis et al., 2005; Ratajczak et al., 2003; Vasyutina et al., 2005; Yusuf et al., 2006). To confirm expression of CXCR4 at the protein level, flow cytometry was used to determine the percentage of CXCR4+ cells in pure cultures of primary mouse myoblasts and myocytes; ~30% of myoblasts were CXCR4+ compared with ~60% of myocytes (Fig. 5A,B). Furthermore, myocytes contained ~two-fold more CXCR4 per cell (Fig. 5C,D), yet myocytes were only 18% larger than myoblasts (Fig. 5E), suggesting that myocytes have a higher density of CXCR4 at the plasma membrane. The increased level of CXCR4 protein in myocytes correlated to the increased mRNA levels of CXCR4 at 24 hours in DM (Fig. 1E). To determine whether CXCR4 and SDF1α are expressed during adult regenerative myogenesis, the percentage of CXCR4+ α7-integrin+ CD31− CD45− myogenic cells was determined at day 3 and day 5 after injury (Fig. 5F,G). From day 3 to day 5, the percentage of myogenic CXCR4+ cells increased from ~45% to 77% (Fig. 5H). In addition, the amount of CXCR4 per muscle cell was increased ~2.5-fold at day 5 compared with day 3 (Fig. 5I), with no change in cell size (data not shown). The percentage of myogenic cells that express CXCR4 in regenerating muscle at day 3 is lower than the 80% CXCR4+ cells observed in freshly isolated quiescent Pax7+ satellite cells on myofibers from uninjured muscle (Cerletti et al., 2008). This discrepancy might be due in part to the marker used for positive selection of myogenic cells in our studies, but is also probably due to modulation of CXCR4-expressing cells by the regenerative process because we observe a 1.7-fold increase in the percentage of CXCR4+ myogenic cells from day 3 to day 7.
Fig. 5.
CXCR4 and SDF-1α are expressed during myogenesis in vitro and in vivo. (A) Primary mouse myoblasts and myocytes were immunostained with antibodies against CXCR4 (APC) in vitro. (B) The percentage of CXCR4+ cells was quantified; a significantly higher percentage of myocytes were CXCR4+. (C) Representative histogram; the level of CXCR4 per cell was also increased between myoblasts and myocytes. (D) Mean fluorescence intensity of CXCR4 per cell; myocytes contained almost twice as much CXCR4 per cell. (E) Myocytes were 18% larger than myoblasts. (F) Mononucleated cells were isolated from gastrocnemius muscles at days 3 and 5 after injury and immunostained with antibodies against CD31 (FITC), CD45 (FITC), α7 integrin (PE) and CXCR4 (APC). Cells were analyzed with the following criteria: CD31+ CD45+, to identify endothelial and immune cells and α7-integrin+ CD31− CD45− for myogenic cells. Day 5 is shown. (G) Myogenic α7-integrin+ CD31− CD45− cells were analyzed for CXCR4 and a representative histogram is shown. (H) The percentage of CXCR4+ myogenic cells; a higher percentage of myogenic cells were CXCR4+ at day 5. (I) The mean fluorescence intensity of CXCR4 per cell was also increased between day 3 and 5; myocytes contained almost twice as much CXCR4 per cell. (J) The level of SDF-1α secreted by primary mouse muscle cells in vitro during myogenesis (24 hours CM) was determined by ELISA. (K) The level of SDF-1α in crushed muscle extract determined by ELISA. The level of SDF-1α was increased at day 3. In all flow cytometry experiments: propidium iodide (PI) was used to remove dead cells from analysis; representative flow plots are shown and isotype controls were used to determine proper gating. Data are means ± s.e.m., n=3 (*P<0.05 compared with Mb, control or 0 days as appropriate).
To validate expression of SDF1-α at the protein level, ELISA assays were performed using control DM and 24 hours CM; significant levels of SDF1-α were detected in CM (Fig. 5J). The levels of SDF1-α in crushed muscle extract, which contains released soluble protein by control and regenerating muscles, were also determined (Bischoff, 1986; Chen and Quinn, 1992). Muscles at day 3 after injury contained significantly higher levels of SDF1-α, compared with uninjured muscles or muscles at day 5 after injury (Fig. 5K). Therefore, SDF1-α might be released under certain conditions after injury. Together, these data demonstrate that CXCR4 and SDF-1α proteins are expressed by primary mouse muscle cells during myogenesis in vitro. As CXCR4 was expressed by mononucleated muscle cells during adult regenerative myogenesis, and SDF-1α was isolated from muscle tissue, this receptor–ligand pair might regulate myogenesis.
The CXCR4–SDF-1α axis is important for proper muscle cell fusion
To examine the role of the CXCR4–SDF-1α axis in myogenesis, we used primary mouse muscle cells in vitro, because direct effects on muscle cells can be analyzed in the absence of other cell types. To determine whether the CXCR4–SDF-1α axis regulates migration during myogenesis, myoblasts and myocytes were allowed to migrate to several concentrations of SDF-1α in Boyden chambers (Fig. 6A). Interestingly, while both cell types were attracted to SDF-1α, myoblasts required a 20-fold higher concentration than myocytes to achieve a similar level of migration. This difference is likely due not only to the greater percentage of CXCR4+ cells in the myocyte population, but also to the increased CXCR4 per myocyte. Thus, SDF-1α affects migration of both myoblasts and myocytes, although myocytes exhibit a greater sensitivity to SDF-1α.
Fig. 6.
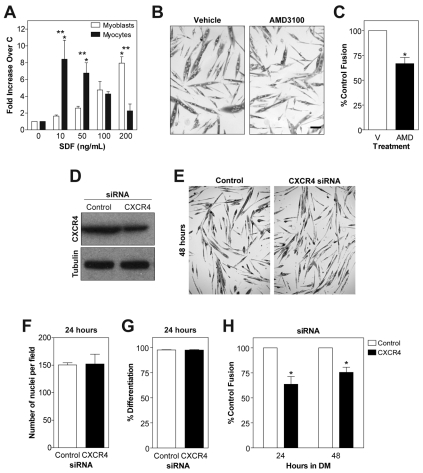
CXCR4 and SDF-1α regulate migration of myoblasts and myocytes, and are necessary for myogenesis. (A) Boyden chamber experiments were performed with primary mouse myoblasts (Mb) and myocytes (Mc) with varying concentrations of SDF-1α. Myoblasts exhibited peak migration to 200 ng/ml, whereas myocytes migrated to 10–50 ng/ml. (B) AMD3100 or vehicle (V) was added to cultures with differentiation media (DM). Cultures were fixed and immunostained for embryonic myosin heavy chain (eMyHC) at 24 hours in DM. Scale bar: 50 μm. (C) Fusion index calculated as the number of nuclei in myotubes divided by the total number of nuclei. Addition of AMD decreased fusion at 24 hours in DM. (D) CXCR4 protein levels were decreased by Cxcr4 siRNA by ~45%. Tubulin was used a loading control. (E) Cells treated with control or Cxcr4 siRNA were placed in differentiation media (DM), fixed and immunostained for embryonic myosin heavy chain (eMyHC) at 48 hours in DM. (F) The total number of nuclei in each field was calculated. No difference between control and Cxcr4 siRNA cultures was observed at 24 hours, indicating that cell survival during differentiation was not affected by Cxcr4 siRNA. (G) Differentiation index calculated as the number of nuclei in eMyHC+ cells divided by the total number of nuclei. No difference was observed, suggesting that terminal differentiation was not affected by Cxcr4 siRNA. (H) Fusion index calculated as the number of nuclei in myotubes divided by the total number of nuclei. Cxcr4 siRNA decreased fusion at both 24 and 48 hours in DM. Data are means ± s.e.m., n=3 (*P<0.05 compared with control or Mb, **P<0.05 compared with Mb at same concentration).
To determine whether CXCR4-dependent processes are necessary for myogenesis, a pharmacological inhibitor of CXCR4, AMD3100 (De Clercq, 2005), was added to cells at the start of differentiation. Nascent myotubes in cultures treated with AMD appeared smaller than vehicle-treated cells at 24 hours in DM (Fig. 6B). However, neither the number of cells per field nor the number of nuclei in differentiated cells was affected (data not shown). Rather, addition of AMD decreased the fusion index, or the total number of nuclei in myotubes, by ~30% compared with the control (Fig. 6C). We also examined myogenesis in vitro in cells containing siRNA to knock down CXCR4. CXCR4 protein levels were decreased by ~45% by Cxcr4 siRNA (Fig. 6D). After 24 or 48 hours in DM, cells were immunostained for eMyHC; at both time points, Cxcr4 siRNA cultures contained smaller myotubes compared with the control (Fig. 6E). This defect in myotube formation was not due to a decrease in the total number of nuclei (Fig. 6F), nor to an affect on differentiation, as measured by the percentage of nuclei found in eMyHC+ cells (Fig. 6G). Rather, Cxcr4 siRNA myocytes exhibited a clear defect in cell fusion (Fig. 6H), because the fusion index was decreased 36% and 24%, at 24 and 48 hours, respectively, in Cxcr4 siRNA cultures (Fig. 6H). Together, these data support the hypothesis that the CXCR4–SDF-1α axis is necessary for proper myogenesis in vitro. The predominant role for CXCR4–SDF-1α during myogenesis might be to regulate the migration of muscle cells, which affects downstream fusion events.
Discussion
Adult regenerative myogenesis is vital for restoring normal myofiber structure after muscle injury. Myogenic progenitor cells must be precisely regulated and positioned in order for proper cell fusion to occur. Using a cell culture model of myogenesis, we demonstrated that a large number of chemokines and chemokine receptors were upregulated during myogenesis when terminally differentiated myocytes were fusing. Differences in migratory behavior were noted between myoblasts and myocytes. These results suggest that regulation of cell migration during myogenesis is complex.
Several chemokines and chemokine receptors we identified were not previously known to be expressed by skeletal muscle cells or tissue (Civatte et al., 2005; De Rossi et al., 2000; Demoule et al., 2009; Hirata et al., 2003; Peterson and Pizza, 2009; Porter et al., 2003; Sachidanandan et al., 2002; Warren et al., 2005; Warren et al., 2004), however, these molecules have known roles in other muscle types. For example, AGTRL1 has protective effects in ischemic heart disease (O'Donnell et al., 2007) and BMP10 regulates hypertropic growth in heart muscle (Chen et al., 2006). Neither of these proteins has identified functions in skeletal muscle but might regulate skeletal muscle growth or repair given their role in smooth and cardiac muscle. Another gene that we found to be expressed during myogenesis, BLR1 (CXCR5), regulates migration of B-cells into ischemia-damaged intestinal tissue through expression of CXCL13 by the damaged areas (Chen et al., 2009), but lacks an identified role during injury repair in skeletal muscle. These results suggest new avenues of research into chemokine-mediated regulation of adult regenerative myogenesis.
A key question is why so many chemokines and chemokine receptors are expressed directly by muscle cells during myogenesis in vitro. As muscle cells are heterogenous (Asakura et al., 2002; Motohashi et al., 2008; Relaix et al., 2005; Tanaka et al., 2009), subpopulations of muscle cells might express a single receptor or ligand. Alternatively, several of these molecules might be expressed by each muscle cell, as occurs in the immune system (Civatte et al., 2005; Porter et al., 2003; Warren et al., 2004). If several receptors are expressed by a single cell, specific chemokine receptors might be used in a spatial-temporal manner. Alternatively, a redundant system might exist, allowing the substitution of one receptor–ligand pair for another. Such a system would allow disruption of a single receptor–ligand pair without serious detriment to myogenesis. Interestingly, our results demonstrate that myocytes did not migrate in response to canonical myoblast migration factors. Instead, myocytes migrated to factors secreted by fusing muscle cells. Thus, regulation of cell migration during different phases of myogenesis is differentially controlled.
The multitude of chemokines and chemokine receptors expressed during myogenesis in vitro might regulate similar or distinct processes. Chemokines regulate cell number at several levels, including survival and proliferation (Miyazaki et al., 2006; Schober and Zernecke, 2007); thus, chemokines expressed early during myogenesis, might regulate myoblast proliferation or survival. Also, because muscle cells must interact directly with one another for terminal differentiation to occur (Krauss et al., 2005), chemokines might also regulate migration of myoblasts. Our data suggest that multiple chemokine receptor–ligand pairs regulate later stages of myogenesis, such as migration and fusion, as these molecules are not expressed at high levels until the majority of cells are terminally differentiated myocytes. Curiously, the expression levels of these molecules were highest during periods of myogenesis in which the myocytes were progressively moving slower, as measured by time-lapse microscopy. Chemokines not only regulate cell velocity, but also directional migration of cells (Kim, 2004). Perhaps chemokines at these later stages of myogenesis are key for positioning myocytes in the correct spatial patterns necessary for cell fusion to occur with other myocytes and with nascent myotubes, rather than acting to enhance cell velocity. Chemokines expressed by muscle cells in vivo might not only have a direct effect on myogenesis, but may also act in a paracrine manner. Chemokines regulate the recruitment of immune cells to damaged tissues (Bleul et al., 1996; Loetscher et al., 1996; Weber et al., 1995), including injured muscle (Robertson et al., 1993); immune cells such as macrophages are crucial for muscle regeneration (Arnold et al., 2007). Therefore, chemokines might regulate myogenesis through several distinct processes.
The investigation of a single receptor–ligand pair, CXCR4 and SDF-1α, indicated that some chemokines identified in this study do regulate migration during myogenesis in vitro. We show that CXCR4 is expressed by both primary mouse myoblasts and myocytes, and its ligand SDF-1α can increase migration of both cell types, albeit at different concentrations. However, despite inhibition of CXCR4 by two different methods, primary muscle cells differentiate similarly to untreated cells, but are unable to undergo fusion as efficiently. Together, these results suggest that CXCR4 is necessary for migration of muscle cells to one another, which is required for normal fusion. Our studies expand on previous CXCR4 studies in the field. The majority of in vitro CXCR4 studies use the immortalized C2C12 mouse muscle cell line (Melchionna et al., 2010; Odemis et al., 2007; Ratajczak et al., 2003). Similarly to our results, the CXCR4–SDF-1α axis enhances migration of C2C12 myoblasts (Odemis et al., 2007; Ratajczak et al., 2003). However, in contrast to our studies, investigations on C2C12 cells suggest that loss of CXCR4 leads to an inhibition of differentiation as measured by decreased expression of differentiation-specific muscle proteins, such as myogenin and/or myosin heavy chain (Melchionna et al., 2010; Odemis et al., 2007). In one study, an almost complete abrogation of muscle cell differentiation was observed with loss of CXCR4, despite the fact that only 15% of C2C12 cells express CXCR4 (Odemis et al., 2007). Differences between primary muscle cells and established cell lines could contribute to some of the differences between our studies and those with C2C12 cells. Interestingly, loss of CD164, a sialomucin that interacts with CXCR4, on the cell surface where it probably functions as a component of a CXCR4 receptor complex (Bae et al., 2008; Forde et al., 2007), also affected migration and myotube formation, but not differentiation of C2C12 cells, similarly to our experiments (Bae et al., 2008). The CXCR4–SDF-1α axis is known to have a role in embryonic muscle development. Most studies that analyze CXCR4 function during embryonic myogenesis in mice, zebrafish and chick suggest that perturbation of CXCR4 signaling alters limb-muscle development mainly as a result of deficiencies in migration of myogenic precursor cells from the somites to the limb buds (Chong et al., 2007; Vasyutina et al., 2005; Yusuf et al., 2006). Since terminal differentiation and fusion occur downstream of migration, defects in these later processes could not be analyzed during embryonic development independently of migration defects. However, one study of embryonic muscle development in Cxcr4-null mice did not observe defects in migration of muscle precursor cells to the limb buds but defects in muscle mass were noted; no mechanism was determined for this loss of muscle mass (Odemis et al., 2005). No studies of the CXCR4–SDF-1α axis have been performed in adult regenerative myogenesis.
CXCR4 is of specific interest to cell-therapy approaches for various muscular disorders. A subset of muscle satellite cells that are CXCR4+ can be engrafted into injured muscle tissue with a high efficiency (Cerletti et al., 2008). As CXCR4 regulates migration of muscle cells both in vitro and in vivo, the increased engraftment might be due to an increased migratory ability of these cells. Furthermore, treatment with SDF-1α enhances migration of myogenic precursors, yielding a positive effect on engraftment of cells into damaged muscle (Galvez et al., 2006). These data suggest that CXCR4–SDF-1α-dependent migration enhances the engraftment of cells into damaged muscle. The large number of chemokine receptors and ligands expressed by muscle cells during myogenesis in vitro suggests further avenues of research to be explored during adult regenerative myogenesis. Further studies of chemokines in vivo might lead to manipulation of these molecules and allow for an increased efficiency of cell-transplantation therapies for various muscle disorders.
Materials and Methods
Animals and muscle injuries
Adult mice between 8 and 12 weeks of age were used and handled in accordance with the institutional guidelines of Emory University. To induce regeneration, gastrocnemius muscles of male C57BL/6 mice were injected with BaCl2 (O'Connor et al., 2007) and collected as described (Abbott et al., 1998).
Primary muscle cell culture, differentiation and fusion assays
Primary myoblasts were derived from the hindlimb muscles of Balb/C mice (Bondesen et al., 2004; Mitchell and Pavlath, 2001) and cultures were >99% myogenic as assessed by MyoD immunostaining (Jansen and Pavlath, 2006). For all experiments 3–5 independent isolates were analyzed. To induce differentiation, primary myoblasts were seeded at a density of 2×105 cells/well on dishes coated with entactin, collagen IV and laminin (E-C-L; Upstate Biotechnology) and switched to differentiation media [DM: DME, 1% insulin-transferrin-selenium-A supplement (Invitrogen)], 100 U/ml penicillin G and 100 μg/ml streptomycin). At indicated time points, cells were immunostained with an eMyHC antibody (F1.652; Developmental Studies Hybridoma Bank) and analyzed as described (Horsley et al., 2001). AMD3100 (Sigma) was dissolved in PBS and used at 10 μM in DM. At least 500 nuclei per condition were analyzed for each assay.
Transfection of primary myoblasts
Stealth RNAi (Invitrogen) was used to knockdown Cxcr4 expression in primary myoblasts. Myoblasts were plated in growth medium (GM; F10, 20% fetal bovine serum, 100 U/ml penicillin G and 100 μg/ml streptomycin) at a density of 8×105 cells per collagen-coated 100 mm plates and after 6 hours, duplexed siRNAs at a final concentration of 27 nM each were used to transfect cells using Lipofectamine 2000 (Invitrogen) in GM according to the manufacturer's instructions. Cells were transfected with either scrambled control or a mixture of three Cxcr4 siRNAs (Invitrogen, ACGAGGUAGAGAAGCAGAUGAAUAUGGCAAUGGAUUGGUGAUCCUGGUCA; ACAGGUACAUCUGUGACCGCCUUUA; CAGUCAUCCUCAUCCUAGCUUUCUU). After 6 hours of incubation, medium containing transfection complexes was replaced by fresh GM. Twenty-four hours after the start of transfection, cells were trypsinized, plated on six-well dishes and differentiated for 24 hours and 48 hours as described for differentiation and fusion assays above. CXCR4 knockdown was assessed by immunoblotting using anti-CXCR4 (Abcam) after 24 hours of transfection. Results represent data from three independent isolates.
Flow cytometry
To analyze CXCR4 expression in vitro by flow cytometry, primary myoblasts were immunostained with anti-CXCR4-APC antibody (1:100; BD Pharmigen) and analyzed on a FACSCalibur (Becton-Dickinson). For analysis of CXCR4 expression during regeneration, mononucleated cells were dissociated from gastrocnemius muscles of mice at the indicated times after BaCl2 injection (n=4 for each time point) and immunostained with antibodies to CD31-FITC (1:100; eBiosciences), CD45-FITC (1:100; BD Biosciences), α7-integrin-PE (1:200; a gift from Fabio Rossi, University of British Columbia, Vancouver, Canada) and CXCR4-APC (1:100; BD Pharmigen). CD31− CD45− cells were analyzed for α7-integrin-PE and CXCR4 expression. For analysis of p21 expression during regeneration, mononucleated cells were dissociated from gastrocnemius muscles of mice at the indicated times after BaCl2 injection (n=10 for each time point), fixed with cold 70% ethanol overnight at −20°C and immunostained with antibodies to CD31-APC (1:100; eBiosciences), CD45-APC (1:100; BD Biosciences), α7-integrin-PE and p21 (1:100; Lifespan Biosciences). To detect p21, cells were incubated with biotin-conjugated donkey anti-goat (1:100; Jackson ImmunoResearch) for 20 minutes, then FITC-conjugated strepavidin (1:100; Jackson ImmunoResearch Lab., Inc.) for 20 minutes. CD31− CD45− cells were analyzed for α7-integrin and p21 expression (n=10 for each time point). For each sample, 10,000 cells were analyzed, and propidium iodide was used to remove dead cells. Isotype controls were used to determine gating. All data analysis was performed using FlowJo v. 6.2.1 (TreeStar).
Immunostaining
Myogenin and eMyHC immunostaining was performed using a VectaStain kit (Vector labs). α7-integrin+ CD31− CD45− muscle cells isolated from gastrocnemius muscles by FACS were plated then fixed in 4% PFA for 10 minutes. Cells were treated with 3% H2O2, biotin-strepavidin blocking kits (Vector), mouse IgG (M.O.M. kit, Vector) and then blocking buffer containing 4% BSA in PBS for 1 hour. Cells were then incubated overnight at 4°C with anti-myogenin (hybridoma supernatant, diluted 1:10 in blocking buffer, F5D; Developmental Studies Hybridoma Bank), anti-eMyHC (hybridoma supernatant, neat, F1.652; Developmental Studies Hybridoma Bank) or appropriate IgG (diluted 1:100 in blocking buffer, Genetex). Following successive washes in PBS with 0.1% BSA, cells were incubated with donkey anti-mouse IgG (Jackson ImmunoResearch) diluted 1:200 in PBS with 4% BSA for 1 hour. Following repeated washes in 0.1% BSA in PBS, the cells were incubated in HRP-conjugated streptavidin (VectorLabs) for 30 minutes followed by visualization with diaminobenzidene (DAB). All immunostaining was performed at room temperature unless stated otherwise. Hybridoma cells were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA, USA.
Real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Life Technologies). All RNA was DNase treated (Invitrogen) and a portion of DNase-treated RNA was reverse transcribed. Real-time PCR was performed, and results were analyzed by using the iCycler iQ Real-Time Detection System and software (Bio-Rad). cDNA (1 μl from each sample) was amplified by using gene-specific primers in a 96-well SABiosciences chemokine array (Chemokines & Receptors PCR Array, Mouse, PAMM-022) and iQ SYBRgreen Supermix (Bio-Rad) in a 25 μl reaction. Samples were incubated at 95°C for 4 minutes, followed by 40 cycles (30 seconds each) of denaturation, annealing, and extension at 95°C, 55°C and 72°C, respectively. SYBRgreen fluorescence was measured at the end of the extension step of each cycle. All reactions were run in triplicate, and PCR product size was verified by melt curve analysis. All samples were normalized using Hypoxanthine guanine phosphoribosyl transferase 1 (HPRT).
Cell-migration assays
Migration of muscle cells was quantified using time-lapse microscopy as described (Jansen and Pavlath, 2006). Briefly, cells were seeded at 2×105 cells per 35 mm dish, and switched to DM for the indicated times before imaging. Images were recorded (QImaging Camera and OpenLab 3.1.4 software) every 5 minutes for 3 hours. Cell velocities were calculated in μm/hour using ImageJ software by tracking the paths of mononucleated cells. Approximately 20 mononucleated cells were tracked for each experiment.
Boyden chamber assays were performed as described (Mylona et al., 2006). Primary myoblasts were seeded on 150-mm plates at low density (9×105 cells/plate) and switched to DM for 24 hours to generate myocytes in the absence of myotube formation. Cells (7.5×104 cells in 200 μl DM) were loaded in the upper wells of the Boyden chamber and incubated at 37°C for 5 hours. Migrated cells were fixed, stained and counted. HGF and PDGF were used at 100 ng/ml in DMEM with 1% BSA, SDF1α at 10-200 ng/ml in DM (Sigma). To prepare conditioned medium (CM), myoblasts were incubated in DM for 24 hours; the medium, which had been ‘conditioned’ with secreted factors, was then collected, filtered (0.45 μm), flash frozen, and stored at −80°C until use.
SDF-1α ELISA assay
SDF-1α was detected using the ELISA Kit for Mouse SDF-1α kit (RayBiotech). Conditioned medium was isolated as above. Crushed muscle extract (CME) was created as described (Chen and Quinn, 1992) using gastrocnemius muscles from C57BL/6 mice (n=10). Briefly, the muscles were dissected, pressed 7–10 times with forceps, pooled, and incubated in TBS (Tris-buffered saline; 20 mM Tris-HCl, pH 7.6, 137 mM NaCl; 1 ml TBS was used for the muscles of each mouse) for 90 minutes at 4°C on a rotator. The extract was centrifuged at 176,000 g for 30 minutes followed by filtration through a 0.2 μm filter and stored at −80°C. Protein concentration was determined using the Bradford assay (Bio-Rad) and equal amounts of protein were used.
Statistics
To determine significance between two groups, comparisons were made using Student's t-tests. Analyses of multiple groups were performed using a one-way or two-way analysis of variance with Bonferroni's post test as appropriate. Statistical analyses were performed using GraphPad Prism 4.0 (GraphPad). For all statistical tests, a confidence interval of P<0.05 was accepted for statistical significance.
Supplementary Material
Acknowledgments
We thank Matthew Randolph for help with control experiments. G.K.P. was supported by grants AR-047314, AR-051372, and AR-052730 from the National Institutes of Health and the Muscular Dystrophy Association. C.A.G. was supported by National Institutes of Health training grant T32-GM08367. L.H.A. and K.K.L. were supported by MDA Development grants. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/18/3052/DC1
References
- Abbott K. L., Friday B. B., Thaloor D., Murphy T. J., Pavlath G. K. (1998). Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell 9, 2905-2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allbrook D. (1981). Skeletal muscle regeneration. Muscle Nerve 4, 234-245 [DOI] [PubMed] [Google Scholar]
- Andres V., Walsh K. (1996). Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 132, 657-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R. K., Chazaud B. (2007). Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A., Seale P., Girgis-Gabardo A., Rudnicki M. A. (2002). Myogenic specification of side population cells in skeletal muscle. J. Cell Biol. 159, 123-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G. U., Gaio U., Yang Y. J., Lee H. J., Kang J. S., Krauss R. S. (2008). Regulation of myoblast motility and fusion by the CXCR4-associated sialomucin, CD164. J. Biol. Chem. 283, 8301-8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini M., Loetscher P., Moser B. (1995). Interleukin-8 and the chemokine family. Int. J. Immunopharmacol. 17, 103-108 [DOI] [PubMed] [Google Scholar]
- Bischoff R. (1986). A satellite cell mitogen from crushed adult muscle. Dev. Biol. 115, 140-147 [DOI] [PubMed] [Google Scholar]
- Bischoff R. (1997). Chemotaxis of skeletal muscle satellite cells. Dev. Dyn. 208, 505-515 [DOI] [PubMed] [Google Scholar]
- Blanco-Bose W. E., Yao C. C., Kramer R. H., Blau H. M. (2001). Purification of mouse primary myoblasts based on alpha 7 integrin expression. Exp. Cell Res. 265, 212-220 [DOI] [PubMed] [Google Scholar]
- Bleul C. C., Fuhlbrigge R. C., Casasnovas J. M., Aiuti A., Springer T. A. (1996). A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J. Exp. Med. 184, 1101-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondesen B. A., Mills S. T., Kegley K. M., Pavlath G. K. (2004). The COX-2 pathway is essential during early stages of skeletal muscle regeneration. Am. J. Physiol. Cell Physiol. 287, C475-C483 [DOI] [PubMed] [Google Scholar]
- Cerletti M., Jurga S., Witczak C. A., Hirshman M. F., Shadrach J. L., Goodyear L. J., Wagers A. J. (2008). Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell 134, 37-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud B., Sonnet C., Lafuste P., Bassez G., Rimaniol A. C., Poron F., Authier F. J., Dreyfus P. A., Gherardi R. K. (2003). Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J. Cell Biol. 163, 1133-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Quinn L. S. (1992). Partial characterization of skeletal myoblast mitogens in mouse crushed muscle extract. J. Cell Physiol. 153, 563-574 [DOI] [PubMed] [Google Scholar]
- Chen H., Yong W., Ren S., Shen W., He Y., Cox K. A., Zhu W., Li W., Soonpaa M., Payne R. M., et al. (2006). Overexpression of bone morphogenetic protein 10 in myocardium disrupts cardiac postnatal hypertrophic growth. J. Biol. Chem. 281, 27481-27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Crispin J. C., Tedder T. F., Dalle Lucca J., Tsokos G. C. (2009). B cells contribute to ischemia/reperfusion-mediated tissue injury. J. Autoimmun. 32, 195-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S. W., Nguyet L. M., Jiang Y. J., Korzh V. (2007). The chemokine Sdf-1 and its receptor Cxcr4 are required for formation of muscle in zebrafish. BMC Dev. Biol. 7, 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civatte M., Bartoli C., Schleinitz N., Chetaille B., Pellissier J. F., Figarella-Branger D. (2005). Expression of the beta chemokines CCL3, CCL4, CCL5 and their receptors in idiopathic inflammatory myopathies. Neuropathol. Appl. Neurobiol. 31, 70-79 [DOI] [PubMed] [Google Scholar]
- Corti S., Salani S., Del Bo R., Sironi M., Strazzer S., D'Angelo M. G., Comi G. P., Bresolin N., Scarlato G. (2001). Chemotactic factors enhance myogenic cell migration across an endothelial monolayer. Exp. Cell Res. 268, 36-44 [DOI] [PubMed] [Google Scholar]
- De Clercq E. (2005). Potential clinical applications of the CXCR4 antagonist bicyclam AMD3100. Mini Rev. Med. Chem. 5, 805-824 [DOI] [PubMed] [Google Scholar]
- De Rossi M., Bernasconi P., Baggi F., de Waal Malefyt R., Mantegazza R. (2000). Cytokines and chemokines are both expressed by human myoblasts: possible relevance for the immune pathogenesis of muscle inflammation. Int. Immunol. 12, 1329-1335 [DOI] [PubMed] [Google Scholar]
- Demoule A., Divangahi M., Yahiaoui L., Danialou G., Gvozdic D., Petrof B. J. (2009). Chemokine receptor and ligand upregulation in the diaphragm during endotoxemia and Pseudomonas lung infection. Mediators Inflamm. 2009, 860565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtermeyer F., Schober S., Poschl E., von der Mark H., von der Mark K. (1996). Specific induction of cell motility on laminin by alpha 7 integrin. J. Biol. Chem. 271, 2071-2075 [DOI] [PubMed] [Google Scholar]
- Forde S., Tye B. J., Newey S. E., Roubelakis M., Smythe J., McGuckin C. P., Pettengell R., Watt S. M. (2007). Endolyn (CD164) modulates the CXCL12-mediated migration of umbilical cord blood CD133+ cells. Blood 109, 1825-1833 [DOI] [PubMed] [Google Scholar]
- Galvez B. G., Sampaolesi M., Brunelli S., Covarello D., Gavina M., Rossi B., Constantin G., Torrente Y., Cossu G. (2006). Complete repair of dystrophic skeletal muscle by mesoangioblasts with enhanced migration ability. J. Cell Biol. 174, 231-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E., Boontheekul T., Mooney D. J. (2006). Designing scaffolds to enhance transplanted myoblast survival and migration. Tissue Eng. 12, 1295-1304 [DOI] [PubMed] [Google Scholar]
- Hirata A., Masuda S., Tamura T., Kai K., Ojima K., Fukase A., Motoyoshi K., Kamakura K., Miyagoe-Suzuki Y., Takeda S. (2003). Expression profiling of cytokines and related genes in regenerating skeletal muscle after cardiotoxin injection: a role for osteopontin. Am. J. Pathol. 163, 203-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., Friday B. B., Matteson S., Kegley K. M., Gephart J., Pavlath G. K. (2001). Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J. Cell Biol. 153, 329-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., Jansen K. M., Mills S. T., Pavlath G. K. (2003). IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell 113, 483-494 [DOI] [PubMed] [Google Scholar]
- Isobe T., Minoura H., Tanaka K., Shibahara T., Hayashi N., Toyoda N. (2002). The effect of RANTES on human sperm chemotaxis. Hum. Reprod. 17, 1441-1446 [DOI] [PubMed] [Google Scholar]
- Jansen K. M., Pavlath G. K. (2006). Mannose receptor regulates myoblast motility and muscle growth. J. Cell Biol. 174, 403-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar K. A., Yi L., Ahmad Y., So L., Rossi F., Pavlath G. K. (2009). Sca-1 expression is required for efficient remodeling of the extracellular matrix during skeletal muscle regeneration. Dev. Biol. 326, 47-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. S., Yi M. J., Zhang W., Feinleib J. L., Cole F., Krauss R. S. (2004). Netrins and neogenin promote myotube formation. J. Cell Biol. 167, 493-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H. (2004). Chemokine-chemokine receptor network in immune cell trafficking. Curr. Drug Targets Immune Endocr. Metabol. Disord. 4, 343-361 [DOI] [PubMed] [Google Scholar]
- Kim C. H. (2005). The greater chemotactic network for lymphocyte trafficking: chemokines and beyond. Curr. Opin. Hematol. 12, 298-304 [DOI] [PubMed] [Google Scholar]
- Krauss R. S., Cole F., Gaio U., Takaesu G., Zhang W., Kang J. S. (2005). Close encounters: regulation of vertebrate skeletal myogenesis by cell-cell contact. J. Cell Sci. 118, 2355-2362 [DOI] [PubMed] [Google Scholar]
- Lafreniere J. F., Mills P., Bouchentouf M., Tremblay J. P. (2006). Interleukin-4 improves the migration of human myogenic precursor cells in vitro and in vivo. Exp. Cell Res. 312, 1127-1141 [DOI] [PubMed] [Google Scholar]
- Lee K. K., Wong C. C., Webb S. E., Tang M. K., Leung A. K., Kwok P. F., Cai D. Q., Chan K. M. (1999). Hepatocyte growth factor stimulates chemotactic response in mouse embryonic limb myogenic cells in vitro. J. Exp. Zool. 283, 170-180 [DOI] [PubMed] [Google Scholar]
- Lluri G., Jaworski D. M. (2005). Regulation of TIMP-2, MT1-MMP, and MMP-2 expression during C2C12 differentiation. Muscle Nerve 32, 492-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluri G., Langlois G. D., Soloway P. D., Jaworski D. M. (2008). Tissue inhibitor of metalloproteinase-2 (TIMP-2) regulates myogenesis and beta1 integrin expression in vitro. Exp. Cell Res. 314, 11-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loetscher M., Gerber B., Loetscher P., Jones S. A., Piali L., Clark-Lewis I., Baggiolini M., Moser B. (1996). Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J. Exp. Med. 184, 963-969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster A. D. (1998). Chemokines-chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338, 436-445 [DOI] [PubMed] [Google Scholar]
- McLennan I. S. (1996). Degenerating and regenerating skeletal muscles contain several subpopulations of macrophages with distinct spatial and temporal distributions. J. Anat. 188, 17-28 [PMC free article] [PubMed] [Google Scholar]
- Melchionna R., Di Carlo A., De Mori R., Cappuzzello C., Barberi L., Musaro A., Cencioni C., Fujii N., Tamamura H., Crescenzi M., et al. (2010). Induction of myogenic differentiation by SDF-1 via CXCR4 and CXCR7 receptors. Muscle Nerve 41, 828-835 [DOI] [PubMed] [Google Scholar]
- Mitchell P. O., Pavlath G. K. (2001). A muscle precursor cell-dependent pathway contributes to muscle growth after atrophy. Am. J. Physiol. Cell Physiol. 281, C1706-C1715 [DOI] [PubMed] [Google Scholar]
- Miyazaki H., Patel V., Wang H., Edmunds R. K., Gutkind J. S., Yeudall W. A. (2006). Down-regulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 66, 4279-4284 [DOI] [PubMed] [Google Scholar]
- Motohashi N., Uezumi A., Yada E., Fukada S., Fukushima K., Imaizumi K., Miyagoe-Suzuki Y., Takeda S. (2008). Muscle CD31(−) CD45(−) side population cells promote muscle regeneration by stimulating proliferation and migration of myoblasts. Am. J. Pathol. 173, 781-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muciaccia B., Padula F., Gandini L., Lenzi A., Stefanini M. (2005a). HIV-1 chemokine co-receptor CCR5 is expressed on the surface of human spermatozoa. Aids 19, 1424-1426 [DOI] [PubMed] [Google Scholar]
- Muciaccia B., Padula F., Vicini E., Gandini L., Lenzi A., Stefanini M. (2005b). Beta-chemokine receptors 5 and 3 are expressed on the head region of human spermatozoon. FASEB J. 19, 2048-2050 [DOI] [PubMed] [Google Scholar]
- Mylona E., Jones K. A., Mills S. T., Pavlath G. K. (2006). CD44 regulates myoblast migration and differentiation. J. Cell Physiol. 209, 314-321 [DOI] [PubMed] [Google Scholar]
- O'Connor R. S., Mills S. T., Jones K. A., Ho S. N., Pavlath G. K. (2007). A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 120, 149-159 [DOI] [PubMed] [Google Scholar]
- O'Donnell L. A., Agrawal A., Sabnekar P., Dichter M. A., Lynch D. R., Kolson D. L. (2007). Apelin, an endogenous neuronal peptide, protects hippocampal neurons against excitotoxic injury. J. Neurochem. 102, 1905-1917 [DOI] [PubMed] [Google Scholar]
- Ocalan M., Goodman S. L., Kuhl U., Hauschka S. D., von der Mark K. (1988). Laminin alters cell shape and stimulates motility and proliferation of murine skeletal myoblasts. Dev. Biol. 125, 158-167 [DOI] [PubMed] [Google Scholar]
- Odemis V., Lamp E., Pezeshki G., Moepps B., Schilling K., Gierschik P., Littman D. R., Engele J. (2005). Mice deficient in the chemokine receptor CXCR4 exhibit impaired limb innervation and myogenesis. Mol. Cell. Neurosci. 30, 494-505 [DOI] [PubMed] [Google Scholar]
- Odemis V., Boosmann K., Dieterlen M. T., Engele J. (2007). The chemokine SDF1 controls multiple steps of myogenesis through atypical PKC{zeta}. J. Cell Sci. 120, 4050-4059 [DOI] [PubMed] [Google Scholar]
- Olguin H. C., Santander C., Brandan E. (2003). Inhibition of myoblast migration via decorin expression is critical for normal skeletal muscle differentiation. Dev. Biol. 259, 209-224 [DOI] [PubMed] [Google Scholar]
- Palumbo R., Sampaolesi M., De Marchis F., Tonlorenzi R., Colombetti S., Mondino A., Cossu G., Bianchi M. E. (2004). Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J. Cell Biol. 164, 441-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. M., Pizza F. X. (2009). Cytokines derived from cultured skeletal muscle cells after mechanical strain promote neutrophil chemotaxis in vitro. J. Appl. Physiol. 106, 130-137 [DOI] [PubMed] [Google Scholar]
- Porter J. D., Guo W., Merriam A. P., Khanna S., Cheng G., Zhou X., Andrade F. H., Richmonds C., Kaminski H. J. (2003). Persistent over-expression of specific CC class chemokines correlates with macrophage and T-cell recruitment in mdx skeletal muscle. Neuromuscul. Disord. 13, 223-235 [DOI] [PubMed] [Google Scholar]
- Ratajczak M. Z., Majka M., Kucia M., Drukala J., Pietrzkowski Z., Peiper S., Janowska-Wieczorek A. (2003). Expression of functional CXCR4 by muscle satellite cells and secretion of SDF-1 by muscle-derived fibroblasts is associated with the presence of both muscle progenitors in bone marrow and hematopoietic stem/progenitor cells in muscles. Stem Cells 21, 363-371 [DOI] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948-953 [DOI] [PubMed] [Google Scholar]
- Robertson T. A., Maley M. A., Grounds M. D., Papadimitriou J. M. (1993). The role of macrophages in skeletal muscle regeneration with particular reference to chemotaxis. Exp. Cell Res. 207, 321-331 [DOI] [PubMed] [Google Scholar]
- Sachidanandan C., Sambasivan R., Dhawan J. (2002). Tristetraprolin and LPS-inducible CXC chemokine are rapidly induced in presumptive satellite cells in response to skeletal muscle injury. J. Cell Sci. 115, 2701-2712 [DOI] [PubMed] [Google Scholar]
- Schober A., Zernecke A. (2007). Chemokines in vascular remodeling. Thromb. Haemost. 97, 730-737 [PubMed] [Google Scholar]
- Schultz E., Gibson M. C., Champion T. (1978). Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J. Exp. Zool. 206, 451-456 [DOI] [PubMed] [Google Scholar]
- Stebler J., Spieler D., Slanchev K., Molyneaux K. A., Richter U., Cojocaru V., Tarabykin V., Wylie C., Kessel M., Raz E. (2004). Primordial germ cell migration in the chick and mouse embryo: the role of the chemokine SDF-1/CXCL12. Dev. Biol. 272, 351-361 [DOI] [PubMed] [Google Scholar]
- Summan M., McKinstry M., Warren G. L., Hulderman T., Mishra D., Brumbaugh K., Luster M. I., Simeonova P. P. (2003). Inflammatory mediators and skeletal muscle injury: a DNA microarray analysis. J. Interferon. Cytokine Res. 23, 237-245 [DOI] [PubMed] [Google Scholar]
- Tanaka K. K., Hall J. K., Troy A. A., Cornelison D. D., Majka S. M., Olwin B. B. (2009). Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell 4, 217-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball J. G. (2005). Inflammatory processes in muscle injury and repair. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R345-R353 [DOI] [PubMed] [Google Scholar]
- Vandercappellen J., Van Damme J., Struyf S. (2008). The role of CXC chemokines and their receptors in cancer. Cancer Lett. 267, 226-244 [DOI] [PubMed] [Google Scholar]
- Vasyutina E., Stebler J., Brand-Saberi B., Schulz S., Raz E., Birchmeier C. (2005). CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 19, 2187-2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. L., O'Farrell L., Summan M., Hulderman T., Mishra D., Luster M. I., Kuziel W. A., Simeonova P. P. (2004). Role of CC chemokines in skeletal muscle functional restoration after injury. Am J. Physiol. Cell Physiol. 286, C1031-C1036 [DOI] [PubMed] [Google Scholar]
- Warren G. L., Hulderman T., Mishra D., Gao X., Millecchia L., O'Farrell L., Kuziel W. A., Simeonova P. P. (2005). Chemokine receptor CCR2 involvement in skeletal muscle regeneration. FASEB J. 19, 413-415 [DOI] [PubMed] [Google Scholar]
- Weber M., Uguccioni M., Ochensberger B., Baggiolini M., Clark-Lewis I., Dahinden C. A. (1995). Monocyte chemotactic protein MCP-2 activates human basophil and eosinophil leukocytes similar to MCP-3. J. Immunol. 154, 4166-4172 [PubMed] [Google Scholar]
- Yao C. C., Ziober B. L., Sutherland A. E., Mendrick D. L., Kramer R. H. (1996). Laminins promote the locomotion of skeletal myoblasts via the alpha 7 integrin receptor. J. Cell Sci. 109, 3139-3150 [DOI] [PubMed] [Google Scholar]
- Yusuf F., Rehimi R., Morosan-Puopolo G., Dai F., Zhang X., Brand-Saberi B. (2006). Inhibitors of CXCR4 affect the migration and fate of CXCR4+ progenitors in the developing limb of chick embryos. Dev. Dyn. 235, 3007-3015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.