Abstract
Our understanding of the mammalian cell cycle is due in large part to the analysis of cyclin-dependent kinase (CDK) 2 and CDK4/6. These kinases are regulated by E and D type cyclins, respectively, and coordinate the G1/S-phase transition. In contrast, little is known about CDK3, a homolog of CDK2 and cell division cycle kinase 2 (CDC2). Previous studies using ectopic expression of human CDK3 suggest a role for this kinase in the G1/S-phase transition, but analysis of the endogenous kinase has been stymied by the low levels of protein present in cells and by the absence of an identifiable cyclin partner. Herein we report the presence of a single point mutation in the CDK3 gene from several Mus musculus strains commonly used in the laboratory. This mutation results in the replacement of a conserved tryptophan (Trp-187) within kinase consensus domain IX with a stop codon. The protein predicted to be encoded by this allele is truncated near the T loop, which is involved in activation by CDK-activating kinase. This mutation also deletes motif XI known to be required for kinase function and is, therefore, expected to generate a null allele. In stark contrast, CDK3 from two wild-mice species (Mus spretus and Mus mus castaneus) lack this mutation. These data indicate that CDK3 is not required for M. musculus development and suggest that any functional role played by CDK3 in the G1/S-phase transition is likely to be redundant with another CDK.
Much of what we know about the mammalian cell cycle comes from an analysis of two prototypical G1/S-phase cyclin-dependent kinase (CDK)-cyclin complexes, CDK2–cyclin E and CDK4/6–cyclin D (refs. 1–7; for reviews, see refs. 8 and 9). These kinases are responsible for activation of the E2F transcriptional program via phosphorylation of the retinoblastoma protein and its family members p107 and p130 (refs. 10 and 11; for reviews, see refs. 12 and 13), and there is evidence that CDK2–cyclin E functions downstream E2F activation to control DNA synthesis and other activities associated with S phase (14–16). There is also evidence of a role for an additional family member CDK3 (17) in the G1/S-phase transition. Human CDK3 is closely related to CDK2 and cell division cycle kinase 2 (CDC2) (76% and 67% identical, respectively). Like CDC2 and CDK2, CDK3 can complement temperature-sensitive mutations in the budding yeast CDC28 gene and interacts with the small CDK subunit CKS1 (17). Dominant negative forms of human CDK3 block cells in G1 phase, and this arrest is not overcome by overexpression of CDK2, suggesting a specialized role for CDK3 in the G1/S-phase transition (18). In addition, expression of dominant negative CDK3 inhibits E2F activity and CDK3 can assemble with E2F-1/DP-1 complexes, suggesting a role for CDK3 in E2F activation (19). Moreover, CDK3–cyclin E complexes can promote S-phase entry in quiescent cells as efficiently as can CDK2–cyclin E complexes (20).
Despite these studies, our understanding of CDK3 and its role in cell division is quite limited. This reflects to a large degree the difficulty in detecting CDK3 in cells and tissues. CDK3 kinase activity has not been reliably detected in extracts from tissue culture cells, and thus its normal kinase activator subunit has not been identified and the timing of its activation has not been reported (17, 18). This low abundance is reflected in extremely low levels of mRNA in tissue Northern blots (17). Consequently, all CDK3 studies thus far have used recombinant human CDK3 or transient overexpression systems (18–20).
We sought to understand CDK3 function by examining the phenotype of mice lacking CDK3. However, during the course of our analysis of the Mus musculus (129Sv) CDK3 gene, we discovered that it contains a point mutation that converts a highly conserved tryptophan (Trp-187) to a premature termination codon. An identical mutation was identified in a M. musculus CDK3 cDNA. Trp-187 is located within kinase signature motif IX and truncation also deletes additional motifs known to be required for kinase activity, likely creating a null allele. In contrast, CDK3 from wild-mice species Mus spretus and Mus mus castaneus lack this mutation. Analysis of several M. musculus mouse strains commonly used in the laboratory revealed that all had the identical CDK3 mutation, consistent with the known genealogy of these strains. These data indicate that CDK3 is not required for M. musculus development or generally for cell division. The absence of a requirement for CDK3 during mouse development likely reflects redundancy with other CDK family members such as CDK2.
Materials and Methods
Isolation and Analysis of Genomic Clones.
A M. musculus (129Sv) genomic library in λ phage (provided by S. Elledge and P. Zhang, Baylor College of Medicine) was screened by using a 700-bp SalI–NotI fragment from a mouse CDK3 cDNA (GenBank accession no. AA607856) containing the extreme C terminus of the coding region (residues 267–304) and 3′ untranslated sequences. Positive clones were converted to plasmids by automatic subcloning. Plasmids were subjected to restriction mapping and the CDK3 gene sequencing by primer extension (primer sequences are available on request from the authors). Exon structure for human and mouse CDK3 were deduced by using the exon identification algorithm fgenes (http://dot.imgen.bcm.tmc.edu:9331/seq-search/gene-search.html). To determine whether the CDK3 gene was present in single copy in the mouse, genomic DNA was isolated from 129Sv embryonic stem cells; digested with EcoRI, BamHI, or BglII; and subjected to Southern blot analysis by using the probe described above. A single hybridizing band was seen with all three restriction enzymes, indicating that CDK3 is a single copy gene (data not shown).
Genomic DNA for wild-mice strains was provided by N. Jenkins (National Cancer Institute) and M. Justice (Baylor College of Medicine) (M. spretus) or was purchased from The Jackson Laboratory (M. m. castaneus), as was FVB/NJ DNA. 129Sv and C57BL/6 DNA was from S. Elledge. B6D2F1 and BALB/c DNA were provided by F. DeMayo and D. Medina (Baylor College of Medicine). For amplification of CDK3 exons 4 and 5, PCRs were performed with CDK3-F7 (5′-GCTTCCCCTGCGCACCTACACCCA) and CDK3-R9 (5′-ACCATCTCTGCAAAGATGCAGCCA) or, alternatively, CDK3-F12 (5′-CCATATGACATCACTGGCGAACTCAGC) and CDK3-R12 (5′-GCTTAGGTGACCACTCTGCCACTG) primers. Reactions (25 μl) were performed by using Expand high-fidelity Taq polymerase (Roche Molecular Biochemicals): 30 cycles of 94°C for 30 sec, 65°C for 30 sec, and 72°C for 45 sec. For primer set F7/R9, a 214-bp fragment was amplified from different mouse strains and directly sequenced by using the F7 primer. For primer set F12/R12, the PCR product (267-bp) was directly sequenced using primer R12.
In Situ Hybridization.
Sense and antisense cRNA probes for CDK3 (700 bp consisting primarily of 3′-untranslated sequences) were generated by using T7 or T3 polymerase and were labeled during synthesis with digoxigenin-labeled UTP as described by the manufacturer (Boehringer Mannheim). Paraffin-imbedded M. musculus embryo sections (Novagen) were dewaxed, rehydrated, digested with proteinase K, acetylated with acetic anhydride, and prehybridized for 3 h at 55°C. Each section was covered with 100 μl of hybridization buffer containing 100 ng of digoxigenin-labeled probe and incubated for 16 h at 55°C. After hybridization, slides were washed and incubated with alkaline phosphatase-conjugated anti-digoxigenin antibodies. Bound antibodies were detected with 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate.
Results and Discussion
Expression of CDK3 During M. musculus Development.
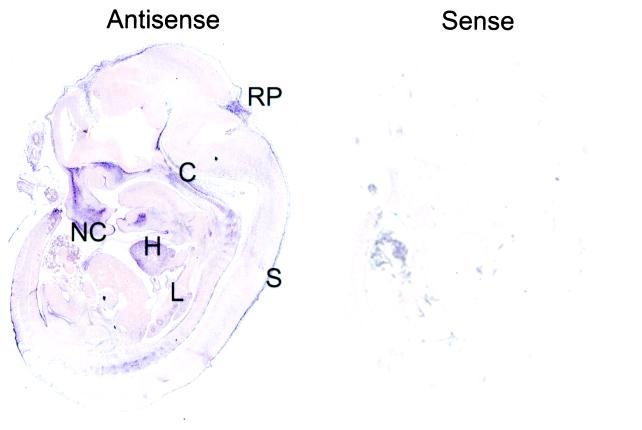
CDK3 mRNA has proven difficult to detect in both tissues and cell lines (17, 18). As such, it is not clear whether it is expressed at very low levels in the vast majority of cell types or is expressed in a cell type-specific manner. Therefore, as an initial step toward understanding CDK3 function, we sought to determine its pattern of expression during mouse development by using in situ hybridization. Although the human CDK3 cDNA is known (17), the mouse cDNA has not been reported. Analysis of the mouse expressed sequence tag (EST) database revealed two sequences derived from a blastocyst cDNA library that were more closely related to human CDK3 than to CDK2 or CDC2 (see below). One of these EST clones (GenBank accession no. AA607856) contained sequences encoding the C terminus of mouse CDK3 (residues 267–304) and the presumptive 3′ untranslated region. This EST was used to generate cRNA probes for hybridization. CDK3 is expressed in a restricted pattern at multiple stages in development (embryonic day 9.5–15.5; Fig. 1 and data not shown). The highest level of CDK3 expression was observed in the heart, lung, nasal cavity, roof-plate of the fourth ventricle, embryonic skin, and cartilage primordium of the basisphenoid and basioccipital bone. The levels of expression throughout much of the embryo were near the level found with the sense probe used as a negative control (Fig. 1). Consistent with the low levels of CDK3 observed in human tissue extracts, extended exposure times were required to detect CDK3 mRNA, indicating that its levels of expression are very low, even in tissues where it is expressed.
Figure 1.
Expression of CDK3 mRNA during M. musculus embryonic development. Embryonic day 12.5 embryos were subjected to in situ hybridization with digoxigenin-labeled antisense and sense probes derived from the 3′ untranslated region of CDK3. Probes were visualized by alkaline phosphatase-conjugated anti-digoxigenin mAbs. S, skin; NC, nasal chamber; RP, roof plate of fourth ventricle; C, cartilage primordium of the basisphenoid and basioccipital bone; L, lung.
Mouse and Human CDK3 Gene Structure.
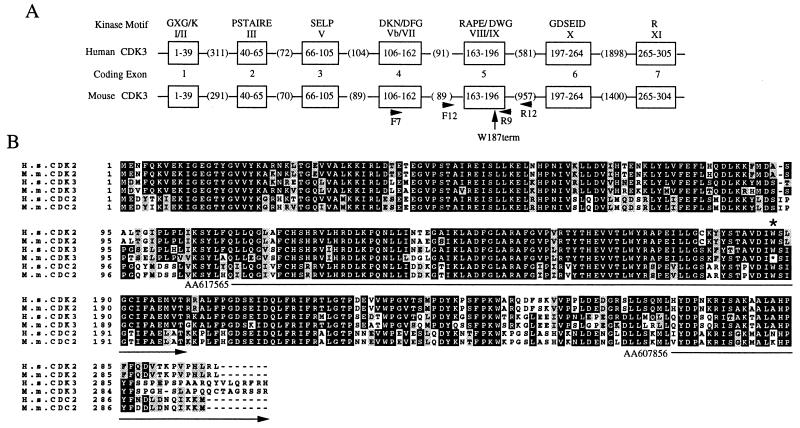
Given this restricted expression pattern, we initiated experiments to delete the CDK3 gene in mice with the expectation that specific roles for CDK3 would be evident in tissues where it is expressed. A partial CDK3 cDNA (GenBank accession no. AA607856) was used to screen a λ genomic library derived from 129Sv mice. Three overlapping clones were identified and the largest clone, containing the entire CDK3 gene, was sequenced. As shown in Fig. 2A, the 4.5-kb mouse CDK3 structural gene contains seven exons. The overall organization of the mouse gene is virtually identical to that of the human gene, as deduced by the sequence of a chromosome 17 derived P1 artificial chromosome clone RP11–685I11 (GenBank accession no. AC040980; Fig. 2A). Southern blotting confirmed that CDK3 is a single copy gene in the 129Sv strain.
Figure 2.
(A) Organization of mouse and human CDK3 genes. The mouse and human CDK3 genes contain seven coding exons (open boxes). The amino acid residues encoded by each exon are indicated within the boxes. The length of introns are indicated in parenthesis. Conserved kinase motifs in each exon are indicated. PCR primers F7 and R9 were used for amplification of exons 4 and 5, including the intervening intron. PCR primers F12 and R12 (located in intron sequences) were used to amplify exon 5 alone. The position of Trp-187 found to be mutated in CDK3 is indicated by the arrow. (B) Alignment of mouse and human CDKs. The sequence of mouse CDK3 was deduced from genomic sequence. The position of the mutation seen in M. musculus CDK3 at the position of Trp-187 is indicated by the asterisk. Identical residues are shown in black and conserved residues are shown in gray. The location of two CDK3 EST clones used in this study are also shown.
Identification of a Premature Termination Codon in the M. musculus CDK3 gene.
The deduced mouse CDK3 protein sequence was found to be 92% identical to human CDK3, including 29 residues spread throughout the protein that are not conserved in CDK2 or CDC2 (Fig. 2B). However, we were surprised to find that the codon for Trp-187 (TGG) in human CDK3 was replaced by a termination codon (TGA) in the mouse protein (Figs. 2B and 3B). Trp-187 is located in conserved kinase motif IX (Asp-Ile-Trp-Ser-Ile-Gly). The Asp, Trp, and Gly residues are identical or conserved in greater than 99% of all protein kinases (21). Truncation at Trp-187 would delete the C-terminal 118 residues of CDK3 and the fully conserved Arg in motif XI (Fig. 2).
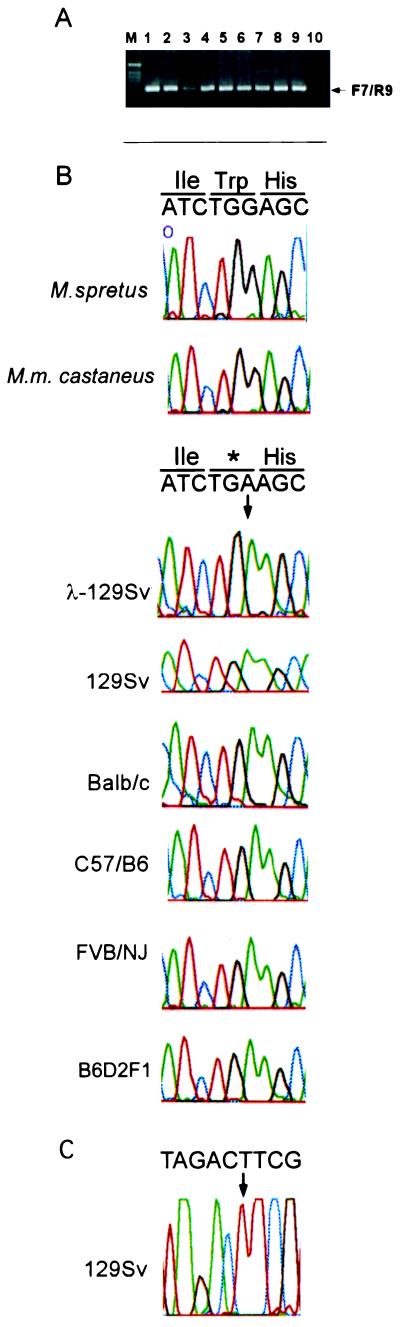
Figure 3.
M. musculus, but not wild-mice strains, contain a premature termination codon in the CDK3 gene. (A) PCR primer pairs F7 and R9 were used to amplify exons 4 and 5 from the CDK3 gene by using total genomic DNA from the indicated strains or from isolated λ clones for CDK3. In all cases, a single PCR product of the expected size (214 bp) was obtained. Lanes: M, DNA ladder; 1, λ-129Sv; 2, 129Sv; 3, C57/B6; 4, BALB/c; 5, FVB/NJ; 6, B6D2F1; 7, M. Spretus; 8, M. castaneus (CASA/RK); 9, M. castaneus (CASA/Ei); 10, negative control. (B) Direct sequencing of PCR products. A portion of the CDK3 sequence flanking the termination mutation is shown, along with the deduced amino acid sequence. The position of the termination codon is indicated by the arrow. The sequences of M. Spretus and M. castaneus contain the expected Trp-187 codon. (C) CDK3 exon 5 was amplified with F12 and R12 primers (Fig. 2A) and sequenced with the reverse R12 primer. The sequence is complementary to that found with the sequences shown in B.
Given this unexpected result, we sought to verify this mutation by direct sequencing of PCR products across exons 4 and 5 derived from 129Sv genomic DNA (Fig. 3 A and B). We found that the sequence derived by this approach was identical to that found with CDK3 genomic clones (Fig. 3B). This rules out the unlikely possibility that CDK3 is heterozygous in this highly inbred strain. To rule out a context-specific sequencing error, we amplified 129Sv genomic DNA with a different set of intron primers flanking exon 5 and sequenced the region in the 3′ → 5′ direction (Figs. 2A and 3C). The sequence observed (Fig. 3C) confirmed the presence of the in-frame stop codon in the M. musculus CDK3 gene and ruled out a context-specific sequencing error.
We next asked whether completely unrelated wild-mice strains also contain this mutation. Genomic DNA from M. spretus and M. m. castaneus (CAST/E1 and CASA/Rk) was used to amplify exons 4 and 5 (Fig. 2A) and PCR products were directly sequenced. The sequences of both wild-mice CDK3 genes were identical to that of the M. musculus strains, except that the codon at position 187 encoded tryptophan (Fig. 2B).
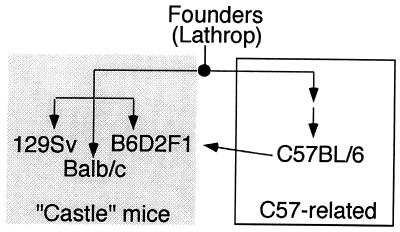
We next examined whether other M. musculus strains, many of which were derived from a small number of founder strains in the early 1900s, also contained this CDK3 mutation. We chose strains in the C57BL group (C57BL/6 and B6D2F1), the “Castle” group (BALB/c and 129Sv), and the Swiss mouse group (FVB/NJ). Genealogical data suggest (22) that strains in the C57BL group and Castle groups have similar origins (Fig. 4) but the precise origins of the Swiss mouse group are not known. Exons 4 and 5 from CDK3 were amplified from genomic DNA (Fig. 3A) and sequenced. As shown in Fig. 3B, all these M. musculus strains contained a termination codon at the position of Trp-187. This result suggests that the mutation was present in the founder mice used to generate C57BL and Castle mice and indicates that FVB/NJ and C57BL strains may have common origins.
Figure 4.
Schematic representation of the genealogy of inbred laboratory mouse strains that contain mutations in CDK3 (adapted from ref. 22). Founder mice bred by A. Lathrop to produce C57-related strains were also used by W. Castle to generate strains that were precursors to 129Sv, BALB/c, and B6D2F1.
Because of the low abundance of CDK3 mRNA in tissues, it has proven difficult to identify cDNA clones, including the use of reverse transcription-coupled PCR (X.Y. and J.W.H., unpublished data). However, we did identify a single M. musculus (B6D2F1) CDK3 EST clone (GenBank accession no. AA617565) that overlaps this region. The sequence of this cDNA also contains a termination codon at the position of Trp-187 in human CDK3 (Fig. 2B). Thus, the mutant CDK3 gene is transcribed and makes an mRNA predicted to encode a protein truncated at Ile-186.
Implications for CDK3 Function.
In this study, we report that several M. musculus strains commonly used in the laboratory contain a premature termination codon in the CDK3 gene. This was achieved by direct sequencing of two distinct PCR products produced from genomic DNA and three independently isolated λ-clones derived from a 129Sv library. This data and the fact that the inbred strains analyzed are more than 99% homozygous argue that these M. musculus strains lack an intact CDK3 gene. This mutation (Trp-187 → Term) results in deletion of two protein kinase motifs (IX and XI) that are highly conserved in serine/threonine kinases and tyrosine kinases and removes a large portion of the structure that interacts with substrates. Therefore, we think it highly likely that this would create a null allele.
There are several lines of evidence that suggests that human CDK3 functions during the G1/S-phase transition. Dominant negative forms of CDK3 arrest cells in G1 phase and this can be overcome by expression of wild-type CDK3, cyclin E, and cyclin D1 but not by CDK2 (18). Moreover, human CDK3 can associate with E2F-1/DP-1 complexes and dominant negative CDK3 blocks E2F activity when overexpressed, suggesting a positive role for CDK3 in the G1/S-phase transition (19). Consistent with this, cyclin E–CDK3 can promote S-phase entry in the absence of serum (20). Thus, although it is clear that CDK3 can promote cell division, our results lead us to conclude that CDK3 is not uniquely required for cell division in the vast majority of cell types in M. musculus. This most likely reflects redundancy between different CDKs in the limited number of cell types where CDK3 is expressed. In particular, CDK2 may be able to substitute for CDK3 activity in vivo. In this regard, we note that CDK3–cyclin E and CDK2–cyclin E give indistinguishable phosphorylation patterns with the retinoblastoma protein in vitro, suggesting that their inherent specificities are similar (23). Although our data indicate that CDK3 is not essential for development as we know it in M. musculus, we cannot exclude the possibility that CDK3 maintains an essential role for the production of particular cell types in organisms, such as humans, in which it remains functional.
Deletion experiments in mice have revealed considerable functional redundancy among various positive and negative cell cycle regulatory components. For example, p107 and p130 appear to play redundant roles in the control of differentiation during mouse development. In isolation, p107 and p130 mutant mice are free of obvious phenotypes but p107−/−;p130−/− mice exhibit neonatal lethality and multiple types of defects in differentiation (24). In other cases, loss of a single cell cycle regulator is not lethal to the organism but produces cell-type-specific effects. For example, CDK4 is not essential for mouse development per se but the mutant animals are much smaller than their wild-type counterparts and there are several tissue-specific abnormalities seen in these mice (25, 26). Thus, although other CDKs, such as CDK6, may be capable of performing the functions of CDK4 in cells where they are coexpressed, tissues where CDK4 is the primary D-type cyclin kinase subunit display clear phenotypic consequences upon loss of CDK4. This effect is mirrored in cyclin D1-deficient mice, which are also much smaller than wild-type animals and also display tissue-specific abnormalities (27). Tissue-specific redundancy is also displayed by the cyclin-dependent kinase inhibitors p21, p27, and p57 (28, 29). Our results emphasize, yet again, the extent to which functional redundancy appears to be used in cell cycle control. Moreover, CDK3 is not the first cell cycle regulator found to be mutated in inbred mouse strains. Of note is the p16INK4a gene, which contains a debilitating point mutation in BALB/c and ABP/Le strains but is wild type and functional in other M. musculus strains (30). Clearly, inactivating mutations of the sort identified herein may have implications for experiments designed to test the function of cell cycle regulators in mice. The mouse single nucleotide polymorphism (SNP) database (31) may help to establish how prevalent loss-of-function mutations are in redundant components of cell cycle machinery in laboratory mouse strains.
Acknowledgments
We thank N. Jenkins, M. Justice, and S. Elledge for reagents. This work was supported by Grants GM54137 and CA58204 from the National Institutes of Health and by the Welch Foundation.
Abbreviations
- CDK
cyclin-dependent kinase
- CDC
cell division cycle kinase
- EST
expressed sequence tag
Footnotes
Data deposition: The M. musculus CDK3 gene sequence reported in this paper has been deposited in the GenBank database (accession no. AA617565).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041596198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041596198
References
- 1.Dulic V, Lees E, Reed S I. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 2.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 3.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 4.Matsushime H, Ewen M E, Strom D K, Kato J Y, Hanks S K, Roussel M F, Sherr C J. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 5.Quelle D E, Ashmun R A, Shurtleff S A, Kato J Y, Bar-Sagi D, Roussel M F, Sherr C J. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 6.Resnitzky D, Reed S I. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtsubo M, Roberts J M. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- 8.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 9.Ekholm S V, Reed S I. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 10.Harbour J W, Luo R X, Dei Santi A, Postigo A A, Dean D C. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 11.Bruce J L, Hurford R K, Jr, Classon M, Koh J, Dyson N. Mol Cell. 2000;6:737–742. doi: 10.1016/s1097-2765(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 12.Harbour J W, Dean D C. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 13.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leng X, Connell-Crowley L, Goodrich D, Harper J W. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 16.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin I, Reed S I, Bartek J. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 17.Meyerson, M., Enders, G. H., Wu, C. L., Su, L. K., Gorka, C., Nelson, C., Harlow, E. & Tsai, L. H. (199) EMBO J.11, 2909–2917. [DOI] [PMC free article] [PubMed]
- 18.Van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann F, Livingston D M. Genes Dev. 1996;10:851–861. doi: 10.1101/gad.10.7.851. [DOI] [PubMed] [Google Scholar]
- 20.Connell-Crowley L, Elledge S J, Harper J W. Curr Biol. 1998;8:65–68. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- 21.Hanks S K, Quinn A M. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 22.Beck J A, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig J T, Festing M F, Fisher E M. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 23.Connell-Crowley L, Harper J W, Goodrich D W. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobrinik D, Lee M H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 25.Rane S G, Dubus P, Mettus R V, Galbreath E J, Boden G, Reddy E P, Barbacid M. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 26.Tsutsui T, Hesabi B, Moons D S, Pandolfi P P, Hansel K S, Koff A, Kiyokawa H. Mol Cell Biol. 1999;19:7011–7019. doi: 10.1128/mcb.19.10.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Wong C, Finegold M, Harper J W, Elledge S J. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Wong C, DePinho R A, Harper J W, Elledge S J. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Ramsay E S, Mock B A. Proc Natl Acad Sci USA. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindblad-Toh K, Winchester E, Daly M J, Wang D G, Hirschhorn J N, Laviolette J P, Ardlie K, Reich D E, Robinson E, Sklar P, et al. Nat Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]