Abstract
The three-dimensional structure of the membrane-bound form of the major coat protein of Pf1 bacteriophage was determined in phospholipid bilayers using orientation restraints derived from both solid-state and solution NMR experiments. In contrast to previous structures determined solely in detergent micelles, the structure in bilayers contains information about the spatial arrangement of the protein within the membrane, and thus provides insights to the bacteriophage assembly process from membrane-inserted to bacteriophage-associated protein. Comparisons between the membrane-bound form of the coat protein and the previously determined structural form found in filamentous bacteriophage particles demonstrate that it undergoes a significant structural rearrangement during the membrane-mediated virus assembly process. The rotation of the transmembrane helix (Q16–A46) around its long axis changes dramatically (by 160°) to obtain the proper alignment for packing in the virus particles. Furthermore, the N-terminal amphipathic helix (V2–G17) tilts away from the membrane surface and becomes parallel with the transmembrane helix to form one nearly continuous long helix. The spectra obtained in glass-aligned planar lipid bilayers, magnetically aligned lipid bilayers (bicelles), and isotropic lipid bicelles reflect the effects of backbone motions and enable the backbone dynamics of the N-terminal helix to be characterized. Only resonances from the mobile N-terminal helix and the C-terminus (A46) are observed in the solution NMR spectra of the protein in isotropic q > 1 bicelles, whereas only resonances from the immobile transmembrane helix are observed in the solid-state 1H/15N-separated local field spectra in magnetically aligned bicelles. The N-terminal helix and the hinge that connects it to the transmembrane helix are significantly more dynamic than the rest of the protein, thus facilitating structural rearrangement during bacteriophage assembly.
Abbreviations used: 6-O-PC, 1,2-di-O-hexyl-sn-glycero-3-phosphocholine; 14-O-PC, 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine; DC, dipolar coupling; DHPC, 1,2-dihexanoyl-sn-glycero-3-phosphocholine; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPG, 1,2-dioleoyl-sn-glycerol-3-[phospho-rac-(1-glycerol)]; CSA, chemical shift anisotropy; HETCOR, heteronuclear chemical shift correlation; HMQC, heteronuclear multiple quantum correlation; HSQC, heteronuclear single quantum correlation; IPAP, in-phase anti-phase; NOESY, nuclear Overhauser effect spectroscopy; PISA, polar index slant angle; RDC, residual dipolar coupling; SLF, separated local field
Introduction
Pf1 is a filamentous bacteriophage that infects Pseudomonas aeruginosa strain Kr. The geometric arrangement of the coat protein subunits on the outside, and the packing of the DNA on the inside differentiate Pf1 as a class II bacteriophage from the better-known class I filamentous bacteriophages (e.g., fd and M13 (1)). In particular, Pf1 has a 1:1 ratio of nucleotides to coat protein subunits, which is consistent with the ordered, symmetrical packing of its DNA, whereas fd has a larger, nonintegral 2.4:1 ratio and its DNA is structurally disordered in the virus particles. Virus assembly is among the most fundamental of biological functions, and in this regard Pf1 is already among the best-characterized systems (1–7).
The major coat protein is of particular interest because during the viral life cycle, it functions both as a membrane protein stored within the inner membrane of the bacterial host before virus assembly occurs, and as a structural protein that forms the bulk capsid in the assembled virus particle. During virus assembly, the basic amino acids in the cytoplasmic C-terminus of the coat protein form electrostatic interactions with the viral DNA, and the protein subunits interact with each other to form the long (1–2 μm) virions. The virus particles are assembled as they are extruded from the bacterial membrane, during which time the coat protein undergoes a large structural rearrangement from L-shaped with a periplasmic N-terminus and cytoplasmic C-terminus in the membrane, to I-shaped in the assembled virus particles, where the coat protein's N-terminus forms the bacteriophage particle's exterior and its C-terminus faces the particle's interior (5–7).
In previous works we determined the three-dimensional (3D) structure of Pf1 coat protein in bacteriophage particles by solid-state NMR spectroscopy (8,9). Fiber diffraction and other physical methods have also been applied to this structural form of the coat protein, and the results have contributed to its description (10–13). However, neither form of the protein has been crystallized. The membrane-bound form of Pf1 coat protein has been investigated primarily by NMR spectroscopy, mainly in micelles by solution NMR, but also in phospholipid bilayers by solid-state NMR (4,6,14,15). In recent solution NMR experiments, we were able to prepare samples of membrane proteins in DHPC micelles with two different types of alignment, which enabled us to determine the 3D structure of the protein in this detergent (16).
Although solution NMR spectroscopy can provide the 3D structure of a membrane protein in micelles, it cannot provide any information about its 3D position (e.g., tilt and rotation of helices in the membrane) within the asymmetric environment of the lipid bilayer membrane. The spatial orientation adopted by membrane proteins in cellular membranes is an intrinsic component of their biological function. However, such directional information is lost in the isotropic micellar samples of membrane proteins required for solution NMR. In contrast, this information is retained in samples where the proteins are embedded in lipid bilayer membranes, such as those that are suitable for solid-state NMR spectroscopy.
Solid-state NMR experiments with lipid bilayer samples that are macroscopically oriented in the magnetic field can provide very precise orientation restraints for structure determination and refinement in environments that closely resemble cellular membranes, and since the direction of alignment is fixed by the sample geometry, they also provide the 3D orientation of the protein in the membrane (17,18). Recently, we showed that this information can also be used to supplement solution NMR structural data to establish a protein's transmembrane orientation (19), provided that similar structural features are present in the two types of samples.
In this work, we combine solution and solid-state NMR orientation restraints to obtain the 3D structure of the Pf1 coat protein within the lipid bilayer membrane. The solution NMR 1H/15N RDCs and solid-state NMR 1H/15N DCs and 15N CSAs, measured for the Pf1 coat protein in weakly aligned micelles or uniaxially aligned lipid bilayers, provide orientation restraints for protein structure determination within the membrane. These results, obtained on the membrane-bound form of the coat protein, complement the atomic-resolution structure of the same protein in intact virus particles, enabling direct comparisons between its two forms and providing insights into the structural rearrangement associated with virus assembly.
Materials and Methods
Sample preparation
Uniformly 15N-labeled and 13C/15N-double-labeled Pf1 coat protein was prepared and purified as described previously (8).
Solution NMR samples were prepared by dissolving 2 mg of the purified protein in 400 μL of 100 mM deuterated DHPC (Cambridge Isotope Laboratories, Andover, MA) containing 10% (v/v) 2H2O at pH 6.7. Weakly aligned samples were prepared either by soaking the micellar sample overnight into a dried 6% polyacrylamide gel with the length of the gel restricted to 22 mm from an initial length of 30 mm, or by adding fd bacteriophage at a final concentration of 28 mg/mL.
Magnetically aligned bilayer samples for solid-state NMR were prepared by dissolving 2–3 mg of pure lyophilized Pf1 coat protein in a solution containing a short-chain lipid, 6-O-PC, which was then added to a dispersion of a long-chain lipid, 14-O-PC. The lipids were obtained from Avanti Polar Lipids (Alabaster, AL). The ratio of long-chain to short-chain lipids is defined by the parameter q. The bicelle sample for q = 3.2 had a lipid concentration of 28% (w/v) and contained 300 mM 14-O-PC in a volume of 180 μL at pH 6.7. For the NMR experiments, a flat-bottomed, 5 mm outer diameter NMR tube (New Era Enterprises, Vineland, NJ) was filled with 160 μL of the bicelle solution.
Glass-aligned bilayer samples for solid-state NMR were prepared by dissolving 3 mg of pure lyophilized coat protein in TFE and 0.1% TFA. The solution was filtered through a 0.22 μm PTFE membrane and dried under nitrogen gas, which was added to an equal volume of DOPC/DOPG (9:1 (w/w), 60 mg total lipid) in chloroform, to ∼400 μL. The solution was spread over 22 glass coverslides (Marienfield GmbH & Co., Lauda-Konigshofen, Germany). After drying was completed, the glass slides were stacked, equilibrated with a saturated ammonium phosphate solution, and sealed in thin polyethylene.
NMR spectroscopy
Solution NMR experiments were performed at 40°C on a Bruker DRX 600 MHz spectrometer equipped with a 5 mm triple-resonance cryogenic probe and a z axis gradient. Heteronuclear NMR experiments were performed on uniformly 15N-labeled or 13C/15N-double-labeled samples with a protein concentration of 1 mM. They included 2D 1H-15N HSQC, 15N-edited HMQC-NOESY with a mixing time of 200 ms, 3D 15N-edited NOESY-HSQC with mixing time of 200 ms, and 3D HNCA. 1H-15N IPAP-HSQC spectra (20) obtained from isotropic and weakly aligned samples were used to measure the 1H-15N RDCs. Heteronuclear 1H-15N NOEs were measured as described previously (21).
Solid-state NMR experiments with magnetically aligned samples were performed at 40°C on Bruker Avance spectrometers operating at 1H resonance frequencies of 700 MHz and 750 MHz. The home-built (1H/15N) and Bruker (1H/15N/13C) probes had 5 mm inner diameter solenoid coils. The one-dimensional (1D) 15N spectra resulted from cross-polarization with a contact time of 1 ms, a recycle delay of 6 s, and an acquisition time of 10 ms. Two-dimensional (2D) SLF spectra were obtained with SAMMY (22). SLF spectra for uniformly 15N-labeled samples had a total of 96 t1 increments and 512 t2 complex points, with 128 scans for each t1 increment. SLF spectra from selectively 15N labeled samples had 48–64 t1 increments. The B1 radiofrequency field strength was 50 kHz, the recycle delay was 6 s, and the acquisition time was 5 ms.
Solid-state NMR experiments with glass-aligned bilayers were performed at 20°C on a Bruker Avance spectrometer, operating at 500 MHz, with a home-built (1H/15N) flat coil probe (22 × 11 × 4 mm). The 1D 15N NMR spectra resulted from cross-polarization with a contact time of 1 ms, a recycle delay of 7 s, and an acquisition time of 10 ms. For each spectrum, 1024–8096 transients were accumulated and an exponential line-broadening of 50–100 Hz was applied.
Resonance assignments
We assigned the solid-state NMR spectra by using selectively labeled samples and the shotgun method (23), which takes advantage of the scattering of each type of amino acid throughout the sequence and the periodic patterns inherent in helical structures. We were able to unambiguously assign 13 out of 21 resonances in the transmembrane helix of Pf1 coat protein from the spectra of the 15N Leu-, 15N Ile-, 15N Val-, and 15N Ala-labeled samples (Fig. 1) because they fall close to the PISA wheel patterns. In contrast, there are two Lys residues (K20 and K45) located in nonhelical regions of the protein (interhelical loop and C-terminus, respectively), and, as expected, their resonances do not fall near an ideal PISA wheel (Fig. 1 F). We made and confirmed these additional resonance assignments by comparing the 15N chemical shift differences between spectra obtained from samples with their bilayer normal aligned perpendicular and parallel to the direction of the magnetic field, as described previously (24).
Figure 1.

2D solid-state NMR 1H/15N SLF spectra of uniformly and selectively 15N-labeled (by residue type) samples of the membrane-bound form of Pf1 coat protein in 14-O-PC/6-O-PC (q = 3.2) bilayers aligned with their normal perpendicular to the magnetic field. The PISA wheel (thin line) for an ideal α-helix with uniform dihedral angles (ϕ = −61°, φ = −45°), tilted 30° from the membrane normal, is superimposed on the experimental spectra. The residue number designates the resonance assignments for each type of labeled amino acid. (A) The data from uniformly 15N-labeled coat protein were described previously (6), and are provided for comparison to the spectra of the selectively labeled samples in (B-F). (B) 15N -valine labeled. (C) 15N-leucine labeled. (D) 15N-isoleucine labeled. (E) 15N-alanine labeled. (F) 15N -lysine labeled.
For example, since the isotropic chemical shifts of two alanine residues (A21 and A46) in nonhelical regions of the protein differ by >6 ppm, we could readily assign their resonances by comparing the 15N chemical shifts observed in spectra from the two alignments. We also took advantage of established patterns of biosynthetic isotopic incorporation. For example, Gly and Ser are readily interconverted during biosynthesis (25); thus, the addition of 15N-Gly to the growth media results in NMR spectra with strong Gly peaks and weak Ser peaks, whereas the addition of 15N-Ser yields spectra with strong Ser and weak Gly peaks. This facilitates resonance assignments by residue type.
We previously described the 3D spectrum of uniformly 15N labeled Pf1 coat protein in magnetically aligned bicelles in the context of demonstrating a new NMR experiment (26). This spectrum was obtained at 900 MHz and contained both 1H/15N HETCOR and 1H-15N SLF planes. Several signals that are overlapped in the 2D spectra are resolved in this 3D spectrum, confirming their frequencies and assignments.
Data analysis and structure calculations
The NMR data were processed using NMRPipe/NMRDraw (27). Molecular structures were analyzed and visualized with PyMOL (28). Structure calculations were performed with XPLOR-NIH (29) using simulated annealing protocols with internal dynamics (30). Dihedral angle restraints were imposed by a quadratic harmonic potential with a force constant of 300 kcal·mol−1·rad−2. The torsion angle database Rama potential (31), with a force constant of 0.02–0.20, was used to select preferred side-chain conformations relative to the backbone dihedral angles. Solution NMR RDC restraints and solid-state NMR DC restraints were imposed using the SANI potential (32), with force constants of 0.2–2.0 kcal·Hz−2 for RDCs, and 1.0–10.0 kcal·kHz−2 for DCs. Solid-state NMR CSA restraints were imposed using the DCSA potential (33) with a force constant of 0.2–2.0 kcal·ppm−2. Other force constants for molecular bonds, angles, and Van der Waals distances were as described previously (34,35).
A starting structure was generated from extended random coil coordinates using a high-temperature simulated annealing protocol (34). The temperature was cooled from 3500°K to 100°K in 25° steps, and the restraints included dihedral angles, the torsion angle database Rama potential, and solution NMR RDCs. The resulting lowest-energy structure was used in a second simulated annealing calculation, which included solid-state NMR DC and CSA restraints. The structure coordinates and NMR data have been deposited in the Protein Data Bank (PDB: 2KSJ). Statistics for the resulting family of 10 lowest-energy structures are reported in Table S1. The ensemble of 10 structures is shown in Fig. S1 of the Supporting Material.
Dihedral angle restraints were derived from TALOS (36) and chemical shift index (37) using the chemical shifts observed in micelle samples, and from dipolar wave (38) and PISA wheel (39,40) analyses of 1H/15N RDCs, 1H/15N DCs, and 15N CSAs. In the dipolar wave analysis, the helical regions of the protein included those residues for which the root mean-square deviation (RMSD) between the experimental values and the data fit to helix was less than the experimental error (0.5 Hz for RDCs and 0.3 kHz for DCs).
For weakly aligned micelle samples, the magnitude (Da) and rhombicity (R) of the alignment tensor were estimated using 2D λ-map (41) analysis of the RDC data from two different alignment media, which yielded values of Da = 11.44 Hz and R = 0.64 for the sample aligned with fd bacteriophage, and Da = 7.74 Hz and R = 0.21 for the sample aligned in compressed gel. For glass-aligned bilayer samples, the membrane normal is parallel to the magnetic field, the order tensor is axially symmetric, and its principal axis coincides with the membrane normal. In this case, values of Da(bilayer) = 10.52 kHz (based on NH bond length of 1.05 Å) and R = 0 were used. For magnetically aligned bicelles, librations about the principal axis of alignment scales the observed frequencies by a factor Szz∼0.8 and Da(bicelle) = Szz × Da(bilayer) (42,43). In this case, values of Da = 8.42 kHz and R = 0 were used.
The traceless values and molecular orientation of the 15N chemical shift tensor are δ11 = −45.7 ppm, δ22 = −62.8 ppm, δ33 = 108.5 ppm, β = 20°, and γ = 11° for non-Gly residues (44), and δ11 = −41.0 ppm, δ22 = −64.0 ppm, δ33 = 105.0 ppm, β = 20°, and γ = 0° for Gly residues (45). Values are reported following the convention |δ33|>|δ22|>|δ11|; the angle β is between δ33 and the NH bond and the angle γ is between δ22 and the axis normal to the peptide plane. The 15N CSA alignment tensor was normalized to the maximum value of the dipolar coupling by setting the XPLOR-NIH DCSA parameter scale to 21,035 Hz. The CSA for each residue was calculated by subtracting the isotropic 15N chemical shift frequency (δiso) from the orientation-dependent chemical shift frequency measured in the solid-state NMR SLF spectra of the aligned protein. Similarly, the CSA values that were back-calculated by simulated annealing were converted to orientation-dependent frequencies by adding δiso. The values of δiso = 119 ppm for non-Gly residues, and δiso = 105 ppm for Gly residues were derived from their respective CSA tensors.
Results and Discussion
Structure of Pf1 coat protein in the lipid bilayer membrane
The structure of the membrane-bound form of Pf1 coat protein was calculated using all of the available restraints measured from solid-state NMR experiments in bicelles and bilayers, as well as solution NMR experiments in micelles (Fig. 2, A and B). Excellent correlations were obtained between the experimental and back-calculated RDCs, DCs, and CSAs when all of these restraints were used in simulated annealing calculations (Fig. S2).
Figure 2.
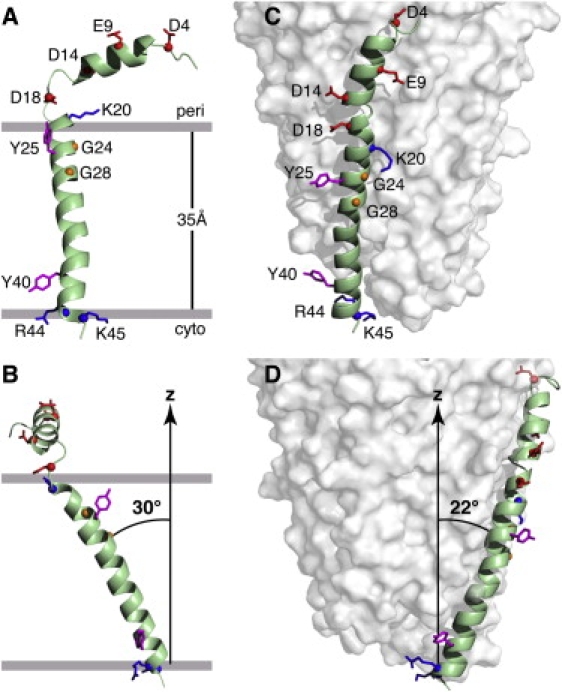
3D structures of the Pf1 coat protein. (A and B) The membrane-bound form of the protein (PDB: 2KSJ). (C and D) The structural form in the intact bacteriophage particle (PDB: 1PJF). The axis of alignment (z) is parallel to the magnetic field and the membrane normal in A and B, and to the long axis of Pf1 bacteriophage in C and D. In the membrane-bound form of the protein, the acidic N-terminus region is exposed to the bacterial periplasmic space (peri) and the basic C-terminal region is exposed to the cytoplasm (cyto). Acidic residues (Asp and Glu) are shown in red, conserved glycine residues in the transmembrane helix are in yellow, basic residues (Arg and Lys) are in blue, and interfacial tyrosines are in pink. Residues R44 and K45 face the cytoplasm in the membrane-bound form and the DNA on the interior of the bacteriophage particles. (B and D) Images obtained by 90° rotation of A and C around the z axis of the images shown in A and C, respectively.
The resulting restraint-based RMSDs reflect experimental errors as well as uncertainties in the NH bond length, the values of the order parameters, and the 15N chemical shift tensor. Given that a common 15N chemical shift tensor was used for all non-Gly residues in the structural analysis, it is remarkable that the back-calculated data reproduce both the DCs and CSAs so well. The favorable RMSDs between experimental and back-calculated data suggest that residue-specific chemical shift tensor variations are minor compared to the spectral manifestation of molecular orientation, indicating that chemical shifts as well as dipolar couplings can be used as restraints for structure determination and refinement. This was previously noted in the solid-state NMR spectra of other membrane proteins (17,19) and is a major factor enabling PISA wheels to be observed in the spectra of oriented proteins.
In lipid bilayers, the coat protein's N-terminal amphipathic helix spans residues 5–15. It aligns parallel to the lipid bilayer surface with hydrophobic residues (A7, V8, A11, I12, and G15) facing the membrane, and its acidic and polar residues (T5, S6, E9, S10, T13, and D14) exposed to the aqueous environment. A three-residue linker (Q16, G17, and D18) connects this N-terminal surface helix to the transmembrane α-helix, which spans residues 23–45 and adopts a tilt of ∼30° from the lipid bilayer normal.
The 30° transmembrane helix tilt of membrane-associated Pf1 is very similar to that of the membrane-associated fd bacteriophage coat protein, whose structure was determined in glass-aligned planar lipid bilayers by solid-state NMR (23). The two aromatic residues in Pf1 (Y25 and Y40) are situated near the periplasmic (Y25) and cytoplasmic (Y40) faces of the bacterial inner membrane, and are likely to play a role in determining transmembrane helix tilt and orientation (46), similarly to Y21, Y24, W26, and F45 in the fd coat protein. In the membrane-bound form of the coat protein, the helix and long-axis rotation are such that the positively charged C-terminal residues (R44 and K45) are exposed to the cytoplasm and positioned to interact and associate with the viral DNA as it is extruded through the membrane. These residues form the positively charged core of the capsid in the bacteriophage particle.
By comparing the membrane-bound form of Pf1 coat protein (Fig. 2, A and B) determined in this study with the previously determined structure (8) of the coat protein incorporated in the bacteriophage particle (Fig. 2, C and D), one can see that a dramatic conformational change occurs during virus assembly. Solid-state NMR chemical shifts and dipolar couplings give accurate measurements of helix tilt angle (between the axis of alignment and helix long axis), helix rotation angle (around the helix long axis), and helix defects (kinks and bulges). These parameters can be derived directly from the solid-state NMR spectra of the membrane-bound and bacteriophage-associated forms of the coat protein (Fig. S3).
In virus particles, the coat protein adopts a long α-helix spanning residues 7–46 that is interrupted by several kinks (8). Of interest, the kink at A29 observed in the structural form is also found in bicelles, as indicated by the deviation of the solid-state NMR dipolar coupling measured for residue A29 from the sinusoidal fitting of the data (Fig. S3). Indeed, the hydrophobic helix of the phage form can be superimposed onto that of the membrane form with resulting RMSDs of 1.12 Å for CA atoms and 2.22 Å for all atoms. The overall orientation of the hydrophobic helix relative to the axis of alignment (i.e., the membrane normal and the bacteriophage long axis) differs significantly between the two forms (Fig. 2, B and D). The helix changes tilt from ∼30° in the membrane to ∼22° in the bacteriophage and undergoes a distinct change (∼160°) in rotation around its long axis (as shown in the corresponding helical wheel projections in Fig. S3), resulting in the basic C-terminal side chains being exposed to the DNA core of the bacteriophage particle.
Dynamics of the N-terminal helix in micelles, bicelles, and bilayers
Our previous solution NMR experiments demonstrated that residues in the coat protein's N-terminal amphipathic α-helix are more mobile in both SDS (15) and DHPC micelles (16). The heteronuclear 1H-15N NOE data (Fig. 3 C), also obtained in DHPC, corroborate the principal structural and dynamic features of the protein, including a relatively rigid transmembrane helix and a mobile interhelical loop and N-terminal helix.
Figure 3.
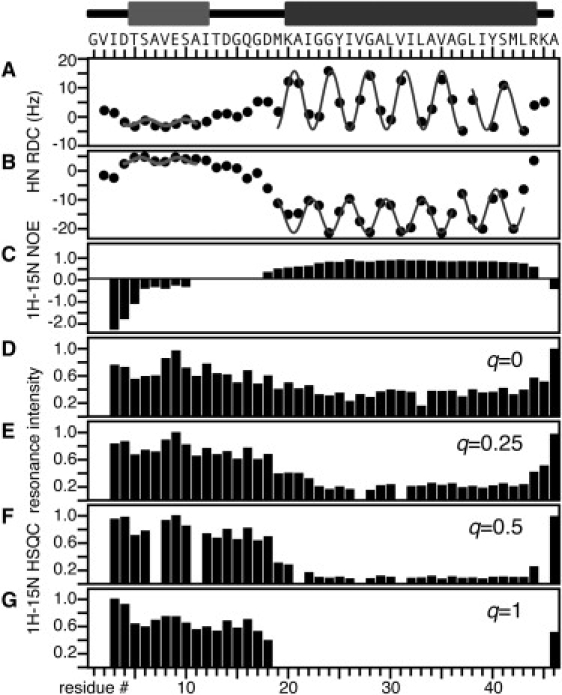
Summary of the experimental data used to characterize the secondary structure and backbone dynamics of the membrane-bound form of Pf1 coat protein in DHPC micelles by solution NMR. The measurements are plotted as a function of residue number for the protein sequence aligned at the top of the figure. Above the sequence is a schematic drawing of the secondary structure; the light gray region is the amphipathic N-terminal helix and the dark gray region is the hydrophobic trans-membrane helix. (A and B) Dipolar waves fit to 1H-15N RDCs. (A) Sample weakly aligned in a stressed polyacrylamide gel. (B) Sample weakly aligned by the presence of fd bacteriophages in the solution (16). (C) 1H/15N heteronuclear NOEs. (D–G). Normalized peak intensities of the amide resonances observed in 2D 1H/15N HSQC spectra samples with different q values for the ratio of DMPC (long chain lipid) to DHPC (short chain lipid) in isotropic bicelles. (D) q = 0. (E) q = 0.25. (F) q = 0.5. (G) q = 1.
Although micelles can be useful model environments for membrane proteins, they suffer from high surface curvature and low lateral pressure in the hydrophobic region, which can obscure important structural and dynamic features of the membrane proteins. Furthermore, their isotropic nature precludes acquisition of information about the global positioning of the protein within the membrane—information that is of paramount importance for understanding functional mechanisms. The solid-state NMR data obtained on the Pf1 coat protein in phospholipid bilayers presented here overcome limitations of the data obtained in micelles, and provide information about its structure and dynamics in the membrane, as well as a precise estimate of the timescale.
Protein-containing bicelles can be used for both solution (47,48) and solid-state (43,49) NMR structural studies of membrane proteins if the parameter q. We previously showed that the membrane-bound form of Pf1 coat protein has the remarkable property of yielding solution NMR spectra in small isotropic bicelles (q = 1) and solid-state NMR spectra in large bicelles (q = 3.2) containing complementary populations of resonances, allowing characterization of the protein's dynamics (6). The q = 1 solution NMR spectrum shows only resonances from the mobile N-terminal helix, whereas the q = 3.2 solid-state NMR spectra show only resonances from residues in the immobile transmembrane helix.
Comparisons of signal intensities observed in the 2D HSQC spectra in micelles and isotropic bicelles over a range of q-values provide insights into the protein's backbone dynamics (Fig. 3, D–G). As the ratio of long- to short-chain lipid increases (from q = 0 to q = 1), the relative resonance intensities of residues 19–45, including the transmembrane helix, decrease systematically and finally disappear. These intensity reductions are not the result of rapid exchange between amide hydrogens, since the transmembrane amide hydrogens are much more resistant to solvent exchange than the N-terminal hydrogens, as shown in our earlier 2H2O exchange experiments (14,50).
Solid-state 15N NMR spectra of uniformly and selectively 15N-labeled Pf1 coat protein in aligned bilayers are compared in Fig. 4. Each amide site contributes a resonance whose frequency is determined by the orientation of the peptide plane relative to the field. Significantly, there is no evidence of underlying powder pattern intensity due to immobile, unoriented polypeptides, or of isotropic intensity from mobile residues. The resonance linewidths in the spectra (Fig. 4, A–C) from glass-aligned bilayer samples are somewhat broader than those obtained from magnetically aligned samples (Fig. 4, D–F). The frequency span of the spectrum in Fig. 4 D is smaller than that in Fig. 4 A because magnetically aligned bilayers have a slightly reduced order parameter of 0.8 (49).
Figure 4.

1D solid-state 15N NMR spectra of the membrane-bound form of Pf1 coat protein in lipid bilayers aligned with their normals parallel to the magnetic field. (A–C) DOPC/DOPG (9/1, w/w) planar lipid bilayers mechanically aligned on glass slides. (D–F) 14-O-PC/6-O-PC (q = 3.2) bilayers magnetically by addition of YbCl3. (A and D) Uniformly 15N-labeled. (B–F) Selectively 15N-labeled (by residue type). (B and E) 15N-isoleucine labeled. (C and F) 15N-threonine labeled. Signals from residues in the amphipathic N-terminal region (I12, T5 and T13) are present in the spectra from samples mechanically-aligned on glass plates (B and C), but are not observed in the spectra from magnetically-aligned samples (E and F). In contrast, signals from the trans-membrane region (I22, I26, I32 and I39) are present in the spectra from both mechanically- and magnetically-aligned samples. The data from the magnetically-aligned sample of the uniformly 15N-labeled protein were described previously (6), and provide for comparison to the spectra of the selectively labeled samples.
Of note, most of the resonance intensity between 70 ppm and 120 ppm present in the spectrum from the glass-aligned sample is missing in the spectrum from the magnetically aligned bicelle sample. This is clearly demonstrated in the spectra from selectively 15N-labeled samples. Pf1 coat protein contains two threonines (T5 and T13) in the N-terminal amphipathic helix. There are two resonances in Fig. 4 C but none in Fig. 4 F, even though careful efforts were made to optimize the experimental parameters. A comparison of the spectra from the 15N-Ile-labeled samples also shows that the signal from I12, next to T13, is present only in the spectrum from the glass-aligned sample (Fig. 4 B versus Fig. 4 E). These differences result from the influence of the dynamics of the N-terminal helix.
Since the dynamics of the N-terminal amphipathic helix prevents its signals from being detected in the solid-state NMR spectra from bicelles (Fig. 4, D–F), the three CSA measurements obtained for T5, I12, and T13 in glass-aligned lipid bilayers were instrumental in defining the helix orientation on the membrane surface. The signals from the N-terminal segment of Pf1 coat protein detected in the solid-state NMR spectra of glass-aligned phospholipid bilayer samples (Fig. 4, A–C) definitively show that the helix lies on the bilayer surface, and provide high-resolution orientation restraints for structure determination.
Both glass-oriented and magnetically oriented lipid bilayers are large, planar, phospholipid membranes. Unlike micelles, they do not suffer from high surface curvature and provide an environment that is close to that of the native membrane. Nevertheless, the slight but significant difference in order parameters between these two systems enables their dynamic features to be characterized very precisely. Taken together, the data in Fig. 4 indicate that that the dynamics of the amphipathic N-terminal helix is sufficient to obliterate the corresponding resonances in the spectra from bicelles. This can be attributed to the factors that contribute to the differences in order parameters between planar glass-aligned planar bilayers (S = 1.0) and magnetically aligned bicelles (S = 0.8), which lead to different NMR timescales for the corresponding solid-state NMR spectra.
For 15N resonances to be observed by 1H-15N cross polarization, the backbone dynamics of the corresponding sites must be slower than the timescale of the 1H-15N dipolar coupling, which is 20 kHz in glass-aligned bilayers with an order parameter of 1.0, and 16 kHz in bicelles with an order parameter of 0.8. The absence of signals from amphipathic helix sites in the spectra from bicelles, together with their presence in the spectra from planar bilayers, indicates that the amphipathic helix experiences backbone dynamics that are intermediate in terms of their NMR behavior—slower than the fully ordered limit, but faster than the reduced timescale. The precise nature of this motion cannot be deduced from the studies presented here, but the results are consistent with the N-terminal helix flicking on and off the membrane surface, as reported previously for the M13 coat protein (51,52).
Implications for the Pf1 bacteriophage particle assembly process
Both Pf1 and fd transmembrane helices are oriented so that the C-terminal positively charged (R44 and K45 in Pf1, and K40, K43, and K44 in fd) and polar (S41 in Pf1, and T36 in fd) residues face the cytoplasmic side of the membrane. This topology may have functional significance for the bacteriophage assembly process, during which the single-stranded phage DNA is extruded through the inner bacterial membrane after being enclosed within a tube of coat protein subunits. The orientation of the coat protein transmembrane helix exposes the charged amino groups of the lysine and arginine side chains to the cytosol, making them available for DNA binding during bacteriophage extrusion.
In contrast, small hydrophobic residues (Gly and Ala) line the transmembrane helix side facing the periplasmic leaflet of the membrane. Some of these residues (G24 and A36) are conserved in the sequences of the coat proteins of Pf1, fd, and M13 bacteriophage, suggesting that they could play a role in mediating helix-helix interactions during viral assembly and in the assembled viral particles (Fig. S4). Indeed, the small-xxx-small motif found in the G24xxxG28 sequence of Pf1 is conserved in the sequences of other filamentous bacteriophage coat proteins (e.g., G23xxxA27 in fd and M13), and was found to be important for stabilizing the interaction between the coat protein subunits in the bacteriophage capsid (8).
In the phage particle (Fig. 5 A), each coat protein subunit at a given position i is closely packed with its neighbors at positions i±5, i±6, and i±11, resulting in extensive hydrophobic interactions that help to stabilize the viral capsid (8). The side chain of R44 is directed inward, as would be expected for a side chain that interacts with DNA, and the charged and polar residues at the N-terminus face the exterior of the phage particle to confer solubility in water. Inspection of the phage-associated form of the protein (8) indicates that the side chains of K20 in subunit i+11, and of D4 in subunit i could be sufficiently close to interact via a salt bridge (Fig. 5 A).
Figure 5.
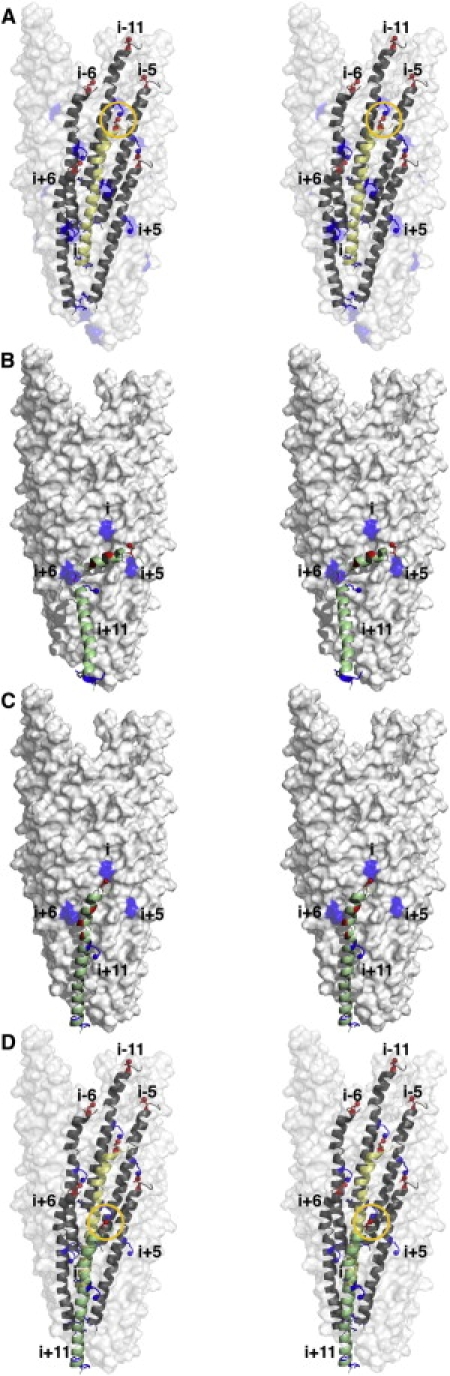
Conformational transition of the Pf1 coat protein from a membrane-bound form to its structural form in bacteriophage particles. Stereo pairs (left, right) are shown for each panel. (A) Molecular surface representation of a 27-subunit repeat of the Pf1 bacteriophage particle showing the interacting subunits (i, i±5, i±6, and i-11). Subunit i is shown in yellow. The yellow circle highlights the proximity of D4 (red) in subunit i, and K20 (blue) in subunit i-11. (B) K20 positive charges (blue) from phage subunits i, i+5, and i+6 are free to interact with a new incoming subunit. These side-chain interactions with the acidic N-terminal region of the membrane-bound form of Pf1 coat protein (green) could guide it to the i+11 subunit position on the growing bacteriophage particle. (C) Additional interactions between the basic C-terminal portion of the coat protein and the bacteriophage DNA, and between neighboring hydrophobic C-terminal helices may trigger the conformational change in the inter-helical loop in the membrane-bound form of the protein and α-helix in the structural form of the protein, upon addition of the i+11 subunit (green) to the growing bacteriophage particle. (D) The newly incorporated i+11 subunit (green) interacts with its neighboring subunits i (yellow), i+5, and i+6.
During phage assembly, as the DNA-protein complex is extruded through the inner bacterial membrane, the unpaired positive charges of K20 on subunits i, i+5, and i+6 could direct the assembly of the next subunit (i+11) and its transformation from the membrane-bound to the phage-bound form by interacting with acidic residues in the N-terminal membrane-associated helix (Fig. 5 B). This electrostatic interaction between membrane-bound coat protein and phage capsid, together with the electrostatic interaction between basic residues in the protein's C-terminus and the viral DNA, could then position the coat protein for further hydrophobic interactions with other capsid-incorporated coat protein subunits, and thus drive its conformational change. The helix-connecting loop itself could then adopt a helical conformation, straightening the coat protein (Fig. 5, B–D).
Conclusions
The Pf1 coat protein is a prime example of a system in which backbone motions are integral to the protein's function—in this case, its ability to assemble into bacteriophage particles from a membrane-inserted state. The dynamics of the backbone in the hinge region of the protein connecting the two helices strongly affects the NMR spectra in various lipid environments. The hinge dynamics exhibited by the amphipathic N-terminal helix moving relative to the transmembrane helix enables the observation of resonances from residues in this helix by solution NMR, interferes with their observation by solid-state NMR in q = 3.2 bicelles, and enables observation of their resonances by solid-state NMR with planar bilayers aligned on glass. This hinge dynamics is also essential for execution of the structural rearrangement that occurs during virus assembly.
Our 3D structures of the Pf1 coat protein determined at the end points of virus assembly (Fig. 2 and Fig. 5) show the conformational changes that occur as the coat protein transitions from the membrane environment to the bacteriophage particle. There are large changes in dynamics and overall orientation for the residues in the N-terminal amphipathic helix as they go from a mobile state, probably flicking on and off the membrane surface, to a more ordered state, oriented nearly parallel to the principal C-terminal hydrophobic helix in the bacteriophage particles. The availability of high-resolution structures with fully defined orientations of both helices for both end points in the membrane and the virus particle provide a better reconstruction of the assembly process compared to our earlier model, which was limited by lack of information about the relative orientations of the helices (6).
Many filamentous bacteriophages have been isolated and characterized. Perhaps the two best studied are fd (class I) and Pf1 (class II). Their coat proteins are small (50 residues of fd and 46 residues of Pf1). We previously determined the 3D structures of both fd and Pf1 in micelles (14–16,53,54), fd in lipid bilayers (23), and the structural forms of both fd and Pf1 in bacteriophage particles (8,9,55). The fd and Pf1 coat proteins are largely α-helical in both the membrane- and bacteriophage-associated forms.
Although the overall structural features are similar, the details of the structure and dynamics of their membrane- and bacteriophage-associated forms are different. Both Pf1 and fd coat proteins have a hinge region connecting the N-terminal amphipathic and C-terminal hydrophobic helices in the membrane. The hinge region of Pf1 coat protein is mobile in both micelles and lipid bilayers (3,14,15,56). In contrast, the hinge of fd coat protein is rigid and structured in lipid bilayers, but exhibits limited mobility in micelles (23,54,57). In Pf1 bacteriophage particles, a few mobile residues are present at the location of the hinge, which disrupts the helix (8). In contrast, the hinge completely disappears in the bacteriophage structure of fd coat protein (55). Of interest, fd coat protein also undergoes a large change in helix rotation from membrane-associated to phage-associated states (23,55), suggesting that the structural and dynamic features observed in the membrane state (e.g., helix tilt, helix rotation, L-shape, and dynamics) are essential for obtaining proper alignment for packing in the filamentous bacteriophages.
The studies presented here are a step toward achieving an atomic-resolution description of the assembly process, and are possible only because of the ability of solid-state NMR to obtain high-resolution spectra and structures from the proteins in their native phospholipid and bacteriophage particle environments. Although additional experiments will be required to provide more details about the assembly process, the structures of the membrane- and phage-associated proteins determined in our studies provide the framework for understanding this transition.
Acknowledgments
This research was supported by grants from the National Institutes of Health. It utilized the NMR facilities at the University of California at San Diego, supported by grants from the National Institutes of Health (P41EB002031 and S10RR23773).
Supporting Material
References
- 1.Caspar D.L., Makowski L. The symmetries of filamentous phage particles. J. Mol. Biol. 1981;145:611–617. doi: 10.1016/0022-2836(81)90549-0. [DOI] [PubMed] [Google Scholar]
- 2.Casjens S., King J. Virus assembly. Annu. Rev. Biochem. 1975;44:555–611. doi: 10.1146/annurev.bi.44.070175.003011. [DOI] [PubMed] [Google Scholar]
- 3.Nambudripad R., Stark W., Makowski L. Membrane-mediated assembly of filamentous bacteriophage Pf1 coat protein. Science. 1991;252:1305–1308. doi: 10.1126/science.1925543. [DOI] [PubMed] [Google Scholar]
- 4.Shon K.J., Kim Y., Opella S.J. NMR studies of the structure and dynamics of membrane-bound bacteriophage Pf1 coat protein. Science. 1991;252:1303–1305. doi: 10.1126/science.1925542. [DOI] [PubMed] [Google Scholar]
- 5.Russel M. Filamentous phage assembly. Mol. Microbiol. 1991;5:1607–1613. doi: 10.1111/j.1365-2958.1991.tb01907.x. [DOI] [PubMed] [Google Scholar]
- 6.Opella S.J., Zeri A.C., Park S.H. Structure, dynamics, and assembly of filamentous bacteriophages by nuclear magnetic resonance spectroscopy. Annu. Rev. Phys. Chem. 2008;59:635–657. doi: 10.1146/annurev.physchem.58.032806.104640. [DOI] [PubMed] [Google Scholar]
- 7.Hemminga M.A., Vos W.L., Stopar D. Viruses: incredible nanomachines. New advances with filamentous phages. Eur. Biophys. J. 2010;39:541–550. doi: 10.1007/s00249-009-0523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiriot D.S., Nevzorov A.A., Opella S.J. Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy. J. Mol. Biol. 2004;341:869–879. doi: 10.1016/j.jmb.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 9.Thiriot D.S., Nevzorov A.A., Opella S.J. Structural basis of the temperature transition of Pf1 bacteriophage. Protein Sci. 2005;14:1064–1070. doi: 10.1110/ps.041220305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas G., Jr., Murphy P. Structure of coat proteins in Pf1 and fd virions by laser Raman spectroscopy. Science. 1975;188:1205–1207. doi: 10.1126/science.1170637. [DOI] [PubMed] [Google Scholar]
- 11.Marvin D.A., Bryan R.K., Nave C. Pf1 Inovirus. Electron density distribution calculated by a maximum entropy algorithm from native fibre diffraction data to 3 A resolution and single isomorphous replacement data to 5 A resolution. J. Mol. Biol. 1987;193:315–343. doi: 10.1016/0022-2836(87)90222-1. [DOI] [PubMed] [Google Scholar]
- 12.Stark W., Glucksman M.J., Makowski L. Conformation of the coat protein of filamentous bacteriophage Pf1 determined by neutron diffraction from magnetically oriented gels of specifically deuterated virions. J. Mol. Biol. 1988;199:171–182. doi: 10.1016/0022-2836(88)90387-7. [DOI] [PubMed] [Google Scholar]
- 13.Goldbourt A., Gross B.J., McDermott A.E. Filamentous phage studied by magic-angle spinning NMR: resonance assignment and secondary structure of the coat protein in Pf1. J. Am. Chem. Soc. 2007;129:2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- 14.Schiksnis R.A., Bogusky M.J., Opella S.J. Structure and dynamics of the Pf1 filamentous bacteriophage coat protein in micelles. Biochemistry. 1987;26:1373–1381. doi: 10.1021/bi00379a025. [DOI] [PubMed] [Google Scholar]
- 15.Lee S., Mesleh M.F., Opella S.J. Structure and dynamics of a membrane protein in micelles from three solution NMR experiments. J. Biomol. NMR. 2003;26:327–334. doi: 10.1023/a:1024047805043. [DOI] [PubMed] [Google Scholar]
- 16.Park S.H., Son W.S., Opella S.J. Phage-induced alignment of membrane proteins enables the measurement and structural analysis of residual dipolar couplings with dipolar waves and λ-maps. J. Am. Chem. Soc. 2009;131:14140–14141. doi: 10.1021/ja905640d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opella S.J., Marassi F.M. Structure determination of membrane proteins by NMR spectroscopy. Chem. Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page R.C., Li C., Cross T.A. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magn. Reson. Chem. 2007;45(S1):S2–S11. doi: 10.1002/mrc.2077. [DOI] [PubMed] [Google Scholar]
- 19.Franzin C.M., Teriete P., Marassi F.M. Structural similarity of a membrane protein in micelles and membranes. J. Am. Chem. Soc. 2007;129:8078–8079. doi: 10.1021/ja0728371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding K., Gronenborn A.M. Sensitivity-enhanced 2D IPAP, TROSY-anti-TROSY, and E.COSY experiments: alternatives for measuring dipolar 15N-1HN couplings. J. Magn. Reson. 2003;163:208–214. doi: 10.1016/s1090-7807(03)00081-8. [DOI] [PubMed] [Google Scholar]
- 21.Bogusky M.J., Leo G.C., Opella S.J. Comparison of the dynamics of the membrane-bound form of fd coat protein in micelles and in bilayers by solution and solid-state nitrogen-15 nuclear magnetic resonance spectroscopy. Proteins. 1988;4:123–130. doi: 10.1002/prot.340040205. [DOI] [PubMed] [Google Scholar]
- 22.Nevzorov A.A., Opella S.J. A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J. Magn. Reson. 2003;164:182–186. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- 23.Marassi F.M., Opella S.J. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 2003;12:403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Angelis A.A., Howell S.C., Opella S.J. Assigning solid-state NMR spectra of aligned proteins using isotropic chemical shifts. J. Magn. Reson. 2006;183:329–332. doi: 10.1016/j.jmr.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Waugh D.S. Genetic tools for selective labeling of proteins with α-15N-amino acids. J. Biomol. NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- 26.Nevzorov A.A., Park S.H., Opella S.J. Three-dimensional experiment for solid-state NMR of aligned protein samples in high field magnets. J. Biomol. NMR. 2007;37:113–116. doi: 10.1007/s10858-006-9121-y. [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.DeLano W.L. PyMOL. 2005 doi: 10.1016/S1359-6446(04)03363-X. www.pymol.org [DOI] [PubMed] [Google Scholar]
- 29.Schwieters C.D., Kuszewski J.J., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 30.Schwieters C.D., Clore G.M. Internal coordinates for molecular dynamics and minimization in structure determination and refinement. J. Magn. Reson. 2001;152:288–302. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 31.Kuszewski J., Gronenborn A.M., Clore G.M. Improvements and extensions in the conformational database potential for the refinement of NMR and X-ray structures of proteins and nucleic acids. J. Magn. Reson. 1997;125:171–177. doi: 10.1006/jmre.1997.1116. [DOI] [PubMed] [Google Scholar]
- 32.Clore G.M., Gronenborn A.M., Tjandra N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- 33.Lipsitz R.S., Tjandra N. 15N chemical shift anisotropy in protein structure refinement and comparison with NH residual dipolar couplings. J. Magn. Reson. 2003;164:171–176. doi: 10.1016/s1090-7807(03)00176-9. [DOI] [PubMed] [Google Scholar]
- 34.Nilges M., Clore G.M., Gronenborn A.M. Determination of three-dimensional structures of proteins from interproton distance data by dynamical simulated annealing from a random array of atoms. Circumventing problems associated with folding. FEBS Lett. 1988;239:129–136. doi: 10.1016/0014-5793(88)80559-3. [DOI] [PubMed] [Google Scholar]
- 35.Chou J.J., Li S., Bax A. Study of conformational rearrangement and refinement of structural homology models by the use of heteronuclear dipolar couplings. J. Biomol. NMR. 2000;18:217–227. doi: 10.1023/a:1026563923774. [DOI] [PubMed] [Google Scholar]
- 36.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 37.Wishart D.S., Sykes B.D. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 38.Mesleh M.F., Veglia G., Opella S.J. Dipolar waves as NMR maps of protein structure. J. Am. Chem. Soc. 2002;124:4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marassi F.M., Opella S.J. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Denny J., Cross T.A. Imaging membrane protein helical wheels. J. Magn. Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay R., Miao X., Valafar H. Efficient and accurate estimation of relative order tensors from λ-maps. J. Magn. Reson. 2009;198:236–247. doi: 10.1016/j.jmr.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders C.R., Hare B., Prestegard J.H. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog. Nucl. Magn. Reson. Spectrosc. 1994;26:421–444. [Google Scholar]
- 43.De Angelis A.A., Jones D.H., Opella S.J. NMR experiments on aligned samples of membrane proteins. Methods Enzymol. 2005;394:350–382. doi: 10.1016/S0076-6879(05)94014-7. [DOI] [PubMed] [Google Scholar]
- 44.Cornilescu G., Bax A. Measurement of proton, nitrogen, and carbonyl chemical shielding anisotropies in a protein dissolved in a dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:10143–10154. [Google Scholar]
- 45.Oas T.G., Hartzell C.J., Drobny G.P. The amide 15N chemical shift tensors of four peptides determined from 13C dipole-coupled chemical shift powder patterns. J. Am. Chem. Soc. 1987;109:5962–5966. [Google Scholar]
- 46.Schiffer M., Chang C.H., Stevens F.J. The functions of tryptophan residues in membrane proteins. Protein Eng. 1992;5:213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- 47.Vold R.R., Prosser R.S., Deese A.J. Isotropic solutions of phospholipid bicelles: a new membrane mimetic for high-resolution NMR studies of polypeptides. J. Biomol. NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 48.Prosser R.S., Evanics F., Al-Abdul-Wahid M.S. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 49.De Angelis A.A., Nevzorov A.A., Opella S.J. High-resolution NMR spectroscopy of membrane proteins in aligned bicelles. J. Am. Chem. Soc. 2004;126:15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 50.Schiksnis R.A., Bogusky M.J., Opella S.J. Secondary structure of filamentous bacteriophage coat protein is preserved in lipid environments. J. Mol. Biol. 1988;200:741–743. doi: 10.1016/0022-2836(88)90485-8. [DOI] [PubMed] [Google Scholar]
- 51.Meijer A.B., Spruijt R.B., Hemminga M.A. Membrane-anchoring interactions of M13 major coat protein. Biochemistry. 2001;40:8815–8820. doi: 10.1021/bi002956s. [DOI] [PubMed] [Google Scholar]
- 52.Meijer A.B., Spruijt R.B., Hemminga M.A. Configurations of the N-terminal amphipathic domain of the membrane-bound M13 major coat protein. Biochemistry. 2001;40:5081–5086. doi: 10.1021/bi002306o. [DOI] [PubMed] [Google Scholar]
- 53.McDonnell P.A., Shon K., Opella S.J. fd coat protein structure in membrane environments. J. Mol. Biol. 1993;233:447–463. doi: 10.1006/jmbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- 54.Almeida F.C., Opella S.J. fd coat protein structure in membrane environments: structural dynamics of the loop between the hydrophobic trans-membrane helix and the amphipathic in-plane helix. J. Mol. Biol. 1997;270:481–495. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- 55.Zeri A.C., Mesleh M.F., Opella S.J. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2003;100:6458–6463. doi: 10.1073/pnas.1132059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park S.H., Loudet C., Opella S.J. Solid-state NMR spectroscopy of a membrane protein in biphenyl phospholipid bicelles with the bilayer normal parallel to the magnetic field. J. Magn. Reson. 2008;193:133–138. doi: 10.1016/j.jmr.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leo G.C., Colnago L.A., Opella S.J. Dynamics of fd coat protein in lipid bilayers. Biochemistry. 1987;26:854–862. doi: 10.1021/bi00377a029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


