Abstract
There is often an interest in knowing, for a given ligand concentration, how many protein molecules have one, two, three, etc. ligands bound in a specific manner. This is a question that cannot be addressed using conventional ensemble techniques. Here, a mathematical method is presented for separating specific from nonspecific binding in nonensemble studies. The method provides a way to determine the distribution of specific binding stoichiometries at any ligand concentration when using nonensemble (e.g., single-molecule) methods. The applicability of the method is demonstrated for ADP binding to creatine kinase using mass spectroscopy data. A major advantage of our method, which can be applied to any protein-ligand system, is that no previous information regarding the mechanism of ligand interaction is required.
Introduction
Ligand-protein interactions are the major driving force for many cellular processes. Determining the equilibrium constants and energetics of such interactions is therefore of fundamental importance. Traditional biochemical methods can provide reliable estimates of affinities, but they cannot reveal distributions of binding stoichiometries, which can be valuable for deciphering reaction mechanisms. However, such distributions can be extracted using the more modern, single-molecule (1) and mass spectroscopy (MS) techniques (2–4). In recent years, MS has proven to be a very valuable tool for characterizing noncovalent interactions (5–13). The power of this approach originates from its high sensitivity, high mass accuracy, low sample requirements and speed of analysis.
Analysis of binding reactions must often take into account the issue of nonspecific binding. Classical biochemical methods can deal with this issue in a fairly straightforward manner (14), but for single-molecule and MS techniques it is a major problem. In the case of MS, nonspecific binding can occur throughout the electrospray process during the desolvation of droplets, leading to an increase in ligand concentration. Consequently, the protein/ligand ratio is altered and the gas-phase measurements no longer reflect the true stoichiometries (15). Several approaches have been suggested for overcoming the problem of nonspecific binding in MS studies (16–21). For example, controlled dissociation of nonspecific gas-phase interactions by blackbody infrared radiation was utilized by Wang et al. (21), but complex dissociation may be induced when the nonspecific binding is strong. Another suggested strategy involves the addition of a reference protein to monitor the appearance of nonspecific complexes (18,19). In this method, there is uncertainty regarding the generality of the approach (20), and the ionization efficiency of the target protein may be compromised, especially in the case of large protein complexes. Finally, van der Rest and co-workers have suggested a mathematical model for distinguishing between specific and nonspecific binding (17), but in their approach, noncooperative binding of the ligand was assumed. Given that nonspecific binding is a problem not only in MS studies, we were motivated to develop a new method for dealing with this issue that is simple, straightforward, and of general applicability.
Theory
Our analysis of specific versus nonspecific binding is general in the sense that it does not assume any particular reaction mechanism and is applicable for studying weak complexes. Two assumptions made are that 1), the number of specific binding sites, Ns, is known, and 2), the nonspecific binding is noncooperative and can be described by a single binding constant. The first step in the analysis involves extracting the value of the nonspecific binding constant from the ratio of the areas of the peaks (referred to as intensities in what follows) corresponding to binding numbers that are larger than Ns. For example, in the case of a dimer with two specific binding sites, we can extract the value of the nonspecific association binding constant, Kn, from the intensities I3 and I4, corresponding to populations with three and four bound ligand molecules, respectively. The ratio I4/I3 for this example is given by
| (1) |
where K1 and K2 are the specific association binding constants, [S] is the free ligand concentration, and [E] is the free protein concentration. The nonspecific binding constant can be evaluated at different [S] values and for different charge states.
Given the value of the nonspecific binding constant from the intensities corresponding to binding numbers that are larger than Ns, we can now determine the values of the specific binding constants from the other peaks in the spectra. The ratio of the intensities I1 and I0, corresponding to populations with one and zero bound ligand molecules, respectively, is given by
| (2) |
Hence, . Likewise, the ratio of the intensities I2 and I0, corresponding to populations with two and zero bound ligand molecules, respectively, is given by
| (3) |
Hence, by combining Eqs. 2 and 3, we get . It can be seen, therefore, that the population of protein species that have ligand bound only to the specific sites can be determined from the appropriate measured intensities and the value of the nonspecific binding constant.
The protein species with ligand bound also to nonspecific sites are considered next. For example, in the case where intensities corresponding to binding numbers from 0 to 3 are observed, the total concentration of species with one ligand bound at a specific site, C1, is equal to:
| (4) |
It can be readily shown that C1/[E] can be expressed as
| (5) |
where the first, second, and third terms in parentheses correspond to K1[S], K1Kn[S]2, and K1Kn2[S]3, respectively. In general, in the case where intensities corresponding to binding numbers from zero to α are observed, one can express C1//[E] as
| (6) |
Likewise, it can be readily shown that one can express C2//[E] as
| (7) |
In general, one can show that CN//[E] is given by:
| (8) |
where N is an integer between zero and the total number of specific binding sites, Ns, (0≤ N ≤ Ns) and α is, as before, the total number of intensity peaks observed (each of which corresponds to a different number of bound ligand molecules, including zero). By definition, IN−1 = 0 when N = 0. It is important to note that, in the above treatment, mixed species with ligand molecules bound to both specific and nonspecific sites are not eliminated but are counted together with other species that have the same number of bound ligands at specific sites. For example, a protein population with, say, one ligand molecule bound to a specific site and two ligand molecules bound to nonspecific sites contributes to I3 but is counted in our above treatment together with the populations of other species that have only one ligand molecule bound to a specific site. In other words, our mathematical treatment makes it possible to correct the detected intensity of each ligand-bound state by 1), subtracting the artificial increase in intensity due to nonspecific binding, and 2), adding the different intensities after subtraction to those that correspond to species with the same number of ligand molecules bound to specific sites (Fig. 1).
Figure 1.
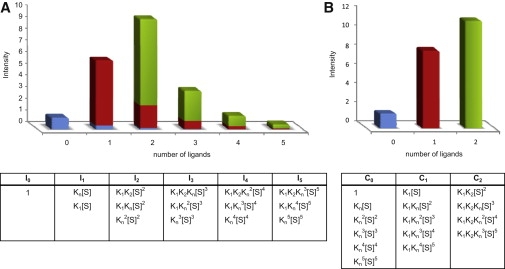
An illustration demonstrating the calculated specific and nonspecific components that contribute to the peak intensities of the 19+ charge state in the case of a solution containing 4 μM CK and 50 μM ADP. (A) Histogram showing the experimentally observed intensities corresponding to the different ligand-bound states. Both specific and nonspecific binding are reflected in each intensity. (B) A histogram generated after correcting for the contribution of nonspecific binding, so that the intensities reflect only specific binding. Listed below each intensity are the factors that contribute to it. The intensities are normalized with respect to I0. The relative contributions of the free protein and protein bound with one or two ligand molecules at the specific sites before (A) and after (B) the correction are indicated in blue, red, and green, respectively.
The final step needed to determine the CN values is to substitute [E] in Eq. 8 with a term that can be calculated. Given that the total protein concentration, CT, is , we can express [E] as , where the terms in the denominator are calculated using Eq. 8. Hence, in general, CN can be calculated from
| (9) |
It can be seen by inspection of Eq. 8 that CN/[E] is related to CN−1/[E], as follows:
| (10) |
where KN is the binding constant of the ligand to the Nth specific site. Hence, the value of KN can be obtained from the slope of a plot of (CN/[E])/(CN−1/[E]) versus [S] for different substrate concentrations by assuming that [S] ≈ [S]total. This assumption is not valid in the case of tight binding. It is important to note, however, that it is not required for determining the distribution of binding stoichiometries.
Materials and Methods
Creatine kinase (CK) from rabbit muscle and adenosine 5′-diphosphate sodium salt (ADP) were purchased from Sigma (St. Louis, MO). CK was further purified on a gel filtration column in 20 mM Tris-HCl (pH 7.5). Before MS analysis, the protein was buffer-exchanged into 250 mM ammonium acetate (pH 7.5) using microbiospin 6 columns (BioRad, Hercules, CA). Titration experiments were performed in the presence of 4 μM CK, 1 mM magnesium acetate and ADP concentrations ranging between 0 and 100 μM. CK/ADP complexes were incubated for 5 min at room temperature before MS analysis.
MS measurements were performed in positive ion mode using a nanoelectrospray ionization quadrupole time-of-flight (ESI-Q-TOF) instrument (Applied Biosystems, Foster City, CA) modified for high mass detection (22,23). Aliquots of 2 μl were electrosprayed from gold-coated borosilicate capillaries prepared in-house as described previously (24). The mass spectrometer was operated at a capillary voltage of 1100 V, declustering potential of 140 V, and second-declustering potential of 15 V. External calibration of the mass spectra was achieved using solutions of 100 mg/ml cesium iodide in water. For each mass spectrum, the peak areas for the free CK and its ligand-bound states were calculated using deconvolution software (peakfit v4, Jandel Scientific, San Rafael, CA).
Results and Discussion
To test our methodology, we applied it to CK homodimer (25) and its ligand ADP. Nanoelectrospray (nanoES) mass spectra were acquired for 4 μM of CK and increasing concentrations of ADP ranging from 0 to 100 μM. Fig. 2 shows the mass spectra of CK acquired at different concentrations of ADP. The CK charge-state distribution is centered at 4550 m/z and the measured mass of the free CK, 85,990 ± 34 Da, is found to be highly consistent with the calculated mass of the homodimer complex (85,960 Da). When titrated with ADP, no change in charge state was obtained, but additional peaks corresponding to the various ligand-bound states appeared (Fig. 2). Due to the relatively small difference in mass between apo and ADP-bound CK (<1%), the binding of the nucleotide does not appear to alter the ionization efficiency, as observed by others (10,17,26).
Figure 2.
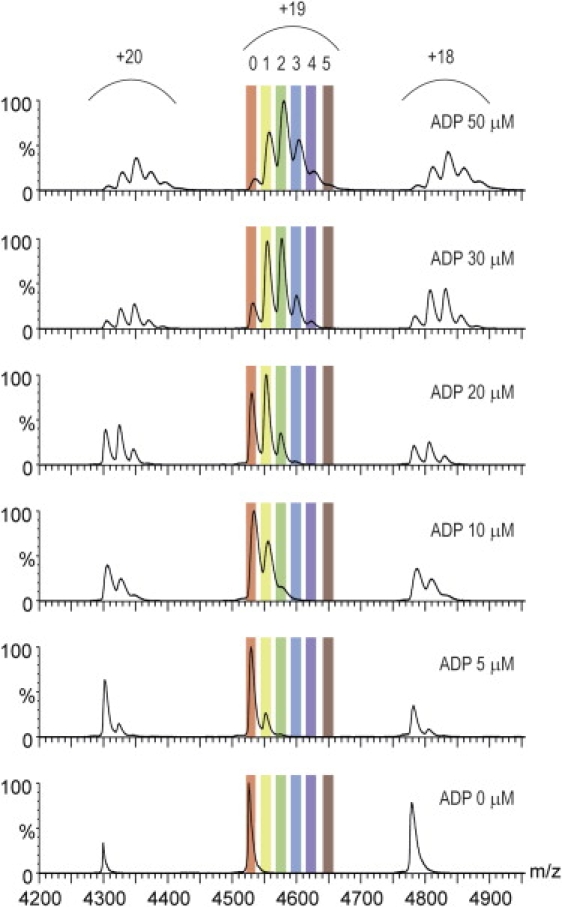
Nanoelectrospray mass spectra of CK obtained at increasing concentrations of ADP. The number of bound ADP molecules is highlighted for the 19+ charge state. The spectra were acquired in the presence of 4 μM CK in 250 ammonium acetate (pH 7).
For each mass spectrum, the peak areas for free CK and its ligand-bound states were calculated using a deconvolution algorithm (Jandel Scientific peakfit v4) (Table S1 in the Supporting Material). In all calculations, it was assumed that the measured peak areas (referred to as In) correlate with the solution concentrations (6,9–13). Initially, we determined the value of Kn by calculating the average value of the I4/I3 and I5/I4 ratios (Table S2) as in Eq. 1. It is important to note that the values of Kn determined from the I4/I3 and I5/I4 ratios are very similar, thus supporting our assumption that the nonspecific binding is not cooperative. Using an average value of 0.007 ± 0.003 μM−1 for Kn and the measured intensities, we next corrected the distribution of binding stoichiometries using Eq. 8 (Table S3). The absolute values of C0, C1, and C2 were then calculated using Eq. 9 (Table 1 and Table S4). Finally, utilizing Eq. 10, we determined the values of the binding (association) constants K1 and K2 to be 0.08 ± 0.002 and 0.02 ± 0.001 μM−1, respectively (Fig. 3). These values correspond to dissociation constants of 12.6 ± 0.3 and 47 ± 2 μM, respectively, which are in very good agreement with previously reported values (17,27–31). In particular, they are in excellent agreement with the respective values of 11.8 ± 1.5 and 48 ± 6 μM for K1 and K2 determined more recently (17). The finding that K1 < K2 is in agreement with the previously reported negative cooperativity of ADP binding to CK (31).
Table 1.
Corrected concentrations of apo CK (C0), and CK with one (C1) and two (C2) specifically bound ADP ligands
| ADP concentration | C0 | C1 | C2 |
|---|---|---|---|
| 0 | 4.00 | 0.00 | 0.00 |
| 5 | 3.28 | 0.72 | 0.00 |
| 10 | 2.31 | 1.38 | 0.31 |
| 15 | 2.28 | 1.51 | 0.21 |
| 20 | 1.71 | 1.81 | 0.48 |
| 30 | 1.09 | 1.89 | 1.02 |
| 40 | 0.87 | 1.71 | 1.42 |
| 50 | 0.29 | 1.50 | 2.22 |
| 100 | 0.13 | 1.41 | 2.46 |
Corrected concentrations were obtained from an average of three charge states in a typical experiment. All values are in μM.
Figure 3.
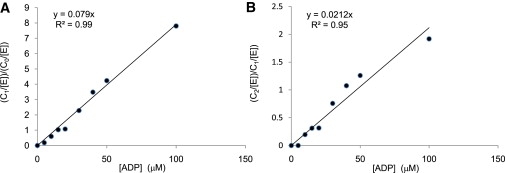
Determination of the values of K1 (A) and K2 (B) by plotting (CN/[E])/(CN−1/(E])) versus [S] and fitting the data using Eq. 10, where [S] is the ADP concentration. Each data point is calculated from the average of the three observed charge states (18+, 19+, and 20+) in four independent experiments.
In summary, the method presented in this article makes it possible to separate specific from nonspecific binding when using nonensemble methods, so that the distribution of specific binding stoichiometries at any ligand concentration can be determined. The method can be applied to data for any ligand-protein system obtained using an experimental approach that can distinguish between populations with different numbers of bound ligand molecules. Major advantages of our method are that no previous information regarding the mechanism of ligand interaction is required and determination of the values of the binding constants is straightforward.
Acknowledgments
This work was supported by grant 153/08 of the Israel Science Foundation (to A.H.) and by funding from the European Research Council (ERC) under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement No. 239679 (to M.S.).
Contributor Information
Michal Sharon, Email: michal.sharon@weizmann.ac.il.
Amnon Horovitz, Email: amnon.horovitz@weizmann.ac.il.
Supporting Material
References
- 1.Jiang Y., Wang Q., Moerner W.E. Hardware-based anti-Brownian electrokinetic trap (ABEL trap) for single molecules: control loop simulations and application to ATP binding stoichiometry in multi-subunit enzymes. Proc. Soc. Photo Opt. Instrum. Eng. 2008;7038:1–12. doi: 10.1117/12.798093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benesch J.L., Ruotolo B.T., Robinson C.V. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 3.Sharon M., Robinson C.V. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 4.van den Heuvel R.H. H., Heck A.J.R. Native protein mass spectrometry: from intact oligomers to functional machineries. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Daneshfar R., Kitova E.N., Klassen J.S. Determination of protein-ligand association thermochemistry using variable-temperature nanoelectrospray mass spectrometry. J. Am. Chem. Soc. 2004;126:4786–4787. doi: 10.1021/ja0316972. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen T.J.D., Roepstorff P., Heck A.J.R. Direct determination of solution binding constants for noncovalent complexes between bacterial cell wall peptide analogues and vancomycin group antibiotics by electrospray ionization mass spectrometry. Anal. Chem. 1998;70:4427–4432. [Google Scholar]
- 7.Li Z., Song F., Anderson K.S. Monitoring enzyme catalysis in the multimeric state: direct observation of Arthrobacter 4-hydroxybenzoyl-coenzyme A thioesterase catalytic complexes using time-resolved electrospray ionization mass spectrometry. Anal. Biochem. 2009;394:209–216. doi: 10.1016/j.ab.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCammon M.G., Robinson C.V. Structural change in response to ligand binding. Curr. Opin. Chem. Biol. 2004;8:60–65. doi: 10.1016/j.cbpa.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Peschke M., Verkerk U.H., Kebarle P. Features of the ESI mechanism that affect the observation of multiply charged noncovalent protein complexes and the determination of the association constant by the titration method. J. Am. Soc. Mass Spectrom. 2004;15:1424–1434. doi: 10.1016/j.jasms.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Sannes-Lowery K.A., Griffey R.H., Hofstadler S.A. Measuring dissociation constants of RNA and aminoglycoside antibiotics by electrospray ionization mass spectrometry. Anal. Biochem. 2000;280:264–271. doi: 10.1006/abio.2000.4550. [DOI] [PubMed] [Google Scholar]
- 11.Wortmann A., Jecklin M.C., Zenobi R. Binding constant determination of high-affinity protein-ligand complexes by electrospray ionization mass spectrometry and ligand competition. J. Mass Spectrom. 2008;43:600–608. doi: 10.1002/jms.1355. [DOI] [PubMed] [Google Scholar]
- 12.Wortmann A., Rossi F., Zenobi R. Determination of zinc to β-peptide binding constants with electrospray ionization mass spectrometry. J. Mass Spectrom. 2005;40:777–784. doi: 10.1002/jms.852. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y., Kirkup C.E., Leary J.A. Characterization of noncovalent protein-ligand complexes and associated enzyme intermediates of GlcNAc-6-O-sulfotransferase by electrospray ionization FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1400–1407. doi: 10.1016/j.jasms.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 14.van Zoelen E.J.J., Kramer R.H., Veerkamp J.H. The use of nonhomologous Scatchard analysis in the evaluation of ligand-protein interactions. Trends Pharmacol. Sci. 1998;19:487–490. doi: 10.1016/s0165-6147(98)01250-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang W., Kitova E.N., Klassen J.S. Influence of solution and gas phase processes on protein-carbohydrate binding affinities determined by nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2003;75:4945–4955. doi: 10.1021/ac034300l. [DOI] [PubMed] [Google Scholar]
- 16.Ayed A., Krutchinsky A.N., Duckworth H.W. Quantitative evaluation of protein-protein and ligand-protein equilibria of a large allosteric enzyme by electrospray ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1998;12:339–344. doi: 10.1002/(SICI)1097-0231(19980415)12:7<339::AID-RCM163>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Daubenfeld T., Bouin A.P., van der Rest G. A deconvolution method for the separation of specific versus nonspecific interactions in noncovalent protein-ligand complexes analyzed by ESI-FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:1239–1248. doi: 10.1016/j.jasms.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Sun J., Kitova E.N., Klassen J.S. Method for stabilizing protein-ligand complexes in nanoelectrospray ionization mass spectrometry. Anal. Chem. 2007;79:416–425. doi: 10.1021/ac061109d. [DOI] [PubMed] [Google Scholar]
- 19.Sun N., Soya N., Klassen J.S. Nonspecific interactions between proteins and charged biomolecules in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2010;21:472–481. doi: 10.1016/j.jasms.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Touboul D., Maillard L., Zenobi R. How to deal with weak interactions in noncovalent complexes analyzed by electrospray mass spectrometry: cyclopeptidic inhibitors of the nuclear receptor coactivator 1-STAT6. J. Am. Soc. Mass Spectrom. 2009;20:303–311. doi: 10.1016/j.jasms.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Wang W., Kitova E.N., Klassen J.S. Blackbody infrared radiative dissociation of nonspecific protein-carbohydrate complexes produced by nanoelectrospray ionization: the nature of the noncovalent interactions. J. Am. Soc. Mass Spectrom. 2005;16:1583–1594. doi: 10.1016/j.jasms.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Chernushevich I.V., Thomson B.A. Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]
- 23.Sobott F., Hernández H., Robinson C.V. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 24.Hernández H., Robinson C.V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 25.Rao J.K.M., Bujacz G., Wlodawer A. Crystal structure of rabbit muscle creatine kinase. FEBS Lett. 1998;439:133–137. doi: 10.1016/s0014-5793(98)01355-6. [DOI] [PubMed] [Google Scholar]
- 26.Loo J.A. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 27.Borders C.L., Jr., Snider M.J., Edmiston P.L. Determination of the affinity of each component of a composite quaternary transition-state analogue complex of creatine kinase. Biochemistry. 2002;41:6995–7000. doi: 10.1021/bi020105+. [DOI] [PubMed] [Google Scholar]
- 28.Burbaum J.J., Knowles J.R. Internal thermodynamics of enzymes determined by equilibrium quench: values of Kint for enolase and creatine kinase. Biochemistry. 1989;28:9306–9317. doi: 10.1021/bi00450a010. [DOI] [PubMed] [Google Scholar]
- 29.Hornemann T., Rutishauser D., Wallimann T. Why is creatine kinase a dimer? Evidence for cooperativity between the two subunits. Biochim. Biophys. Acta. 2000;1480:365–373. doi: 10.1016/s0167-4838(00)00098-4. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin A.C. The interaction of 8-anilino-1-naphthalenesulfonate with creatine kinase. Evidence for cooperativitiy of nucleotide binding. J. Biol. Chem. 1974;249:1445–1452. [PubMed] [Google Scholar]
- 31.Price N.C., Hunter M.G. Non-identical behaviour of the subunits of rabbit muscule creatine kinase. Biochim. Biophys. Acta. 1976;445:364–376. doi: 10.1016/0005-2744(76)90090-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


