Abstract
The sculpting of membranes into dynamic, curved shapes is central to intracellular cargo trafficking. Though the generation of membrane curvature during trafficking necessarily involves both lipids and membrane-associated proteins, current mechanistic views focus primarily on the formation of rigid cages and curved scaffolds by protein assemblies. Here we report on a different mechanism for the control of membrane deformation, unrelated to the imposition of predefined curvature, involving modulation of membrane material properties: Sar1, a GTPase that regulates vesicle trafficking from the endoplasmic reticulum, lowers the rigidity of the lipid bilayer membrane to which it binds. In vitro assays in which optically trapped microspheres create controlled membrane deformations revealed a monotonic decline in bending modulus as a function of Sar1 concentration, down to nearly zero rigidity, indicating a dramatic lowering of the energetic cost of curvature generation. This is the first demonstration that a vesicle trafficking protein lowers the rigidity of its target membrane, leading to a new conceptual framework for vesicle biogenesis.
Introduction
Cellular membranes are quasi-two-dimensional materials that bend and curve in three-dimensional space. Control of membrane curvature is crucial in many cellular contexts, such as lamellipodial and filopodial extensions at the edges of motile cells, cleavage furrow development during cell division, and the construction of transport vesicles that traffic cargo between membranous organelles (1–3). For many of these systems, the proteins associated with curvature have been identified and a large and growing body of knowledge describes their structures and biochemical interactions. Much less is known, however, about the mechanics of curvature generation—how specific proteins modulate forces and energies to sculpt intracellular membranes into dynamic, curved forms. Existing mechanistic views of transport vesicle formation have focused largely on the creation of scaffolds by rigid macromolecular assemblies, motivated by proteins such as clathrin (4) and Sec13 and Sec31 (5) that form the edges of polyhedral cages, and by stiff BAR domains (6,7) that bend membranes to conform to their crescentlike shapes. The possibility of protein-mediated alteration of underlying geometry-independent membrane material properties such as rigidity, however, has been neglected, although lipid membrane mechanics are a key determinant of the energetics of curvature generation and can couple to protein structures in a wide variety of ways (1–3,8–10). We report here that Sar1, a GTPase that regulates the assembly of coat protein II (COPII) transport vesicles, lowers the mechanical rigidity of the lipid bilayer membrane to which it binds. This is the first discovery of membrane softening for any trafficking protein.
The well-studied COPII complex mediates trafficking from the endoplasmic reticulum (ER) (11–13). Of the five COPII coat proteins, only Sar1, a 21.5 kDa G-protein of the Ras superfamily, binds directly to the ER lipid bilayer membrane. Sar1's 23 N-terminal amino acids form an amphipathic α-helix that is exposed when the protein is GTP-bound, allowing the hydrophobic hemicylinder to insert into the lipid bilayer (14–16). Sar1-GDP binds membranes with lower affinity (14). The recruitment of Sar1-GTP to the ER is the crucial first step in vesicle formation, after which the remaining coat proteins, the two heterodimer pairs Sec23/Sec24 and Sec13/Sec31, assemble. Sec23/Sec24 recruits cargo and accelerates GTP hydrolysis by Sar1. Sec13/Sec31 plays a scaffolding role, as the dimer is capable in itself of cage formation in vitro (5).
Sar1 with bound GTP or nonhydrolyzable GTP analogs has been shown to generate curvature and tubulation in vitro from liposomes with compositions mimicking that of the ER, visualized with electron microscopy (15,16). This tubulation was found to require the N-terminal helix, being absent in truncated Sar1 that bound to membranes via engineered interactions with lipid headgroups (15,16). Curvature generation was interpreted in the context of the bilayer couple mechanism (17): helix insertion increases the area of one bilayer leaflet relative to the other; the area differential forces curvature. This process should be especially pronounced in systems such as liposomes, where the constraint of a small bounded volume means that a small asymmetry in leaflet areas leads to significant deviations from a spherical shape. In an amorphous, large-area structure such as the ER, it is more difficult to envision mechanisms by which a global differential leaflet area caused by Sar1 insertion enables the local curvature generation required for trafficking, especially as Sar1 has no known tendencies to cluster or oligomerize. While Sar1 may employ the bilayer couple mechanism, explorations of other mechanisms, which need not be mutually exclusive, are warranted.
Materials and Methods
Lipid composition
Lipid membranes were composed of the “Major Mix” mixture as in Matsuoka et al. (14), modified to include fluorescent probes and biotinylated lipids. See additional Methods in the Supporting Material for details.
Sar1 expression
Proteins were expressed and purified using protocols slightly modified from Lee et al. (15) and Barlowe et al. (18). See additional Methods in the Supporting Material for details.
Sample preparation
Dried lipid films were hydrated with 0.2–0.5 ml HKM buffer (20 mM HEPES-KOH, pH 6.8, 160 mM potassium acetate, 1 mM MgCl2), yielding multilayered membrane stacks. Sar1p or Δ23-Sar1, 100 μM GMPPNP (Sigma-Aldrich, St. Louis, MO) and 4 mM EDTA were incubated together in an Eppendorf tube for 5 min, after which they were added to the chamber containing membranes and buffer. Nucleotide uptake by Sar1p was verified by tryptophan fluorescence experiments (19,20). After ∼5 min, a few microliters of a suspension of 4.8-μm-diameter streptavidin-coated silica microspheres (Bangs Laboratories, Fishers, IN) were added to the chamber. See additional Methods in the Supporting Material for details.
Optical trapping and particle tracking
Microspheres were trapped with a home-built optical trap setup using a 671-nm, 120-mW diode laser (model No. RS71-100PS; Meshtel, AKA Intelite, Genoa, NV). Microsphere images were captured with bright-field microscopy using a Model No. pco.1200 camera (Cooke, Romulus, MI) at 100 frames per s. Particle positions were determined using home-built tracking software that employs well-established algorithms (21,22) with ∼10 nm precision. See additional Methods in the Supporting Material for details.
Drag coefficient
The drag coefficient, b, is determined independently for each tethered microsphere following the general approach described in Sainis et al. (23). See additional Methods in the Supporting Material for details.
Tether images and radii
Fluorescence images of membrane tethers were captured with an ORCA-ER charge-coupled device camera (Hamamatsu, Hamamatsu City, Japan). See additional Methods in the Supporting Material for details.
Electrostatic calculations
Electrostatic potential maps for Sar1 lacking the terminal helix were calculated using PBEQ-Solver (24) applied to the structure from Bi et al. (25). (PDB ID 1M2O) and visualized with PyMol (http://www.pymol.org/).
Experimental Design
We hypothesized that Sar1's helix insertion alters the material character of the membrane, specifically its bending modulus (or rigidity, κ), which determines the energetic cost of curvature (1–3). We measured rigidity and its modulation by Sar1 using a membrane tether-pulling assay that recapitulates relevant aspects of ER morphology in an in vitro setting. In both cell-free and live-cell contexts, membrane deformation by an external unidirectional force can draw out cylindrical membrane tethers (26–30) (Fig. 1, A and B). Tether mechanics are well understood (26–32): the mechanical energy associated with the one-dimensional curvature of a tether of radius R and length L is proportional to the tether area and hence to the product RL, giving rise to a length-independent contractile force, f. Measurement of both f and R reveals the membrane rigidity; a derivation is provided as Supporting Material. In brief: the bending energy per unit area of a membrane, ɛ, is a function of the local principle radii of curvature (R1 and R2), the bending modulus, the Gaussian curvature modulus (κG), and the spontaneous curvature (c0) (33,34):
The spontaneous curvature, at which the bracketed term is minimal, gives the geometry of the lowest energy configuration of the unperturbed membrane. For lipid bilayers with identical leaflet composition, c0 = 0 (2,3). Membrane inclusions, such as inserted proteins, can change the membrane rigidity directly by altering κ (e.g., by altering lipid packing or thinning the membrane) (10,27,28,35,36), or indirectly by coupling to the local curvature and reducing the effective rigidity with respect to deformations (9). The curvature-mediated interaction can be accounted for by an additional energetic term proportional to the local curvature c = R1−1 + R2−1, the inclusion density ϕ, and a coupling constant, Λ. (Additional terms describing self-interactions among inclusions are described in the Supporting Material.) For a membrane tether of radius R,
and the total energy, E, is the sum of contributions from the bending energy, the inclusion-curvature coupling, the elastic energy given by the product of the tether area and the membrane tension, σ, and the mechanical work given by the product of f and L. Minimizing E, it follows that
| (1) |
The functional relationship between the measurable force and radius values reveals κ and Λϕ. In the absence of coupling between the inclusions and the local curvature (i.e., Λ = 0), fR = 2πκ; we will denote the rigidity in this case, in which the role ascribed to the proteins is to directly alter membrane rigidity, as
The tether energy function can also be analyzed to provide σΛ=0, a measure of the sum of the membrane tension and the (indistinguishable) interaction energy of the proteins; details are provided in the derivation provided as Supporting Material.
Figure 1.
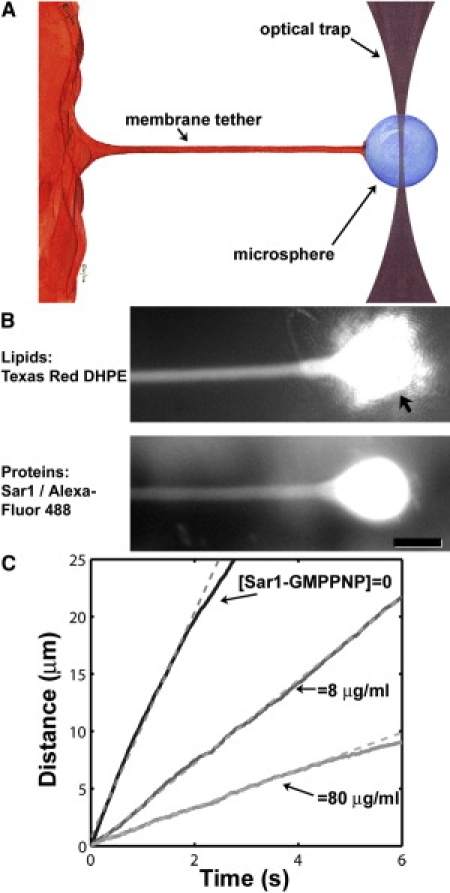
(A) Schematic: Pulling a membrane-anchored microsphere with an optical trap generates a cylindrical membrane tether. Upon release of the microsphere, the tether retracts to lower its curvature-associated mechanical energy. (B) A membrane tether, pulled by an optically trapped bead. (Top) Fluorescence image of Texas Red DHPE labeling the lipid membrane. (Arrow) The bead and the trapping laser light. (Bottom) Fluorescence image of Alexa Fluor 488-labeled Sar1 from the same tether. (Bar) 10 μm. (C) Tether retraction. The bead position after its release from the optical trap is plotted for three representative tethers of similar radii (R = 0.45–0.55 μm) and microsphere drag coefficients (b = 0.07–0.11 μN s/m), in the presence or absence of Sar1-GMPPNP. (Dashed lines) Linear fits to the initial trajectory.
We created, deformed, and mechanically characterized lipid membranes incubated with varying concentrations of Sar1 from the yeast S. cerevisiae (Sar1p). Details of compositions and procedures are given in Materials and Methods. In brief: Lipid films of the desired compositions were dried on glass coverslips and hydrated in HKM buffer. The resulting multilayer lipid bilayer sheets had a large surface area (∼106 μm2), like the ER and unlike liposomes. Again, in contrast to liposomes, the lack of strong geometric constraints on area enables low tension in our system (37). As in earlier liposome-based studies (14,15), we used lipid compositions rich in phosphoethanolamine and phosphatidylinositol lipids. A small fraction of fluorescent-labeled lipids (0.5 mol % Texas Red DHPE) allowed membrane visualization by fluorescence microscopy. Lipids with biotin-conjugated headgroups (1 mol % biotin-cap-DPPE) allowed binding of 5-μm-diameter streptavidin-coated polystyrene beads that served as handles for tether pulling. Membranes were incubated with Sar1p, EDTA, and the nonhydrolyzable GTP analog GMP-PNP (14,15).
After incubation with streptavidin-coated beads, individual microspheres were manipulated manually using a home-built optical trap to pull a membrane tether tens or hundreds of microns in length. The tether is pulled parallel to the substrate, beyond the edge of the hydrated membrane sheets, and is therefore clearly visible against the lipid-free coverslip background. The trap was then turned off, leading to immediate retraction of the tether, dragging the bead with it; retraction lowers the area of curved membrane and the associated bending energy. The beads were imaged and tracked via high-speed video microscopy, yielding the tether speed, v. The tether force, f, equilibrates with the drag force on the bead, bv, where b is the drag coefficient. The drag coefficient was experimentally determined for each tether by analyzing the probability distribution of position fluctuations of the bead (23), described further in Materials and Methods. This approach provides b independent of the nature of dissipation in the system, which may be a combination of hydrodynamic drag of the bead through the surrounding fluid or viscosity within the lipid bilayer (38). The tethered bead trajectory reveals the tether force (f = bv). Notably, this approach is intrinsically insensitive to properties of the optical trap, being applied when the trap is off and relying only on fundamental statistical mechanical behaviors to quantify forces (23). Tether radii were measured from fluorescence images.
Results
Upon the release of the bead from the optical trap, the tethers exhibited a constant-velocity regime of bead retraction, indicating a constant, length-independent tether force as expected. Fluorescence images of a tether are shown in Fig. 1 B. The mean R was 0.6 μm with a standard deviation (SD) of 0.2 μm for >300 tethers examined (see Methods in the Supporting Material for details of the radius determination). Three representative trajectories from tethers with similar radii are plotted in Fig. 1 C, with 0, 8, and 80 μg/mL Sar1-GMPPNP, the final value being similar to the concentration employed in liposome deformation assays (15). At long times after release, the microsphere velocities often slowed; one would expect the bead trajectory to be nontrivially related to the flexible tether's Brownian dynamics. We therefore focus on the initial constant velocity regime, in which the trajectory and the tether mechanics are simply related as described above. For the hundreds of tethers examined, the mean drag coefficient was b = 0.085 μN s/m, with an SD of 0.032 μN s/m—approximately twice the Stokes drag of a 4.8-μm-diameter bead in an infinite three-dimensional aqueous space. The drag coefficient shows a slight rise as a function of the solution concentration of Sar1p ([Sar1p]), which may indicate that the protein increases the interleaflet viscosity (Fig. S1) (38).
We find that Sar1p-GMPPNP lowers membrane rigidity in a concentration-dependent manner. In Fig. 2 A we plot κΛ=0 as a function of the solution concentration of Sar1p ([Sar1p]), which shows a pronounced decline at both physiological and low ionic strengths (1× and 0.05× HKM, 180 and 9 mM, respectively). The values of κΛ=0, also provided as Table S1, are determined by averaging fR(2π)−1 over all tethers pulled at a given protein and buffer concentration, regardless of tether radius. This is by construction: κΛ=0 is precisely the rigidity if there is no curvature coupling, and hence if fR(2π)−1 is independent of R. The more general case will be considered below. We note here, however, that partitioning the tethers into subsets with radii less than and greater than the median R results in similar values for κΛ=0 for each subset, suggesting that any curvature coupling is small (Fig. S2 provided in the Supporting Material).
Figure 2.
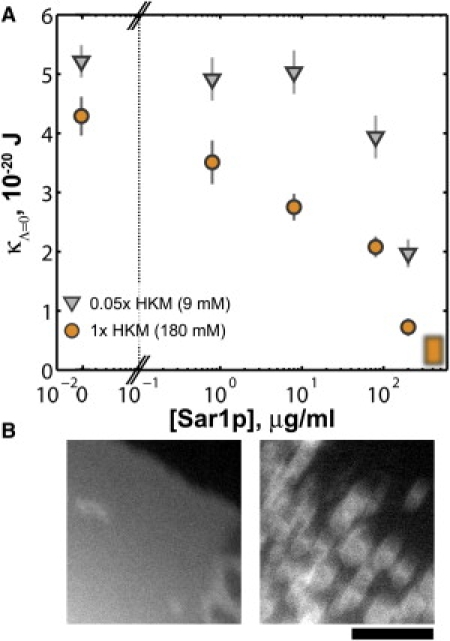
(A) Membrane rigidity, κΛ=0, as a function of Sar1p-GMPPNP concentration. The blurred box is drawn at a concentration at which spontaneous membrane vesiculation is observed, indicating near-zero rigidity. (Circles and triangles) Data from experiments at 1× (180 mM) and 0.05× (9 mM) HKM buffer, respectively. (Error bars) Standard errors of the means. (B) Spontaneous membrane disintegration. Fluorescence images of a region of membrane before (left) and after (right) the addition of Sar1p-GMPPNP at 400 μg/mL. (Bar) 10 μm. See also Movie S1, provided as Supporting Material.
At very high [Sar1p], ≈400 μg/mL, we observed, in the absence of any optical manipulation, spontaneous disintegration of the membranes into liposomes or small tubules (Fig. 2 B; Movie S1). This indicates that the curvature energy was smaller than the ambient thermal energy, implying that, roughly, κ < kBT = 0.4 × 10−20 J, where kB is Boltzmann's constant and T is the absolute temperature. Tethers pulled in the absence of Sar1p, but with GMPPNP present at the same concentration as above, showed κΛ=0 = (5.5 ± 0.4) × 10−20 J (N = 49, 0.05× HKM), equal within uncertainties to the value for lipid-only membranes, κΛ=0 = (5.2 ± 0.3) × 10−20 J (N = 34, 0.05× HKM), indicating that nucleotide is not responsible for the membrane softening indicated in Fig. 2 A. The mean concentration of Sar1p in S. cerevisiae is roughly 5 μg/mL (39), and so is spanned by the range of in vitro concentrations examined.
The reduction in κΛ=0 induced by Sar1p could, in principle, be due to a reduction in the true membrane rigidity, κ, or a positive value of Λ, i.e., an energetic preference for convex local curvature. Force measurements from tethers spanning a range of radii allow separation of κ and Λϕ (Eq. 1). We first apply this analysis to Sar1p lacking the N-terminal helix, denoted Δ23-Sar1p. As in Lee et al. (15), the helix is replaced by a hexahistidine tag, enabling strong binding to membranes that include a small fraction (5 mol %) of nickel-chelating lipids. (At solution concentrations of 80 μg/mL, we find that Δ23-Sar1p binds membranes with a surface density ∼2× greater than wild-type Sar1p.) With Δ23-Sar1p, the product of f and R shows a linear dependence on R with a strongly positive slope (Fig. 3 A), 80 μg/mL Δ23-Sar1p-GMPPNP, 0.05× HKM. This indicates a negative (concave) curvature coupling on the part of the protein, consistent with its inability to generate buds in liposome deformation experiments (15). With 400 μg/mL Δ23-Sar1p, we observe no spontaneous vesiculation of the membrane, in contrast to wild-type Sar1p, implying an energetic barrier to (convex) vesicle formation by the helixless protein. Neither the Ni-chelating lipids nor the hexahistidine tag in itself alters the membrane rigidity or induces significant curvature coupling: for protein-free membranes with Ni-chelating lipids, κΛ=0 = (5.1 ± 0.3) × 10−20 J and Λϕ = −0.3 ± 2.6 J/μm (N = 30, 0.05× HKM). For such membranes incubated with a hexahistidine peptide at molar concentrations equal to that of 80 μg/mL Δ23-Sar1p, κΛ=0 = (4.5 ± 0.4) × 10−20 J and Λϕ = −1.7 ± 1.8 J/μm (N = 17, 0.05× HKM). In contrast, Δ23-Sar1p gives Λϕ = −9.9 ± 1.1 J/μm.
Figure 3.
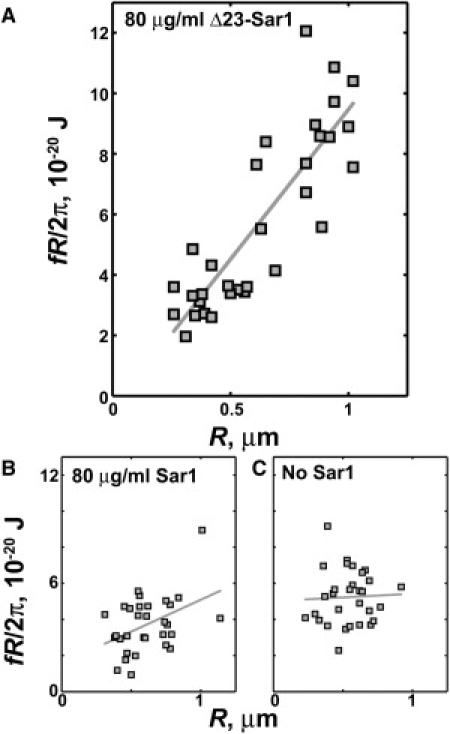
(A) The product of the tether retraction force (f) and radius (R) plotted as a function of R, for 80 μg/mL Δ23-Sar1-GMPPNP, which lacks the amphipathic α-helix of wild-type Sar1. A positive slope indicates a negative coupling between the protein and the local curvature, or equivalently a concave effective spontaneous curvature. (B) fR versus R for 80 μg/mL Sar1p-GMPPNP, and (C) fR versus R for membranes with no Sar1, with the same plot axes as panel A. The data plotted are from experiments in 0.05× HKM buffer. (Solid lines) Linear fits.
Tethers formed with GMPPNP-bound wild-type Sar1p exhibit a much weaker dependence of fR on R (Fig. 3 B, 80 μg/mL Sar1p-GMPPNP, 0.05× HKM), and hence demonstrate little curvature coupling. This disproves the hypothesis that Sar1p imposes particular curvatures on the membranes to which it binds (15). The magnitude of Λϕ is so small as to be barely resolvable, and shows a weak progression to increasingly negative values as a function of Sar1p concentration (Fig. S3). The curvature coupling can be considered as an effective spontaneous curvature for the protein-bound membrane ceff = Λϕ/(2κ) (9) (derived in Supporting Material). Throughout the range of [Sar1p] examined, the values of ceff are negative and satisfy |ceff| < 0.5 μm−1. This corresponds to a preferred radius of curvature R0 = c0−1 > 2 μm, much larger than any of the characteristic dimensions of transport vesicles and hence unlikely to determine vesicle morphology. It follows from Λ (or ceff) being negative that the true rigidity, κ, is smaller than κΛ=0 and the steep decline in κΛ=0 observed as a function of [Sar1p] (Fig. 2) reflects a reduction in κ. Moreover, the dependence of fR on R allows direct determination of κ (Eq. 1). While subject to large uncertainties, κ shows the same sharp decline with [Sar1p] as does κΛ=0 (Fig. 4). The membrane softening is similar at ionic strengths of 180 and 9 mM, corresponding to a factor-of-5 difference in Debye screening length.
Figure 4.
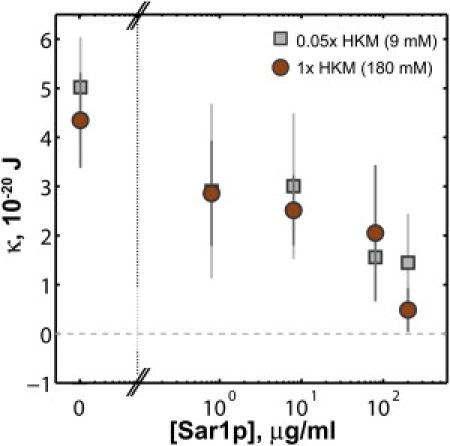
Membrane rigidity, κ, as a function of Sar1p-GMPPNP concentration. The values and uncertainties are determined from a linear fit of fR versus R (Eq. 1).
A 23-amino-acid peptide corresponding to N-terminal Sar1p helix domain appears in itself capable of lowering membrane rigidity. The peptide requires 5% dimethylsulfoxide (DMSO) added to the buffer for solubility; the DMSO alone lowers κΛ=0 by ∼40%. Relative to the rigidity with DMSO and without the peptide, κΛ=0 is reduced by a factor of 0.61 ± 0.15 at a peptide concentration of 0.2 μM in 0.05× HKM buffer (Fig. S4) and induces membrane disintegration at concentrations >≈5 μM. For comparison, full-length Sar1p lowers κΛ=0 by ∼0.6× at a concentration at ∼5 μM (100 μg/mL) (Fig. 2 A). We caution against drawing conclusions regarding the magnitude of the membrane softening of the peptide compared to the full-length protein, as their membrane-binding affinities may be dissimilar, and note simply that the data show that the helix is sufficient to lower bilayer rigidity.
The effective membrane tension, σΛ=0, shows a decline with [Sar1p], especially at high ionic strength (1× HKM) (Fig. S5). As noted earlier, both the true membrane tension and the (indistinguishable) interaction energy of the proteins contribute to σΛ=0. Tension in lipid bilayer membranes may in general be near zero (37) and moreover, unlike rigidity, it depends on extrinsic factors such as surface area and bounded volume that are unlikely to be constant between samples. The decrease of σΛ=0 as a function of protein concentration is likely an indication, therefore, of repulsive interactions among Sar1p molecules.
The membrane tethers are likely to be single-bilayer shells, because the protein-coated microspheres whose manipulation pulls the tethers are bound only to the outermost surface of the membrane sheet. Although we lack a direct assay for potential multilamellar structure in the tethers, fluorescence imaging (as in Fig. 1 B) allows an assessment of whether there is appreciable polydispersity in structure. The background-subtracted fluorescence intensity integrated over a cross-section of the tether should be proportional to the circumference of the tether and to the number of layers in the cylindrical shell. The intensity normalized by tether radius, therefore, is proportional to the multilamellarity. In practice, differences in fluorescence illumination intensity and photobleaching will contribute to differences in tether intensity. Nonetheless, we found that only 8% of all the tethers have normalized intensities >1 SD above the mean intensity. Moreover, on every individual day on which experiments were conducted, during which variation in illumination intensity would be minimal, the ratio of the standard deviation of the integrated intensity to the mean was <1. We conclude, therefore, that the vast majority of tethers form a homogenous population.
Discussion
Our measurements reveal that Sar1p dramatically lowers the rigidity of lipid bilayer membranes, the first such discovery for a vesicle-trafficking protein. This membrane softening does not coincide with the imposition of strong local curvature, and is a profoundly different mode of action than those typically ascribed to intracellular curvature-associated proteins, such as the construction of rigid scaffolds (4,6,7,40) or the sensing of local geometry (41,42). Sar1 alters the mechanical properties of the membrane to which it binds. We suggest that this synergistic activity on the part of both membrane and protein facilitates coat formation by the rest of the COPII proteins, especially the rigid, cage-forming Sec13/Sec31 heterodimer (5), by locally lowering the energetic cost of membrane deformation, proportional to rigidity. We speculate that this may be a common tactic among proteins with membrane-inserting motifs, such as myristoylated Arf (43,44) and proteins with N-BAR domains, the latter of which feature an amphipathic helix together with a curved, rigid form (7,45). The widespread occurrence of N-BAR proteins at various curved intracellular surfaces, not only those whose radius conforms to the rigid BAR crescent (45), is puzzling in the context of scaffolding, yet is sensible when viewed as related to local membrane softening. We suggest that the lack of significant curvature generation by Sar1p, i.e., the small Λϕ, is a structural consequence of the positive (convex) Λ expected from the bilayer insertion of the N-terminal helix (15,17) being counteracted by the negative (concave) Λ induced by the rest of the protein (Δ23-Sar1p, Fig. 3 A). It should be kept in mind that our finding of small Λϕ means only that Sar1 does not necessarily couple strongly to curvature; spatial constraints may still enable it to contribute to membrane bending independent of other COPII coat proteins. Particular geometries (as in the case of liposome-binding (15,16)) or interactions with other proteins or lipids (e.g., that localize it to particular regions of the ER membrane), could spatially enhance asymmetries in leaflet areas and generate curvature via the bilayer couple mechanism noted earlier.
Membrane softening is a known mode of action for several small peptides (10,46–49) and has been ascribed both to coupling to local curvature (i.e., nonzero Λ), and to direct modulation of κ by membrane thinning or increased lipid disorder. It will be interesting to further examine Sar1p's N-terminal helix alone and compare its membrane interactions with the behaviors of other, similarly-sized peptides. Physically meaningful comparisons between different macromolecules, however, will require quantification of the two-dimensional concentrations of the proteins—in general, a challenging task (50).
Our studies examine the equilibrium material properties of lipid bilayers with Sar1p. Cellular membranes are nonequilibrium systems, however, and several recent studies have shown that active membrane proteins that harness energy fluxes from nucleotide hydrolysis cycles, incident light, and other sources to generate molecular motion can change membrane tension and rigidity (51–54). This raises the intriguing possibility of nonequilibrium effects beyond those explored here, especially when mediated by the full COPII complex, which employs proteins that regulate Sar1's nucleotide exchange and GTP hydrolysis rates (11–13).
Acknowledgments
We thank Christopher Harland, Tyler Campbell, and Gregory Tietjen for experimental assistance, Pete von Hippel, Jennifer Curtis, and Kirsten Bacia for useful discussions, Karen Kallio and Jim Remington for graciously assisting with protein expression, and Bob Lesch and Randy Schekman for generously providing bacteria for Sar1p and Δ23-Sar1p expression.
R.P. acknowledges support from the Alfred P. Sloan Foundation. R.P. and A.F.L. acknowledge support from the M. J. Murdock Trust.
Supporting Material
References
- 1.McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 3.Parthasarathy R., Groves J.T. Curvature and spatial organization in biological membranes. Soft Matter. 2007;3:24–33. doi: 10.1039/b608631d. [DOI] [PubMed] [Google Scholar]
- 4.Pearse B.M., Smith C.J., Owen D.J. Clathrin coat construction in endocytosis. Curr. Opin. Struct. Biol. 2000;10:220–228. doi: 10.1016/s0959-440x(00)00071-3. [DOI] [PubMed] [Google Scholar]
- 5.Stagg S.M., Gürkan C., Balch W.E. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 6.Frost A., Perera R., Unger V.M. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peter B.J., Kent H.M., McMahon H.T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 8.Phillips R., Ursell T., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leibler S. Curvature instability in membranes. J. Phys. 1986;47:507–516. [Google Scholar]
- 10.Huang H.W. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta. 2006;1758:1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Lee M.C., Miller E.A. Molecular mechanisms of COPII vesicle formation. Semin. Cell Dev. Biol. 2007;18:424–434. doi: 10.1016/j.semcdb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Gürkan C., Stagg S.M., Balch W.E. The COPII cage: unifying principles of vesicle coat assembly. Nat. Rev. Mol. Cell Biol. 2006;7:727–738. doi: 10.1038/nrm2025. [DOI] [PubMed] [Google Scholar]
- 13.Lee M.C., Miller E.A., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka K., Orci L., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 15.Lee M.C., Orci L., Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 16.Bielli A., Haney C.J., Aridor M. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J. Cell Biol. 2005;171:919–924. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheetz M.P., Singer S.J. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc. Natl. Acad. Sci. USA. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barlowe C., d'Enfert C., Schekman R. Purification and characterization of SAR1p, a small GTP-binding protein required for transport vesicle formation from the endoplasmic reticulum. J. Biol. Chem. 1993;268:873–879. [PubMed] [Google Scholar]
- 19.Antonny B., Madden D., Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- 20.Antonny B., Huber I., Cassel D. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J. Biol. Chem. 1997;272:30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- 21.Duda R.O., Hart P.E. Use of the Hough transformation to detect lines and curves in pictures. Commun. ACM. 1972;15:11–15. [Google Scholar]
- 22.Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Coll. Interf. Sci. 1996;179:298–310. [Google Scholar]
- 23.Sainis S.K., Germain V., Dufresne E.R. Statistics of particle trajectories at short time intervals reveal fN-scale colloidal forces. Phys. Rev. Lett. 2007;99:018303. doi: 10.1103/PhysRevLett.99.018303. [DOI] [PubMed] [Google Scholar]
- 24.Jo S., Vargyas M., Im W. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 2008;36(Web Server issue):W270–W275. doi: 10.1093/nar/gkn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bi X., Corpina R.A., Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 26.Waugh R.E., Hochmuth R.M. Mechanical equilibrium of thick, hollow, liquid membrane cylinders. Biophys. J. 1987;52:391–400. doi: 10.1016/S0006-3495(87)83227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai J., Sheetz M.P. Mechanical properties of neuronal growth cone membranes studied by tether formation with laser optical tweezers. Biophys. J. 1995;68:988–996. doi: 10.1016/S0006-3495(95)80274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raucher D., Sheetz M.P. Characteristics of a membrane reservoir buffering membrane tension. Biophys. J. 1999;77:1992–2002. doi: 10.1016/S0006-3495(99)77040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borghi N., Brochard-Wyart F. Tether extrusion from red blood cells: integral proteins unbinding from cytoskeleton. Biophys. J. 2007;93:1369–1379. doi: 10.1529/biophysj.106.087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorre B., Callan-Jones A., Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derényi I., Jülicher F., Prost J. Formation and interaction of membrane tubes. Phys. Rev. Lett. 2002;88:238101. doi: 10.1103/PhysRevLett.88.238101. [DOI] [PubMed] [Google Scholar]
- 32.Powers T.R., Huber G., Goldstein R.E. Fluid-membrane tethers: minimal surfaces and elastic boundary layers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2002;65:041901. doi: 10.1103/PhysRevE.65.041901. [DOI] [PubMed] [Google Scholar]
- 33.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 34.Lipowsky R., Sackmann E. Structure and Dynamics of Membranes. Vol. 1. Elsevier; New York: 1995. [Google Scholar]
- 35.Huang H.W. Elasticity of lipid bilayer interacting with amphiphilic helical peptides. J. Phys. II. 1995;5:1427–1431. doi: 10.1016/S0006-3495(95)80418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J., Tieleman D.P., Tristram-Nagle S. Alamethicin in lipid bilayers: combined use of x-ray scattering and MD simulations. Biochim. Biophys. Acta. 2009;1788:1387–1397. doi: 10.1016/j.bbamem.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jähnig F. What is the surface tension of a lipid bilayer membrane? Biophys. J. 1996;71:1348–1349. doi: 10.1016/S0006-3495(96)79336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans E., Yeung A. Hidden dynamics in rapid changes of bilayer shape. Chem. Phys. Lipids. 1994;73:39–56. [Google Scholar]
- 39.Gygi S.P., Rochon Y., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimada A., Niwa H., Yokoyama S. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 41.Ramamurthi K.S., Lecuyer S., Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bigay J., Casella J.F., Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 44.Beck R., Sun Z., Wieland F. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. USA. 2008;105:11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh T., De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 46.Hackl W., Seifert U., Sackmann E. Effects of fully and partially solubilized amphiphiles on bilayer bending stiffness and temperature dependence of the effective tension of giant vesicles. J. Phys. II (France) 1997;7:1141–1157. [Google Scholar]
- 47.Vitkova V., Méléard P., Bivas I. Alamethicin influence on the membrane bending elasticity. Eur. Biophys. J. 2006;35:281–286. doi: 10.1007/s00249-005-0019-5. [DOI] [PubMed] [Google Scholar]
- 48.Tristram-Nagle S., Nagle J.F. HIV-1 fusion peptide decreases bending energy and promotes curved fusion intermediates. Biophys. J. 2007;93:2048–2055. doi: 10.1529/biophysj.107.109181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouvrais H., Méléard P., Ipsen J.H. Softening of POPC membranes by magainin. Biophys. Chem. 2008;137:7–12. doi: 10.1016/j.bpc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Galush W.J., Nye J.A., Groves J.T. Quantitative fluorescence microscopy using supported lipid bilayer standards. Biophys. J. 2008;95:2512–2519. doi: 10.1529/biophysj.108.131540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manneville J.B., Bassereau P., Prost J. Activity of transmembrane proteins induces magnification of shape fluctuations of lipid membranes. Phys. Rev. Lett. 1999;82:4356–4359. [Google Scholar]
- 52.El Alaoui Faris M.D., Lacoste D., Bassereau P. Membrane tension lowering induced by protein activity. Phys. Rev. Lett. 2009;102:038102. doi: 10.1103/PhysRevLett.102.038102. [DOI] [PubMed] [Google Scholar]
- 53.Girard P., Prost J., Bassereau P. Passive or active fluctuations in membranes containing proteins. Phys. Rev. Lett. 2005;94:088102. doi: 10.1103/PhysRevLett.94.088102. [DOI] [PubMed] [Google Scholar]
- 54.Betz T., Lenz M., Sykes C. ATP-dependent mechanics of red blood cells. Proc. Natl. Acad. Sci. USA. 2009;106:15320–15325. doi: 10.1073/pnas.0904614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


