Abstract
This work explores the effect of long-range tertiary contacts on the distribution of residual secondary structure in the unfolded state of an α-helical protein. N-terminal fragments of increasing length, in conjunction with multidimensional nuclear magnetic resonance, were employed. A protein representative of the ubiquitous globin fold was chosen as the model system. We found that, while most of the detectable α-helical population in the unfolded ensemble does not depend on the presence of the C-terminal region (corresponding to the native G and H helices), specific N-to-C long-range contacts between the H and A-B-C regions enhance the helical secondary structure content of the N terminus (A-B-C regions). The simple approach introduced here, based on the evaluation of N-terminal polypeptide fragments of increasing length, is of general applicability to identify the influence of long-range interactions in unfolded proteins.
The study of structural and dynamic characteristics of unfolded proteins has gained tremendous momentum in recent years (1,2). The discovery of intrinsically disordered proteins in living cells has led to a deeper appreciation for the nontrivial biological role of unstructured states in substrate binding specificity, transport, regulation, and disease. Partly or fully disordered proteins under physiologically relevant conditions constitute ∼30% of all eukaryotic proteins (3). The energy-weighted ensemble character of a protein's unfolded state at equilibrium is best described by the term “statistical coil” (4), although “random coil” has historically been more commonly used. Although the most physiologically relevant unfolded state is the one populated in vivo at neutral pH, unfolded states generated in vitro under denaturing conditions are a more directly accessible alternative. Furthermore, this state is the starting condition of most in vitro refolding studies. Its detailed comprehension is therefore an important gateway toward a better understanding of both the unfolded ensemble and the pathways of in vitro protein folding.
The secondary structure distribution of unfolded proteins has been characterized, in some cases, even at atomic resolution. However, very little is known about its origin. Recent experimental studies have detected the presence of both residual secondary structure and specific native and nonnative long-range interactions in the unfolded state (5–7). There is, however, an urgent need to clarify whether the residual secondary structure typically detected in the unfolded ensemble arises from local contacts or is a consequence of long-range interactions (i.e., contacts among residues far apart in sequence). The two aspects of this issue, which have clear implications for unfolded chain dynamics and protein folding, are illustrated in Fig. 1 a.
Figure 1.
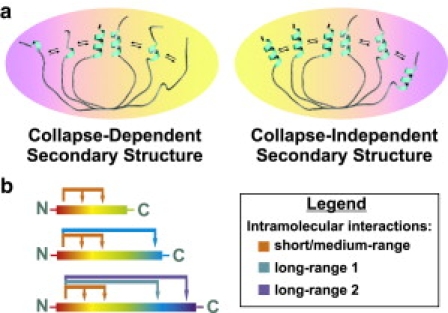
(a) General cartoon showing that chain collapse and the resulting long-range contacts may (image, left) or may not (image, right) be responsible for the residual secondary structure often observed in unfolded proteins. (Arrows) Transitions among different conformations. (b) Scheme illustrating the chain elongation strategy adopted here.
This work tackles the above question by what we consider a novel approach. In short, the evolution of backbone secondary structure of the unfolded ensemble is monitored by NMR at atomic resolution upon comparing unfolded N-terminal protein fragments of increasing length (Fig. 1 b). This procedure enables pinpointing the specific contributions of the C-terminal regions to the residual secondary structure in the N-terminal portion of the chain. Sperm whale apomyoglobin (apoMb), a very well-studied member of the ubiquitous globin fold, was selected as the target model chain. We found that, overall, most of the detectable α-helical conformation is populated irrespective of fragment length. However, specific long-range interactions involving the last (∼30) C-terminal residues enhance the secondary structure content of the N-terminal region.
The acid unfolded (pH 2.3) state of full-length apoMb, here denoted as (1-153)apoMb, was characterized at atomic resolution (5) and shown to populate partial helical conformation for the amino acids corresponding to the A, D/E, and H helices. In addition, paramagnetic spin labeling led to the identification of long-range interactions involving residues corresponding to the native A and G-H helices and medium-range interactions within the A-B-C and G-H regions (6,7). Such interactions, due to their transient nature and to extensive conformational averaging in the unfolded state, are undetectable by long-range nuclear Overhauser effects (5).
Here, we focus on unfolded (pH 2.4) 13C-15N-labeled (1-77), (1-119), and (1-153)apoMb. The (1-77)apoMb fragment lacks long-range interactions involving the G and H regions whereas (1-119)apoMb lacks contacts involving the H region. Previous data (8,9) show that all three unfolded apoMb chains have overall dimensions consistent with expanded polymers in a good solvent according to Flory's scaling law (10) and all three chains fit the definition of random coil (10,11). Resonance assignments were carried out by triple-resonance methods, and chemical shift analysis was performed on the Cα, C′, HN, and N nuclei. The Cα NMR chemical shift deviations from random coil reference values, denoted as secondary chemical shifts (SCS), are employed as selective reporters of backbone secondary structure (12).
Fig. 2 a shows that the Cα SCS values for the same residues in each fragment are mostly independent of chain length. Therefore, overall, the distribution of residual secondary structure in the unfolded ensemble does not vary significantly upon progressive extension of the polypeptide chain from 77 amino acids to full-length protein. However, a more detailed comparison provided by the difference between Cα secondary chemical shifts (ΔCα SCS) among individual chain lengths (Fig. 2 b) indicates that specific long-range interactions modify the secondary structure of the N-terminal portion of the chain.
Figure 2.
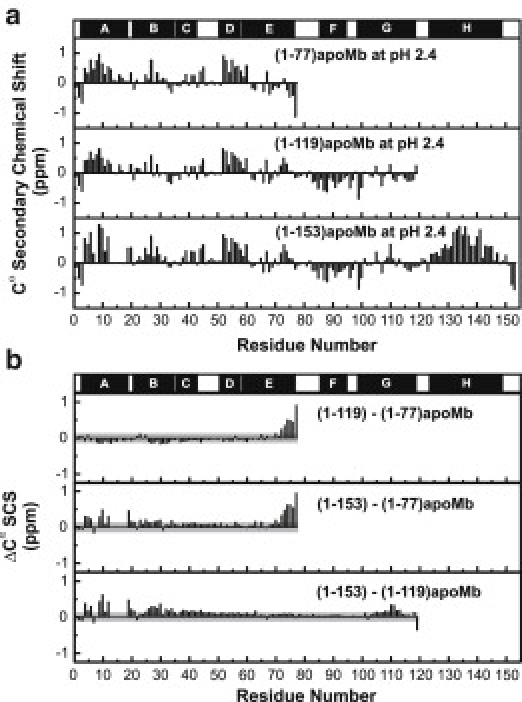
(a) Cα secondary chemical shifts (SCS) and (b) differences among the Cα secondary chemical shifts (ΔCα SCS) of (1-77), (1-119), and (1-153)apoMb at pH 2.4 and 25°C in 5 mM CD3COOH, 5% D2O. Native apoMb helices (solid bars) are mapped above the graph. (Gray-shaded regions) ΔCα SCSs falling within experimental error, assessed from backbone assignments on two independent samples (see the Supporting Material).
The Cα SCSs for the residues corresponding to the native A, B, and C helices (i.e., amino acids 3–42) are nearly identical in (1-77) and (1-119)apoMb (Fig. 2 b). Thus, the helicity of this portion of the polypeptide does not vary in the presence and absence of long-range interactions involving the G region. Such contacts are known to exist in full-length unfolded apoMb (6,7). However, they cannot be present in (1-77)apoMb. In essence, either long-range contacts involving the G residues do not affect secondary structure (in the A-B-C region), or the population experiencing long-range contacts with G is small and undetectable. The former scenario is more likely, given that 1), the overall populations interacting with either G or H regions are similar (3.7 and 3.6%, respectively) (7); and 2), the effect of interactions with H region is explicitly detectable (see below). However, caution should be exercised, considering that the estimated populations (7) do not explicitly take into account the potential distance-dependence of the long-range contacts.
The slightly negative ΔCα SCS in the A-B-C region may reflect some small (i.e., within experimental error) second-order effects (see the Supporting Material). The carbonyl carbon SCSs (C′ SCS) are also effective reporters of secondary structure, particularly in the unfolded state (5). The trends followed by C′ SCS and ΔC′ SCS (Fig. 3) parallel those of Cα SCS (Fig. 2). HN and N SCSs, known to be less diagnostic of secondary structure, are reported in the Supporting Material.
Figure 3.
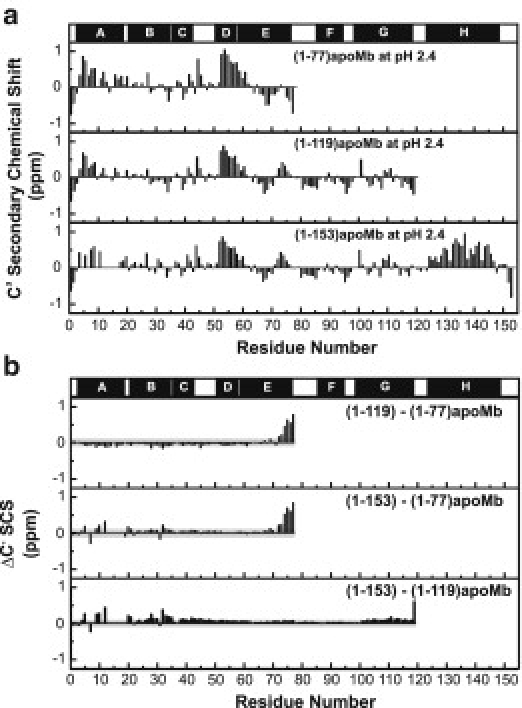
(a) C′ SCS and (b) ΔC′ SCS of (1-77), (1-119), and (1-153)apoMb at pH 2.4 and 25°C in 5 mM CD3COOH, 5% D2O. The native apoMb helices are mapped above the graph. (Gray-shaded regions) ΔC′ SCS values' experimental errors, determined as described in the legend for Fig. 2.
The ΔCα SCS values of residues 74–77 fall far outside the estimated propagated error (∼±0.1 ppm) and are consistently positive (Fig. 2 b). We attribute them to end-effects due to the proximity of (1-77)apoMb's residues 74–77 to the C-terminus.
The two lower panels of Fig. 2 b highlight the effect of the H region, which is the major finding of this work. Clearly, the C-terminal residues corresponding to the native H helix enhance the residual helicity of the amino acids in the A-B-C cluster, from 5 to 12% (see Fig. S7 in the Supporting Material). This result demonstrates that specific long-range contacts are capable of increasing local chain helicity in the unfolded state.
The H-region-dependent enhancement in secondary structure in the A-B-C region goes hand-in-hand with selective line-broadening beyond detection in the A region (Fig. S8). Hence, the observed tertiary contact-driven increase of secondary structure in the A region appears linked to the establishment of slow dynamic processes (on the microsecond-to-millisecond timescale; see also Yao et al. (5)).
Residues in the D, E, and F regions are known not to experience medium- and long-range interactions with other portions of the chain (6,7). Despite the absence of such contacts, Cα SCS reveal the presence of local clusters spanning 6–12 residues. Therefore, Flory's isolated pair hypothesis (10), stating that backbone dihedral angles are independent on those of the neighboring residues, does not apply here. Our results agree with the computational prediction of more-extended local backbone biases (13,14) in the unfolded ensemble, and with the fact that apoMb Cα SCSs depend on the position of each residue type (see Fig. S9).
The long-range ΔCα SCS effects observed here may either be due to small populations of compact species experiencing a large increase in helicity (due to A-B-C to H contacts) or to a small increase in helical content by relatively larger populations of semicompact species. A combination of the above effects is also possible.
In summary, this work shows that only specific long-range interactions between the N-terminal A-B-C cluster and the C-terminal H region lead to detectable increased helicity in the N-terminal region. Thus, some long-range interactions lead to enhanced secondary structure. The resulting collapsed species are likely important, as they may provide preferential kinetic escape routes to the native state, upon switching to folding conditions.
Acknowledgments
This work was funded by the National Science Foundation (MCB-0544182 and MCB-0951209), the Research Corporation (Research Innovation Award to S.C.), and the Milwaukee Foundation (Shaw Scientist Award to S.C.). The National Institutes of Health (S10-RR13866-01) supported the NMR instruments.
Footnotes
Senapathy Rajagopalan's present address is The Methodist Hospital Research Institute, Diabetes Research, 6565 Fannin St., F8-060, Houston, TX 77030.
Eric C. Fulmer's present address is Faculty of Earth and Life Sciences, VU University Amsterdam, De Boelelaan 1085, 1081 HV Amsterdam, The Netherlands.
Ye-Jin Eun's present address is Department of Biochemistry, University of Wisconsin-Madison, 433 Babcock Dr., Madison, WI 53706.
Supporting Material
References and Footnotes
- 1.Dyson H.J., Wright P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 2.McCarney E.R., Kohn J.E., Plaxco K.W. Is there or isn't there? The case for (and against) residual structure in chemically denatured proteins. Crit. Rev. Biochem. Mol. Biol. 2005;40:181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]
- 3.Fink A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Vila J.A., Ripoll D.R., Scheraga H.A. Unblocked statistical-coil tetrapeptides and pentapeptides in aqueous solution: a theoretical study. J. Biomol. NMR. 2002;24:245–262. doi: 10.1023/a:1021633403715. [DOI] [PubMed] [Google Scholar]
- 5.Yao J., Chung J., Dyson H.J. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 2001;40:3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 6.Lietzow M.A., Jamin M., Wright P.E. Mapping long-range contacts in a highly unfolded protein. J. Mol. Biol. 2002;322:655–662. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- 7.Felitsky D.J., Lietzow M.A., Wright P.E. Modeling transient collapsed states of an unfolded protein to provide insights into early folding events. Proc. Natl. Acad. Sci. USA. 2008;105:6278–6283. doi: 10.1073/pnas.0710641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurt N., Rajagopalan S., Cavagnero S. Effect of hsp70 chaperone on the folding and misfolding of polypeptides modeling an elongating protein chain. J. Mol. Biol. 2006;355:809–820. doi: 10.1016/j.jmb.2005.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gast K., Damaschun H., Damaschun G. Compactness of protein molten globules: temperature-induced structural changes of the apomyoglobin folding intermediate. Eur. Biophys. J. 1994;23:297–305. doi: 10.1007/BF00213579. [DOI] [PubMed] [Google Scholar]
- 10.Flory P.J. Wiley; New York: 1969. Statistical Mechanics of Chain Molecules. [Google Scholar]
- 11.Kohn J.E., Millett I.S., Plaxco K.W. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart D.S., Sykes B.D., Richards F.M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 13.Fitzkee N.C., Rose G.D. Reassessing random-coil statistics in unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran H.T., Wang X., Pappu R.V. Reconciling observations of sequence-specific conformational propensities with the generic polymeric behavior of denatured proteins. Biochemistry. 2005;44:11369–11380. doi: 10.1021/bi050196l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


