Abstract
Background
Five isolates (JS09GY2, JS09GY3, JS09GY4, JS09GY5, and JS09GY6) of avian leukosis virus subgroup J (ALV-J) were isolated from six infected commercial layer flocks displaying both hemangioma and myeloid leukosis (ML), which shared the same parental line, in China in 2009.
Results
All six of the commercial layer chickens examined showed hemangiomas on their body surface or feet. Some developed hemangiomas in their internal organs, causing hepatorrhexis and blood loss. Histopathologically different stages of hemangiomas with ML in the liver, heart, and spleen, were observed. Five viral isolates were obtained from infected DF1 cells incubated with the spleen tissue or serum of the birds from the six flocks. By full genome sequences analysis, a 19-nucleotide repeat sequence was identified in the primer binding site (PBS)-leader region of isolates JS09GY3 and JS09GY6, located between sites 249 and 250 according to the sequence of reference strain HPRS103, and also present in Rous sarcoma virus strain Schmidt–Ruppin B (RSV-SRB), Rous associated virus type 1 (RAV-1), and Rous associated virus type 2 (RAV-2). The predicted Gp85 proteins of isolates JS09GY2, JS09GY3, JS09GY5, and JS09GY6 were highly variable. Interestingly, the E elements of these four examined isolates showed a key deletion at site 30, which produced a new c-Ets-1 binding site. An 11-bp insertion was also found in the E element of isolate JS09GY3 located between bp 66 and 67 according to the sequence of reference strain HPRS103, while almost all previously reported Chinese strains showed an almost identical deletion of 127 bp in the same region.
Conclusions
Five ALV-J isolates were obtained from six field infected commercial layer chickens. Coexistence of hemangioma and ML were observed in these infected cases both macro- and microscopically. Complete proviral genome sequences of two isolates (JS09GY3 and JS09GY6) and the partial sequences of the other two isolates (JS09GY2 and JS09GY5) were determined. The isolates were found to be recombinants of ALV-J with a PBS-leader sequence originating from other retroviruses. The Gp85 protein with an amino acid deletion, a contiguous 11-bp insertion mutation in the E element, and a novel binding site, were noted in the proviral genomes.
Keywords: Recombinant avian leukosis virus, Subgroup J, Layer chickens, Hemangioma, Myeloid leukosis
Background
Exogenous avian leukosis viruses (ALVs) from chickens have been subclassified into different subgroups (A, B, C, D, and J) based on their host range, viral envelope interference, and cross-neutralization patterns (Payne and Fadly 1997). Avian leukosis virus subgroup J (ALV-J) was isolated from meat-type chickens in the late 1980s in UK (Payne et al. 1991). Since its occurrence ALV-J infection has emerged worldwide and causes economic losses associated with mortality, delayed growth, and tumor development including myelocytomas, erythroblastosis, hemangiomas, nephromas, and sarcomas in broilers (Payne 1999). ALV-J has a broad host range and all lines of chickens tested are susceptible to infection (Payne et al. 1992). Myeloid leukosis (ML) caused by ALV-J in commercial layer flocks has been reported in China (Xu et al. 2004).
The E element of ALV-J has been investigated and appears to play a role in tumorigenesis (Chesters et al. 2006a). However, various substantial deletions (>50%) have been reported in the E element of ALV-J proviruses isolated from breeding stocks and broiler chickens between 1988–2007 (Zavala et al. 2007) and all eight ALV-J strains isolated from field cases of meat-type chickens between 1999 and 2001 in China had an almost identical deletion of 127 bp in the E element (Cui et al. 2003). No significant insertion mutations in the E element have been reported since the prototype strain was isolated. Clinically, tumors associated with ALV-J infection are primarily expressed as ML (Arshad et al. 1997). Whereas, the proportion of hemangioma in all tumor cases associated with ALV-J is only 3% (Payne 1999). Most reported cases of ALV-J infection in layer chickens in China to date have been associated with ML, characterized by pathological lesions or molecular detection, rather than complete genome analysis (Xu et al. 2004; Wang and Cui 2008).
Here we investigated the characteristics of ALV-J infection and isolated and analyzed viruses from different commercial layer flocks sharing the same parental line in China in 2009. The complete or partial proviral genome sequences were determined and compared with reference sequences. Interestingly, hemangioma accompanied by ML was observed both macro- and microscopically. Significant insertion mutations in the E element, and a high degree of genetic variation in the gp85 gene in the viral genome were detected. These results will contribute towards our understanding of the infection process of ALV-J in layer chickens and may shed light on the multiformity of tumor types induced by ALV-J in recent years.
Results
Epidemiology of the parental line flock
To study the origin and epidemiology of ALV-J, chickens in the parental line were tested for shedding of virus group-specific antigen p27 and antibodies against ALV-J by an enzyme-linked immunosorbent assay (ELISA), as shown in Table 1. Various levels of p27 antigen shedding were found in all four chicken age groups (5, 12, 36 and 60-week-old), but no antibody for ALV-J was presented in chickens in the younger age groups (5-week-old and 12-week-old). Whereas, in the older age groups (36-week-old and 60-week-old), the percentage of antibodies specific for subgroup J reached 14% and 31%, respectively.
Table 1.
ELISA test for ALVs antigen and ALV-J antibody in the parental line chickens
| Age (Weeks) | ♀ (positive rates) | ♂ (positive rates) | Status for p27 shedding and ALV-J antibody (♀ + ♂) | ||
|---|---|---|---|---|---|
| p27 | ALV-J Ab | p27 | ALV-J Ab | ||
| 5 | 5/20 | 0/20 | 2/8 | 0/8 | S + A- (25%) |
| 12 | 1/20 | 0/20 | 0/8 | 0/8 | S + A- (3.5%) |
| 36 | 6/20 | 4/20 | 0/8 | 0/8 | S + A- (21%), S-A + (14%) |
| 60 | 1/21 | 8/21 | 2/8 | 1/8 | S + A- (10%), S-A + (31%) |
The letter “S” indicated p27 shedding; “A” indicated antibodies of ALV-J
Six birds from the six commercial flocks were also examined for ALV antigens and antibodies against ALV-J. All six chickens were positive for p27 shedding and serum ALV antigen and negative for ALV-J antibody (data not shown). These results indicated the possible presence of immune tolerance for ALVs in these chickens.
Clinical signs and histopathology of samples from the commercial layer chickens
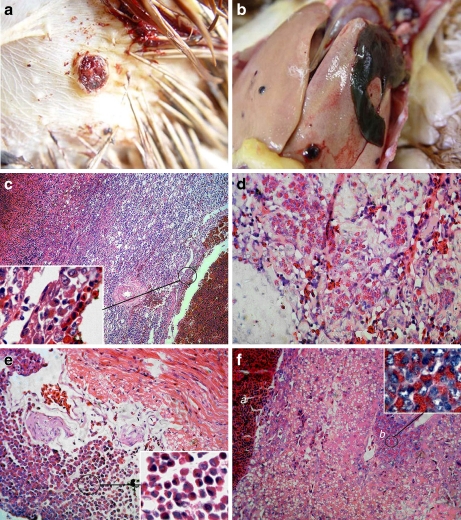
The six birds used in this study were termed JS09GY1–JS09GY6, according to the viruses isolated from them. Infected birds showed hemophthisis symptoms and hemangiomas with a diameter of 0.4–1 cm on or under the skin of the abdominal region (Fig. 1a, showing the hemangioma in bird JS09GY2), the feet, the cervical region, or the extremitas inferior. Gross lesions around hemangiomas (0.2 to 3.5 cm in diameter) were also observed in the liver, spleen, kidney, intestinal tract, and mesenterium. Hepatorrhexis (Fig. 1b, JS09GY3), and blood loss in the spleen and kidney caused by hemangiomas, occurred in three cases, respectively.
Fig. 1.
Gross lesions and Histopathology (haematoxylin and eosin staining). (a) Hemangioma on the body surface. JS09GY2 case was showed. (b) Hepatorrhexis in JS09GY3 case caused by hemangioma in liver. (c) Spleen of JS09GY6 showing different staged hemangiomas. Myeloid cells aggregations were found around the hemangiomas (H&E stain, 200×). (d) Myeloid cells in the interstitium of the hemangioma of JS09GY2 (H&E stain, 400×). (e) Myeloid cells aggregation in heart of JS09GY5 (H&E stain, 400×). (f) Hemangioma and ML tumor coexisted in the liver of JS09GY3 (H&E stain, 400×), the tumor cells in the ML were observed (inserted, 1,000×); (a) Hemangioma, (b) ML
Four (JS09GY2, JS09GY3, JS09GY5, and JS09GY6) of the six chickens (66.7%) showed the coexistence of hemangioma and ML by histopathology examination, while the other two chickens showed simple hemangiomas. Different stages of hemangioma were observed on body surfaces and internal organs including the liver, kidney, and spleen (Fig. 1c, JS09GY6). Hemangiomas were similar in appearance but the tumors on the body surface were dermatic in structure. The parietal layer of the hemangioma was a pultaceous, inordinate sponge-like structure composed of connective tissue and vascular endothelial cells. Poikilocytes and myeloid cell aggregation were found around the hemangiomas (Fig. 1d, JS09GY2). ML was the other main tumor type observed. These tumor cells were focal, eosinophilic, granulocytic, large, with round or elliptical nuclei located on one side. Focal infiltrations of myeloid cells at different proliferative and differentiation stages were seen in the heart (Fig. 1e, JS09GY5) and spleen. Many ML tumor cells gathered around blood vessels in the liver. Specifically, hemangioma and ML coexisting in the liver tissue was observed in one case (Fig. 1f, JS09GY3).
In the four cases, in which two types of tumors were observed, the degree of development of both hemangioma and ML was not coincidental. In two cases, hemangioma was the main tumor type. Different developmental stages of hemangiomas were found within one bird, whereas, ML tumors were localized and the accrementition focuses were recently formed. Another two cases showed severe ML with extensive tissue distribution, but the mature hemangiomas in these cases were relatively slight and almost all hemangiomas were accompanied by myeloid cell infiltration into the interstitium.
Detection of virus in commercial chickens
ALV-J specific RT-PCR using the primer set H5/H7 (Smith et al. 1998) and RNA isolated from the liver of infected birds as template, gave a fragment of about 545 bp (pol–env) for all birds. And the other primer set H5/AD1 for subgroup A–E got no product from the same samples. These result confirmed infection with ALV-J in these birds and ruled out other ALV subgroups infection in these chickens (data not shown).
Isolation and identification of viruses from commercial chickens
Five ALV-J isolates, designated JS09GY2, JS09GY3, JS09GY4, JS09GY5 and JS09GY6, were obtained from the spleen tissue of different field cases after two passages of the infected DF1 cells and were confirmed by an indirect fluorescence assay (IFA) with monoclonal antibody JE9 (data not shown), which is specific for subgroup J ALV (Qin et al. 2001). Viruses were also isolated from infected DF1 cells inoculated with the serum from the five commercial chickens and viremia was therefore confirmed in these field cases.
Comparison of the proviral DNA sequence
Complete genome analysis
Two ALV-J isolates (JS09GY3: GenBank accession number GU982308, and JS09GY6: GenBank accession number GU982310) from DF1 cells inoculated with the spleen tissues, both associated with hemangioma and ML, were used for full genome sequencing. The 3′ noncoding region of the env gene of isolates JS09GY2 (GenBank accession number GU982307) and JS09GY5 (GenBank accession number GU982309) was also sequenced.
The genomes of isolates JS09GY3 and JS09GY6 were 7661 and 7653 nucleotides, respectively, with a genetic organization typical of ALV-J, as determined using the NCBI tool BLAST (http://blast.ncbi.nlm.nih.gov/). The complete genome nucleotide sequences of these two isolates shared 99.6% identity to each other, and 95.1% to 95.3% nucleotide identity to the four reference ALV-J isolates (HPRS103, ADOL-7501, SD07LK1, and NX0101). The genomes of isolates JS09GY3 and JS09GY6 were most closely related to the prototype strain HPRS103.
When compared with the ALV-J reference strains and Rous sarcoma virus strain Schmidt- Ruppin B (RSV-SRB), the predicted Gag amino acid sequence similarities of isolates JS09GY3 and JS09GY6 to the reference strains ranged from 96.0% to 98.1%, whereas, those of the Pol protein ranged from 97.7% to 99.0%. The percentage identity of the gp37 nucleotide sequence of the four isolates (JS09GY2, JS09GY3, JS09GY5, and JS09GY6) to the prototype strain HPRS103 ranged from 93.1% to 93.2%. The direct repeat 1 (DR1) sequences of the four isolates were 95.7% identical to that of strain HPRS103.
Novel recombinant ALV-J viruses
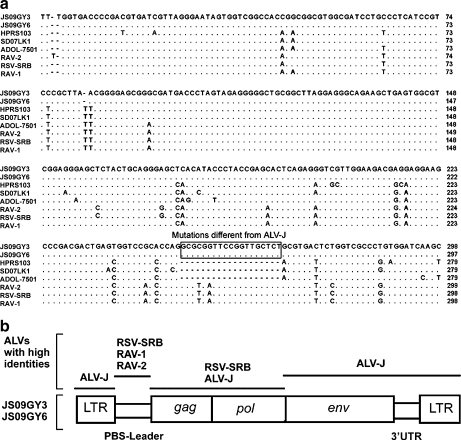
Special insertions of 19 nucleotide repeat sequences (5′-GCGCG GTTCC GGTTG CTCT-3′) located between bp 249 and 250 of the primer binding site (PBS)-leader sequence according to strain HPRS103, were also identified in isolates JS09GY3 and JS09GY6. By BLASTN analysis, these insertion sequences were also identified in the leader region of RSV-SRB (GenBank: AF052428, Rous sarcoma virus (strain Schmidt- Ruppin), Rous sarcoma virus (strain Schmidt-Ruppin B)), Rous associated virus type 1 (RAV-1, GenBank: M62407, avian leukosis virus, Rous-associated virus type 1), and Rous associated virus type 2 (RAV-2, GenBank: K02374, avian leukosis virus, Rous-associated virus type 2), but were absent in all of the available ALV-J reference strains (Fig. 2a). Notably, the nucleotide percentage identity of the PBS-leader sequence of the two isolates to the corresponding sequences from RSV-SRB, RAV-1, and RAV-2 reached 93% while nucleotide identity to strain HPRS103 was only 89.2%. These results suggested that isolates JS09GY3 and JS09GY6 originated from recombination between ALV-J and other strains of ALSV (Avian Leukosis and Sarcoma Virus) such as RSV-SRB, RAV-1, or RAV-2 (Fig. 2b). However, the recombination mechanism is unclear.
Fig. 2.
The isolates were ALV-J recombinant viruses with PBS-Leader sequences similar to other retroviruses. (a) Alignment analysis of the sequences of the PBS-Leader of the two isolates (JS09GY3 and JS09GY6) and reference sequences of HPRS103 (GenBank: Z46390), SD07LK1 (GenBank: FJ216405), ADOL-7501 (GenBank: AY027920), RAV-2 (GenBank: K02374), RAV-1 (GenBank: M62407) and RSV-SRB (GenBank: AF052428). Sequences identical to JS09GY3 are showed as dot (.) and the deleted sequences are represented by dashed line (-). (b) Sequence comparison of the two isolates (JS09GY3 and JS09GY6) with other retroviruses. Bottom boxes, structure of the JS09GY3 and JS09GY6 genome; lines above, viral sequences showing high identity with those regions of JS09GY3 and JS09GY6
Genetic variations in the E element, env gene, and U3 region
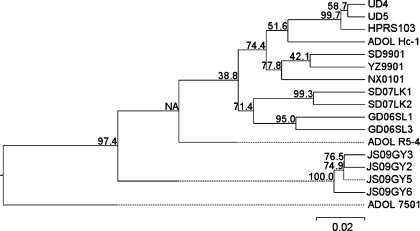
When aligned, significant sequence heterogeneity in the gp85 gene of the four ALV-J isolates was observed. Mutations were distributed mainly in the hypervariable regions of the isolates, notably with deletion of an amino acid (Lys, at site 179 of the Gp85 protein of strain HPRS103) in the protein encoded by the hr1 region (data not shown). The predicted amino acid sequences of Gp85 from the four isolates clustered in a distinct group in the phylogenetic tree (Fig. 3). These sequences were relatively closely related to the Gp85 predicted amino acid sequence of strain HPRS103 with 89.3–90.4% similarity, whereas they showed less similarity, 86.8–91.5%, to the corresponding sequences from other reference strains. Most Chinese strains from meat-type chickens and local “yellow” chickens were 90.4–95.2% identical to strain HPRS103 in the entire Gp85 protein sequence. The signal peptide sequences of the four isolates were absolutely identical to those in strain HPRS103, while most other reference strains showed various mutations in this region.
Fig. 3.
Phylogenetic relationship of the predicted amino acid sequences of the entire gp85 in JS09GY2, JS09GY3, JS09GY5, and JS09GY6 with reference strains. HPRS103 (GenBank: Z46390), YZ9901 (GenBank: AY897222), SD9901 (GenBank: AY897220), NX0101 (GenBank: AY897227), SD07LK1 (GenBank: FJ216405, from infected layer chicken), SD07LK2 (GenBank: ABX83206, from infected layer chicken), GD06SL1 (GenBank: ABM26973, from local “yellow” chicken), GD06SL3 (GenBank: ABM52966, from local “yellow” chicken), UD4 (GenBank: AAG45097), UD5 (GenBank: AAG45098), ADOL-7501 (GenBank: AAK58476), ADOL-Hc-1 (GenBank: AAC96313), and ADOL-R5-4 (GenBank: AAD25920). Numbers denote the bootstrap confidence interval for 1000 replicates in which a certain branch occurred
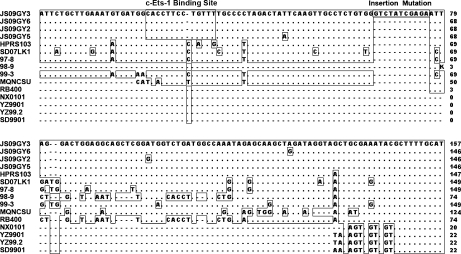
Surprisingly, an 11-bp (5′-GTCTA TCGAG A-3′) insertion (between bp 66 and 67 of the E element according to strain HPRS103) was noted in the intact E element of isolate JS09GY3. No such significant insertions were found in the same region of the available ALV-J reference strains or ASV strains using the BLASTN program (Fig. 4). The partial homoplastic sequence (5′-CTATC GAGA-3′) was found in the insertion encoding transmembrane segment of the gp37 gene in all ALV-J strains analyzed. Besides the insertion sequence in isolate JS09GY3, alignment of the E elements of the four isolates revealed high sequence conservation, with 95.6% identity to strain HPRS103. Despite the conservation of sequence observed in isolates from layer chickens, most Chinese reference strains (86%, 127/147) from meat-type chickens exhibited substantial deletions in the E element. These results suggested that conservation of the E element corresponded to the host breed. To confirm the mutation in the E element in isolate JS09GY3, the liver tissue DNA from the infected chicken was used as a template to amplify the E element and the same sequence was obtained.
Fig. 4.
DNA alignment of the nucleotide sequences of the E elements of the four isolates and reference strains. JS09GY3, JS09GY6, JS09GY2, JS09GY5, HPRS103 (GenBank: Z46390), YZ9901 (GenBank: AY897222), YZ9902 (Cui et al. 2003), SD9901 (GenBank: AY897220), SD0001 (GenBank: AY897223), SD0002 (GenBank: AY897224), SD0101 (GenBank:AY897225), HN0001 (GenBank: AY897219), NX0101 (GenBank: AY897227), SD07LK1 (GenBank: FJ216405), RB400 (GenBank: AY689439), 97-8 (GenBank: AY689456), 98-9 (GenBank: AY689464) and 99-3 (GenBank: AY689468). Sequences identical to HPRS103 are showed as dot (.) and the deleted sequences are represented by dashed lines (-). The insertion region and the special binding site are represented in panes
Motif analysis revealed that a key deletion mutation (at site 30 according to strain HPRS103) in the E element of the four isolates created a distinct binding site for c-Ets-1 (p54), located at site 22 to 34 bp according to the sequence of strain HPRS103. The U3 regions were the most variable in the long terminal repeat (LTR) of the isolates. The U3 sequences of JS09GY2, JS09GY3, JS09GY5, and JS09GY6 showed just 91.6, 92.4, 92.6, and 91.6% identity to that of strain HPRS103, respectively. The U3 region of isolate JS09GY5 showed a contiguous deletion mutation located between bp 9 to 18, according to the sequence of strain HPRS103. The putative binding sites were highly conserved including the Y box, CCAAT-like box, PRE box, and CarG-like box. A special binding site for transcription factor 11 (TCF11) in the U3 region was observed in isolate JS09GY3 (located at bp 104 to 116), JS09GY5 (located at bp 96 to 108), and JS09GY6 (located at bp 104 to 116), which is known to mediate regulation of tumor growth and metastasis (Berg et al. 2007; Wei et al. 2003).
Discussion
This is the first study to report the full genome sequences of ALV-J isolates showing recombination with a leader sequence from other retroviruses and insertion in the E element from different commercial layer flocks. The commercial layer chickens developed two tumor phenotypes, hemangioma and ML, induced by ALV-J but the corresponding parental line birds showed no symptoms of ALV-J infection, except positive ALV-J antibodies in the serum of birds within the older age groups. As previously reported, the genetic differences among the lines of white leghorn chickens were found to influence the response of chickens to infection with ALV-J (Williams et al. 2004) and differences in the type of tumors may be attributed to differences in the genetics of the commercial layer flock (Mays et al. 2006). The potential positive effect of the host genetic background on the clinical symptoms and tumor phenotypes induced by ALV-J may explain the differences in clinical appearance of commercial chickens and their parental line birds infected with ALV-J. However, the infection conditions of the commercial chickens and the parental birds were different. The absence of antibodies against ALV-J accompanied by p27 shedding and viremia, proven by virus isolation from serum, reflected immune tolerance in the examined commercial chickens. This may indicate that vertical transmission or horizontal transmission occurred at an early age in these chickens. Whereas, in the parental line flock, the ev21 gene carried in the slow feathering female chickens may positively effect the p27 shedding rates and positive p27 shedding could not indicate ALV-J infection due to the lack of virus isolation from the birds. Therefore, it was only the ALV-J antibodies detected in the older age groups that could indicate infection in the parental line flock. It was suggested that the absence of ALV-J infection symptoms in the parental line could be due to the presence of antibodies and the higher resistance of older chickens to the tumors induced by ALV-J (Mays et al. 2006). As previously reported, it was thought that short-term direct or indirect contact with infected chickens at hatching resulted in high rates of horizontal transmission of ALV-J (Witter and Fadly 2001). It is possible that a minority of infected birds within the parental chickens transmitted the virus vertically to commercial offsprings, which in turn transferred ALV-J to other chickens horizontally at an early age. These horizontal and vertical transmissions resulted in immunotolerance to ALV-J in the commercial chickens and tumors developed at a high rate. Interestingly, most field cases of ALV-J infection in commercial layer chickens in China, including our findings, were reported in commercial brown layer chickens, indicating that this chicken line is relatively susceptible to ALV-J infection (Xu et al. 2004).
One of the main characteristics of retroviruses is genomic instability, resulting in rapid evolution (Nichol 1996). Recombination has been detected among exogenous viruses, between exogenous and endogenous viruses, and exogenous viruses and non-homologous cellular genes (Svoboda et al. 1986). The possible recombinant pattern of the isolates in our study (Fig. 2b) was different from that reported previously, suggesting that the 3′UTR upstream of the DR1 is a hot-spot for recombination between ALV-J and other exogenous viruses (Gingerich et al. 2002; Hatai et al. 2008; Lupiani et al. 2006). The ALSV 5′ leader sequence affects the packaging efficiency of genomic RNA (Donze and Spahr 1992; Sonstegard and Hackett 1996). Thus, the reported recombination event in our study may affect viral translation and duplication. The relationship between tumor multiplicity and recombination has not yet been examined functionally. According to the DNA sequence, the two isolates originating from different commercial flocks with the same parental flock, showed the same recombination model and extremely high similarity at the nucleotide level (>99%). This indicated that the viruses shared the same ancestor and that recombination with other retroviruses may have occurred in the parental line, or even in the grandparental flock. Taking into account that isolates JS09GY3 and JS09GY6 were from two commercial layer flocks located in different districts, in addition to the fact that all flocks were vaccinated with different commercial vaccines, this seems to exclude the possibility that vaccination resulted in ALV-J infection.
Previous studies have shown that the Gp85 protein is primarily responsible for receptor binding (Rong et al. 1997) and cell type oncogenicity of ALV (Chesters et al. 2002). Some reports suggested that another unique sequence of ALV-J, the E element, may play an important role in oncogenesis and pathogenesis of ALV-J in certain genetic lines of chickens (Hue et al. 2006a; Chesters et al. 2006b). Interestingly, the Gp85 protein sequences of the four isolates showed significant variation from that of strain HPRS103, while the corresponding regions in Chinese strains from meat-type chickens were more conserved (Cui et al. 2003). Phylogenetic examination of the Gp85 protein (Fig. 3) revealed that ALV-J had evolved to generate fairly distinct viral lineages corresponding to the origin of the host breed in China. On the other hand, the E elements in the compared isolates from infected layer chickens showed no deletions and the JS09GY3 isolate showed an insertion, while reference isolates from meat-type chickens showed various deletions even up to >86% (Hue et al. 2006b; Zavala and Jackwood 2007). In contrast with strains from meat-type chickens, hypervariable gp85 gene sequences and conserved E element sequences were found to coexist in isolates from infected layer chickens in this study. The proposed correlation between mutations in the viral genome and the host breeds reported in this study requires further functional analysis and proviral sequence analysis of the common strains in meat-type chickens. It is possible that ALV-J isolates in China have acquired some mutations in their genomes. Since the E element was supposed to form the largest and most stable hairpin structure in the viral genome (Schwartz et al. 1983) and contribute to oncogenesis of ALV-J, more research is required to determine whether the insertion affects virus replication and tumor type in commercial layer chickens.
Motif analysis of the E elements of the four isolates (JS09GY2, JS09GY3, JS09GY5, and JS09GY6) showed a specific binding site for c-Ets-1, which is associated with vascular endothelial cell differentiation (Dorsey et al. 2002; Gilles et al. 1996; Goldberg et al. 1994). Furthermore, a special binding site TCF11 was contained within the U3 regions of the isolates. These binding sites may contribute to the tumor type observed clinically.
Conclusions
Following the report by Xu et al. (2004), this study investigated the infection of ALV-J in commercial layer chickens from different flocks in China. In contrast to previous studies in which only ML was reported, two tumor phenotypes of hemangioma and ML were found to be induced by ALV-J in chickens in this study, as determined by histopathological examination. The presence of ALV-J antibodies revealed ALV-J infection in the clinically asymptomatic parental flock. Five isolates were obtained from the separately infected DF1 cells incubated with homogenates of spleen tissues or serum from the six commercial layer chickens. The viruses were identified by IFA detecting the Gp85 protein of ALV-J. The complete and partial genome sequences of the isolates were determined. Sequences analysis revealed that the isolates were recombinants of ALV-J containing the PBS-leader region of other retrovirus. The E element of JS09GY3 exhibited 11 nucleotide repeat sequence insertions, while most reported Chinese strains showed an almost identical 127 nucleotide deletion in the same region. The Gp85 protein of the four ALV-J isolates displayed one amino acid deletion and clustered in a distinct group in the phylogenetic tree.
The results presented here will contribute towards our understanding of the infection process of ALV-J in layer chickens in China and may shed light on the tumor phenotypes induced by ALV-J in recent years. Further investigations are required to study the relationship between mutations within the viral genome and the tumor types induced by ALV-J.
Methods
Background of the samples
Six commercial layer flocks (150-days-old) in Jiangsu, China, comprising about 3000–4000 chickens each, experienced production depression and elevated mortality in 2009. Approximately 20–30% of birds showed hemangioma on their body surface or internal organs, and the mortality associated with blood loss caused by hemangioma reached 15–20%. The six flocks shared the same batch of parent-breeding chickens and had no contact history with other flocks. One bird from each of the six commercial flocks was examined in this study. Serum and cloacal swabs samples were used to test for the p27 antigen and ALV-J antibodies, spleen and serum samples were used for virus isolation, and various tissues were used in histopathological examination.
The 12,000 breeding chickens in the common parental line flock of the six commercial layer flocks were introduced from a clinically asymptomatic grand parental-breeding flock in Beijing, China. This parental flock showed no clinically symptoms but had a short period of low hatching-rate (about 80%). The slow feathering maternal line and fast feathering paternal origin were contained in this parental breeding flock. The four different batches of chickens in this parental line were of different ages: 5-weeks-old, 12-weeks-old, 36-weeks-old, and 60-weeks-old. A total of 81 blood/cloacal swabs and 32 samples from the maternal line and paternal origin, respectively, from these four different batches, were tested for ALV-J infection by ELISA.
Group-specific antigen p27 in samples of either cloacal swabs from parental line chickens, or in cloacal swabs and serum from the six commercial chickens were detected by ELISA using the Avian Leukosis Virus Antigen Test Kit (IDEXX Laboratories, Westbrook, ME, U.S.A.). ALV-J antibodies in serum were detected using and Avian Leukosis Virus Antibody Test Kit - subgroup J (IDEXX Laboratories).
Necropsy and histopathology
The six commercial layer chickens used in this study were submitted by the owners to the laboratory. Clinical signs and gross lesions were examined. Tissues including the hemangioma, liver, spleen, kidney, heart, pancreas, muscular stomach, and duodenum, were fixed in 15% formalin, embedded in paraffin, sectioned into 5 μm slices and stained using haematoxylin and eosin for histopathology.
Viral genetic examination
Viral RNA extracted using the Multisource Total RNA Miniprep Kit (Axygen Biotechnology (Hangzhou) Limited, Hangzhou, China) from liver samples was examined by reverse transcription polymerase chain reaction (RT-PCR) with primers H5/H7 specific for ALV J subgroup and primer pair H5/AD1 for other ALV A–E subgroups using the PCR procedure performed according to the previously described method (Smith et al. 1998).
Virus isolation and identification
Samples of the spleen and serum from the six chickens were inoculated onto a monolayer of the chicken embryo fibroblast cell line (DF1 cells) which was resistant to subgroup E ALVs, to isolate the ALV-J. Cells were passaged every 7–10 days for two passages. The presence of ALV-J in infected cells was confirmed by an IFA with ALV-J specific monoclonal antibody JE9 (Qin et al. 2001).
PCR and sequencing of the full-length proviral DNA
DNA was extracted from the infected DF1 cells according to the method described by Sambrook et al. (2001). Briefly, after digestion, DF1 cell DNA was extracted with phenol and chloroform/isoamylalcohol, and precipitated with 70% ethanol. Primers were designed according to the prototype ALV strain HPRS103 (GenBank: Z46390) and were used to amplify the 5′LTR-gag, gag–pol, and pol–env-3′ noncoding regions of the proviral genome. The LTR–gag fragments (located at bp 1 to bp 2554 according to strain HPRS103) were obtained using the designed forward primer F1 5′-GGCAG CTGTG TAGTG TTATG CAATA CTCTT-3′ and the reverse primer R1 5′-GCATG GGAAT TCCCC CTCCTA-3′. The PCR to amplify the gag–pol gene fragments (bp 2543 to bp 5263 of strain HPRS103) were carried out with the forward primer F2 5′-CGAAT TCCCA TGCGA AAATCT-3′ and the reverse primer R2 5′-CTTGA TCATC CTTTT GGGTG ATGT-3′. The forward primer F3 5′-AGGTC GACCC CCGGT TAAGA TACGA AT-3′ and reverse primer R3 5′-GAATT CTGAA GCCAT CCGCT TCATG CAGGT-3′ were used to amplify the fragment of the pol–env-3′ noncoding regions (bp 5104 to bp 7841 of strain HPRS103). PCR amplifications were carried out with LA Taq polymerase (Takara Bio Inc., Dalian, China) and the following cycle conditions for all three reactions: 95°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 50°C for 1 min, and 72°C for 3 min. A final extension step was conducted at 72°C for 10 min. PCR products were ligated into the pGEM-T Easy cloning vector (Promega, USA). DNA sequences from the positive clones were determined by a local company (Invitrogen Biological Company, Shanghai, China). For all reactions, the isolation of DNA and PCR amplification were carried out at least twice to avoid PCR errors.
DNA sequence analysis
Reference strain sequences obtained from GenBank are described in the legends of the table and figures respectively. Phylogenetic analysis was performed using MegAlign in the DNASTAR program (Madison, WI, USA). After sequence alignment using the Jotun Hein or Clustal W algorithm of MegAlign module provided in DNASTAR program. Bootstrap analyses were carried out to evaluate statistical reliability based on 1000 re-samplings of the data set. Motif analysis of the transcription control sequences were carried out using the internet-based MOTIF-sequence search in the Kyoto encyclopedia of genes and genomes site (KEGG) (http://motif.genome.jp) (Kanehisa and Goto 2000).
Acknowledgements
The research was supported by the Joint Funds of the National Natural Science Foundation of China and Guangdong province (Grant No. U0831002), the R&D Special Foundation for Public Welfare Industry (No. 200803019) and the Major Basic Research of Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 07KJA23021).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Author contributions
Dr Qin supervised all of the research. Xiaoping Wu and Dr. Qin were responsible for the histopathological examination, virus detection isolation and sequence analysis. Haiyu Shen and Pingping Wang were responsible for the ELISA tests. Dr. Jin and Dr. Qian was responsible for sample collection, virus detection and isolation. Dr Eltahir gave some disccusion and revised the manuscript. All authors read and approved the manuscript.
Contributor Information
Xiaoping Wu, Email: pingping19781015@yahoo.cn.
Kun Qian, Email: qiankun@yzu.edu.cn.
Aijian Qin, Phone: +86-514-87979224, FAX: +86-514-87979217, Email: aijian@yzu.edu.cn.
Haiyu Shen, Email: ming521066@sina.com.
Pingping Wang, Email: wpp85@qq.com.
Wenjie Jin, Email: wenjiejin1@yzu.edu.cn.
Yassir Mohammed Eltahir, Email: yassrtahr@yahoo.com.
References
- Arshad SS, Howes K, Barron GS, Smith LM, Russell PH, Payne LN. Tissue tropism of the HPRS-103 strain of J subgroup avian leukosis virus and of a derivative acutely transforming virus. Vet Pathol. 1997;34(2):127–137. doi: 10.1177/030098589703400205. [DOI] [PubMed] [Google Scholar]
- Berg DT, Gupta A, Richardson MA, O'Brien LA, Calnek D, Grinnell BW. Negative regulation of inducible nitric-oxide synthase expression mediated through transforming growth factor-beta-dependent modulation of transcription factor TCF11. J Biol Chem. 2007;282(51):36837–36844. doi: 10.1074/jbc.M706909200. [DOI] [PubMed] [Google Scholar]
- Chesters PM, Howes K, Petherbridge L, Evans S, Payne LN, Venugopal K. The viral envelope is a major determinant for the induction of lymphoid and myeloid tumours by avian leukosis virus subgroups A and J, respectively. J Gen Virol. 2002;83(Pt 10):2553–2561. doi: 10.1099/0022-1317-83-10-2553. [DOI] [PubMed] [Google Scholar]
- Chesters PM, Smith LP, Nair V. The E (XSR) element contributes to the oncogenicity of avian leukosis virus (subgroup J) J Gen Virol. 2006;87:2685–2692. doi: 10.1099/vir.0.81884-0. [DOI] [PubMed] [Google Scholar]
- Chesters PM, Smith LP, Nair V. E (XSR) element contributes to the oncogenicity of Avian leukosis virus (subgroup J) J Gen Virol. 2006;87(Pt 9):2685–2692. doi: 10.1099/vir.0.81884-0. [DOI] [PubMed] [Google Scholar]
- Cui Z, Du Y, Zhang Z, Silva RF. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis. 2003;47(4):1321–1330. doi: 10.1637/6085. [DOI] [PubMed] [Google Scholar]
- Donze O, Spahr PF. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11(10):3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey JF, Cunnick JM, Mane SM, Wu J. Regulation of the Erk2-Elk1 signaling pathway and megakaryocytic differentiation of Bcr-Abl (+) K562 leukemic cells by Gab2. Blood. 2002;99(4):1388–1397. doi: 10.1182/blood.V99.4.1388. [DOI] [PubMed] [Google Scholar]
- Gilles F, Raes MB, Stehelin D, Vandenbunder B, Fafeur V. The c-ets-1 Proto-oncogene Is a New Early-Response Gene Differentially Regulated by Cytokines and Growth Factors in Human Fibroblasts. Exp Cell Res. 1996;222:370–378. doi: 10.1006/excr.1996.0046. [DOI] [PubMed] [Google Scholar]
- Gingerich E, Porter RE, Lupiani B, Fadly AM. Diagnosis of myeloid leukosis induced by a recombinant avian leukosis virus in commercial white leghorn egg laying flocks. Avian Dis. 2002;46(3):745–748. doi: 10.1637/0005-2086(2002)046[0745:DOMLIB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Goldberg Y, Treier M, Ghysdael J, Bohmann D. Repression of AP-1-stimulated transcription by c-Ets-1. J Biol Chem. 1994;269(24):16566–16573. [PubMed] [Google Scholar]
- Hatai H, Ochiai K, Nagakura K, Imanishi S, Ochi A, Kozakura R, Ono M, Goryo M, Ohashi K, Umemura T. A recombinant avian leukosis virus associated with fowl glioma in layer chickens in Japan. Avian Pathol. 2008;37(2):127–137. doi: 10.1080/03079450801898815. [DOI] [PubMed] [Google Scholar]
- Hue D, Dambrine G, Denesvre C, Laurent S, Wyers M, Rasschaert D. Major rearrangements in the E element and minor variations in the U3 sequences of the avian leukosis subgroup J provirus isolated from field myelocytomatosis. Arch Virol. 2006;151(12):2431–2446. doi: 10.1007/s00705-006-0811-2. [DOI] [PubMed] [Google Scholar]
- Hue D, Dambrine G, Denesvre C, Laurent S, Wyers M, Rasschaert D. Major rearrangements in the E element and minor variations in the U3 sequences of the avian leukosis subgroup J provirus isolated from field myelocytomatosis. Arch Virol. 2006;151:2431–2446. doi: 10.1007/s00705-006-0811-2. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiani B, Pandiri AR, Mays J, Hunt HD, Fadly AM. Molecular and biological characterization of a naturally occurring recombinant subgroup B avian leukosis virus with a subgroup J-like long terminal repeat. Avian Dis. 2006;50(4):572–578. doi: 10.1637/7656-053006R.1. [DOI] [PubMed] [Google Scholar]
- Mays JK, Pandiri AR, Fadly AM. Susceptibility of various parental lines of commercial white leghorn layers to infection with a naturally occurring recombinant avian leukosis virus containing subgroup B envelope and subgroup J long terminal repeat. Avian Dis. 2006;50(3):342–347. doi: 10.1637/7493-121505R.1. [DOI] [PubMed] [Google Scholar]
- Nichol S. RNA viruses. Life on the edge of catastrophe. Nature. 1996;384(6606):218–219. doi: 10.1038/384218a0. [DOI] [PubMed] [Google Scholar]
- Payne A: The emergence of myeloid leukosis in meat-type chickens. Proceedings of 99 international conference and exhibition on veterinary poultry 1999:l–10.
- Payne LN, Fadly AM, editors. Leukosis/sarcoma group. Ames IA: Iowa State University Press; 1997. [Google Scholar]
- Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72(Pt 4):801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- Payne LN, Howes K, Gillespie AM, Smith LM. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol. 1992;73(Pt 11):2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- Qin A, Lee LF, Fadly A, Hunt H, Cui Z. Development and characterization of monoclonal antibodies to subgroup J avian leukosis virus. Avian Dis. 2001;45(4):938–945. doi: 10.2307/1592872. [DOI] [PubMed] [Google Scholar]
- Rong L, Edinger A, Bates P. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J Virol. 1997;71(5):3458–3465. doi: 10.1128/jvi.71.5.3458-3465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW: Preparation and analysis of eukaryotic genomic DNA. In: Molecular Cloning: A Laboratory Manual Edited by JS, DW R, vol. 1. New York Cold Spring Harbor Laboratory Press; 2001: 6.11–16.14.
- Schwartz DE, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Smith EJ, Williams SM, Fadly AM. Detection of avian leukosis virus subgroup J using the polymerase chain reaction. Avian Dis. 1998;42(2):375–380. doi: 10.2307/1592488. [DOI] [PubMed] [Google Scholar]
- Sonstegard TS, Hackett PB. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70(10):6642–6652. [PMC free article] [PubMed] [Google Scholar]
- Svoboda J, Dvorak M, Guntaka RV, Geryk J. Transmission of (LTR, v-src, LTR) without recombination with a helper virus. Virol. 1986;153(2):314–317. doi: 10.1016/0042-6822(86)90035-8. [DOI] [PubMed] [Google Scholar]
- Wang H, Cui Z. The Identification and Sequence Analysis of ALV-J Isolated from Layers. Chinese Journal of Virology. 2008;24(5):369–375. [PubMed] [Google Scholar]
- Wei D, Richardson EL, Zhu K, Wang L, Le X, He Y, Huang S, Xie K. Direct demonstration of negative regulation of tumor growth and metastasis by host-inducible nitric oxide synthase. Cancer Res. 2003;63(14):3855–3859. [PubMed] [Google Scholar]
- Williams SM, Reed WM, Bacon LD, Fadly AM. Response of white leghorn chickens of various genetic lines to infection with avian leukosis virus subgroup J. Avian Dis. 2004;48(1):61–67. doi: 10.1637/7052. [DOI] [PubMed] [Google Scholar]
- Witter RL, Fadly AM. Reduction of horizontaltransmission of avian leukosis virus subgroup J in broiler breeder chickens hatched and reared in small groups. Avian Pathol. 2001;30:641–654. doi: 10.1080/03079450120092134. [DOI] [PubMed] [Google Scholar]
- Xu B, Dong W, Yu C, He Z, Lv Y, Sun Y, Feng X, Li N, Lee LF, Li M. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol. 2004;33(1):13–17. doi: 10.1080/03079450310001636237a. [DOI] [PubMed] [Google Scholar]
- Zavala SC G, Jackwood MW. Molecular epidemiology of avian leukosis virus subgroup J and evolutionary history of its untranslated region. Avian Dis. 2007;51(4):942–953. doi: 10.1637/0005-2086(2007)51[942:MEOALV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Zavala G, Cheng S, Jackwood MW. Molecular epidemiology of avian leukosis virus subgroup J and evolutionary history of its 3′ untranslated region. Avian Dis. 2007;51(4):942–953. doi: 10.1637/0005-2086(2007)51[942:MEOALV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]