Abstract
Objective:
In addition to the main multiple sclerosis (MS) major histocompatibility complex (MHC) risk allele (HLA DRB1*1501), investigations of the MHC have implicated several class I MHC loci (HLA A, HLA B, and HLA C) as potential independent MS susceptibility loci. Here, we evaluate the role of 3 putative protective alleles in MS: HLA A*02, HLA B*44, and HLA C*05.
Methods:
Subjects include a clinic-based patient sample with a diagnosis of either MS or a clinically isolated syndrome (n = 532), compared to subjects in a bone marrow donor registry (n = 776). All subjects have 2-digit HLA data. Logistic regression was used to determine the independence of each allele's effect. We used linear regression and an additive model to test for correlation between an allele and MRI and clinical measures of disease course.
Results:
After accounting for the effect of HLA DRB1*1501, both HLA A*02 and HLA B*44 are validated as susceptibility alleles (pA*02 0.00039 and pB*44 0.00092) and remain significantly associated with MS susceptibility in the presence of the other allele. Although A*02 is not associated with MS outcome measures, HLA B*44 demonstrates association with a better radiologic outcome both in terms of brain parenchymal fraction and T2 hyperintense lesion volume (p = 0.03 for each outcome).
Conclusion:
The MHC class I alleles HLA A*02 and HLA B*44 independently reduce susceptibility to MS, but only HLA B*44 appears to influence disease course, preserving brain volume and reducing the burden of T2 hyperintense lesions in subjects with MS.
GLOSSARY
- BPF
= brain parenchymal fraction;
- CIS
= clinically isolated demyelinating syndrome;
- EDSS
= Expanded Disability Status Scale;
- KIR
= killer immunoglobulin receptor;
- MHC
= major histocompatibility complex;
- MS
= multiple sclerosis;
- MSSS
= Multiple Sclerosis Severity Score;
- NK
= natural killer;
- SNP
= single nucleotide polymorphism.
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the CNS that has a component of genetic susceptibility. The locus with the strongest association with susceptibility is the major histocompatibility complex (MHC), and this association has been well-validated in a multitude of studies.1 The main risk haplotype for MS within the MHC is tagged by the HLA DRB1*1501 allele.1 However, there is mounting evidence that other haplotypes may also contribute to MS susceptibility in European populations.2–5 In particular, 3 class I MHC alleles have been proposed to be associated with susceptibility to MS after accounting for the effect of HLA DRB1*1501: HLA A*02, HLA B*4402, and HLA C*05.2,6–10 The most recent class I MHC allele, HLA B*4402, was discovered by imputing HLA alleles from single nucleotide polymorphism (SNP) genotype data and is consistent with a recent report that the HLA Bw4 group of HLA B alleles (which includes HLA B4402) is associated with reduced susceptibility to MS.8,11
While imputation of SNP data provides good estimates of HLA type in many cases, the effectiveness of this method is allele-specific. SNP data can localize an effect, but there is, as yet, not a comprehensive set of surrogate SNPs for each MHC class I or II allele.12 Thus, we turned to a novel, independent dataset of class I and II MHC typing data to validate the respective roles of these 3 MHC class I alleles. Our new data are of 2-digit resolution, meaning that, for example, HLA B*4402 is one of several alleles captured under the HLA B*44 rubric used in the HLA typing data that we have collected in the subjects from the Partners MS Center in Boston.
Here, we replicate the existence of HLA DRB1*1501-independent MHC class I susceptibility haplotypes and refine the relative roles of HLA A*02, HLA B*44, and HLA C*05. While the HLA C*05 allele appears to be secondary to HLA B*44, HLA A*02 is an independent risk allele. Secondarily, we extend our analysis to explore the phenotypic associations of the protective HLA A*02 and HLA B*44 alleles in subjects with MS.
METHODS
Human subjects.
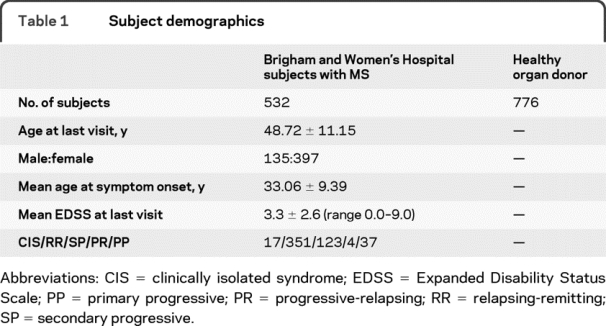
All subjects with MS met the revised McDonald diagnostic criteria13 or had a clinically isolated demyelinating syndrome (CIS); the latter subjects have a history of a single episode of inflammatory demyelination documented by a neurologist and had 2 or more periventricular or ovoid hyperintense T2 lesions of >3 mm on MRI.14 The control subjects consist of a population of 776 anonymous samples from unrelated subjects consecutively recruited as volunteer donors for a bone marrow registry by the C.W. Bill Young Department of Defense Marrow Donor Program. Because of the recruitment setting, individuals are unlikely to be related and are likely to originate from different areas of the United States. The MS and control subjects are self-identified as non-Latino, non-Asian, non–African American subjects of European ancestry. The demographic profile of both sets of subjects is outlined in table 1.
Table 1 Subject demographics
Standard protocol approvals, registrations, and patient consents.
The study was approved by the institutional review board of Partners Healthcare and written informed consent was obtained from all patients participating in the study.
Clinical and HLA typing data.
Clinical data were obtained from the Partners MS Center Oracle relational database. At each clinic visit, the Expanded Disability Status Scale (EDSS) score is measured on each patient.15 Using the EDSS and disease duration recorded at the most recent clinic visit, the Multiple Sclerosis Severity Score (MSSS) of each subject was calculated using the Global MSSS table provided by Roxburgh and colleagues.16 A total of 374 patients had an EDSS and recorded disease duration of 30 years or less and contributed to the MSSS analysis. Details on the generation of the HLA typing data are available in appendix e-1 on the Neurology® Web site at www.neurology.org.17,18
Radiographic data.
The details of the Partners MS Center's collection of volumetric data have been previously described.19 In short, routine clinical MRI scans were obtained on a 1.5-Tesla magnetic resonance system (Signa, GE Medical Systems, Milwaukee, WI) using a standard birdcage quadrature coil as part of routine clinical practice at the Partners MS Center.20 Each brain is segmented using an automated template-driven segmentation pipeline termed TDS+.21,22 No healthy control subjects had radiographic data. A total of 378 patients (2,067 scans) with MHC class I information had radiographic data and recorded disease duration less than 40 years. A detailed description of our data extraction and quality control pipeline is available in appendix e-1. The final dataset consisted of 375 patients with 1,933 scans.
Statistical analysis.
The allele frequencies for HLA DRB1*1501, HLA A*02, HLA B*44, and HLA C*05 were calculated by tabulating the total number of alleles of each category. Genetic association was assessed using a standard χ2 test completed on a 2 × 2 contingency table for each HLA allele tested. The association between other alleles at each loci was also considered using the same approach. In order to address the potential for association due solely to linkage disequilibrium, multiple logistic regression models were used to estimate the association between HLA A*02, HLA B*44, and HLA C*05 and susceptibility controlling for HLA DRB1*1501 and the other loci. In each case, the number of alleles was used as the predictor in the model. Interactions between alleles on the log odds scale were investigated using the logistic regression model. In addition, the association between the secondary loci and susceptibility was investigated in HLA DRB1*1501−/− patients. For comparison of clinical characteristics in the patients with MS, a Kruskal-Wallis test was used to compare the MSSS across the 3 genotype groups. For comparison of age at onset and radiographic characteristics (brain parenchymal fraction [BPF] and log-transformed T2 lesion volume), we compared the characteristics at last visit using linear regression with an additive effect of each allele. Multivariate models for the radiographic characteristics were also investigated to control for age, disease duration, and gender. For longitudinal MRI follow-up, the change over time in BPF and T2 lesion volume was investigated using a random intercept and slope model. For this approach, an additive effect of each allele was used as a predictor. To assess whether an allele had a significant effect on the change with time, the interaction between the allele and study time was investigated. The model also included baseline age, disease duration, and gender as potential confounders. To minimize heterogeneity due to disease course and biologic age, we also completed a restricted analysis among patients under age 65 years and within the first 10 years of disease (calculated from the onset of first symptom). All statistical analysis was completed in the statistical package R (http://www.R-project.org).
RESULTS
This is the first assessment of the MHC in our collection of subjects with MS from the Partners MS Center. When the subjects with MS or CIS (n = 532) were compared to healthy organ donors (n = 776), the expected increased frequency of the HLA DRB1*1501 allele among MS subjects was observed (table 2) (p = 2.44 × 10−12). Additional associations were observed at several MHC class I alleles including the 3 alleles of interest: HLA A*02, HLA B*44, and HLA C*05. Unlike our earlier study in subjects from the United Kingdom,2 significant associations were not observed with HLA DRB1*01 either before or after taking the effect of HLA DRB1*1501 into account and only a marginal evidence of association was observed with HLA DRB1*03 after accounting for HLA DRB1*1501 in these subjects from the United States (tables e-1 and e-2). Given our data, these alleles were not included in further modeling, and we focused on the class I alleles since they displayed much more robust evidence of association. No other DRB1 alleles were considered in the modeling since HLA A*02 and HLA B*44 were the first and second most associated allele among the tested loci after the effect of HLA DRB1*1501 was taken into account.
Table 2 Associations with loci of interest
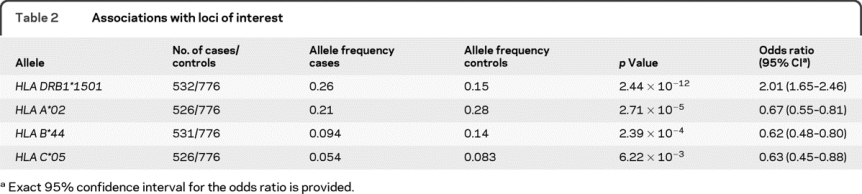
To address whether the 3 HLA alleles of interest have independent effects, multiple logistic regression was used. After accounting for the effect of HLA DRB1*1501, all 3 alleles remained associated with MS susceptibility (table 3). Given this residual association, a set of additional regression analyses examined the effect of each of the three alleles in the presence of the other alleles (table 3). This series of analyses demonstrated that the effect of the HLA C*05 allele was secondary to that of the HLA B*44 allele and HLA A*02 allele since no significant residual association was observed with HLA C*05 in the larger models. No significant gene/gene interactions were observed in these analyses. This confirms our earlier analysis that suggested the secondary role of HLA C*05 relative to HLA B*44 using SNP data in a different sample set.8 These analyses also examined the relative roles of the HLA A*02 and HLA B*44 alleles in the presence of the other allele. In our dataset, these 2 associations appeared to be distinct since a residual association with MS susceptibility remained for (1) HLA A*02 in the presence of HLA DRB1*1501 and HLA B*44 and (2) HLA B*44 in the presence of HLA DRB1*1501 and HLA A*02 (table 3). No significant gene/gene interaction between HLA B*44 and HLA A*02 was observed in this analysis. Thus, our study suggests the existence of 2 separate MHC class I associations, confirming both the recent report concerning HLA B*44 and the published associations with HLA A*02.
Table 3 Associations of each loci in regression analyses of all subjects (n = 1,308)
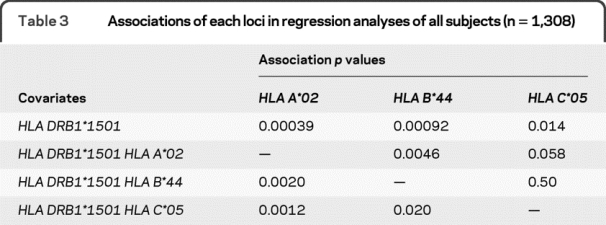
To confirm these results using a different approach, we also analyzed our data after limiting the analysis to those subjects who were HLA DRB1*1501−/−. Table e-3 reports the results of these analyses, which mirror the results of the primary analysis. In particular, HLA A*02 and HLA B*44 had independent effects while HLA C*05 appeared to be have no residual effect in the presence of HLA B*44.
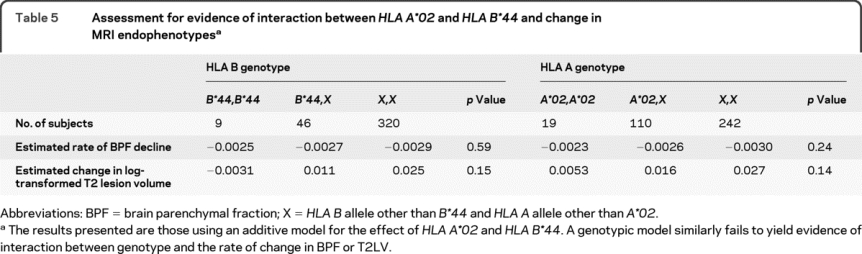
Given the existence of 2 MHC class I loci associated with MS susceptibility, the association between these loci and MS outcomes was examined. In our cross-sectional analyses, HLA A*02 did not have a significant association with either our clinical measures—the age at symptom onset and MSSS—or measures derived from MRI—BPF and T2 hyperintense lesion volume at last visit (table 4). On the other hand, HLA B*44 was associated with both MRI measures at last visit in both univariate (p = 0.03 for each measure, table 4) and multivariate analyses (p = 0.02 for BPF and p = 0.03 for T2 lesion volume). HLA B*44 was associated with higher BPF and lower T2 lesion volume, which is consistent with the protective effect of this allele in terms of susceptibility. No significant association between HLA B*44 and age at onset or MSSS was observed (table 4). In longitudinal analyses, neither allele was significantly associated with the rate of change in BPF or T2 lesion volume, but the direction of the effect was toward a protective effect for each allele (table 5). This suggests that subjects with an HLA B*44 allele have a slower rate of brain atrophy and T2 hyperintense lesion accumulation than other subjects with MS.
Table 4 Assessment of association between HLA A*02 and HLA B*44 genotypes and clinical/demographic/radiologic outcomes
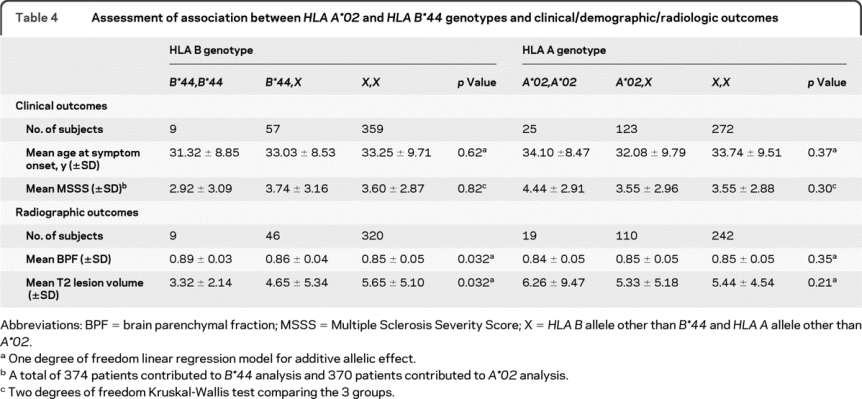
Table 5 Assessment for evidence of interaction between HLA A*02 and HLA B*44 and change in MRI endophenotypes
A comprehensive assessment of evidence of association between HLA A, B, C, and DRB1 alleles and MS susceptibility is provided in tables e-1 and e-2. Consistent with previous reports, the relatively uncommon HLA DRB1*14 allele displayed modest evidence of a protective effect in terms of MS susceptibility.3,23 In addition, there is evidence of further heterogeneity in the MHC locus, with 3 suggestive associations appearing in the analysis where the effects of HLA A*02, HLA B*44, and HLA DRB1*1501 are accounted for (full model p) (tables e-1 and e-2): HLA A*03, univariate (no covariates) p = 0.8, full model p = 0.001; HLA C*03, univariate p = 0.0002, full model p = 0.002; and HLA DRB1*11, univariate p = 0.06, full model p = 0.003. Given the 50 alleles tested in this secondary analysis, study-wide significance is established as a full model p < 0.001. Of note, we saw a nonsignificant trend for a higher frequency of an HLA DRB1*1501/*08 genotype in subjects with MS (data not shown); this genotype combination has previously been proposed as a high-risk genotype in relation to MS susceptibility.3
DISCUSSION
In our focused analysis of the MHC class I region, we confirm that this chromosomal segment is associated with MS independently of the HLA DRB1*1501 association. Specifically, we validate for the first time the earlier observation concerning the role of the HLA B*44 allele and confirm the role of the HLA A*02 allele first identified in Scandinavian populations by demonstrating their association with MS susceptibility in an independent cohort of subjects with MS from the Partners MS Center. The results are consistent with other reports validating the role of the HLA A*02 allele and implicating the HLA Bw4 group of alleles.7,9,11,24 Further, we enhance the state of the field by demonstrating that these 2 effects are independent of one another and that the previously described association of the HLA Cw*05 allele with susceptibility to MS2 is likely to be due to linkage disequilibrium with HLA B*44.
In our dataset, both HLA A*02 and HLA B*44 have substantial effects on susceptibility (ORA*02 = 0.67 and ORB*44 = 0.62). These effects are larger than that of the non-MHC risk alleles discovered to date25 and will play an important role as we assemble a comprehensive map of the genetic architecture of MS in coming years. Given the MHC's allelic heterogeneity, long-range linkage disequilibrium, and the large effect size of HLA DRB1*1501, it is not surprising that loci with effects of this magnitude are only being validated now. In fact, we suspect that additional susceptibility loci will be found within the MHC as part of future investigations in larger sample sets. While the 2 MHC class I alleles discussed here are clearly associated with susceptibility to MS, we should remember that they exist on haplotypes that extend over long physical distances and encompass many different polymorphisms. Thus, we cannot say that HLA A*02 and HLA B*44 are causal variants at this time; they simply represent the best markers of independent protective haplotypes within the MHC. It is also possible that multiple alleles in strong linkage disequilibrium with either allele could be required to achieve the protective effects that we have validated.
The association of MHC class I alleles with susceptibility to MS highlights the role of certain cell types, such as CD8+ and natural killer (NK) cells, in the onset of MS. These cell types harbor receptors for MHC class I, and these findings fit well with the reported dysfunction of CD8+ and NK cells in early stages of demyelinating disease.26 Recently, in a large-scale scan of peripheral blood cell populations, we identified and validated a reduction in the frequency of a CD56+ NK cell population in untreated subjects who either had relapsing-remitting MS or a clinically isolated demyelinating syndrome.27 Many other investigators have assessed the role of NK cells in MS, and, while some early studies were negative, the preponderance of the evidence available to date suggests that NK cell frequency is reduced in MS and that they may be dysfunctional.28 The convergence of these immunologic and genetic data strongly argue for a comprehensive assessment of the complex interactions between MHC class I molecules and their various receptors in subjects with MS, both at the genetic and functional level. An early effort in this direction used HLA Bw4 and explored the role of killer immunoglobulin receptors (KIR) in MS susceptibility, including KIR3DL1, which is a receptor for this group of HLA B alleles.11 While KIR alleles did not appear to be associated in this study of moderate size, modest effects typical of inflammatory disease susceptibility alleles cannot yet be excluded and will require the careful assessment of allele combinations between KIR and MHC class I alleles.
Interestingly, a protective effect of HLA B*44 has been previously reported in subjects with CD95/TNFRSF6 mutations resulting in autoimmune lymphoproliferative syndrome type 1a. Subjects with such mutations who also bear an HLA B*44 allele were less likely to develop the full spectrum of autoimmune lymphoproliferative syndrome clinical features such as lymphadenopathy, splenomegaly, autoimmunity, and lymphoma.29 Furthermore, HLA B*44 homozygotes have also been reported to have lower NK cell activity, offering one suggestion as to the mechanism by which HLA B*44 may reduce MS susceptibility.30
While HLA A*02 and HLA B*44 are clearly validated as protective haplotypes in this and other studies,9,11,24 we see evidence that only the HLA B*44 haplotype affects volumetric MRI measures (BPF and T2 hyperintense lesion volume): on average, subjects with MS with HLA B*44 alleles had a larger brain volume and a lower lesion burden than subjects with MS who lacked this allele. Our secondary analysis of longitudinal data suggests that these correlations may arise from a reduction in the rates of brain atrophy and T2 lesion volume accumulation. Thus, the difference in brain volume at a subject's last MRI is less likely to be due to differences in baseline brain volume. However, further work is needed to confirm our observations relating these MS outcome measures and HLA B*44 as well as an independent report of associations between HLA A*02 and measures of MS disease course that we did not validate in our sample set.31 Overall, MS susceptibility alleles appear to have a limited role in MS disease severity so far,32–35 and dedicated studies are needed to explore the genetic architecture of disease course in MS.
As suggested by our comprehensive assessment of MHC alleles in table e-1, additional risk-associated and protective alleles, particularly in the HLA C locus, may well exist, and dedicated efforts to further explore this critical region are therefore warranted. Such efforts that include-high resolution HLA typing of multiple loci are needed to supplement the ongoing large SNP-based efforts: so far, HLA types are, for the most part, imperfectly tagged by SNP markers and will be needed to identify the causal allele or alleles in each locus.
AUTHOR CONTRIBUTIONS
Statistical analysis was conducted by Dr. Brian Healy.
DISCLOSURE
Dr. Healy receives research support from Merck Serono. Dr. Liguori and D. Tran report no disclosures. Dr. Chitnis has served as a consultant for Biogen Idec, Teva Pharmaceutical Industries Ltd., and EMD Serono, Inc.; served on a speakers' bureau for EMD Serono, Inc.; and receives research support from the NIH (NINDS K08 NS 047669-01 [PI]) and the National Multiple Sclerosis Society. Dr. Glanz receives research support from Merck Serono, the NIH (NINDS 5R01NS055083-03 [coinvestigator]), and the National Multiple Sclerosis Society. C. Wolfish reports no disclosures. Dr. Gauthier has received funding for travel or speaker honoraria from Biogen Idec and EMD Serono, Inc.; and has received research support from EMD Serono, Inc. Dr. Buckle has served on scientific advisory boards for Acorda Therapeutics Inc., Biogen Idec, EMD Serono, Inc., Bayer Schering Pharma, Teva Pharmaceutical Industries Ltd., and Pfizer Inc.; and has served on speakers' bureaus for and received speaker honoraria from Biogen Idec, EMD Serono, Inc., Bayer Schering Pharma, Teva Pharmaceutical Industries Ltd., and Pfizer Inc. Dr. Houtchens has served on scientific advisory boards for and received speaker honoraria from EMD Serono, Inc., Biogen Idec, and Teva Pharmaceutical Industries Ltd. L. Stazzone has received funding for travel and speaker honoraria from EMD Serono, Inc. Dr. Khoury has served on scientific advisory boards for EpiVax and Repligen Corporation; serves as a Section Editor for Clinical Immunology; has received speaker honoraria from Repligen Corporation and EMD Serono, Inc.; has received research support from Biogen Idec, the NIH (R01 AI071448 [PI], R01 AI067472 [PI], and R56 AI058680-05 [PI]), the National Multiple Sclerosis Society, and the Fidelity Foundation; and her spouse receives research support from Genzyme Corporation. Dr. Hartzmann is an employee of the US Navy and has received intramural research funding from the Office of Naval Research and Naval Medical Research Center. Dr. Fernandez-Vina serves on editorial advisory boards for Immunogenetics and Human Immunology and serves as a consultant for the National Marrow Donor Program. Dr. Hafler serves on scientific advisory boards and as medico-legal consultant for, and has received speaker honoraria from, Allozyne, Inc., Eisai Inc., and Xceed Molecular Inc.; and receives research support from the NIH (NINDS R37 NS024247 [PI], NINDS POI NS038037, NINDS ROI NS049477, U19 AI0703S2 [PI], POI AI073748, and P01 AI045757) and the National Multiple Sclerosis Society. Dr. Weiner has served/serves on scientific advisory boards for and received speaker honoraria from Biogen Idec, Genentech, Inc., EMD Serono, Inc., and Teva Pharmaceutical Industries Ltd.; serves on the editorial boards of Clinical Immunology, Multiple Sclerosis, the Journal of Immunology, and the Journal of Autoimmunity; and receives research support from the NIH (NINDS P01 NS 038037-06A2 [Program Director], NIAID R01 AI43458 [PI], and R01 AG027437 [PI]), and the National Multiple Sclerosis Society. Dr. Guttmann has served on a scientific advisory board for Tibotec Therapeutics/Johnson & Johnson; holds patents re: Versatile stereotactic device and methods of use and overlay of tinted images for visualizing change in serial radiologic images; and has received/receives research support from Teva Pharmaceutical Industries Ltd., the NIH (R01 AG022092-01 [PI of subcontract], HRCA PO1AG004390 [PI of subcontract], NIH R01 NS036524-05 [PI of subcontract], R01 NS055083–01A1 [coinvestigator], NCRR UL1 RR025758-01 [coinvestigator], 2P41RR013218 Supplement [coinvestigator], 2U01AI063623-06 [coinvestigator], NCRR P41 RR013218 [coinvestigator]), the Maurice Pechet Foundation, and the National Multiple Sclerosis Society. Dr. De Jager has served on scientific advisory boards for Teva Pharmaceutical Industries Ltd. and Biogen Idec; serves on the editorial board of the Journal of Neuroimmunology; has received speaker honoraria from Biogen Idec and The Jackson Laboratory; has served as a consultant for Merck Serono; and receives research support from the NIH (ARRA RC2 AG036650 [PI], R01 NS067305 [PI], ARRA RC2 AG036547 [coinvestigator], ARRA RC2 GM093080 [coinvestigator], ARRA RC2 NS070340 [coinvestigator], and R01 AG036042 2008-R01 AG15819 [coinvestigator]), and the National Multiple Sclerosis Society.
Supplementary Material
Address correspondence and reprint requests to Dr. Philip L. De Jager, Program in Translational NeuroPsychiatric Genomics, Department of Neurology, Brigham and Women's Hospital, 77 Avenue Louis Pasteur, NRB 168c, Boston, MA 02115 pdejager@rics.bwh.harvard.edu
Supplemental data at www.neurology.org
*These authors contributed equally to this work.
Study funding: Supported by the National Multiple Sclerosis Society. P.L.D. is a Harry Weaver Neuroscience Scholar Award Recipient of the National MS Society (NMSS). D.A.H. is a Jacob Javits Scholar of the NIH.
Disclosure: Author disclosures are provided at the end of the article.
Received November 25, 2009. Accepted in final form May 3, 2010.
REFERENCES
- 1.Hauser SL, Oksenberg JR. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 2006;52:61–76. [DOI] [PubMed] [Google Scholar]
- 2.Yeo TW, De Jager PL, Gregory SG, et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol 2007;61:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcellos LF, Sawcer S, Ramsay PP, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet 2006;15:2813–2824. [DOI] [PubMed] [Google Scholar]
- 4.Harbo HF, Lie BA, Sawcer S, et al. Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens 2004;63:237–247. [DOI] [PubMed] [Google Scholar]
- 5.Chao MJ, Barnardo MC, Lincoln MR, et al. HLA class I alleles tag HLA-DRB1*1501 haplotypes for differential risk in multiple sclerosis susceptibility. Proc Natl Acad Sci USA 2008;105:13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogdell-Hahn A, Ligers A, Gronning M, Hillert J, Olerup O. Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens 2000;55:140–148. [DOI] [PubMed] [Google Scholar]
- 7.Brynedal B, Duvefelt K, Jonasdottir G, et al. HLA-A confers an HLA-DRB1 independent influence on the risk of multiple sclerosis. PLoS One 2007;2:e664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rioux JD, Goyette P, Vyse TJ, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA 2009;106:18680–18685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergamaschi L, Leone MA, Fasano ME, et al. HLA-class I markers and multiple sclerosis susceptibility in the Italian population. Genes Immun 2010;11:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burfoot RK, Jensen CJ, Field J, et al. SNP mapping and candidate gene sequencing in the class I region of the HLA complex: searching for multiple sclerosis susceptibility genes in Tasmanians. Tissue Antigens 2008;71:42–50. [DOI] [PubMed] [Google Scholar]
- 11.Lorentzen AR, Karlsen TH, Olsson M, et al. Killer immunoglobulin-like receptor ligand HLA-Bw4 protects against multiple sclerosis. Ann Neurol 2009;65:658–666. [DOI] [PubMed] [Google Scholar]
- 12.de Bakker PI, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006;38:1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 2001;50:121–127. [DOI] [PubMed] [Google Scholar]
- 14.Miller D, Barkhof F, Montalban X, Thompson A, Filippi M. Clinically isolated syndromes suggestive of multiple sclerosis, part 2: non-conventional MRI, recovery processes, and management. Lancet Neurol 2005;4:341–348. [DOI] [PubMed] [Google Scholar]
- 15.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 1983;33:1444–1452. [DOI] [PubMed] [Google Scholar]
- 16.Roxburgh RH, Seaman SR, Masterman T, et al. Multiple Sclerosis Severity Score: using disability and disease duration to rate disease severity. Neurology 2005;64:1144–1151. [DOI] [PubMed] [Google Scholar]
- 17.Tu B, Mack SJ, Lazaro A, et al. HLA-A, -B, -C, -DRB1 allele and haplotype frequencies in an African American population. Tissue Antigens 2007;69:73–85. [DOI] [PubMed] [Google Scholar]
- 18.Cao K, Chopek M, Fernandez-Vina MA. High and intermediate resolution DNA typing systems for class I HLA-A, B, C genes by hybridization with sequence-specific oligonucleotide probes (SSOP). Rev Immunogenet 1999;1:177–208. [PubMed] [Google Scholar]
- 19.Gauthier SA, Mandel M, Guttmann CR, et al. Predicting short-term disability in multiple sclerosis. Neurology 2007;68:2059–2065. [DOI] [PubMed] [Google Scholar]
- 20.Meier DS, Guttmann CR. MRI time series modeling of MS lesion development. Neuroimage 2006;32:531–537. [DOI] [PubMed] [Google Scholar]
- 21.Wei X, Warfield SK, Zou KH, et al. Quantitative analysis of MRI signal abnormalities of brain white matter with high reproducibility and accuracy. J Magn Reson Imaging 2002;15:203–209. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Meier D, Polgar-Turcsanyi M, Karkocha P, Bakshi R, Guttmann CR. Multiple sclerosis medical image analysis and information management. J Neuroimaging 2005;15:103S–117S. [DOI] [PubMed] [Google Scholar]
- 23.Ramagopalan SV, Morris AP, Dyment DA, et al. The inheritance of resistance alleles in multiple sclerosis. PLoS Genet 2007;3:1607–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva AM, Bettencourt A, Pereira C, et al. Protective role of the HLA-A*02 allele in Portuguese patients with multiple sclerosis. Mult Scler 2009;15:771–774. [DOI] [PubMed] [Google Scholar]
- 25.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 2009;41:776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFarland HF, Martin R. Multiple sclerosis: a complicated picture of autoimmunity. Nat Immunol 2007;8:913–919. [DOI] [PubMed] [Google Scholar]
- 27.De Jager PL, Rossin E, Pyne S, et al. Cytometric profiling in multiple sclerosis uncovers patient population structure and a reduction of CD8low cells. Brain 2008;131:1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segal BM. The role of natural killer cells in curbing neuroinflammation. J Neuroimmunol 2007;191:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacek MM, Schaffer AA, Davis J, et al. HLA B44 is associated with decreased severity of autoimmune lymphoproliferative syndrome in patients with CD95 defects (ALPS type Ia). Clin Immunol 2006;118:59–65. [DOI] [PubMed] [Google Scholar]
- 30.Dubey DP, Alper CA, Mirza NM, Awdeh Z, Yunis EJ. Polymorphic Hh genes in the HLA-B(C) region control natural killer cell frequency and activity. J Exp Med 1994;179:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivadinov R, Weinstock-Guttman B, Zorzon M, et al. Gene-environment interactions between HLA B7/A2, EBV antibodies are associated with MRI injury in multiple sclerosis. J Neuroimmunol 2009;209:123–130. [DOI] [PubMed] [Google Scholar]
- 32.Okuda DT, Srinivasan R, Oksenberg JR, et al. Genotype-Phenotype correlations in multiple sclerosis: HLA genes influence disease severity inferred by 1HMR spectroscopy and MRI measures. Brain 2009;132:250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baranzini SE, Wang J, Gibson RA, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet 2009;18:767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smestad C, Brynedal B, Jonasdottir G, et al. The impact of HLA-A and -DRB1 on age at onset, disease course and severity in Scandinavian multiple sclerosis patients. Eur J Neurol 2007;14:835–840. [DOI] [PubMed] [Google Scholar]
- 35.Sombekke MH, Lukas C, Crusius JB, et al. HLA-DRB1*1501 and spinal cord magnetic resonance imaging lesions in multiple sclerosis. Arch Neurol 2009;66:1531–1536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


