Abstract
Although the importance of peptidoglycan recognition proteins (PGRPs) in detecting bacteria and promoting immunity is well recognized in Drosophila melanogaster and other insect species, such a role has not yet been experimentally established for PGRPs in the tobacco hornworm, Manduca sexta. In this study, we purified M. sexta PGRP1 from the baculovirus-insect cell expression system, tested its association with peptidoglycans and intact bacteria, and explored its possible link with the prophenoloxidase activation system in larval hemolymph. Sequence comparison suggested that PGRP1 is not an amidase and lacks residues for interacting with the carboxyl group of meso-diaminopimelic acid-peptidoglycans (DAP-PGs). M. sexta PGRP1 gene was constitutively expressed at a low level in fat body, and the mRNA concentration became much higher after an injection of Escherichia coli. Consistently, the protein concentration in larval plasma increased in a time-dependent manner after the immune challenge. Purified recombinant PGRP1 specifically bound to soluble DAP-PG of E. coli but not to soluble Lys-type PG of Staphylococcus aureus. In addition, this recognition protein completely bound to insoluble PGs from Micrococcus luteus, Bacillus megaterium and Bacillus subtilis, whereas its association with the bacterial cells was low even though their peptidoglycans are exposed on the cell surface. After PGRP1 had been added to plasma of naïve larvae in the absence of microbial elicitor, there was a concentration-dependent increase in prophenoloxidase activation. Phenoloxidase activity, as usual, increased after the plasma was incubated with peptidoglyans or bacterial cells. These increases became more prominent when insoluble M. luteus or B. megaterium PG or soluble E. coli PG and PGRP1 were both present. Statistic analysis suggested a synergistic effect caused by interaction between PGRP1 and these PGs. Taken together, these results indicated that PGRP1 is a member of the M. sexta prophenoloxidase activation system, which recognizes peptidoglycans from certain bacteria and initiates the host defense response. The unexplained difference between the purified PGs and intact bacteria clearly reflects our general lack of understanding of PGRP1-mediated recognition and how it leads to proPO activation.
Keywords: pattern recognition, hemolymph protein, melanization, insect immunity, serine protease pathway
1. Introduction
Arthropods rely solely on their innate immunity to fight off invading pathogens (Gillespie et al., 1997; Lavine and Strand, 2002; Lemaitre and Hoffmann, 2007). This system is elicited by conserved molecular patterns associated with microbes, such as peptidoglycan (PG), lipopolysaccharide, and β-1,3-glucan. Peptidoglycan, a cell wall component of bacteria, is a high molecular weight polymer consisting of unbranched glycan strands cross-linked by short stem peptides (Vollmer et al., 2008). The glycan strands are composed of alternating β-1,4-linked N-acetylglucosamine and N-acetylmuramic acid. The stem peptides attached to N-acetylmuramic acid residues contain a lysine or meso-diaminopimelic acid (DAP) at the third position, which connects these peptides directly or indirectly through a peptide cross-bridge. Lys-type PGs are found in most Gram-positive bacteria, whereas DAP-PGs are present in Gram-negative bacteria and the Gram-positive genera Bacillus and Clostridium. Different bacteria vary in stem peptide, cross-bridge, and physical properties (e.g. thickness, elasticity, porosity) of their peptidoglycans.
Peptidoglycan recognition proteins (PGRPs) are immunity-related molecules conserved from insects to humans (Royet and Dziarski, 2007; Chaput and Boneca, 2007). The founding member of this protein family was isolated from the silkworm, Bombyx mori (Yoshida et al., 1996). The carboxyl-terminal PG-binding domain of B. mori PGRP is approximately 165 residues long and homologous to lysozyme of bacteriophage T7 (Ochiai and Ashida, 1999; Kang et al., 1998). Due to the lack of key catalytic residues in T7 lysozyme, the silkworm protein does not cleave an amide bond between N-acetylmuramic acid and L-alanine of the stem peptide. Its binding of Lys-type PG from Micrococcus luteus, however, triggers a serine proteinase cascade that leads to prophenoloxidase (proPO) activation and melanin formation. In Drosophila, PGRP-SA and -SD bind Lys-PG to induce Toll pathway; PGRP-SA is also involved in proPO activation; PGRP-LC and -LE bind DAP-PG fragments and induce Imd pathway; PGRP-LC stimulates phagocytosis (Charroux et al., 2009). On the other hand, Drosophila PGRP-SB, -SC, and -LB hydrolyze the amide bond between N-acetylmuramic acid and L-alanine in the stem peptide to reduce the amount of immunogenic muropeptide and down-regulate the immune responses (Royet and Dziarski, 2007). In Tenebrio molitor, clustered PGRP-SA detects Lys-type PG and activates the proPO cascade and Toll pathway (Park et al., 2007).
Three-dimensional structures of PG-binding domains in the Drosophila and human PGRPs reveal an α/β mixed fold similar to that of T7 lysozyme (Kim et al., 2003; Reiser et al., 2004; Guan et al., 2004 and 2005; Lim et al., 2006; Cho et al., 2007; Leone et al., 2008). This general fold consists of a central β-sheet and three peripheral α-helices, parts of which form the walls and bottom of a PG-binding groove found in most PGRPs. The groove is composed of His, Tyr, His, Thr and Cys/Ser residues that correspond to His17, Tyr46, His122, Lys128 and Cys130 of T7 lysozyme (Mellroth et al., 2003). Some of these residues are responsible for Zn2+-binding and N-acetylmuramoyl-L-alanine amidase activity of T7 lysozyme and hydrolytic PGRPs. Structural analysis of free or muropeptide-bound PGRPs, along with site-directed mutagenesis and binding assays, suggests that certain residues in the binding groove are responsible for the differential recognition of Lys- and DAP-PGs (Guan et al., 2004; Lim et al., 2006; Cho et al., 2007).
In M. sexta, cDNA cloning disclosed a 19 kDa protein most similar in sequence to B. mori PGRP (identity: 54%) (Yu et al., 2002; Zhu et al., 2003). While its protein level in larval plasma increased after an injection of M. luteus, immunological function of M. sexta PGRP1 remains unclear. In fact, unpublished data indicated that addition of PGRP1 to plasma failed to enhance melanization triggered by M. luteus (Kanost et al., 2004). To explore its possible role in bacteria recognition and initiation of proPO activation, we expressed M. sexta PGRP1 in insect cells and tested its binding to soluble and insoluble PGs from various bacteria and to intact bacterial cells. By correlating the binding data with proPO activation upon exposure to these elicitors, we confirmed the unusual binding properties of M. sexta PGRP1 and established its role as a sensor of the proPO activation system. Implications of our findings regarding recognition of bacteria are discussed as well.
2. Materials and methods
2.1. Insect rearing, bacterial challenge, and hemolymph collection
M. sexta eggs were purchased from Carolina Biological Supply and larvae were reared on an artificial diet (Dunn and Drake, 1983). Day 2, 5th instar larvae were injected with formaldehyde-killed Escherichia coli (2 × 108 cells/larvae). Hemolymph was collected from a cut proleg of the immune challenged larvae 6, 12, and 24 h later. For use as a control at 0 h, hemolymph was also collected from day 2, 5th instar naïve larvae. After centrifugation at 5000g for 5 min, plasma samples from the naïve and induced insects were aliquoted and stored at −80 °C.
2.2. Detection of PGRP1 in hemolymph
The control and induced plasma samples were analyzed by mixing 2 μl of plasma with 6 μl of 20 mM Tris-HCl, pH 8.0 and 4 μl of 5 × SDS sample buffer. After incubation at 95 °C for 5 min, 7 μl of the mixture was separated by 15% SDS-PAGE, transferred to a nitrocellulose membrane, and reacted with 1:2000 diluted PGRP1 polyclonal antiserum kindly provided by Dr. Michael Kanost at Kansas State University. Antibody-antigen complexes were detected using alkaline phosphatase conjugated to goat-anti-rabbit IgG, 5-bromo-4-chloro-3-indolyl phosphate, and nitro-blue tetrazolium (Bio-Rad).
2.3. Reverse transcription-PCR analysis of PGRP1 mRNA levels
Hemocyte and fat body total RNA was prepared from the naïve and induced 5th instar larvae (day 3) using Micro-to-Midi Total RNA Purification System (Life Technologies). The RNA (2 μg), oligo(dT) (0.5 μg) and dNTPs (1 μl, 10 mM each) were mixed with diethylpyrocarbonate-treated H2O in a final volume of 12 μl, denatured at 65 °C for 5 min, and quickly chilled on ice for 3 min. M-MLV reverse transcriptase (1 μl, 200 U/μl) (Life Technologies), 5 × buffer (4 μl), 0.1 M dithiothreitol (2 μl), and RNase OUT (1 μl, 40 U/μl) (Life Technologies) were added to the denatured RNA sample (12 μl) for first strand cDNA synthesis at 37 °C for 50 min. M. sexta ribosomal protein S3 mRNA was used as an internal control to normalize the cDNA samples using specific primers j501 (5′-GCCGTTCTTGCCCTGTT) and j504 (5′-CGCGAGTTGACTTCGGT). Primers j297 (5′-CACATTGAGTACCTTACCCGTCCA) and j298 (5′-GAACGAAGATCCGATGTCCAGTC) were used to amplify M. sexta PGRP1 cDNA under conditions empirically chosen to avoid saturation: 30 cycles of 94 °C, 30 s; 50 °C, 30 s; 72 °C, 30 s in a duplex PCR reaction. Relative levels of PGRP1 mRNA in the samples were estimated by 1.5% agarose gel electrophoresis.
2.4. Construction of recombinant baculovirus for PGRP1 expression
M. sexta PGRP1 cDNA (obtained from Dr. Michael Kanost at Kansas State University) was amplified using PCR primer j285 (5′-GGAATTCACTGCAACGTCGTC) and j288 (5′-CTCGAGGTCTTTATATTCGGACAC). The PCR product was cloned into pGEM-T (Promega) and completely verified by DNA sequencing. The 528 bp EcoRI-XhoI fragment was retrieved from the plasmid by restriction digestion and directionally inserted into the same sites of pMFH6 (Lu and Jiang, 2008) to generate plasmid PGRP1/pMFH6, which allows efficient secretion of the recombinant protein into medium using the honeybee mellitin signal peptide and purification on a Ni2+-nitrilotriacetic (NTA) agarose column via the carboxyl-terminal hexahistidine tag. In vivo transposition of the expression cassette, selection of bacterial colonies carrying the recombinant bacmid, and isolation of bacmid DNA were carried out according to manufacturer’s protocols (Life Technologies). The initial viral stock (V0) was obtained by transfecting Spodoptera frugiperda Sf9 cells with a bacmid-CellFECTIN mixture, and its titer was improved through serial infections. The V6 viral stock, containing the highest level of baculovirus (1~2 × 108 pfu/ml), was stored at −70 °C for further experiments.
2.5. Expression and purification of M. sexta PGRP1
Sf9 cells (2.0 × 106 per ml) in 100 ml of Sf-900™ III serum-free medium (Life Technologies) were infected with the baculovirus stock at a multiplicity of infection of 5~10 and grown at 27 °C for 96 h with gentle agitation (100 rpm). After centrifugation at 5000g for 10 min, the culture supernatant was collected and mixed with an equal volume of distilled water and once again centrifuged at 22100g for 20 min. The cleared supernatant was applied to a dextran sulfate-Sepharose column (5 ml) equilibrated with buffer A (1 mM benzamidine in 10 mM potassium phosphate, pH 6.4). Following a washing step with 25 ml of buffer A, bound proteins were eluted with a linear gradient of 0–1.0 M NaCl in buffer A (30 ml). Based on the result of immunoblot blot analysis, fractions containing M. sexta PGRP1 were pooled and applied to a Ni2+-NTA agarose column (1 ml) (Qiagen), equilibrated with buffer B (50 mM potassium phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.005% Tween-20). After washing with buffer B (5 ml), bound proteins were eluted from the column with a linear gradient of 10–100 mM imidazole in buffer B (20 ml). Finally, tightly bound proteins were eluted with 5 ml of buffer B containing 250 mM imidazole. All the purification steps were carried out at 4 °C. After electrophoretic analysis, PGRP1 fractions were combined and concentrated using Amicon ultracentrifugal 5K MWCO filter device (Millipore). Buffer exchange with 20 mM Tris-HCl, pH 7.5, 50 mM NaCl was performed on the same device.
2.6. Plate assay of PGRP1 binding to soluble peptidoglycans
Soluble PGs (InvivoGen) from E. coli and Staphylococcus aureus were used as ligands in a binding assay. Each ligand (2 μg/well) was applied to wells in a 96-well microplate, air dried overnight at room temperature, and fixed to the wells at 60 °C for 30 min. After blocking with 200 μl of 1 mg/ml bovine serum albumin (BSA) in Tris buffered saline (TBS: 137 mM NaCl, 3 mM KCl, 25 mM Tris-HCl, pH 7.6) at 37 °C for 2 h and after washing with 200 μl TBS four times (5 min each), PGRP1 (300 ng) in 50 μl TBS containing 0.1 mg/ml BSA was added to the wells and incubated at room temperature for 3 h. To test binding specificity, a competition experiment was performed in parallel by pre-incubating PGRP1 (300 ng) with 200 μg of the ligand at room temperature for 30 min. The mixture was then added to the ligand-coated wells and incubated at room temperature for 3 h. Following a washing step with TBS, 100 μl of 1:1000 diluted anti-(His)5 monoclonal antibody (Bio-Rad) in TBS containing 0.1 mg/ml BSA was added to the wells and incubated at 37 °C for 2 h. After washing with 200 μl of TBS four times, 100 μl of 1:1500 diluted goat-anti-mouse IgG conjugated to alkaline phosphatase (Bio-Rad) in TBS containing 0.1 mg/ml BSA was added to the wells and incubated at 37 °C for 2 h. After washing with TBS four times and 0.5 mM MgCl2, 10 mM diethanolamine once, 50 μl aliquots of p-nitrophenyl phosphate (1.0 mg/ml in the diethanolamine buffer) were added to the wells and absorbance at 405 nm was monitored in the kinetic mode on a VersaMax microplate reader (Molecular Devices).
2.7. Isolation of insoluble peptidoglycans from Gram-positive bacteria
M. luteus, S. aureus, Bacillus megaterium, and Bacillus subtilis were separately grown in Luria-Bertani medium (2.0 L) overnight at 37 °C with shaking. These bacteria, separated from the medium by centrifugation at 2000g for 20 min, were resuspended in 100 ml saline (0.85% NaCl) and heated at 100 °C for 20 min. The cells were washed twice with saline (50 ml each), once with water (50 ml), and three times with acetone (50 ml each) by complete resuspension and centrifugation. After drying at 37 °C for 8 h, peptidoglycans were extracted from the bacteria according to a modified protocol (Tsuchiya et al., 1996). Briefly, dried cells (10 g) were stirred in 300 ml of 10% trichloroacetic acid in a boiling water bath for 20 min. After centrifugation at 10000g for 30 min, the pellet was washed three times with 250 ml water and once with 250 ml of buffer C (100 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 1 mM CaCl2). Bacteria resuspended in 100 ml buffer C were incubated with 30 mg of bovine trypsin at 37 °C for 24 h with gentle agitation. After the enzyme was inactivated with 1 mM phenylmethanesulfonyl fluoride at 37 °C for 30 min, treated cells were centrifuged at 10000g for 30 min and washed with 200 ml of H2O ten times. Each time the precipitate was completely resuspended by sonication and, after the final wash, lyophilized and stored at −20 °C.
2.8. Binding of PGRP1 to insoluble peptidoglycans
One mg of insoluble PG was mixed with 10 μl of PGRP1 (3 μg) and 40 μl of buffer D (20 mM Tris-HCl, pH 8.0, 20 mM NaCl). After incubation for 2 h at 4 °C with mixing, the mixture was centrifuged at 16000g for 15 min. The supernatant was treated with 5 × SDS sample buffer and analyzed as unbound fraction. The pellet was washed 3 times with 200 μl of buffer D each and then boiled with 20 μl of 2 × SDS sample buffer for 5 min to obtain bound fraction. The total, unbound, and bound samples (equivalent to 7 μl of 1:5 diluted PGRP1) were separated by 15% SDS-PAGE followed by immunoblot analysis using 1:2000 diluted anti-(His)5 monoclonal antibody and goat-anti-mouse IgG conjugated to alkaline phosphatase.
2.9. Binding of PGRP1 to bacterial cells
A single bacterial colony was grown in LB medium overnight at 37 °C and subcultured in 4 ml of the same medium until OD600 was close to 0.8 (ca. 1.6 × 108 cells/ml). After centrifugation at 1000g and washing with buffer D twice, the bacteria were resuspended in 40 μl of buffer D. Purified PGRP1 (10 μl, 3 μg) was added to the cell suspension and incubated for 2 h at 4 °C with mixing. After centrifugation at 4000g for 15 min, the supernatant was treated with 5 × SDS sample buffer and analyzed as unbound fraction. The cell pellet was washed 3 times with 200 μl of buffer D, suspended with 20 μl of 2 × SDS sample buffer, and heated at 95 °C for 5 min to obtain the bound fraction. The total, unbound, and bound fractions (equivalent to 7 μl of 1:5 diluted PGRP1) were analyzed as described in Section 2.8.
2.10. Role of PGRP1 in proPO activation
Plasma from day 3, fifth instar naïve larvae was diluted ten times with buffer E (20 mM Tris-HCl, pH 8.0, 1 mM CaCl2, 0.001% Tween-20). Five μl of diluted sample was mixed with 19 μl of buffer E and incubated at 25 °C for 10 min. PO activity was determined using dopamine as a substrate on a microplate reader (Jiang et al., 2003). Plasma samples with low PO activity were pooled and stored in 10 μl aliquots at −80 °C. Aliquots of the 1:10 diluted plasma (5 μl) with buffer E (1 μl) or purified recombinant PGRP1 (1 μl, 0.2 μg/μl) were separately incubated with 1 μl of buffer E (negative control) or one of the following elicitors to investigate PGRP1-enhanced proPO activation: insoluble PGs from M. luteus, S. aureus, B. megaterium and B. subtilis (1 μg), soluble PGs (1 μg) from S. aureus and E. coli, curdlan (10 μg), and 2 × 105 cells of M. luteus, S. aureus, B. megaterium, B. subtilis and E. coli. The controls were mixtures of diluted plasma with buffer E, elicitor, or PGRP1, and the test was diluted plasma mixed with elicitor and PGRP1. The total volume of the four mixtures in each elicitor group was adjusted to 24 μl with buffer E and incubated at 25 °C for 1 h prior to the PO activity assay.
3. Results
3.1. Structure features, expression pattern, and recombinant production of M. sexta PGRP1
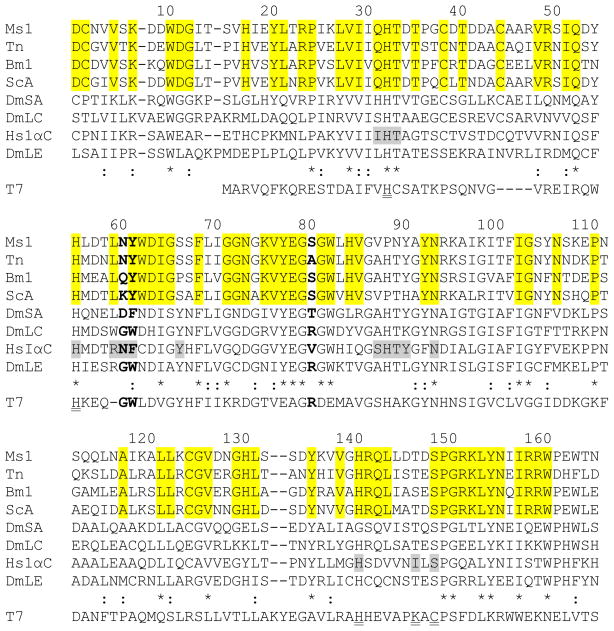
M. sexta PGRP1 was initially identified as a bacteria-induced pattern recognition receptor in plasma (Yu et al., 2002; Zhu et al., 2003). The open reading frame encodes a 192-residue protein sequence, including a 20-residue signal peptide. The mature protein has a calculated molecular mass of 19,319 Da. No potential N- or O-linked glycosylation sites are found in the sequence. The conserved PGRP domain is located between residues 22–164. Sequence alignment with other PGRPs revealed that M. sexta PGRP1 has a serine residue (Ser148) in the position equivalent to Cys130 in T7 lysozyme (Fig. 1). This common feature of receptor-type PGRPs suggests that M. sexta PGRP1 is not an amidase (Mellroth et al., 2003). Asn60 and Tyr61of M. sexta PGRP1 correspond to Asn and Phe of human PGRP-1αC, Asp and Phe of Drosophila PGRP-SA, as well as Gly and Trp of Drosophila PGRP-LC and -LE (Fig. 1). The Asn/Asp-Phe and Gly-Trp are responsible for specific binding to Lys- and DAP-type PGs, respectively (Guan et al., 2004; Kaneko et al., 2004). Additionally, Ser80 of M. sexta PGRP1 is in a position equivalent to Arg254 in Drosophila PGRP-LE, which interacts with the carboxyl group of DAP-PGs (Fig. 1) (Lim et al., 2006). This Arg residue is conserved in all known Drosophila and human PGRPs that recognize DAP-PGs, but not always found in PGRPs that bind Lys-PGs (Onoe et al., 2007).
Fig. 1. Multiple sequence alignment of PGRP sequences and T7 lysozyme.
Amino acid sequences of M. sexta PGRP1 (Ms1, AF413068), T. ni PGRP (Tn, AAC31820), B. mori PGRP (Bm1, BAA77209), S. cynthia PGRP-A (ScA, BAF03522), D. melanogaster PGRP-SA (DmSA, NP_572727), -LC (DmLC, NP_729468.2), -LE (DmLE, NP_573078), H. sapiens PGRP-IαC (HsIαC, AAK72484), and T7 lysozyme (T7, NP_0419731) are aligned. Residue numbers of mature M. sexta PGRP1 are indicated on top of its sequence. Identical and similar residues in PGRPs are marked with “*” and “:”, respectively. Residues corresponding to Asn236 and Phe237 of human PGRP-IαC and Arg254 of Drosophila PGRP-LE are shown in bold. The muramyl tripeptide-interacting residues in human PGRP-IαC are shaded gray, residues identical in the lepidopteran PGRPs are highlighted, and residues for Zn2+-binding and amidase activity in T7 lysozyme are double underlined.
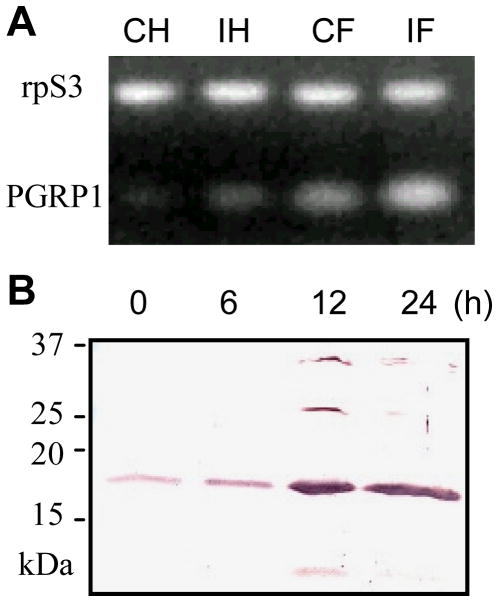
The PGRP1 gene was constitutively expressed at a low level in fat body, and its transcripts became more abundant after an immune challenge with E. coli (Fig. 2). The mRNA level was much lower in hemocytes from naïve larvae, which also increased after the bacteria injection. The PGRP1 protein is present in the plasma of naïve larvae and its concentration dramatically increased at 12 h after the injection of E. coli. These data indicated that constitutive expression of M. sexta PGRP1 is induced in a time-dependent manner by E. coli, as well as by M. luteus in the previous experiments (Yu et al., 2002; Zhu et al., 2003).
Fig. 2. Induced expression of M. sexta PGRP1 gene.
(A) RT-PCR analysis of total RNA samples from control hemocytes (CH) and fat body (CF) of naïve larvae and induced hemocytes (IH) and fat body (IF) of larvae at 24 h after injection with E. coli. M. sexta ribosomal protein S3 (rpS3) transcripts were normalized for the analysis of PGRP1 mRNA. (B) Immunoblot analysis of plasma samples from larvae at 0, 6, 12, 24 h after injection with E. coli. Diluted PGRP1 antiserum was used as the first antibody.
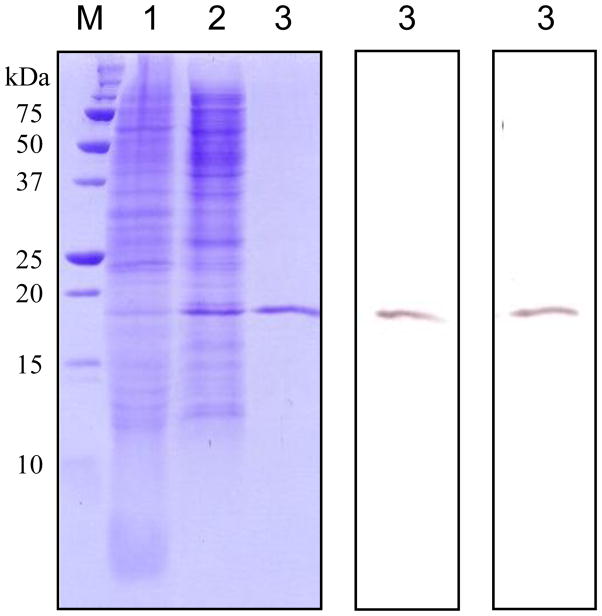
We expressed M. sexta PGRP1 in a baculovirus-insect cell system. The recombinant protein in the medium reached the highest level at 72–84 h after infection. PGRP1 was directly captured by cation exchange chromatography on a dextran sulfate-Sepharose column and further purified by affinity chromatography on a Ni-NTA column. From 100 mL conditioned medium, we obtained 0.15 mg of PGRP1. The purified recombinant protein migrated as a single band at 19 kDa position, which was recognized by anti-(His)5 and PGRP1 antibodies (Fig. 3).
Fig. 3. Isolation of M. sexta PGRP1 from baculovirus-infected Sf9 cells.
Left panel, Coomassie blue staining; central panel, immunoblot analysis using anti-(His)5 as the first antibody; right panel, immunoblot analysis using diluted PGRP1 antiserum as the first antibody. Conditioned cell culture medium (lane 1, 10 μl), proteins eluted from dextran sulfate-Sepharose (lane 2, 10 μl), and affinity-purified PGRP1 from Ni2+-NTA agarose (lane 3, 10 μl) were separated by 15% SDS-PAGE, along with pre-stained molecular weight standards (M) with their sizes indicated on the left.
3.2. Differential binding of M. sexta PGRP1 to microbial cells and cell wall components
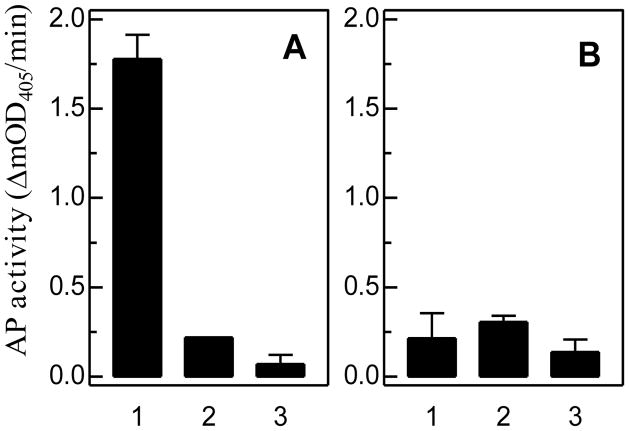
To measure the relative binding of PGRPs to Lys- and DAP-PGs, we developed an ELISA-based, semiquantitative binding assay. Contrary to the prediction based on sequence comparison (Fig. 1), this test showed that M. sexta PGRP1 binds soluble DAP-PG from E. coli (Fig. 4A) more than soluble Lys-PG from S. aureus (Fig. 4B). The binding was specific for the former since pre-incubation of PGRP1 with excess amount of E. coli PG eliminated its binding to the ligand immobilized on the ELISA plate. In contrast, the association of PGRP1 with Lys-PG of S. aureus was nonspecific, as shown by the competition experiment. The total binding was not significantly higher than the negative control of BSA.
Fig. 4. Binding of M. sexta PGRP1 to soluble peptidoglycans from E. coli (A) and S. aureus (B).
As described in Section 2.6, the purified recombinant PGRP1 was incubated with soluble PG immobilized on a 96-well microplate. The binding was detected via ELISA and alkaline phosphatase activity is shown as mean ± SEM (n=3). Binding without a competitor (#1), with excess soluble PG as competitor (#2), and the negative control of BSA (#3).
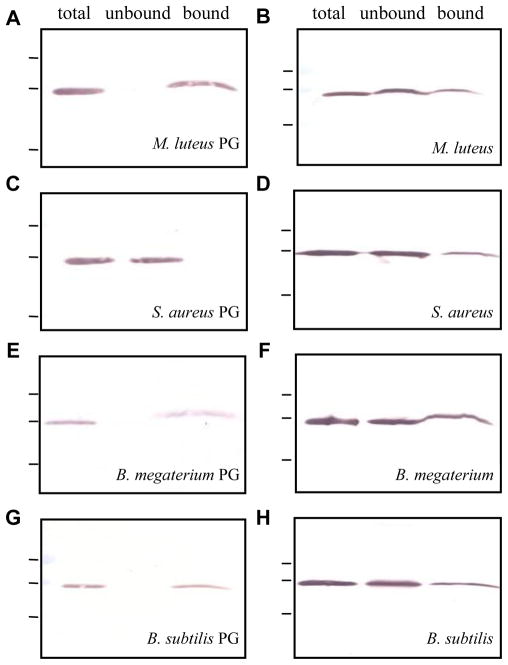
M. sexta PGRP1 showed complete binding to insoluble PGs from M. luteus (Lys-type), B. megaterium (DAP-type), and B. subtilis (DAP-type) (Fig. 5, A, E, and G). Consistent with the ELISA results, recombinant PGRP1 did not show any binding to insoluble PG from S. aureus (Lys-type) (Fig. 5C). When PGRP1 was incubated with M. luteus, S. aureus, B. megaterium and B. subtilis cells (Fig. 5, B, D, F, and H), partial binding was observed in each case. Although PGs are located on the surface of Gram-positive bacteria and enough cells were used, a majority of PGRP1 was detected in the flow-through fractions, indicating that other cell surface structures (e.g. techoic acids, lipoteichoic acids, and proteins) may interfere with the binding. In contrast, low complexity and high accessibility of insoluble PGs purified from M. luteus, B. megaterium, and B. subtilis may have contributed to the complete binding.
Fig. 5. Binding of M. sexta PGRP1 to insoluble PGs (A, C, E, G) or cells (B, D, F, H) of different bacteria.
As described in Sections 2.8 and 2.9, binding assays were performed using the purified PGRP1 and PGs or whole bacteria. The total, unbound, and bound fractions were separated by SDS-PAGE followed by immunoblot analysis using anti-(His)5 as the first antibody.
3.3. Role of M. sexta PGRP1 in the proPO activation system
To test a possible role of M. sexta PGRP1 in the proPO activation system, we added purified PGRP1 to the cell-free hemolymph from naïve larvae and observed a concentration-dependent increase in proPO activation in the absence of bacterial elicitor (Fig. 6). In contrast, no such response was detected after we added BSA to the plasma.
Fig. 6. PO activity increase in control hemolymph from naïve larvae after adding purified M. sexta PGRP1.
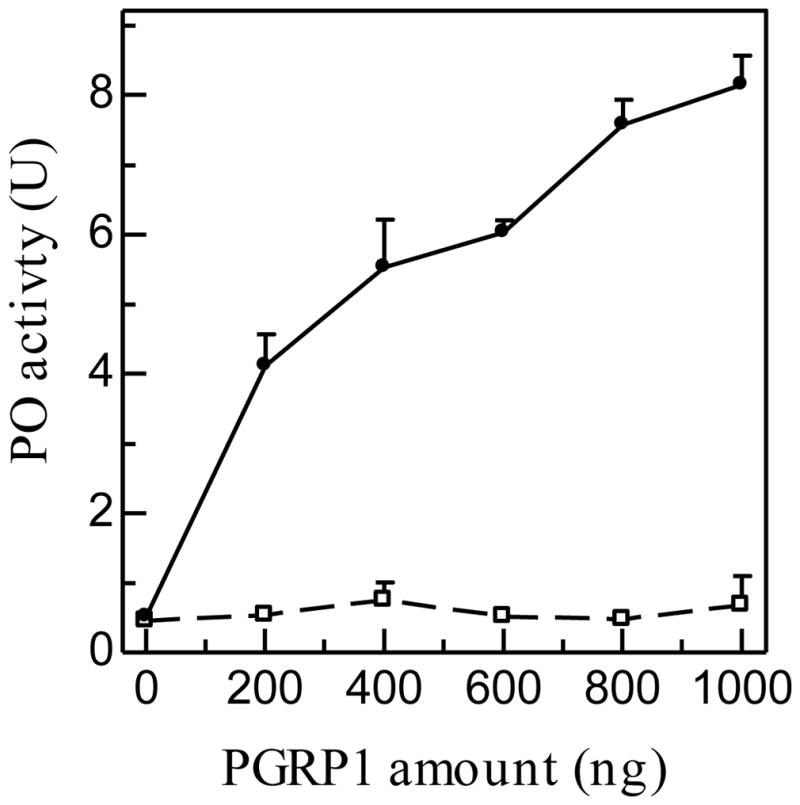
Diluted plasma (5 μl) was incubated at room temperature with 0.2 to 1.0 μg of BSA ( ) or purified recombinant PGRP1 (
) or purified recombinant PGRP1 ( ) in a final volume of 24 μl. PO activity was measured after 60 min and plotted as mean ± SEM (n = 3) against amount of PGRP1 added.
) in a final volume of 24 μl. PO activity was measured after 60 min and plotted as mean ± SEM (n = 3) against amount of PGRP1 added.
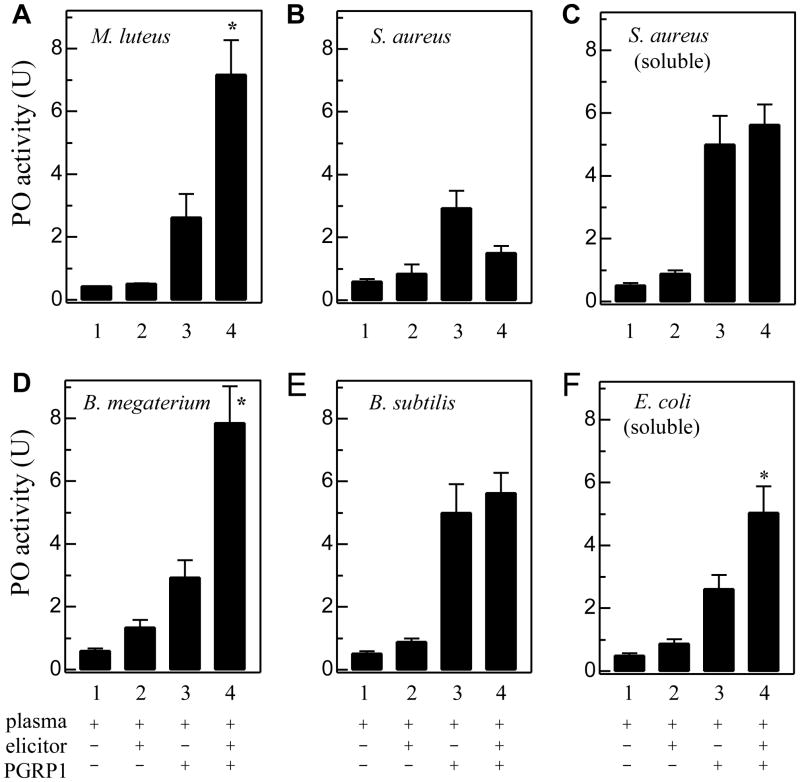
How may exogenous PGRP1 enhance proPO activation triggered by various PGs? To answer this question, we incubated diluted plasma with PGs in the absence or presence of recombinant PGRP1. Compared with the control of plasma only, small but significant increase (#2 - #1) was observed after insoluble PG from M. luteus, B. megaterium, or B. subtilis (Fig. 7, A, D and E) or curdlan (Fig. S1) was added, indicating that the proPO activation system was functional. Such increase (#2 - #1, representing PG-plasma interaction) was not as large as that in the mixtures of plasma and PGRP1 (#3 - #1, representing exogenous PGRP1-plasma interaction). When PG, PGRP1, and diluted plasma were present simultaneously, the PO activity increase (#4 - #1) was significantly higher than the sum of the components [(#2 – #1) + (#3 – #1)] in the following three cases: insoluble PG from M. luteus (Lys-type) and B. megaterium (DAP-type) and soluble PG from E. coli (DAP-type) (Fig. 7, A, D, and F). This synergistic effect, likely caused by specific interaction of these PGs and exogenous PGRP1 (Fig. 4A and Fig. 5, A, E, and G), was not observed in the #4 reactions containing insoluble or soluble PG from S. aureus (Fig. 7, B and C). Consistent with that, the recombinant PGRP1 did not bind to S. aureus PGs (Fig. 4B and Fig. 5C). Neither did M. sexta PGRP1 synergistically enhance proPO activation triggered by curdlan (Fig. S1), an insoluble form of β-1,3-glucan not binding to PGRP1 (data not shown). Insoluble PG from B. subtilis (DAP-type) did cause an increase in proPO activation (#4 - #1), but it was statistically insignificantly higher than the sum of (#2 – #1) and (#3 – #1) (Fig. 7E).
Fig. 7. Enhancement of proPO activation in plasma from naïve larvae by peptidoglacans in the absence or presence of M. sexta PGRP1.
As described in Section 2.10, PGs of M. luteus (A), S. aureus (B), S. aureus (C, soluble), B. megaterium (D), B. subtilis (E), and E. coli (F, soluble) were separately incubated with plasma and purified PGRP1 for 60 min at room temperature. PO activity (#4) was measured and plotted as mean ± SEM (n = 3), along with those of the controls (#1, plasma only; #2, plasma and PG; #3, plasma and PGRP1). Since interaction of plasma factors with elicitor (#2 - #1) and co-presence of plasma and exogenous PGRP1 (#3 - #1) both lead to proPO activation, an interaction of elicitor and exogenous PGRP1 in plasma (#4 - #1) is expected to increase proPO activation to a level significantly higher than the sum of the two components [i.e. (#2 - #1) + (#3 - #1)]. To detect a possible synergistic effect of elicitor-PGRP1 interaction, the PO activity changes represented by (#4 - #1) and (#2 + #3 – 2×#1) are compared using unpaired t-test. An asterisk (*) on #4 indicates that (#4 - #1) is significantly higher (p < 0.05).
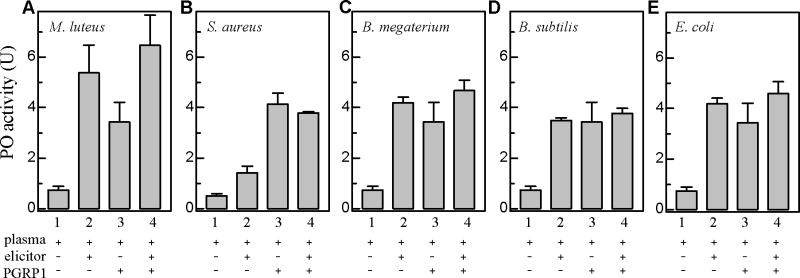
We have further examined a possible synergistic enhancement of proPO activation caused by interaction between exogenous PGRP1 and whole bacteria. M. luteus, S. aureus, B. megaterium, B. subtilis, and E. coli cells all stimulated the melanization response by interacting with plasma factors (Fig. 8, #2 vs. #1). The elevated PO activity was comparable to that caused by exogenous PGRP1 interacting with plasma proteins (#2 vs. #3). After bacteria, PGRP1, and plasma had been incubated for 1 h at 25 °C, the PO activity increase (#4 - #1) was lower than the sum of the two components (#2 + #3 – 2×#1). The lack of synergism coincided with the incomplete binding of PGRP1 to these bacteria (Fig. 6).
Fig. 8. Activation of proPO in plasma from naïve larvae by bacterial cells with or without M. sexta PGRP1.
M. luteus (A), S. aureus (B), B. megaterium (C), B. subtilis (D), and E. coli (E) cells were individually incubated with plasma and purified PGRP1 for 60 min at room temperature. PO activity (#4) was measured and plotted as mean ± SEM (n = 3), along with those of the controls (#1, plasma only; #2, plasma and cells; #3, plasma and PGRP1). Since interaction of plasma factors with cells (#2 - #1) and co-presence of plasma and exogenous PGRP1 (#3 - #1) both lead to proPO activation, an interaction of bacteria and exogenous PGRP1 in plasma (#4 - #1) is expected to increase proPO activation at a level significantly higher than the sum of the two components [(#2 - #1) + (#3 - #1)] in an unpaired t-test. Comparisons of the PO activity changes, however, provide no support for any synergistic effect caused by bacteria-PGRP1 interaction.
4. Discussion
Insects have evolved a battery of proteins to recognize characteristic molecular patterns of microbes and promote defense responses (Gillespie and Kanost, 1997; Steiner et al., 2004). In M. sexta, immulectins, β-1,3-glucan recognition proteins, and other plasma factors participate in the immune surveillance (Jiang, 2008; Ragan et al., 2008). M. sexta PGRP1 is a hemolymph protein induced upon bacterial injection (Yu et al., 2002; Zhu et al., 2003). Although it is 54% identical in sequence to the B. mori PGRP (a sensor of the silkworm proPO activation cascade), addition of M. sexta PGRP1 to plasma failed to enhance proPO activation stimulated by M. luteus cells (Kanost et al., 2004). Knockdown of PGRP1 expression did not have any effect on cellular immunity but increased the susceptibility of larvae to Photorhabdus asymbiotica (Eleftherianos et al., 2007). To elucidate its immunological function, we carried out this study using recombinant PGRP1 and a variety of microbial elicitors. The biochemical analysis provided evidence that M. sexta PGRP1 functions as a recognition molecule for Lys-PG of M. luteus and DAP-PGs and that the specific binding to PGs triggers the proPO activation cascade.
4.1. Differences in binding to Lys-PGs from M. luteus and S. aureus
M. sexta PGRP1 bound insoluble Lys-PG of M. luteus (Fig. 5A) but not that of S. aureus (Fig. 5C). Nor did specific binding occur after the protein was incubated with soluble PG of S. aureus (Fig. 4B). This is consistent with the observation that M. sexta PGRP1 in induced plasma did not bind to S. aureus cells (Ragan, 2008) although, in the absence of other proteins, the recombinant PGRP1 slightly associated with the bacteria (Fig. 5D). The lack of binding also coincided with the result that PGRP1 did not further enhance proPO activation triggered by S. aureus cells or its PG (Fig. 7, B and C; Fig. 8B). Similarly, Drosophila PGRP-LC mediates the IMD pathway activation by M. luteus PG but not by S. aureus PG (Kaneko et al., 2005). S. aureus PG is highly cross-linked by a pentaglycine bridge, whereas M. luteus uses a branched pentapeptide (identical to its stem-peptide) for the cross-bridge (Shokman and Barrett, 1983). The bridge structure, low cross-linking, and high accessibility of glycan strand of M. luteus PG appear to be responsible for the PGRP binding and proPO activation. S. cynthia ricini PGRP-A binds to linear M. luteus PG with high affinity, but little binding was detected with cross-linked PG from the same species (Onoe et al., 2007).
4.2. Binding to DAP-PGs from E. coli, B. megaterium, and B. subtilis
M. sexta PGRP1 also recognized insoluble DAP-PGs of B. megaterium and B. subtilis (Fig. 5, E and G). While DAP-PGs are exposed on the surface of Gram-positive genera Bacillus and Clostridium, they are located underneath the outer membrane of Gram-negative bacteria. Thus, we tested soluble DAP-PG purified from E. coli and detected specific binding (Fig. 4A). PGRP-A of the eri-silkworm has a similar specificity by binding to Bacillus licheniformis and M. luteus PGs (Onoe et al., 2007). In Drosophila, recombinant PGRP-SA, -SC1b, -LC, and -LF bind to both Lys- and DAP-PGs in vitro (Mellroth et al., 2003 and 2005; Persson et al., 2007).
The modification of glycan strand, composition of stem peptide, type and extent of cross-linking, and interference of other cell surface components all seem to affect associations between PGs and their recognition proteins (Royet and Dziarski, 2007). M. sexta PGRP1 binds to one milligram of insoluble DAP-PGs (equivalent to those in 1.0 × 109 bacteria) far more completely than to 6.4 × 108 intact cells (Fig. 5, E through H) and the binding to B. megaterium cells was more complete than to B. subtilis, probably due to a lower degree of cross-linking in B. megaterium PG (Bricas et al., 1967) and deacetylation of glucosmine in B. subtilis PG (Atrih et al., 1999). These structural differences may also be responsible for the differential stimulation of melanization by PGs from the two Bacillus species (Fig. 7, D and E).
In the beetle Holotrichia diomphalia, PGRP binds to β-1,3-glucan (a polysaccharide from fungal cell wall) and activates proPO system (Lee et al., 2004). To test if M. sexta PGRP1 does that, we incubated larval plasma with curdlan in the absence or presence of the recombinant protein. The coexistence of insoluble β-1,3-glucan and PGRP1 failed to synergistically enhance proPO activation (Fig. S1). This result is consistent with the lack of curdlan binding by M. sexta PGRP1 (data not shown).
4.3. Structural basis for the binding specificity of M. sexta PGRP1
Both mammalian and insect PGRPs contain about sixteen residues that contact PG (Guan et al., 2004). Five of them (His208, His231, Tyr242, His264, and Asn269 in human PGRP-1αC) form a nearly contiguous patch on the groove floor, which correspond to His32, His55, Ser66, Pro88 and Asn93 in M. sexta PGRP1 (Fig. 1). Asn236 and Phe237 of human PGRP-1αC form several van der Waals bonds with the side chain of lysine in Lys-PG (Guan et al., 2004). Sequence variations at these positions affect the ability of some PGRPs to discriminate Lys- and DAP-PGs (Steiner, 2004; Royet and Dziarski, 2007; Charroux et al., 2009). For instance, Drosophila PGRP-SA which recognizes Lys-PG contains Asp96 and Phe97 at the two sites (Fig. 1). In contrast, Drosophila PGRP-LC and -LE which recognize DAP-PG have Gly and Trp residing at the corresponding positions (Kaneko et al., 2004). M. sexta PGRP1 and Trichoplusia ni PGRP, with Asn60 and Tyr61 located in the same sites (Fig. 1), recognize Lys-PG from M. luteus and DAP-PG from B. megaterium (Kang et al., 1998). It has also been suggested Arg254 in Drosophila PGRP-LE interacts with the carboxyl group of DAP-PG (Lim et al., 2006). This Arg is conserved in Drosophila and human PGRPs which recognize DAP-PG, but not always present in PGRPs that recognize Lys-PG. M. sexta PGRP1 and S. cynthia PGRP-A have a serine residue at the corresponding position (Fig. 1). Since both PGRPs recognize Lys- and DAP-type PGs (Onoe et al., 2007), this Arg residue seems dispensable for binding to DAP-PG.
4.4. Recognition of PGs by PGRP1 and stimulation of proPO activation
A connection between PGRP1 and the proPO cascade is supported by the observation that adding PGRP1 to the larval plasma concentration-dependently enhanced proPO activation in the absence of a microbial elicitor (Fig. 6). An increase in spontaneous melanization was observed when PGRP-LE was over-expressed in Drosophila (Takehana et al., 2002). Constitutively present in hemolymph at a low level (Fig. 2), PGRPs probably form clusters only on pathogen surface and transmit the invasion signal to other molecules (Park et al., 2007). Supplementation of recombinant PGRPs increased their plasma concentrations and may have triggered pathogen-independent self-association of the pattern recognition molecules, a process favoring interactions with other components of the proPO activation system.
The correlation between binding characteristics of M. sexta PGRP1 and proPO activation further supported the protein’s role as a biosensor of the cascade (Table 1). There was a complete binding of M. sexta PGRP1 to three insoluble PGs: M. luteus and B. megaterium PGs interacted with exogenous PGRP1 and synergistically enhanced proPO activation; a similar increase caused by B. subtilis PG was statistically insignificant. When purified PGs were replaced by bacteria in these reactions, we did not observe any synergistic enhancement. This is consistent with the previous finding that supplementation of PGRP1 to larval plasma did not boost proPO activation after exposure to M. luteus (Kanost et al., 2004; Ragan, 2008). The complex structure of cell surface may have interfered the binding of PGRP1 to the bacteria. PGRP1 did not bind to soluble PG of S. aureus or enhance proPO activation triggered by S. aureus. In contrast, specific binding of PGRP1 to soluble PG isolated from E. coli led to a synergistic increase in PO activity.
Table 1.
Relationship between binding of exogenous PGRP1 and increase in proPO activation
| Lys-type PG | DAP-type PG | ||||
|---|---|---|---|---|---|
| M. luteus | S. aureus | B. megaterium | B. subtilis | ||
| bacterial cells | binding | * | * | * | * |
| proPO activation | − | − | − | − | |
| insoluble PG | binding | ** | no | ** | ** |
| proPO activation | + | − | + | +/− | |
| soluble PG | binding | S. aureus | no | E. coli | ** |
| proPO activation | − | + | |||
partial binding;
complete binding;
+ or −: synergistic enhancement of proPO activation.
4.5. Concluding remarks
We have produced M. sexta PGRP1 as a soluble 19 kDa protein in insect cells and confirmed its induced expression in larvae. The recombinant protein specifically binds to Lys-PG from M. luteus and DAP-PGs from Gram–negative bacteria (e.g. E. coli) and Gram-positive Bacilli. This unusual binding specificity is probably related to Asn60, Tyr61 and Ser80 in the protein. The recognition is affected by glycan structure, stem peptide, and cross-linking of PGs, as well as by other cell wall components. Significant and specific binding of PGs by PGRP1 correlates with synergistic enhancement in the proteolytic activation of proPO in plasma. PGRP1 is a component of the M. sexta proPO system, but we do not understand precisely how PG-PGRP1 recognition leads to proPO activation.
Supplementary Material
Curdlan was incubated with plasma and purified PGRP1 for 1 h at 25 °C. PO activity (#4) was measured and plotted as mean ± SEM (n = 3), along with those of the controls (#1, plasma only; #2, plasma and curdlan; #3, plasma and PGRP1). Since interaction of plasma factors with curdlan (#2 - #1) and co-presence of plasma and exogenous PGRP1 (#3 - #1) both trigger proPO activation, an interaction of cudlan and exogenous PGRP1 in plasma (#4 - #1) would increase proPO activation at a level significantly higher than the sum of the two components [(#2 - #1) + (#3 - #1)]. Unpaired t-test, however, fails to show a synergistic effect.
Acknowledgments
We thank Drs. Kanost, Dillwith, and the anonymous reviewers for their critical comments on the manuscript. Dr. Michael Kanost at Kansas State University provided useful regents for this work. We would like to thank Xiufeng Zhang for repeating the experiment of proPO activation by microbial cells. The study was supported by National Institutes of Health Grant GM58634. The article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450.
The abbreviations used are
- PG and PGRP
peptidoglycan and its recognition protein
- DAP
meso-diaminopimelic acid
- PO and proPO
phenoloxidase and its precursor
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- BSA
bovine serum albumin, NTA, nitrilotriacetic acid
- TBS
Tris buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atrih A, Bacher G, Allmaier G, Williamson MP, Foster SJ. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP5 in peptidoglycan maturation. J Bacteriol. 1999;181:3956–3966. doi: 10.1128/jb.181.13.3956-3966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricas E, Ghuysen JM, Dezélée P. The cell wall peptidoglycan of Bacillus megateriumKM. I Studies on the stereochemistry of α,α′-diaminopimelic acid. Biochemistry. 1967;6:2598–2607. doi: 10.1021/bi00860a043. [DOI] [PubMed] [Google Scholar]
- Chaput C, Boneca IG. Peptidoglycan detection by mammals and flies. Microbes Infect. 2007;9:637–647. doi: 10.1016/j.micinf.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 2009;11:631–636. doi: 10.1016/j.micinf.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Cho S, Wang Q, Swaminathan CP, Hesek D, Lee M, Boons GJ, Mobashery S, Mariuzza RA. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc Natl Acad Sci USA. 2007;104:8761–8766. doi: 10.1073/pnas.0701453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J Invert Pathol. 1983;41:77–85. [Google Scholar]
- Eleftherianos I, Gökçen F, Felföldi G, Millichap PJ, Trenczek TE, ffrench-Constant RH, Reynolds SE. The immunoglobulin family protein hemolin mediates cellular immune responses to bacteria in the insect Manduca sexta. Cell Microbiol. 2007;9:1137–1147. doi: 10.1111/j.1462-5822.2006.00855.x. [DOI] [PubMed] [Google Scholar]
- Gillespie JP, Kanost MR, Trenczek T. Biological mediators of insect immunity. Ann Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- Guan R, Roychowdhury A, Ember B, Kumar S, Boons GJ, Mariuzza RA. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc Natl Acad Sci USA. 2004;101:17168–17173. doi: 10.1073/pnas.0407856101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Roychowdury A, Ember B, Kumar S, Boons GJ, Mariuzza RA. Crystal structure of a peptidoglycan recognition protein (PGRP) in complex with a muramyl tripeptide from Gram-positive bacteria. J Endotoxin Res. 2005;11:41–46. doi: 10.1179/096805105225006713. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J Biol Chem. 2003;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Science. 2008;15:53–66. [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Golenbock D, Silverman N. Peptidoglycan recognition by the Drosophila Imd pathway. J Endotoxin Res. 2005;11:383–389. doi: 10.1179/096805105X76823. [DOI] [PubMed] [Google Scholar]
- Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc Natl Acad Sci USA. 1998;95:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Kim MS, Byun M, Oh BH. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol. 2003;4:787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- Lee MH, Osaki T, Lee JY, Baek MJ, Zhang R, Park JW, Kawabata S, Soderhall K, Lee BL. Peptidoglycan recognition proteins involved in 1,3-β-D-glucan-dependent prophenoloxidase activation system of insect. J Biol Chem. 2004;279:3218–3227. doi: 10.1074/jbc.M309821200. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Leone P, Bischoff V, Kellenberger C, Hetru C, Royet J, Roussel A. Crystal structure of Drosophila PGRP-SD suggests binding to DAP-type but not lysine-type peptidoglycan. Mol Immunol. 2008;45:2521–2530. doi: 10.1016/j.molimm.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan-recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- Lu Z, Jiang H. Expression of Manduca sexta serine proteinase homolog precursors in insect cells and their proteolytic activation. Insect Biochem Mol Biol. 2008;38:89–98. doi: 10.1016/j.ibmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278:7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Håkansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci USA. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A pattern recognition protein for peptidoglycan: cloning the cDNA and the gene of the silk worm, Bombyx mori. J Biol Chem. 1999;274:11854–11858. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- Onoe H, Matsumoto A, Hashimoto K, Yamano Y, Morishima I. Peptidoglycan recognition protein (PGRP) from eri-silkworm, Samia cynthia ricini: protein purification and induction of the gene expression. Comp Biochem Physiol. 2007;147:512–519. doi: 10.1016/j.cbpb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Park J, Kim C, Kim J, Je B, Roh K, Kim S, Lee H, Ryu J, Lim J, Oh B, Lee W, Ha N, Lee B. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci USA. 2007;104:6602–6607. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C, Oldenvi S, Steiner H. Peptidoglycan recognition protein LF: A negative regulator of Drosophila immunity. Insect Biochem Mol Biol. 2007;37:1309–1316. doi: 10.1016/j.ibmb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Ragan EJ. PhD dissertation. Kansas State University; 2008. Immune-related protein complexes and serpin-1 isoforms in Manduca sexta plasma. [Google Scholar]
- Reiser JB, Teyton L, Wilson IA. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution. J Mol Biol. 2004;340:909–917. doi: 10.1016/j.jmb.2004.04.077. [DOI] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nature. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- Shockman GD, Barrett JF. Structure, function, and assembly of cell walls of Gram-positive bacteria. Ann Rev Microbiol. 1983;37:501–527. doi: 10.1146/annurev.mi.37.100183.002441. [DOI] [PubMed] [Google Scholar]
- Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev. 2004;198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x. [DOI] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurta S. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Asahi N, Suzuoki F, Ashida M, Matsuura S. Detection of peptidoglycan and β-glucan with silkworm larvae plasma test. FEMS Immunol Med Microbiol. 1996;15:129–134. doi: 10.1111/j.1574-695X.1996.tb00063.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem. 1996;271:13854–13860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Zhu Y, Ma C, Fabrick JA, Kanost MR. Pattern recognition proteins in Manduca sexta plasma. Insect Biochem Mol Biol. 2002;32:1287–1293. doi: 10.1016/s0965-1748(02)00091-7. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem Mol Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Curdlan was incubated with plasma and purified PGRP1 for 1 h at 25 °C. PO activity (#4) was measured and plotted as mean ± SEM (n = 3), along with those of the controls (#1, plasma only; #2, plasma and curdlan; #3, plasma and PGRP1). Since interaction of plasma factors with curdlan (#2 - #1) and co-presence of plasma and exogenous PGRP1 (#3 - #1) both trigger proPO activation, an interaction of cudlan and exogenous PGRP1 in plasma (#4 - #1) would increase proPO activation at a level significantly higher than the sum of the two components [(#2 - #1) + (#3 - #1)]. Unpaired t-test, however, fails to show a synergistic effect.