Abstract
Previous complementation and mapping of mutations that change the usual yellow color of the Zygomycete Phycomyces blakesleeanus to white or red led to the definition of two structural genes for carotene biosynthesis. We have cloned one of these genes, carRA, by taking advantage of its close linkage to the other, carB, responsible for phytoene dehydrogenase. The sequences of the wild type and six mutants have been established, compared with sequences in other organisms, and correlated with the mutant phenotypes. The carRA and carB coding sequences are separated by 1,381 untranslated nucleotides and are divergently transcribed. Gene carRA contains separate domains for two enzymes, lycopene cyclase and phytoene synthase, and regulates the overall activity of the pathway and its response to physical and chemical stimuli from the environment. The lycopene cyclase domain of carRA derived from a duplication of a gene from a common ancestor of fungi and Brevibacterium linens; the phytoene synthase domain is similar to the phytoene and squalene synthases of many organisms; but the regulatory functions appear to be specific to Phycomyces.
Carotenoids are tetraterpenoid pigments synthesized by photosynthetic organisms and many bacteria and fungi and acquired by animals with their diet. The attractive colors and the beneficial health effects of β-carotene and other carotenoids have stimulated interest in their chemical and biological production.
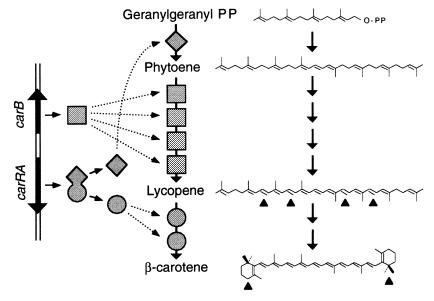
The production of β-carotene by the Zygomycete Phycomyces blakesleeanus has been investigated for over half a century (1). Three enzyme activities are required (Fig. 1): phytoene synthase joins two molecules of geranylgeranyl pyrophosphate to form phytoene; phytoene dehydrogenase introduces four double bonds to yield lycopene; and lycopene cyclase shapes the ψ acyclic ends of lycopene as β rings. This scheme is maintained in all carotenogenic organisms, with some modifications. For example, oxygen atoms may be introduced in the carotenes to form xanthophylls.
Figure 1.
Genes, enzymes, and chemical reactions for carotene biosynthesis in Phycomyces. Two molecules of geranylgeranyl pyrophosphate are converted to one molecule of β-carotene by phytoene synthase (diamonds), four copies of phytoene dehydrogenase (squares), and two copies of lycopene cyclase (circles). The scheme on the left represents the genes that code for these enzymes. The chemical changes are marked with arrowheads.
The yellow color of wild-type Phycomyces is modified by mutations to red, white, or various gradations of yellow. The red mutants accumulate lycopene, and the white mutants either accumulate phytoene or lack all carotenes, or at least do not have enough for a visible color (2, 3). The genetic analysis by complementation, recombination, and reversion of the red and white mutants identified two closely linked genes, carB and carRA (4–6). Their products, and possibly those of other genes, are organized into an enzyme complex working as an assembly chain in which the four dehydrogenations are catalyzed by four identical units of phytoene dehydrogenase (7, 8) and the two cyclizations are catalyzed by two identical units of lycopene cyclase (9, 10).
Gene carRA has two distinct domains (5). Domain R, proximal with respect to the translation start, is characterized by the red carR mutants and is responsible for lycopene cyclase. The distal domain A is characterized by carA mutants, which normally are white and have small amounts of β-carotene, but, in the presence of retinol, produce substantial amounts of β-carotene and become yellow (11). The carRA mutants with null phenotype for both domains are white and do not respond to retinol (5).
The concentration of β-carotene in the cells depends on environmental factors. Synergisms and differential effects on the mutants allow the classification of these factors in four groups with separate mechanisms of action. Blue light increases the carotene content in the wild type more than 10-fold. This response is defective in mutants of many genes (12). Sexual activity, mediated by trisporates, increases the carotene content more than 5-fold (13, 14) and retinol and dimethyl phthalate more than 40-fold (11, 15). Some “insensitive” mutants were isolated because of their limited response to retinol, but they turned out to be equally defective in their responses to the other activators (11, 14, 15); the mutation in one of these mutants, strain S119, is very closely linked to a carA mutation; the one in strain S144 is unlinked (6) and defines gene carI. Under standard conditions, both are light yellow and have just a little less β-carotene than the wild type.
The carotene content is increased permanently by recessive mutations at gene carS (13), distant from the carB–carRA gene cluster on the same chromosome (6). The carS mutants are insensitive to retinol, but sensitive to trisporates and dimethyl phthalate (14, 15). Other variations in carotene content are caused by mutations in the carC, carD, and carF regulatory genes (16–18), none of which is known to be close to the structural genes.
The gene for phytoene dehydrogenase, carB, was cloned (19) because of its similarity to al-1, the gene from Neurospora crassa with the same function (20). We have cloned and sequenced gene carRA from Phycomyces by taking advantage of its close linkage with carB and have correlated the sequences of several Phycomyces mutants with their phenotypes and with homologous sequences from various organisms.
Materials and Methods
Strains and Culture Conditions.
The P. blakesleeanus Bgff. wild-type NRRL1555 (used unless otherwise stated) and the mutant strains used in this work are listed in Table 1. In the strain designations, NRRL stands for the U.S. Department of Agriculture laboratory in Peoria, IL; C for the collection of the former Prof. Max Delbrück at the California Institute of Technology (Pasadena), and S for our collection. The mutants were obtained after treatments with the mutagen N-methyl-N′-nitro-N-nitrosoguanidine (21). Phycomyces was cultured and handled as described (22); standard conditions are 4 days on minimal agar at 22°C. Escherichia coli DH5α was used for the multiplication of plasmids.
Table 1.
Strains of P. blakesleeanus used in this work with the nucleotide changes in their carRA gene and the predicted amino acid changes
| Strain | Mutation | Main carotene | Nucleotide change | Amino acid change | ||
|---|---|---|---|---|---|---|
| NRRL1555 | none | β-carotene | None | None | ||
| C9 | carR21 | Abundant lycopene | 908 | C → T | 215 | P → S |
| C11 | carR23 | Abundant lycopene | 229 | G → A | 77 | E → K |
| C6 | carRA12 | Traces of lycopene | 400 | G → A | See text | |
| S92 | carRA91 | Traces of lycopene | 177 | G → A | 59 | W → stop |
| C2 | carA5 | Scarce β-carotene | 1710 | C → T | 481 | P → L |
| S119 | carA113 | β-carotene | 1659 | T → C | 465 | I → T |
See ref. 21 for additional details.
DNA Isolation and Manipulation.
Genomic DNA was isolated as described (23). Cleaner genomic DNA preparations (more sensitive to restriction enzymes) are obtained from sporangiophores than from mycelium (24). For Southern blots, genomic DNA (1–4 μg) was digested overnight with restriction enzymes, separated by electrophoresis in an agarose gel (8 g/liter), transferred to a nylon membrane, hybridized with probes labeled with digoxigenin-11-dUTP, and detected with Lumigen PPS following the manufacturer's recommendations (Roche, Mannheim, Germany). Phycomyces cDNA was obtained with the TimeSaver cDNA kit and mRNA from vegetative mycelia isolated with the QuickPrep mRNA Purification kit (both from Amersham Pharmacia).
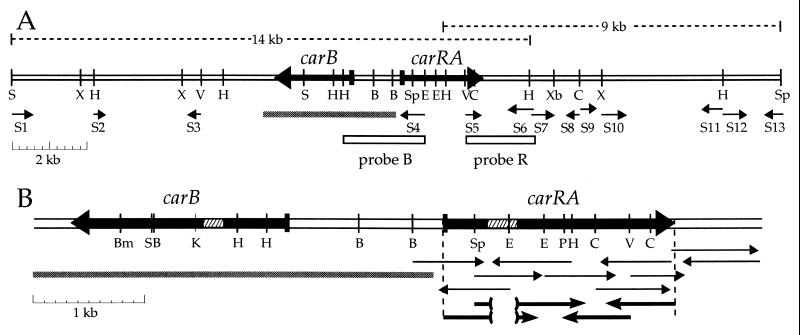
A lambda 2001 gene library was constructed by inserting Phycomyces DNA partially digested with Sau3A into the BamHI site of the vector and packaged into phage particles with the Gigapack Gold Kit (Stratagene). The library, consisting of about 7,000 independent clones, was multiplied and screened for hybridization with a 2.2-kb Phycomyces DNA fragment (probe B in Fig. 2A) that contains the promoter and part of the coding sequence of gene carB. Probe B was cut with HindIII from the 5-kb EcoRI fragment that includes the whole carB gene (19). A positive clone contained a 13-kb insert, a SacI site in the insert, and two other SacI sites in the polylinker sequence, flanking the insertion site. Two DNA fragments of about 5.7 kb and 7.8 kb, respectively, were cut from the chimeric lambda molecule by SacI digestion, subcloned in the SacI site of vector Bluescript KS(+), and characterized by restriction and partial sequencing. The location of gene carB was deduced by comparison of the restriction fragments with those expected from the known carB gene sequence.
Figure 2.
Structural genes for carotene biosynthesis in Phycomyces. (A) A DNA region that contains genes carB and carRA. Probes B and R were used to screen a lambda library and a partial plasmid library and isolate the 14-kb and the 9-kb fragments, respectively. Sequencing reactions are indicated by the thin arrows S1 through S13. The gray segment represents the DNA sequence published by Ruiz-Hidalgo et al. (19). (B) A 6494-bp DNA sequence that contains genes carB and carRA. Hatched segments represent introns and white segments represent intergenic sequences. Thin arrows represent genomic sequencing reactions, and thicker arrows below represent cDNA sequencing reactions. The target sites of restriction endonucleases are: B, BglII; Bm, BamHI; C, ClaI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SacI; Sp, SpeI; V, EcoRV; X, XhoI; and Xb, XbaI.
A 1.4-kb EcoRV–SacI fragment of the 13-kb insert (probe R in Fig. 2A) was found to hybridize with a single 9-kb EcoRI–SpeI genomic DNA fragment. A partial genomic library of 6- to 10-kb EcoRI–SpeI fragments of Phycomyces genomic DNA in Bluescript KS(+) was constructed and screened with probe R. This led to the isolation of a positive clone with a 9-kb insert.
The PCR mixtures contained about 5 ng genomic DNA, 0.2 mM of each of the four deoxynucleoside triphosphates, 1 μM of each primer, and 0.5 μl of the DNA polymerase mixture Expand High Fidelity (Roche) in a final volume of 50 μl. After overlaying them with light mineral oil, they were incubated once at 95°C for 3 min, 35 times at 95°C for 1 min, 55°C for 1 min, and 68°C for 3 min, and once at 68°C for 10 min in a GeneAmp 2400 programmable thermocycler (Perkin–Elmer/Cetus). The cDNA to be sequenced was multiplied with primers 5′-TTCTTTATTCCTGTGACTTTCCACGATCA-3′ and 5′-GCGCGCATGCTGACTTATATGGAAGT-3′. For sequencing the mutant alleles, this last primer was replaced by 5′-CAGTACTAAGCACCATTCAC-3′.
DNA was sequenced by Medigene (Martinsried, Germany) using the ABI377 automatic sequencer (Perkin–Elmer). Mutant alleles were sequenced twice from independent PCR products. For other DNA manipulations and technical details, see ref. 25.
Sequence Analyses.
Alignments were facilitated by the CLUSTAL W 1.6 and X 1.63B computer programs (26–28). Hydrophobicity profiles were obtained with the Expert Protein Analysis System (ExPASy; Swiss Institute of Bioinformatics, Geneva, Switzerland). The search for protease cleavage sites (29) was carried out with the signalp 1.1 (Technical University of Denmark, Lyngby) and the psort ii (Institute for Medical Science, University of Tokyo, Tokyo) computer programs.
Results
DNA Flanking the carB Gene of Phycomyces.
The very close linkage of genes carRA and carB led us to expect both to be present simultaneously in small DNA fragments of the Phycomyces genome. A Phycomyces lambda 2001 gene library was screened for hybridization with a DNA fragment (probe B in Fig. 2A) that contains gene carB and its promoter. A clone with a 14-kb DNA insert was isolated and characterized to obtain a restriction map and the location of the carB gene.
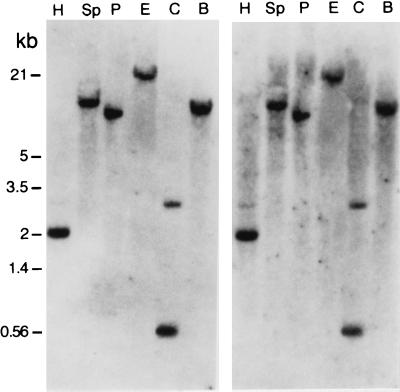
A fragment from one of the ends of the 14-kb DNA insert (probe R in Fig. 2A) hybridized with single fragments of Phycomyces genomic DNA cut with various restriction enzymes, even at low stringency conditions (Fig. 3). This indicated that the Phycomyces genome contains a single copy of the probe R sequence. This probe was used to isolate a 9-kb fragment of genomic DNA, which extended to over 20 kb the available DNA that includes and surrounds gene carB.
Figure 3.
Hybridization of probe R (Fig. 2A) with Phycomyces genomic DNA fragments cut with several restriction enzymes (abbreviated as in Fig. 2) and separated by electrophoresis. The two pictures differ in the stringency of the test (washed at 65°C, Left, and 42°C, Right).
The whole region was explored by cloning and sequencing 13 subfragments (Fig. 2A). S4 and S5 caught our attention and led us to sequence completely a segment of 3,100 bp, where a putative new gene was found (Fig. 2B). This gene was identified as gene carRA by sequencing the same fragment from some mutants that had been used to define gene carRA, as described below.
The sequences S2, S3, and S11 had low proportions of GC base pairs, as is usual in the intergenic sequences of Phycomyces (30), and contained abundant stop codons in all possible reading frames. Several putative genes seem to be present in the region, but they do not seem to be related to carotene biosynthesis. One of them, closely similar to α-β hydrolases (well known fungal lipases), would be transcribed toward the right overlapping sequences S12 and S13. Another one, very similar to an unknown gene from Drosophila melanogaster and, to a lower extent, to genes for mucins from several animals, seem to be present in sequence S10. Three additional ORFs were found in sequences S1, S6-S7, and S8–S9.
The Wild-Type carRA Sequence.
Gene carRA was composed of two ORFs with a total of 1,806 bp, interrupted by a 265-bp intron. The sequence and the precise limits of the intron were defined by sequencing DNA fragments obtained by PCR from a cDNA sample (Fig. 2B). The putative CarRA protein would have 602 aa. Codon usage in gene carRA is similar to that of other Phycomyces genes, with a marked preference for pyrimidines, and particularly for C, in the third position of the codons.
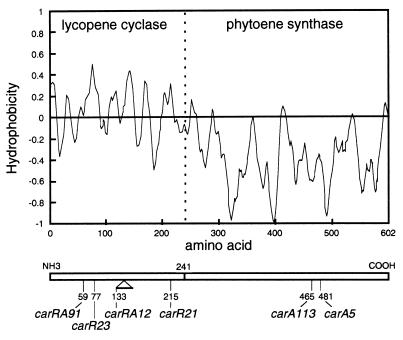
The polarity profile of CarRA (Fig. 4) defined two different domains. Domain R, covering 40% of the protein from its amino end, would be hydrophobic and transmembranal; the other one, domain A, covering the rest to the carboxyl end, would be hydrophilic. The frontier between both domains lies approximately at the motif AHAIV (amino acid residues 239–243), which is a putative protease cleavage site that could split the polypeptide between residues 241 and 242.
Figure 4.
Hydrophobicity index of the predicted CarRA protein sequence. The vertical dotted line marks the location of the putative protease cleavage site AHAIV. The sites of the mutations in six color mutants are indicated below.
DNA Sequences in carRA Mutants.
The carRA sequence was obtained from six independent color mutants that were known or suspected to carry mutations in gene carRA, according to previous genetic analyses (4–6). Three of these mutants are albino, two are red, and one is light yellow. All of them carried mutations in the newly cloned gene (Table 1).
Strains C6 and S92 are albino carRA mutants, with very little lycopene, no β-carotene, and insensitive to retinol. The mutation carRA91 in strain S92 replaced a TGA stop with the normal tryptophan at codon 59. In strain C6, the first base of the intron, guanine, was replaced by adenine. Both changes in the sequence of the gene amply justify the phenotypes of the respective mutants.
Strains C9 and C11 are red carR mutants, deficient in lycopene cyclase, rich in lycopene, and insensitive to retinol. In each, an amino acid of the R domain was replaced by a very different amino acid (proline by cysteine, glutamate by lysine). Critical sites for lycopene cyclase activity therefore must lie in the R domain of the CarRA protein.
The two other strains had single amino acid substitutions in the A domain of the CarRA protein. The albino phenotype of strain C2 is the result of a proline to leucine replacement. Strain S119, isolated because of its weak response to chemical activators, was suspected to have a mutated gene carRA only because of tight linkage with the carA5 mutation (6). The finding of an isoleucine to threonine substitution in the A domain confirms this suspicion and justifies calling the mutation carA113.
Sequence Comparisons.
Gene carRA from P. blakesleeanus is similar to genes from other fungi (Fig. 5) in the general structure of the predicted product (an amino-terminal hydrophobic domain and a carboxyl-terminal hydrophilic domain) and in the actual sequence. Particularly well conserved are the sites of mutations that we have analyzed, with the obvious exception of the one at the intron. The putative protease cleavage site AHAIV that would split the Phycomyces translation product after residue 241 is not conserved, but the other gene products contain alternative cleavage sites. For example, the Xanthophyllomyces product may be split at LSACD after residue 248 and the Neurospora gene product, at LWAVD after residue 190.
Figure 5.
Alignment of the protein sequences Pb, Mc, Nc, and Xd predicted from the sequences of genes carRA from Phycomyces blakesleeanus, al-2 (ref. 31; GenBank accession no. L27652) from Neurospora crassa, crtYB (ref. 32; accession no. AJ133646) from Xanthophyllomyces dendrorhous (syn. Rhodomyces dendrorhous and Phaffia rhodozyma), and carRP (ref. 33; accession no. AJ250827) from Mucor circinelloides. The line marks the putative cleavage site in the Phycomyces sequence at the approximate limit of the lycopene cyclase and phytoene synthase domains. The black circles locate the mutations in six Phycomyces color mutants. The arrowheads indicate the presence of introns in the gene sequence.
The Phycomyces R domain retains the traces of an old duplication. This is seen by comparing the sequences before and after the location of the intron (Fig. 6). These approximate halves are related to each other (27 positions in the corresponding polypeptide sequences are occupied by identical amino acids) and even more so to gene product CrtYc from Brevibacterium linens (31 and 38 identical amino acids, respectively). The Phycomyces R domain is similar to those of other fungi (130, 62, and 56 identical amino acids with those of Mucor, Xanthophyllomyces, and Neurospora, respectively; Fig. 5), but bears little or no relationship to the lycopene cyclases from plants.
Figure 6.
Alignment of two parts of the lycopene cyclase domain of the CarRA protein from Phycomyces and proteins CrtYc and CrtYd from Brevibacterium linens (ref. 34; GenBank accession no. AF139916). CarRA(1) and CarRA(2) represent the 133-aa sequence that precedes the intron location and the next 128 aa, respectively. The black circles locate missense mutations in red mutants of Phycomyces.
The Phycomyces domain A coincides in 211, 120, and 100 amino acids with the corresponding domains of Mucor, Xanthophyllomyces, and Neurospora, respectively (Fig. 5), in 74–98 amino acids with nine phytoene synthases (Agrobacterium aurantiacum, Rhodobacter sphaeroides, Synechococcus PCC7942, Synechocystis sp, Spirulina platensis, Dunaliella bardawil, Arabidopsis thaliana, Lycopersicon esculentum, and Zea mays) and in 69–79 amino acids with six squalene synthases (Candida utilis, Capsicum annuum, Solanum tuberosum, Zea mays, Leishmania major, and Homo sapiens). Phytoene and squalene synthases catalyze exactly the same chemical changes on different substrates.
Discussion
We have cloned a gene from the Phycomyces wild type and six color mutants often used to investigate the genetics and other aspects of carotene biosynthesis. The sequences of the six color mutants clearly identified the gene as the carRA gene previously defined by complementation and recombination (4–6). Our molecular characterization of gene carRA confirms the results of classical geneticists, to such an extent that the best way to state our conclusions is to copy theirs (5): “The A and R functions are determined by contiguous DNA segments, cotranscribed to a single mRNA, and cotranslated to a single polypeptide. The R segment is proximal to the gene's beginning (5′ of the mRNA and NH2 end of the polypeptide). Phenotype R would arise from missense mutations in the R segment; phenotype AR from nonsense or frameshift mutations in the R segment.”
In fact, the two carR mutants carry missense mutations in the R domain. These are far apart on the primary structure, but both affect the PLEE tetrapeptide that is repeated in the two halves of the R domain (Fig. 6). The carR21 mutant protein suffers from a drastic loss of enzyme activity, but integrates in the enzyme aggregate as effectively as the wild-type protein (9).
The carRA91 mutation introduces a premature TGA stop codon in the R domain and would result in a short, presumably inactive, polypeptide with only 58 aa. The traces of lycopene found in mutant mycelia require some phytoene synthase activity, which may result from a low natural level of TGA suppression. It is also possible that squalene synthase processes phytoene to a certain extent.
The carRA12 mutation in strain C6 modifies the first nucleotide of the critical GU dinucleotide splice site at the 5′ end of the intron. Because the intron contains a stop codon in-phase with the reading frame of the gene, the excision failure would result in a polypeptide with 149 aa, 16 of them in a tail unrelated to the original sequence. The traces of lycopene in the mutant might result from residual excision of the intron or from the rare, in-phase excision of another nucleotide segment. Red revertants were obtained from strain C6, and the mutation in one of them was very closely linked to the original mutation (5). This is likely to result from the creation of a new intron excision site, so that the R activity is lost, but the gene is read through to produce a correct phytoene synthase.
The missense mutation carA5 in strain C2 results in a 10- to 15-fold reduction of the β-carotene content with respect to the appropriate controls. Under standard conditions, strain C2 contains 4 μg/g dry mass, vs. 62 in the wild type (35), and similar reductions are found under sexual stimulation, in the presence of trisporic acids, retinol, and dimethyl phthalate, and after introduction of a carS mutation in the genetic background (11, 14, 15, 36). Other carA mutants exhibit their own reduction levels. This means that the mutations impair to a certain extent the catalytic activity of the protein, regardless of the regulatory interactions.
The carA5 mutation has other pleiotropic effects that can hardly be ascribed to the reduced phytoene synthase activity. This mutation, even in heterokaryosis, inhibits substrate transfer in the cyclization of lycopene to β-carotene and renders these reactions more susceptible to chemical inhibition (37). This defect may be relatively trivial, because it is corrected by the mere addition of detergent to the medium. Additionally, the mutant lacks photoinduction of carotene synthesis (12) and has a severe photomorphogenetic defect (38). These two defects cannot be attributed to the carotene shortage, because they occur in carA carS double mutants that contain more β-carotene than the wild type.
Phenotypes such as that of strain C2 cannot result from severe mutations in domain A that would cause an irreparable loss of phytoene synthase activity. Such mutants would have the null AR phenotype. In fact, all mutants induced by a frameshift-inducing agent presented the null phenotype (5).
S119 strain has the same phenotype as S144, whose mutation carI131 affects an unlinked gene (6). We propose that mutation carA113 marks the binding site of the carI gene product, which mediates the stimulatory effects of light, sexual activity, retinol, and dimethyl phthalate. The carS gene product mediates only the effect of retinol and its analogues, probably by interacting with the carI gene product, although both could interact with phytoene synthase.
The results from classical genetics (5) suggested that “the polypeptide is split into separate R and A proteins,” unless “the A function is mediated by a diffusible product.” It is thus interesting to note the presence of a putative protease cleavage site at about the limit between domains R and A in Phycomyces and other fungi.
The three enzyme activities that catalyze the seven chemical reactions needed for the synthesis of β-carotene in Phycomyces depend on the expression of two adjacent genes, carRA and carB, which are transcribed divergently. The advantage of this set-up would be the joint regulation of the whole pathway from a single promoter region. This arrangement is not conserved: the homologous genes of Fusarium fujikuroi are adjacent, but transcribed in the same direction (M. M. Prado and J.A., unpublished results); those of Neurospora are linked, but separated by other genes (39).
Gene carRA is remarkably similar (Fig. 5) to homologous genes from representatives of three major classes of Fungi, Zygomycetes (Mucor), Ascomycetes (Neurospora), and Basidiomycetes (Xanthophyllomyces). Gene al-2 from Neurospora, defined by albino mutations and responsible for phytoene synthase (40), was the first to be cloned and sequenced (31). A cyclase activity has been shown to be present in the amino-terminal domain of the al-2 gene product by the recent isolation of reddish mutants of Neurospora that carry mutations in that domain (J.A. and T. Schmidhauser, unpublished observations). Verdoes et al. (32) were the first to show, by functional complementation in E. coli, that lycopene cyclase and phytoene synthase are coded by a single gene from Xanthophyllomyces, which they called crtYB.
Sequence comparisons suggest that the lycopene cyclase domain of the fungal genes was formed by a duplication of a small gene in a common ancestor of the fungi and Brevibacterium. The lycopene cyclase activity of this bacterium (34) is the result of the joint action of the products of genes crtYc and crtYd, which are contiguous, related by their sequence, and probably formed by a different duplication. The phytoene synthase domain of the fungal genes seems to derive from the common ancestor of all present phytoene synthases and squalene synthases.
The activation of the carotene pathway by light and chemicals is not universal. Light is an activator in Phycomyces and Neurospora, but not in Xanthophyllomyces (41) or Blakeslea (42). As far as they have been tested, the best chemical activators for Phycomyces have little or no effect on Ascomycetes, Basidiomycetes, bacteria, and plants. In view of the conservation of the A domain, including the amino acids that are changed in Phycomyces mutants, it would not be surprising if other organisms used the same protein domain for their own regulatory purposes.
Acknowledgments
We thank Prof. A. P. Eslava, Universidad de Salamanca, Salamanca, Spain, for the kind gift of the Phycomyces DNA fragment that contains the carB gene, Prof. L. M. Corrochano, Universidad de Sevilla, for the Phycomyces cDNA, and D. Pérez del Camino for technical assistance. This work was financed by the European Union (Contract FAIR PL96-1633), the Spanish Government (Grant DGES PB96-1336), and Junta de Andalucía (Group 3038).
Footnotes
Data deposition: The nucleotide sequence that contains gene carRA has been deposited in the GenBank database (accession no. AJ278287).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021555298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021555298
References
- 1.Cerdá-Olmedo E. In: Phycomyces. Cerdá-Olmedo E, Lipson E D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1987. pp. 199–222. [Google Scholar]
- 2.Heisenberg M, Cerdá-Olmedo E. Mol Gen Genet. 1968;102:187–195. doi: 10.1007/BF00385973. [DOI] [PubMed] [Google Scholar]
- 3.Meissner G, Delbrück M. Plant Physiol. 1968;43:1279–1283. doi: 10.1104/pp.43.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ootaki T, Crafts-Lighty A, Delbrück M, Hsu W J. Mol Gen Genet. 1973;121:57–70. doi: 10.1007/BF00353693. [DOI] [PubMed] [Google Scholar]
- 5.Torres-Martínez S, Murillo F J, Cerdá-Olmedo E. Genet Res. 1980;36:299–309. doi: 10.1017/s0016672300019911. [DOI] [PubMed] [Google Scholar]
- 6.Roncero M I G, Cerdá-Olmedo E. Curr Genet. 1982;5:5–8. doi: 10.1007/BF00445733. [DOI] [PubMed] [Google Scholar]
- 7.Eslava A P, Cerdá-Olmedo E. Plant Sci Lett. 1974;2:9–14. [Google Scholar]
- 8.Aragón C M G, Murillo F J, de la Guardia M D, Cerdá-Olmedo E. Eur J Biochem. 1976;63:71–75. doi: 10.1111/j.1432-1033.1976.tb10208.x. [DOI] [PubMed] [Google Scholar]
- 9.De la Guardia M D, Aragón C M G, Murillo F J, Cerdá-Olmedo E. Proc Natl Acad Sci USA. 1971;68:2012–2015. doi: 10.1073/pnas.68.9.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candau R, Bejarano E R, Cerdá-Olmedo E. Proc Natl Acad Sci USA. 1991;88:4936–4940. doi: 10.1073/pnas.88.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eslava A P, Alvarez M I, Cerdá-Olmedo E. Eur J Biochem. 1974;48:617–623. [Google Scholar]
- 12.Bejarano E R, Avalos J, Lipson E D, Cerdá-Olmedo E. Planta. 1991;183:1–9. doi: 10.1007/BF00197560. [DOI] [PubMed] [Google Scholar]
- 13.Murillo F J, Cerdá-Olmedo E. Mol Gen Genet. 1976;148:19–24. doi: 10.1007/BF00268541. [DOI] [PubMed] [Google Scholar]
- 14.Govind N S, Cerdá-Olmedo E. J Gen Microbiol. 1986;132:2775–2780. [Google Scholar]
- 15.Bejarano E R, Parra F, Murillo F J, Cerdá-Olmedo E. Arch Microbiol. 1988;150:209–214. [Google Scholar]
- 16.Revuelta J L, Eslava A P. Mol Gen Genet. 1983;192:225–229. [Google Scholar]
- 17.Salgado L M, Bejarano E R, Cerdá-Olmedo E. Exp Mycol. 1989;13:332–336. [Google Scholar]
- 18.Mehta B J, Salgado L M, Bejarano E R, Cerdá-Olmedo E. Appl Environ Microbiol. 1997;63:3657–3661. doi: 10.1128/aem.63.9.3657-3661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Hidalgo M J, Benito E P, Sandmann G, Eslava A P. Mol Gen Genet. 1997;253:734–744. doi: 10.1007/s004380050378. [DOI] [PubMed] [Google Scholar]
- 20.Schmidhauser T J, Lauter F R, Russo V E A, Yanofsky C. Mol Cell Biol. 1990;10:5064–5070. doi: 10.1128/mcb.10.10.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerdá-Olmedo E. Methods Enzymol. 1985;110:220–243. [Google Scholar]
- 22.Cerda-Olmedo E. In: Phycomyces. Cerdá-Olmedo E, Lipson E D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1987. pp. 337–339. [Google Scholar]
- 23.Giordano W, Avalos J, Fernandez-Martín R, Cerdá-Olmedo E, Domenech C. Microbiology. 1999;145:2997–3002. doi: 10.1099/00221287-145-10-2997. [DOI] [PubMed] [Google Scholar]
- 24.Weinkove D, Poyatos J A, Greiner H, Oltra E, Avalos J, Fuksansky L, Barrero A F, Cerdá-Olmedo E. Fungal Genet Biol. 1998;25:196–203. doi: 10.1006/fgbi.1998.1098. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins D G, Thompson J D, Gibson T J. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Avalos J, Corrochano L M, Brenner S. Gene. 1996;174:43–50. doi: 10.1016/0378-1119(96)00304-6. [DOI] [PubMed] [Google Scholar]
- 31.Schmidhauser T J, Lauter F R, Schumacher M, Zhou W, Russo V E A, Yanofsky C. J Biol Chem. 1994;269:12060–12066. [PubMed] [Google Scholar]
- 32.Verdoes J C, Krubaski P, Sandmann G, van Ooyen A J J. Mol Gen Genet. 1999;262:453–461. doi: 10.1007/s004380051105. [DOI] [PubMed] [Google Scholar]
- 33.Velayos A, Eslava A P, Iturriaga E A. Eur J Biochem. 2000;267:5509–5519. doi: 10.1046/j.1432-1327.2000.01612.x. [DOI] [PubMed] [Google Scholar]
- 34.Krubaski P, Sandmann G L. Mol Gen Genet. 2000;263:423–432. doi: 10.1007/s004380051186. [DOI] [PubMed] [Google Scholar]
- 35.Bejarano E R, Cerdá-Olmedo E. Phytochemistry. 1989;28:1623–1626. [Google Scholar]
- 36.Salgado L M, Cerdá-Olmedo E. Curr Genet. 1992;21:67–71. [Google Scholar]
- 37.Murillo F J, Torres-Martínez S, Aragón C M G, Cerdá-Olmedo E. Eur J Biochem. 1981;119:511–516. doi: 10.1111/j.1432-1033.1981.tb05637.x. [DOI] [PubMed] [Google Scholar]
- 38.Corrochano L M, Cerdá-Olmedo E. J Photochem Photobiol. 1990;6:325–335. [Google Scholar]
- 39.Perkins D D, Radford A, Sachs M A. The Neurospora Compendium. San Diego: Academic; 2001. [Google Scholar]
- 40.Harding R W, Turner R V. Plant Physiol. 1981;68:745–749. doi: 10.1104/pp.68.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An G-H, Johnson E A. Antonie van Leeuwenhoek. 1990;57:191–203. doi: 10.1007/BF00400151. [DOI] [PubMed] [Google Scholar]
- 42.Sutter R P. J Gen Microbiol. 1970;64:215–221. doi: 10.1099/00221287-64-2-215. [DOI] [PubMed] [Google Scholar]