Abstract
Infertility is a worldwide reproductive health problem, affecting men and women roughly equally. Mouse genetic studies demonstrate that more than two hundred genes specifically or predominantly regulate fertility. However, few genetic causes of infertility in humans have been identified. Here we focus on the regulation of male fertility by X-linked germ cell-specific genes. Previous genomic studies reveal that the mammalian X chromosome is enriched for genes expressed in early spermatogenesis. Recent genetic studies in mice show that X-linked germ cell-specific genes, such as Akap4, Nxf2, Taf7l, and Tex11, indeed play important roles in regulation of male fertility. Moreover, we find that the Taf7l Tex11 double mutant males exhibited much more severe defects in meiosis than either single mutant, suggesting that these two X-linked genes regulate male meiosis synergistically. The X-linked germ cell-specific genes are particularly attractive in the study of male infertility in humans. Because males are hemizygous for X-linked genes, loss-of-function mutations in the single copy X-linked genes, unlike in autosomal genes, would not be masked by a normal allele. The genetic studies of X-linked germ cell-specific genes in mice have laid a foundation for mutational analysis of their human orthologues in infertile men.
Keywords: male fertility, infertility, X chromosome, meiosis, spermatogenesis
Male infertility
An estimated 15% of couples are affected by infertility worldwide (Matzuk and Lamb, 2002). Males and females contribute roughly equally to infertility. Studies in various model organisms have provided great insights into the molecular and genetic pathways that regulate fertility (de Rooij and de Boer, 2003; Matzuk and Lamb, 2002). In particular, genetic studies in mice have identified more than 200 genes that are specifically or preferentially involved in the regulation of fertility, providing potential molecular targets for contraception and facilitating genetic studies of infertility in humans (Matzuk and Lamb, 2002). Major known causes of male infertility in humans include Y chromosome deletions and chromosomal abnormalities such as Klinefelter syndrome (47, XXY), which together account for about 25% of infertility in otherwise healthy men (Reijo et al, 1996; Van Assche et al, 1996). Therefore, the majority (~75%) of cases of male infertility in humans are idiopathic (of unknown origin) and the underlying causes are believed to be genetic. However, to date, efforts to uncover point mutations in single genes that contribute to human spermatogenic failure have been largely unsuccessful (Matzuk and Lamb, 2002; Nuti and Krausz, 2008). The slow progress in human studies could be attributed to the possibility that even though a great number (hundreds if not thousands) of genes specifically regulate fertility, the contribution of mutations in each gene is likely to be small. Therefore, the likelihood to find a causative mutation in one particular gene in infertile men is expected to be extremely low.
Enrichment and deficiency of germ cell-specific genes on the X chromosome
The transcription status of the X chromosome changes dramatically during male germ cell development. In mammals, sex chromosomes and autosomes are transcriptionally active in mitotically dividing spermatogonia and early meiotic (pre-pachytene) spermatocytes. During meiosis, sex chromosomes undergo meiotic sex chromosome inactivation (MSCI) and are thus transcriptionally silenced, while autosomes are actively transcribed (Handel et al, 1994; Solari, 1974; Turner et al, 2005). In post-meiotic germ cells, sex chromosomes remain transcriptionally repressed (Namekawa et al, 2006; Turner et al, 2006). Genomic studies have shown that germ cell-specific genes are not randomly distributed in the genome, and that, in particular, the unique hemizygous and transcription status of the X chromosome has shaped its germ cell-specific gene content (Khil et al, 2004; Namekawa et al, 2006; Turner et al, 2006; Wang et al, 2001). In summary, the X chromosome is enriched for genes expressed prior to meiosis but is deficient in spermatogenesis genes expressed meiotically and post-meiotically.
However, two recent studies added provocative episodes to the saga of the mammalian X chromosome (Mueller et al, 2008; Song et al, 2009). Many X-linked microRNAs are expressed at the pachytene stage, when MSCI occurs (Song et al, 2009). The escape from MSCI silencing by X-linked microRNAs suggests that they may contribute to MSCI itself or regulate post-transcriptional regulation of autosomal mRNAs during the meiotic and post-meiotic stages. Thirty-three multicopy gene families (a total of about 273 X-linked genes) are expressed predominantly in post-meiotic germ cells (Mueller et al, 2008). The expression level of these X-linked multicopy gene family is comparable to that of autosomal genes, suggesting that multiplication of gene copy numbers evolved to counteract transcriptional repression of the X chromosome in post-meiotic germ cells.
The importance of the X chromosome in mammalian spermatogenesis was first suggested by its enrichment of germ cell-specific genes expressed in early spermatogenesis. A systematic genomic screen identified 36 such genes from mouse spermatogonia, the majority of which are single-copy genes (Wang et al, 2001). Nearly one third of these genes maps to the X chromosome, suggesting that the X chromosome plays a preeminent role in early spermatogenesis. In retrospect, the number of germ cell-specific genes expressed at the spermatogonial stage is rather small (at least 36), compared to the large number of germ cell-specific genes (at least 350) expressed at the meiotic and post-meiotic stages (Fujii et al, 2002; Griswold, 1998; Schultz et al, 2003). To date, genetic studies of half of these 36 spermatogonially expressed genes by targeted inactivation have demonstrated that they play important roles in many aspects of spermatogenesis (Wang and Pan, 2007). Here we focus on the genetic studies of four X-linked germ cell-specific genes (Akap4, Nxf2, Taf7l, and Tex11) in mouse, as they may represent “hotspots” for mutations causing infertility in men. As these are single-copy genes and males are hemizygous for the X chromosome, mutations in these X-linked genes, unlike autosomal recessive mutations, would not be masked by a wild type allele. Therefore, it is more likely to identify causative point mutations in X-linked genes than in autosomal genes in infertile men. We will review the genetic studies of these X-linked genes in mice and humans, describe the synergistic regulation of male meiosis by two X-linked genes (Taf7l and Tex11), and discuss the opportunities and challenges in the study of male infertility in humans.
Akap4 is essential for sperm motility and male fertility
Cyclic AMP (cAMP) functions as a second messenger in the signal transduction pathway that regulates sperm motility. The cAMP-dependent protein kinase (PKA) is compartmentalized through its interaction with a family of A-kinase anchoring proteins (AKAPs). AKAP4 is the most abundant protein in the fibrous sheath, a cytoskeletal structure in the principle piece of sperm flagellum (Carrera et al, 1994). AKAP4 interacts with AKAP3, another sperm-specific AKAP (Brown et al, 2003). In Akap4-deficient mice, sperm count was not reduced, but sperm were immotile, resulting in male infertility (Miki et al, 2002). In Akap4 mutant sperm, fibrous sheath-associated proteins such as glycolytic enzymes (GAPDS) and AKAP3 were either absent or reduced in abundance. Therefore, AKAP4 is required for structural and functional integrity of the fibrous sheath. Mutations ablating the AKAP4 function could cause dysplasia of the fibrous sheath (DFS) in infertile men.
Nxf2 regulates male meiosis and maintenance of spermatogonial stem cells
In eukaryotes, bulk mRNAs are actively transported from the nucleus to the cytoplasm by a family of nuclear mRNA export factors (NXF). NXF1 is a house-keeping gene evolutionarily conserved from yeast to humans and is responsible for the nuclear export of bulk mRNAs (Kang and Cullen, 1999; Katahira et al, 1999). In mouse, four Nxf genes have been identified: Nxf1, Nxf2, Nxf3, and Nxf7 (Sasaki et al, 2005; Tan et al, 2005). Among these Nxf genes, Nxf2 is specifically expressed in germ cells in the testis (Wang et al, 2001). NXF2 localization in male germ cells exhibits distinct patterns: it is nuclear in spermatogonia, but localizes to the nuclear periphery (envelope) in early spermatocytes (Lai et al, 2006; Wang and Pan, 2007). NXF2 is associated with several proteins such as FMR1 (Fragile X mental retardation syndrome 1), KIF17 (a cytoplasmic motor protein), and MAP1B (a microtubule-associated protein), suggesting that NXF2 might regulate mRNA stability or trafficking (Lai et al, 2006; Takano et al, 2007; Tretyakova et al, 2005).
Inactivation of Nxf2 in mice demonstrated that it plays a dual function in spermatogenesis: progression of meiosis and maintenance of spermatogonial stem cells (Pan et al, 2009). In a mixed genetic background, about one third of Nxf2-deficient mice exhibited meiotic arrest, while the remaining mutant males had apparently normal spermatogenesis. On the C57BL/6J inbred background, Nxf2-deficient males exhibited reduced sperm count, impaired sperm motility, decreased spermatogonial proliferation, and age-dependent loss of spermatogonia, resulting in male infertility or subfertility.
Disruption of Taf7l causes reduced sperm production
TFIID, a highly conserved general transcription factor, is required for transcription of protein-coding genes by RNA polymerase II. TFIID consists of TBP (TATA-binding protein) and at least 12 TAFs (TBP-associated proteins) (Hochheimer and Tjian, 2003; Veenstra and Wolffe, 2001). Testis-specific TAFs have been identified in Drosophila, mouse, and human (Hiller et al, 2004; Hiller et al, 2001; Wang et al, 2001; Wang and Page, 2002). Interestingly, testis-specific TAFs in Drosophila are required for meiotic progression and male fertility (Lin et al, 1996). In mammals, Taf7l is a testis-specific paralogue of Taf7, which is ubiquitously expressed (Pointud et al, 2003; Wang et al, 2001). Taf7l is the ancestral gene of Taf7, since the Taf7l coding region is interrupted by 12 introns but the Taf7 coding region is intronless. Thus, Taf7 appears to have originated from the X-linked Taf7l by retroposition (Cheng et al, 2007). Taf7l is expressed in various types of germ cells, including spermatogonia, spermatocytes, and spermatids (Pointud et al, 2003). Moreover, biochemical studies demonstrate that TAF7L replaces TAF7 in the TFIID complex in male germ cells (Pointud et al, 2003).
Disruption of Taf7l in mice caused a significant reduction in sperm count and sperm motility. Taf7l-deficient sperm exhibited morphological defects in tails. The testis weight of Taf7l−/Y mice was consistently reduced by more than 10%. As a result, Taf7l-deficient males were subfertile (Cheng et al, 2007). Microarray profiling showed that the abundance of six genes decreased in Taf7l-deficient testes by more than two fold. These studies suggested that even though deficiency of TAF7L might be compensated in part by TAF7, TAF7L has evolved a specialized function in transcription in male germ cells.
TEX11 is the first X-linked meiosis factor
Meiotic sex chromosome inactivation (MSCI) leads to the hypothesis that meiosis-specific factors are rarely if ever encoded by the sex chromosomes. This perception was countered when TEX11 was found to be essential for male meiosis (Yang et al, 2008). Tex11 was originally identified as an X-linked germ cell-specific gene of unknown function (Wang et al, 2001). The mouse Tex11 gene spans about 224 kb and thus is one of the largest genes in the mammalian genome. The only known domain in TEX11 is a tetratricopeptide repeat (TPR) protein-protein interaction domain, which is present in proteins that form multimeric complexes such as chaperones (Blatch and Lassle, 1999). TEX11 forms distinct foci on meiotic chromosomes in both spermatocytes and oocytes, suggesting that it might be a meiosis-specific factor (Yang et al, 2008). TEX11 colocalizes with recombination-related proteins. In our study, we generated Tex11-null mice by deleting 27 (out of 30) exons (Yang et al, 2008). Interestingly, the Tex11-null males were sterile, while the Tex11-null females were subfertile. Further analyses revealed that Tex11 is essential for male meiosis and it plays two distinct functions in meiosis: promotion of chromosomal synapsis and regulation of crossover formation. TEX11 interacts with SYCP2, which is an integral component of the synaptonemal complex (Yang et al, 2006; Yang et al, 2008). These studies suggest that TEX11 might provide a physical link between chromosomal synapsis and crossover formation.
Intriguingly, in another study, only the exon 3 of Tex11 was deleted in the mutant mice (Adelman and Petrini, 2008). The exon 3 deletion mutant males and females had normal fertility, suggesting that this mutant allele is not null (Adelman and Petrini, 2008). By yeast two-hybrid assay, TEX11 was found to interact with NBS1, a component of the Mre11 complex. Although it remains to be confirmed by co-immunoprecipitation from testicular extracts or co-immunolocalization on meiotic chromosomes, the potential association between TEX11 and NBS1 is consistent with the role of TEX11 in crossover formation.
While Tex11 is conserved in vertebrates, it also has sequence homologues (SPO22/ZIP4) in Arabidopsis and budding yeast (Chelysheva et al, 2007; Tsubouchi et al, 2006). However, TEX11 sequence homologues were not found in fission yeast, fly, or worm. SPO22 localizes as foci on yeast meiotic chromosomes (Tsubouchi et al, 2006). Like mouse TEX11, yeast SPO22 promotes both synaptonemal complex polymerization (a.k.a. chromosomal synapsis) and crossover formation. Mutation in Arabidopsis SPO22 reduces crossover formation but does not prevent chromosomal synapsis (Chelysheva et al, 2007). Taken together, these studies demonstrate that the function of TEX11/SPO22 in meiosis is evolutionarily conserved among diverse organisms.
Synergistic regulation of male meiosis by Taf7l and Tex11
We attempted to test whether Taf7l and Tex11 function synergistically in spermatogenesis for two reasons. First, they exhibit the same developmental expression pattern in male germ cells (Wang et al, 2005). Second, both genes are germ cell-specific and map to the X chromosome, suggesting that they might have co-evolved to regulate spermatogenesis. We crossed Taf7l and Tex11 mutant (knockout) alleles to the same X chromosome to generate double mutant (Taf7l−/Y Tex11−/Y) males.
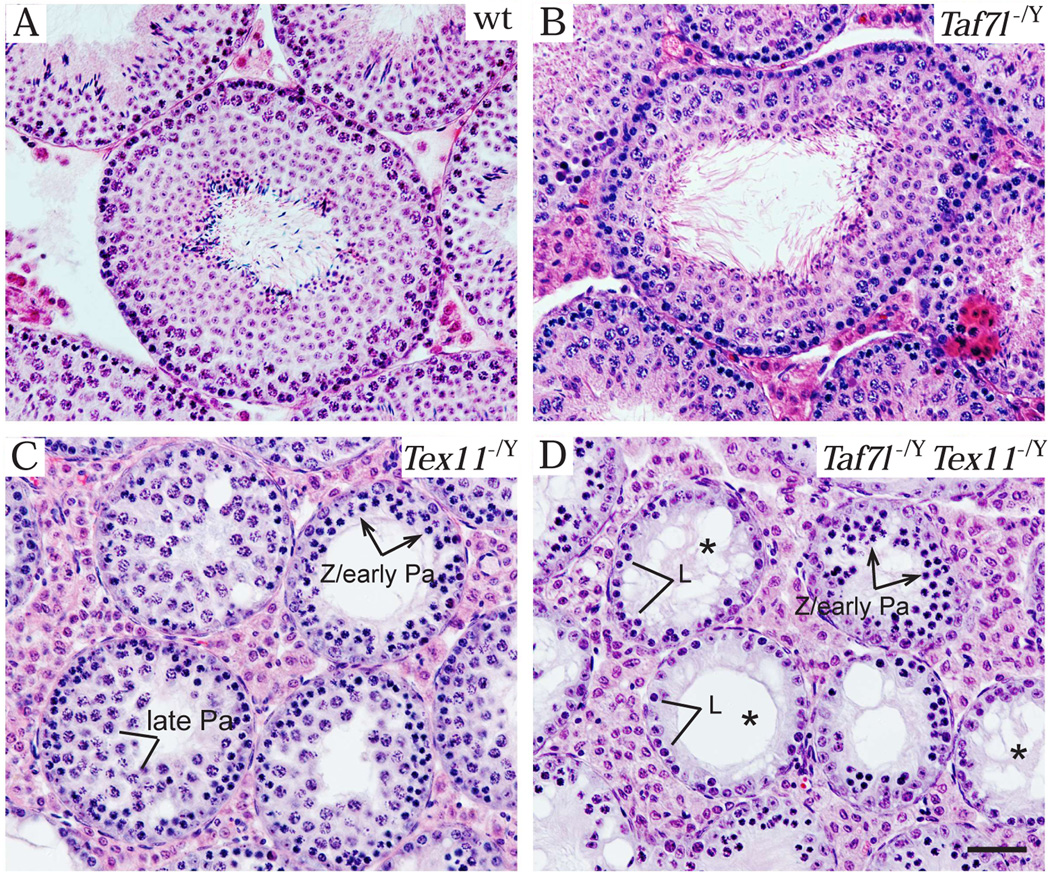
While Taf7l-deficient testes exhibited no spermatogenic block (Fig. 1B), Tex11-deficient testes displayed meiotic arrest (Fig. 1C) (Cheng et al, 2007; Yang et al, 2008). Interestingly, meiotic defects in the Taf7l Tex11 double mutant males were much more severe than that in the Tex11 single mutant. Late pachytene spermatocytes were abundant In Tex11-deficient seminiferous tubules (Fig. 1C), but were rare or absent in Taf7l Tex11 double mutant (Fig. 1D). As a result, tubules with a single layer of germ cells (pre-leptotene or leptotene spermatocytes) were prevalent in the double mutant testes (Fig. 1D) but not in Tex11 single mutant testes (Fig. 1C). The most advanced germ cells in the double mutant appeared to be zygotene or early pachytene spermatocytes (Fig. 1D).
Figure 1.
The Taf7l Tex11 double mutant mice exhibited more serve defects in male meiosis. In this new study, we generated Taf7l Tex11 double mutant mice. Histological analysis was performed on testes from 6-month-old mice of four genotypes: wild type (A), Taf7l−/Y (B), Tex11−/Y (C), and Taf7l−/Y Tex11−/Y double mutant (D). Asterisks in (D) indicate the seminiferous tubules with a single layer of germ cells. Mice were maintained and used for experimentation according to the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania. Abbreviations: L, leptotene spermatocyte; Z, zygotene spermatocyte; early Pa, early pachytene spermatocyte; late Pa, late pachytene spermatocyte. Scale bar, 50 µm.
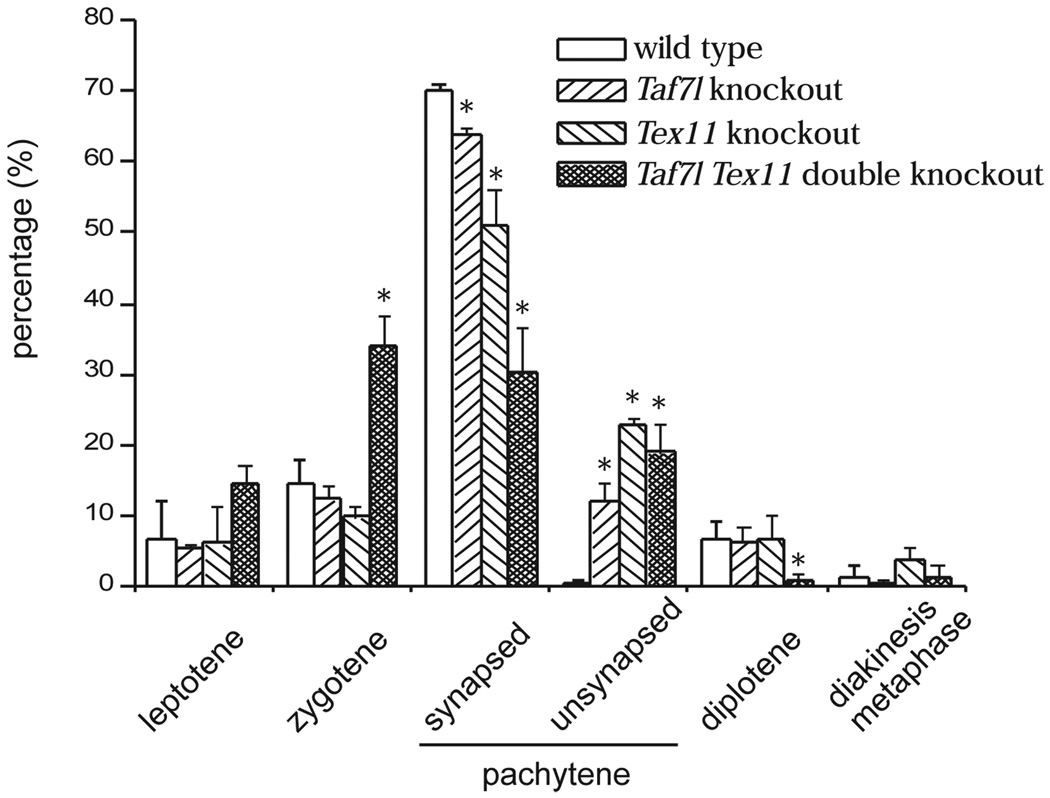
We then determined the percentage of each stage of spermatocytes by spread analysis (Fig. 2). This analysis showed that Taf7l Tex11 double mutant spermatocytes, unlike either single mutant cells, rarely progressed to the diplotene stage. Strikingly, the percentage of zygotene spermatocytes (35%) in the double mutant was significantly higher than that (~10%) in either single mutant. In addition, the percentage of pachytene spermatocytes with normal synapsis was dramatically reduced in the double mutant. These data were in agreement with the lack of late pachytene spermatocytes in the double mutant revealed by histological analysis. These studies suggested that Taf7l and Tex11 regulate male meiosis synergistically at the zygotene and early pachytene stages.
Figure 2.
Early meiotic defects in Taf7l Tex11 double mutant male mice. Surface spread analysis was performed on spermatocytes from adult (2-month-old) mice of four genotypes: wild type, Taf7l−/Y, Tex11−/Y, and Taf7l−/Y Tex11−/Y double mutant. Spread spermatocyte nuclei were immunostained with antibodies against SYCP1 (component of synaptonemal complex central element) and SYCP2 (component of synaptonemal complex lateral elements) as previously described (Yang et al, 2006). Three mice/genotype were analyzed. At least 100 spermatocytes from each mouse were scored and were categorized into leptotene, zygotene, pachytene, diplotene, and diakinesis/metaphase spermatocytes, according to the immunostaining patterns of SYCP1 and SYCP2. Pachytene spermatocytes were further divided into synapsed (all chromosomes were synapsed) and unsynapsed (some chromosomes were unsynapsed). The percentage of each category of spermatocytes (average ± SD) was plotted. *Values were statistically significant in comparison with wild type or other genotypes (p < 0.05) by Student’s t test.
Infertility studies from mice to men: opportunities and challenges
A large number (> 200) of mouse mutants with infertility or subfertility as a major defect have been generated (Matzuk and Lamb, 2002). The number of infertile mouse mutants will increase dramatically and thus provide a plethora of candidate fertility factors for humans. In stark contrast, little progress has been made in identifying the genetic cause (single gene mutations) of infertility in humans (Matzuk and Lamb, 2002; Nuti and Krausz, 2008). As discussed earlier, causative mutations in a given gene are likely to occur at an extremely low frequency. The X chromosome is of particular interest in the genetic study of human male infertility. Because males have only one X chromosome, mutations in an X-linked gene would not be masked by a normal allele and thus would manifest in males.
Mutation screening in infertile men had been performed for several X-linked genes such as Akap4, Taf7l, and Nxf2. However, causative mutations remain elusive. Mutation screening and immunolocalization studies in nine men with dysplasia of fibrous sheath (DFS) did not identify defects in AKAP4 and AKAP3 (Turner et al, 2001). In a case report, one infertile men with DFS appeared to be negative for AKAP4 in sperm tails and contain partial intragenic deletions in the AKAP3 and AKAP4 genes, however, further molecular analyses were necessary to define the genomic deletions (Baccetti et al, 2005). Mutation screening in the TAF7L gene were reported in two human studies involving 25 and 16 azoospermic patients (no sperm in semen) respectively (Akinloye et al, 2007; Stouffs et al, 2006). In both reports, the sequence changes in TAF7L were found in both azoospermic and fertile men, and thus were not causative (Akinloye et al, 2007; Stouffs et al, 2006; Tuttelmann et al, 2007). Screening of 65 azoospermic men with Sertoli cell-only syndrome did not identify mutations in NXF2 (Stouffs et al, 2008).
The failure to identify causative mutations in X-linked genes (as well as in autosomal genes) in infertile men highlights the challenges in the genetic studies of human male infertility (Matzuk and Lamb, 2002; Nuti and Krausz, 2008). First, given that hundreds, if not thousands, of genes specifically regulate male fertility, the contribution of mutations in a single gene to infertility in men would be extremely small (< 1%). Therefore, it would be necessary to screen a large number (at least a hundred) of infertile patients in the future. Second, the phenotype of knockout mice serves as an informative guide in selecting infertile patients for mutation screening. The mutation screening of X-linked germ cell-specific genes (Taf7l and Nxf2) was performed before the study of the mouse mutants and thus only included azoospermic men. Given that inactivation of either Taf7l or Nxf2 in mice causes reduced sperm count, future mutation screening in these two genes needs to focus on oligozoospermic (reduced sperm count) rather than azoospermic men. In contrast, mutation screening of human TEX11 gene in azoospermic men with maturation arrest would be more appropriate, since Tex11 is essential for male meiosis in mice (Yang et al, 2008). Third, the ultimate challenge has been and will remain how to distinguish between a causative mutation and a polymorphism. Traditional pedigree-based linkage analysis is not applicable to the genetic study of fertility, as infertility leads to no offspring. Biochemical and molecular biological studies would likely yield some insights into the effects of a given mutation on protein functions, but would stop short of being a definitive proof of causality. Therefore, despite the possible complications of generating mouse models of any human disease, we propose that modeling human male infertility by generation of knockin mice with analogous mutations found in infertile men will be the most vigorous approach to test whether a sequence variant (mutation) is the cause of infertility in humans.
Acknowledgments
We thank C. Höög for anti-SYCP1 antibody. This work is supported by an NIH/NIGMS grant RO1GM076327 (PJW).
References
- Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4:e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinloye O, Gromoll J, Callies C, Nieschlag E, Simoni M. Mutation analysis of the X-chromosome linked, testis-specific TAF7L gene in spermatogenic failure. Andrologia. 2007;39:190–195. doi: 10.1111/j.1439-0272.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- Baccetti B, Collodel G, Estenoz M, Manca D, Moretti E, Piomboni P. Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum Reprod. 2005;20:2790–2794. doi: 10.1093/humrep/dei126. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Brown PR, Miki K, Harper DB, Eddy EM. A-kinase anchoring protein 4 binding proteins in the fibrous sheath of the sperm flagellum. Biol Reprod. 2003;68:2241–2248. doi: 10.1095/biolreprod.102.013466. [DOI] [PubMed] [Google Scholar]
- Carrera A, Gerton GL, Moss SB. The major fibrous sheath polypeptide of mouse sperm: structural and functional similarities to the A-kinase anchoring proteins. Dev Biol. 1994;165:272–284. doi: 10.1006/dbio.1994.1252. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG, de Boer P. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet Genome Res. 2003;103:267–276. doi: 10.1159/000076812. [DOI] [PubMed] [Google Scholar]
- Fujii T, Tamura K, Masai K, Tanaka H, Nishimune Y, Nojima H. Use of stepwise subtraction to comprehensively isolate mouse genes whose transcription is up-regulated during spermiogenesis. EMBO Rep. 2002;3:367–372. doi: 10.1093/embo-reports/kvf073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- Handel MA, Park C, Kot M. Genetic control of sex-chromosome inactivation during male meiosis. Cytogenet Cell Genet. 1994;66:83–88. doi: 10.1159/000133672. [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Lai D, Sakkas D, Huang Y. The fragile X mental retardation protein interacts with a distinct mRNA nuclear export factor NXF2. RNA. 2006;12:1446–1449. doi: 10.1261/rna.94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4 Suppl:s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol. 2002;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online. 2008;16:504–513. doi: 10.1016/s1472-6483(10)60457-9. [DOI] [PubMed] [Google Scholar]
- Pan J, Eckardt S, Leu NA, Buffone MG, Zhou J, Gerton GL, McLaughlin KJ, Wang PJ. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116:1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Takeda E, Takano K, Yomogida K, Katahira J, Yoneda Y. Molecular cloning and functional characterization of mouse Nxf family gene products. Genomics. 2005;85:641–653. doi: 10.1016/j.ygeno.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari AJ. The behavior of the XY pair in mammals. Int Rev Cytol. 1974;38:273–317. doi: 10.1016/s0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffs K, Tournaye H, Van der Elst J, Liebaers I, Lissens W. Is there a role for the nuclear export factor 2 gene in male infertility? Fertil Steril. 2008;90:1787–1791. doi: 10.1016/j.fertnstert.2007.08.071. [DOI] [PubMed] [Google Scholar]
- Stouffs K, Willems A, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. The role of the testis-specific gene hTAF7L in the aetiology of male infertility. Mol Hum Reprod. 2006;12:263–267. doi: 10.1093/molehr/gal020. [DOI] [PubMed] [Google Scholar]
- Takano K, Miki T, Katahira J, Yoneda Y. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Res. 2007;35:2513–2521. doi: 10.1093/nar/gkm125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Zolotukhin AS, Tretyakova I, Bear J, Lindtner S, Smulevitch SV, Felber BK. Identification and characterization of the mouse nuclear export factor (Nxf) family members. Nucleic Acids Res. 2005;33:3855–3865. doi: 10.1093/nar/gki706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, Felber BK. Nuclear export factor family protein participates in cytoplasmic mRNA trafficking. J Biol Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell. 2006;10:809–819. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- Turner RM, Musse MP, Mandal A, Klotz K, Jayes FC, Herr JC, Gerton GL, Moss SB, Chemes HE. Molecular genetic analysis of two human sperm fibrous sheath proteins, AKAP4 and AKAP3, in men with dysplasia of the fibrous sheath. J Androl. 2001;22:302–315. [PubMed] [Google Scholar]
- Tuttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M. Gene polymorphisms and male infertility--a meta-analysis and literature review. Reprod Biomed Online. 2007;15:643–658. doi: 10.1016/s1472-6483(10)60531-7. [DOI] [PubMed] [Google Scholar]
- Van Assche E, Bonduelle M, Tournaye H, Joris H, Verheyen G, Devroey P, Van Steirteghem A, Liebaers I. Cytogenetics of infertile men. Hum Reprod. 1996;11 Suppl 4:1–24. doi: 10.1093/humrep/11.suppl_4.1. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP. Gene-selective developmental roles of general transcription factors. Trends Biochem Sci. 2001;26:665–671. doi: 10.1016/s0968-0004(01)01970-3. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Pan J. The role of spermatogonially expressed germ cell-specific genes in mammalian meiosis. Chromosome Res. 2007;15:623–632. doi: 10.1007/s10577-007-1141-2. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC. Functional substitution for TAFII250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet. 2002;11:2341–2346. doi: 10.1093/hmg/11.19.2341. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173:497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22:682–691. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]