Abstract
Genomic imprinting is an epigenetic phenomenon affecting a small number of genes which leads to differential expression from the two parental alleles. Imprinted genes are known to regulate fetal growth and a ‘kinship’ or ‘parental conflict’ model predicts that paternally- and maternally-expressed imprinted genes promote and inhibit fetal growth, respectively. In this review we examine the role of imprinted genes in postnatal growth and metabolism, with an emphasis on the GNAS/Gnas locus. GNAS is a complex imprinted locus with multiple oppositely imprinted gene products, including the G protein α-subunit Gsα which is expressed primarily from the maternal allele in some tissues and the Gsα isoform XLαs which is expressed only from the paternal allele. Maternal, but not paternal, Gsα mutations lead to obesity in Albright hereditary osteodystrophy. Mouse studies show that this phenomenon is due to Gsα imprinting in the central nervous system leading to a specific defect in the ability of central melanocortins to stimulate sympathetic nervous system activity and energy expenditure. In contrast mutation of paternally-expressed XLαs leads to opposite metabolic effects in mice. While these findings conform to the ‘kinship’ model, the effects of other imprinted genes on body weight regulation do not conform to this model.
Keywords: G protein, genomic imprinting, obesity, pseudohypoparathyroidism, Prader-Willi syndrome, Angelmans syndrome
Introduction
Genomic imprinting is an epigenetic phenomenon affecting a small number of genes which results in partial or complete suppression of gene expression from one parental allele 1. The primary biochemical imprint ‘marks’ (differences in DNA methylation and/or histone modification) which distinguish the two parental alleles are erased in primordial germ cells, reestablished during oogenesis or spermatogenesis, and maintained in all somatic tissues throughout development. All imprinted genes have one or more regions in which the maternal and paternal allele are differentially methylated. In many cases DNA methylation is associated with silencing of the affected allele as the methylation occurs within the gene’s promoter region. In other cases DNA methylation may be present on the active allele, particularly when the methylation silences a cis-acting negative regulatory element for the gene promoter or when the methylation occurs on a promoter for an interfering RNA.
The most widely accepted hypothesis for why imprinting occurs is the ‘kinship’ or ‘parental conflict’ hypothesis which predicts that paternally and maternally transmitted alleles promote and inhibit fetal growth, respectively 2, 3. This model assumes that father wants to allocate finite resources to his offspring while it is in the mother’s interest to conserve resources over multiple litters that she will bear throughout life. In addition to their effects on fetal growth, it is clear that imprinted genes also have important roles in postnatal energy metabolism and in many cases obesity is associated with altered function of imprinted genes. Several human genetic disorders affecting imprinted genes lead to obesity and population studies have identified several chromosomal regions associated with parent-of-origin effects on body weight regulation in humans 4–7. In this review we will summarize the what is presently known about the role of imprinted genes in the development of obesity and regulation of energy balance, with a particular focus on the GNAS/Gnas locus.
Organization and Gene Products of GNAS/Gnas
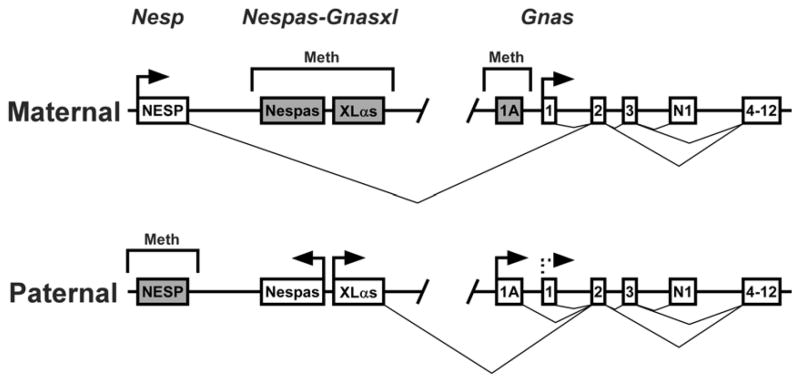
GNAS and the mouse ortholog Gnas are located within syntenic regions at 20q13.2-13.3 and distal chromosome 2, respectively and have similar overall organization and imprinting patterns 8. GNAS/Gnas generates multiple gene products through the use of alternative promoters and first exons that splice onto a common exon (exon 2, Fig. 1). The most downstream alternative promoter generates transcripts encoding the ubiquitously expressed G protein α-subunit Gsα that couples many receptors for hormones, neurotransmitters and other extracellular signals to adenylyl cyclase 9. Gsα is required for receptor-stimulated cAMP production and most of its downstream effects are mediated via cAMP, which in turn activates signaling molecules such as protein kinase A (PKA) and cAMP-regulated guanine nucleotide exchange factors (cAMP-GEFs). PKA is a serine/threonine kinase which acutely activates many metabolic processes such as lipolysis, gluconeogenesis, and glycogenolysis through direct phosphorylation of enzymes and other factors involved in intermediary metabolism. In addition PKA has more chronic effects on gene expression by phosphorylation of transcription factors such as the cAMP response element binding protein (CREB) 10. cAMP-GEFs are highly expressed in neuroendocrine cells and may lead to activation of the ras-like protein Rap1. There is some evidence that Gsα may also stimulate other effectors besides adenylyl cyclase and may also be activated by receptors outside of the seven-transmembrane G protein-coupled receptor family 8.
Fig. 1.
Organization and imprinting of the GNAS/Gnas locus. The maternal (top) and paternal (bottom) alleles of GNAS/Gnas are depicted with regions of methylation shown above and splicing patterns shown below for each allele. The regions designated Nesp, Nespas-Gnasxl, and Gnas in mice are indicated at the top. Active promoters are indicated with horizontal arrows while inactive promoters are indicated with shaded boxes. First exons for transcripts encoding NESP55 (NESP), Nespas, XLαs, and Gsα (exon 1) are shown along with downstream exons 3–13 and the alternative neural-specific terminal exon N1 (exons 4–13 are shown as a single box). The dashed horizontal line at the paternal Gsα promoter indicates that transcription is suppressed due to imprinting in a tissue-specific manner. For clarity only the first exon for Nespas is shown. The figure is not drawn to scale.
The Gsα coding region spans 13 exons (12 exons in mice) and generates two long and two short forms of Gsα protein by alternative splicing of exon 3 9, 11. The Gsα promoter resides within a unmethylated CpG island (GC-rich region with a high frequency of CpG dinucleotides) 9, 12–14. Although its promoter is not methylated, Gsα is imprinted in a tissue-specific manner, being expressed primarily from the maternal allele in several tissues, including pituitary, thyroid, renal proximal tubules, gonads, and the paraventicular nucleus of the hypothalamus, but is biallelically expressed in most tissues 15–23. Gsα also appears to be imprinted in neonatal brown adipose tissue (BAT) 24, but is not imprinted in adult BAT or white adipose tissue (WAT) 25–27. Temporal changes in Gnas regulation appear to occur in adipose tissue in the early postnatal period, as the alternative Gsα isoform XLαs is expressed in neonatal BAT and WAT but this expression is lost within 1–2 weeks 28. Splicing to an alternative terminal exon (N1) located between Gsα exons 3 and 4 generates a neural-specific truncated splice variant of unknown function (GsαN1) 29.
The most upstream alternative promoter (referred to as Nesp in the mouse; Fig. 1) generates transcripts for the neuroendocrine-specific protein of 55 kDa (NESP55) 13, 30, 31. NESP55 is a chromogranin-like protein located within large dense core granules of secretory cells and is presumed to be a cosecretagogue 32 which is totally unrelated to the G protein α-subunit family. It is primarily expressed in neuroendocrine tissues, including the adrenal medulla, pituitary, hypothalamus, and other midbrain and brainstem regions 32–35. The entire NESP55 coding sequence is within its specific upstream exon and Gsα exons 2–13 are within the 3′ untranslated region of NESP55-specific transcripts 30, 32. NESP55 is only transcribed from the maternal allele and its promoter region is methylated only on the paternal allele 13, 30, 36. Imprinting of NESP55 in mice is not established until after implantation, suggesting that this region is not a primary imprinting control region for the GNAS/Gnas locus 14 but is rather its imprinting is dependent on the promoter for the antisense transcript Nespas 24(see Fig. 1 and below).
Downstream of the Nesp promoter is another alternative promoter (referred to as Gnasxl in the mouse, Fig. 1) that generates transcripts encoding the Gsα isoform XLαs 12, 13, 31, 37, 38. Gnasxl is oppositely imprinted to Nesp, being transcriptionally active only on the paternal allele and methylated on the maternal allele 12, 13, 36. XLαs has a long amino-terminal extension encoded by its specific first exon while the remainder of the protein encoded by exons 2–13 is identical to Gsα. XLαs has been shown to be capable of mediating receptor signaling in a similar fashion as Gsα in cultured cells 39. XLαs has a more restricted tissue distribution than Gsα, being primarily expressed in neuroendocrine tissues including the brain, adrenal medulla, pars intermedia of the pituitary, stomach, heart, kidney and pancreatic islets 37, 40, 41, 42. Mouse brain regions where XLαs is expressed include the hypothalamus, locus coeruleus, and the laterodorsal tegmental, hypoglossal, trigeminal, ambiguous, medullary reticular, raphe obscurus and facial nuclei 42. XLαs is expressed in mouse adipose tissue in the early postnatal period but its expression is lost within two weeks after birth 28.
High levels of a neural-specific truncated form of XLαs (XLN1) are generated by alternative splicing of XLαs to alternative terminal exon N1 40 which has been proposed to be potential dominant negative regulator of Gsα function 28, 42. The XLαs first exon has been shown to also generate the unrelated protein Alex from a second open reading frame, and this protein has been reported to directly interact with the XLαs protein 38, 43, 44. Polymorphisms within the XLαs exon 1 may generate alternative forms of Alex and XLαs which fail to interact with each other 44.
Just upstream of the XLαs promoter and within the same differentially methylated region is the Nespas promoter which generates paternally expressed antisense transcripts that traverse the NESP55 exon 36, 45–47. Within the Nespas promoter region is a primary DNA methylation imprint mark in which methylation is established during oogenesis and maintained throughout development 48. A mouse model in which the Nespas promoter region was deleted confirmed that Nespas is the imprint control center for Nesp and probably is important for normal imprinting throughout the Gnas locus 24.
The exon 1A promoter region (also referred to as exon A/B in humans) located just upstream of Gsα exon 1 (Fig. 1) is within a differentially methylated region which is a primary imprint control region as methylation on the maternal allele is established during oogenesis 14, 49, 50. Exon 1A generates untranslated transcripts from the paternal allele of unknown function. 14. In pseudohypoparathyroidism type 1B, a form of renal parathyroid hormone resistance, maternal-specific methylation of the exon 1A region is absent due to either a failure to establish or maintain the methylation in this region 50–53. Based upon this observation it has been suggested that tissue-specific Gsα imprinting (suppression of the paternal Gsα promoter) is due to the presence of one or more negatively regulatory cis-acting elements within the exon 1A differentially methylated region that are both tissue-specific and methylation-sensitive (and therefore does not suppress Gsα expression on the methylated maternal allele). Evidence for this model comes from mice in which paternal deletion of the exon 1A region resulted in reversal of Gsα imprinting with biallelic expression of Gsα in all tissues that were examined 54, 55.
Evidence for a Role of GNAS in Weight Regulation in Humans
The most direct evidence for an important role of GNAS in the regulation of energy balance comes from patients with Albright hereditary osteodystrophy (AHO), a congenital disorder characterized by the presence of short stature, shortening of various long bones in the hands and feet (brachydactyly), subcutaneous ossifications, rounded face, and neurobehavioral abnormalities, which is caused by heterozygous GNAS mutations which disrupt Gsα expression or function 56. Patients in whom the mutation is present on the maternal GNAS allele also develop resistance to various hormones, including parathyroid hormone, thyrotropin, growth hormone-releasing hormone, and gonadotropins, a condition known as pseudohypopararthyroidism type 1A (PHP1A) while patients with mutations on the paternal allele develop the features of AHO without multihormone resistance, a condition referred to as pseudopseudohypoparathyroidism (PPHP) 15, 56. This is the result of Gsα imprinting in specific hormone target tissues: mutation of the active maternal Gsα allele leads to severe Gsα deficiency and hormone resistance while mutation of the inactive paternal allele has little effect on Gsα expression or hormone sensitivity 17, 18, 20–22.
Similar to the parent-of-origin effect on the development of hormone resistance, it has recently been shown that the development of obesity only occurs with maternal, but not paternal, Gsα mutations (PHPIA but not PPHP) 57. The obesity generally occurs very early (within the first year) and tends to be severe in early childhood, similar to other monogenic forms of obesity (e.g. those secondary to melanocortin 4 receptor [MC4R] mutations).
Few studies have been performed in humans examining the underlying mechanism for the obesity in PHPIA. Although one study showed evidence that adipocytes from PHP1A patients have reduced lipolytic responsiveness to epinephrine due to reduced levels of Gsα/cAMP signaling 58, the adipose tissue is unlikely to be involved in this parent-of-origin effect on energy balance as studies have shown no evidence for Gsα imprinting in adipose tissue 25–27. This same study 58 did show PHP1A patients to have extremely low circulating norepinephrines, even when compared to similarly obese children without PHP1A, suggesting that the defect may lie in central nervous system (CNS) regulation leading to low sympathetic nervous system activity and metabolic rates. Although food intake has not been systematically examined in these patients, one recent case was reported in which severe obesity developed in the first year of life in the absence of hyperphagia, suggesting that low energy expenditure rates may be the major contibutor to obesity in this disorder 59. There is some evidence that growth hormone deficiency due to growth hormone releasing hormone resistance in the pituitary may contribute to obesity in some cases 60. Although the incidence of comorbid metabolic abnormalities such as insulin resistance and diabetes in PHP1A has not been systematically examined, recently a case of severe insulin resistance in a young PHP1A patient has been reported 61.
In addition to the monogenic obesity resulting from clear loss-of-function mutations, two single nucleotide polymorphisms have been identified within the GNAS locus that have been associated with altered body weight or response to weight loss regimens. In one study in German women with polycystic ovarian syndrome, the common T393C polymorphism within the GNAS coding region (which does not alter the protein sequence) was associated with increased obesity and insulin resistance, although it should be noted that no such association was noted in their larger control population 62. A functional polymorphism within the Gsα promoter region (G vs. A at nucleotide −1211 relative to the Gsα translational start site) was shown to be associated with differences in binding of the transcriptional factor upstream stimulatory factor 1 (USF-1), Gsα expression levels, lipolytic rates in adipocytes, and weight loss on short-term calorie restriction, but was not associated with adiposity at baseline 63, 64.
Gsα Imprinting Underlies the Parent-of-Origin Metabolic Effects of GNAS Mutations
The initial GNAS knockout line that was developed harbored a large insertion mutation within exon 2 (E2−) 17 (the metabolic phenotypes of various Gnas knockout are summarized in Table 1). Homozygotes were embryonically lethal. Interestingly heterozygous E2− mice showed opposite changes in energy metabolism depending on whether the mutation was present on the maternal (E2m−/+) or paternal (E2+/p−) allele 65. E2m−/+ mice developed obesity with increased lipid accumulation in BAT and WAT and proportionally increased serum leptin levels. This obesity was primarily associated with reduced resting and total energy expenditure levels, with no evidence for hyperphagia. These effects seemed to be due to reduced activation of the sympathetic nervous system, as these mice had reduced urine excretion of norepinephrine and its metabolites, normal metabolic and biochemical responsiveness to a β3-adrenergic agonist which only activates adipose tissue, and normal energy expenditure at thermoneutral temperature (30°C) when sympathetic activity is minimized 65. Despite the presence of obesity, E2m− had very mild hypolipidemia and slightly improved glucose tolerance and insulin sensitivity with increased insulin-stimulated glucose uptake in isolated skeletal muscles 65, 66. In contrast, E2+/p− mice had a severely lean phenotype with markedly increased sympathetic nervous system activity, glucose tolerance, and insulin sensitivity that is the result of XLαs deficiency (see below).
Table 1.
Metabolic Phenotypes of Gnas Knockout Models
| Model | Gene Defect | Transcripts Disrupted | Metabolic Phenotype | References |
|---|---|---|---|---|
| E2m−/+ | Gsα exon 2 insertion (maternal) |
|
|
17, 65, 66 |
| E1m−/+ | Gsα exon 1 deletion (maternal) |
|
|
25, 26, 55, 69 |
| Oed | Gsα exon 6 Val159Glu point mutation (maternal) |
|
Similar phenotype to E2m/+ | 68 |
| E2+/p− | Gsα exon 2 insertion (paternal) |
|
|
17, 65, 66, 93 |
| E1+/p− | Gsα exon 1 deletion (paternal) |
|
|
25, 26 |
| Sml | Gsα exon 6 Val159Glu point mutation (maternal) |
|
Similar to E1+/p− | 68 |
| Gnasxl+/p− | XLαs exon 1 deletion |
|
|
28, 42 |
| NESP55 KO | Nesp deletion |
|
|
67 |
| mBrGsKO | CNS-specific Gsα exon 1 deletion (maternal) |
|
|
23 |
| pBrGsKO | CNS-specific Gsα exon 1 deletion (both alleles) |
|
Normal phenotype | 23 |
| LGsKO | Liver-specific Gsα exon 1 deletion (both alleles) |
|
|
72 |
| MGsKO | Skeletal muscle-specific Gsα exon 1 deletion (both alleles) |
|
|
73 |
| βGsKO | β cell-specific Gsα exon 1 deletion (both alleles) |
|
|
70 |
Although NESP55 deficiency could potentially contribute to the E2m−/+ metabolic phenotype as it is a maternally-expressed GNAS gene product which includes exon 2 in its mRNA, this is not likely to be the case, as exon 2 is not within the NESP55 coding region and NESP55 transcripts are still present in E2m−/+ mice (T.X., L.S.W., unpublished results). Moreover mice with specific disruption of NESP55 expression do not develop metabolic abnormalities 67. NESP55 is unlikely to significantly contribute to regulation of energy balance in humans as PHP1B patients with NESP55 deficiency due to imprinting defects involving the NESP55 promoter leading to promoter methylation on both parental alleles do not appear to be prone to obesity 50.
Studies in Gsα-specific knockout mice in which Gsα exon 1 was deleted (E1−) have confirmed that the obesity observed in E2m−/+ mice is due to loss of expression of Gsα from the maternal Gnas allele 25, 26. Like E2m−/+ mice, E1m−/+ mice develop obesity associated with normal food intake but reduced sympathetic nervous system activity and energy expenditure levels in the presence of normal thyroid function. Unlike E2m−/+ mice, E1m−/+ mice have a phenotype more reminiscent of metabolic syndrome, in that in addition to obesity E1m−/+ mice develop glucose intolerance, insulin resistance, and hyperlipidemia 26. Mice with a single base missense mutation (V159E) within Gsα exon 6 on the maternal allele (known as Oed mice) also developed obesity with reduced energy expenditure 68. As the Oed mutation disrupts Gsα function without affecting expression of the neural-specific truncated Gsα isoform GsαN1, this is consistent with loss of Gsα rather than loss of GsαN1 as the cause of the obesity that occurs secondary to maternal GNAS mutations. Unlike E1m−/+ mice, Oed mice did not develop glucose intolerance or hypertriglyceridemia, although this difference between the two lines may be due to the fact that they were on different genetic backgrounds.
In contrast to E1m−/+ mice, E1+/p− mice with the paternal Gsα mutation developed a very minimal increase in adiposity and minimal glucose intolerance and insulin resistance 26. Therefore the development of obesity in E1− mice mimics the pattern of inheritance observed in AHO patients, in whom obesity is much more prominent in PHP1A (who have maternal Gsα mutations) than in PPHP patients (who have paternal Gsα mutations) 57. These similar observations in both Gsα knockout models and AHO patients strongly implicates tissue-specific Gsα imprinting as being a required for the metabolic consequences of maternal Gsα mutations, with obesity resulting from severe Gsα deficiency in one or more metabolically active tissues due to mutation of the maternal Gsα allele and suppressed Gsα expression from the paternal allele due to imprinting. To further test this model female E1− mice were mated with males harboring a deletion of the 1A imprinting control region for Gsα 69. The metabolic phenotype in E1m−/+ offspring was completely reversed in those mice which also harbored the paternal 1A− deletion, and therefore had reversal of tissue-specific Gsα imprinting, thus confirming that the maternal Gsα metabolic phenotype is dependent on tissue-specific Gsα imprinting. In addition, mice with only the paternal 1A− deletion had reduced body growth (both lean and fat mass) and hyperactivity, without any measurable changes in food intake or energy expenditure rates 69.
The GNAS Metabolic Phenotype is Due to Gsα Imprinting in the Central Nervous System
Gsα is ubiquitously expressed and therefore any tissue involved in regulation of energy balance and glucose metabolism is a potential candidate as being the site where Gsα imprinting produces the parent-of-origin metabolic effects of Gsα mutations. Liver, adipose tissue, and muscle are not likely candidates as Gsα expression is not affected by imprinting in these tissues 8, 17, 25, 27, 65. Moreover mice with loss of Gsα expression in liver, muscle, adipose tissue, or pancreatic β cells do not develop the metabolic phenotype observed in mice with maternal Gsα mutations in the germline 70–73.
Recent work shows that Gsα imprinting in the CNS underlies the GNAS metabolic phenotype 23. Mice with CNS-specific disruption of maternal Gsα allele (mBrGsKO mice) which were generated by mating female Gsα E1-floxed mice with male Nestin-cre mice developed severe obesity starting at age 5–6 weeks which was at least initially not associated with hyperphagia, but rather with lower sympathetic nervous system activity, energy expenditure rates, and activity levels and greater metabolic efficiency (weight gain/calorie intake). In contrast mice with CNS-specific disruption of the paternal Gsα allele (pBrGsKO mice) maintained a normal phenotype. The effects in mBrGsKO mice were not due to maternal gestational effects as the mothers were Nestin-cre− and therefore had normal Gsα expression levels and metabolic phenotype. Thyroid hormone levels were also normal in mBrGsKO mice.
In addition to abnormal energy balance, mBrGsKO mice also developed hyperglycemia, glucose intolerance, and insulin resistance, while glucose metabolism and insulin sensitivity remained normal in pBrGsKO mice. The abnormalities in glucose metabolism and insulin sensitivity in mBrGsKO mice began prior to the development of obesity and therefore CNS-specific Gsα deficiency has a primary effect on peripheral glucose metabolism independent of obesity.
Overall, these findings in BrGsKO mice show that the metabolic phenotype generated by germline maternal Gsα mutations can be reproduced by loss of maternal Gsα expression in the CNS alone and therefore Gsα imprinting in the CNS underlies this parent-of-origin effect. Initial studies have shown that Gsα undergoes imprinting in the paraventricular nucleus of the hypothalamus, an area known to be important for central regulation of metabolism 23. In contrast, there was no evidence for Gsα imprinting in the nucleus of the solitary tract, another site that is important for metabolic regulation.
Gsα Mutation Leads to Loss of Stimulation of Energy Expenditure by Central Melanocortins
The central melanocortin pathways work downstream of leptin to promote negative energy balance by inhibiting food intake and stimulating sympathetic nervous system activity and energy expenditure 74–76. Proopiomelanortin neurons which synthesize and release melanocortins are located in the arcuate nucleus of the hypothalamus where they are stimulated by leptin. These neurons have axonal projections to the paraventricular nucleus and other CNS sites where they synapse with downstream neurons which express the melanocortin receptors MC3R and MC4R. These receptors are classic seven-transmembrane G protein-coupled receptors which activate Gsα/cAMP pathways. MC4R deficiency leads to obesity in both mice and humans 75, 77, 78 and MC4R receptors mediate the acute effects of melanocortins on food intake and energy expenditure 79, 80. In addition, MC4R also directly regulates glucose metabolism and peripheral insulin sensitivity 74, 81, 82. MC3R deficiency leads to only minor changes in adiposity 75.
In many respects mBrGsKO mice have a similar metabolic phenotype to MC4R knockout mice and therefore the primary defect leading to the metabolic consequences of maternal Gsα mutation may be resistance to central melanocortin action through loss of MC4R/Gsα signalling. In fact the ability of a central melanocortin agonist to acutely stimulate energy expenditure was significantly impaired in mBrGsKO mice 23. This finding is consistent with the reduction in sympathetic nervous activity and energy expenditure measured at baseline in mBrGsKO, E1m−/+, E2m−/+, and Oed mice 23, 26, 65, 68, 69, the absence of diet-induced thermogenesis in mBrGsKO mice (which is known to be dependent on MC4R signalling) 23, 83, 84, and the reduction of heart rate and diastolic blood pressure in mBrGsKO mice in the setting of severe obesity 23. Reduced heart rate and blood pressure was also observed in obese patients and mice with MC4R mutations 85, 86. Loss of MC4R-mediated stimulation of sympathetic nervous system activity and energy expenditure rates may also underlie the obesity observed in PHP1A patients, as these patients were also shown to have low circulating levels of norepinephrine 58.
Although MC4R-mediated effects on sympathetic nervous system activity and energy expenditure were impaired in mBrGsKO mice, the ability of the same melanocortin agonist to acutely inhibit food intake was maintained in these same mice (as well as in E1m−/+ mice) 23. This finding is consistent with fact that the onset of obesity in mBrGsKO mice is not associated with hyperphagia and and in at least one child it was documented that early weight gain occurred in the absence of hyperphagia 59.
Mice with maternal Gsα mutation had some of the same features as those of MC4R mutations (obesity, reduced sympathetic nervous system activity and energy expenditure, and impaired glucose metabolism), while several features associated with MC4R mutation were absent (hyperphagia, increased linear growth). MC4R signalling stimulates the expression of Sim1, a transcription factor expressed in the paraventricular nucleus which mediates some of the physiological actions of central melanocortins 87. Sim1 mutations in humans and in mice leads to effects that are reciprocal to those of Gsα mutations, namely obesity due primarily to hyperphagia and increased linear growth, with no effects on energy expenditure or glucose metabolism 88–92. Moreover, Sim1 haploinsufficient mice had an impaired anorexic response to administered melanocortins while the ability of melanocortins to stimulate energy expenditure in these mice were normal 88, a pattern opposite to what is observed in mBrGsKO mice. It is therefore possible that melanocortins mediate their physiological effects through multiple pathways downstream of MC4R: a Gsα-dependent pathway which mediates the effects on sympathetic nervous system activity, energy expenditure, and glucose metabolism, and a Gsα-independent pathway through Sim1 that mediates the effects on food intake and linear growth. Further studies are required to determine whether this model is correct, and if so, what are the signals between MC4R and Sim1 (another G protein such as Gqα or a G protein-independent signaling mechanism).
XLαs Has Metabolic Effects Opposite to Those of Gsα
E2+/p− mice with the E2− mutation on the paternal allele showed a metabolic phenotype opposite to that of E2m−/+ mice 65. Neonatal E2+/p− mice had poor suckling and most died within hours of birth. Adult E2+/p− mice were severely lean with markedly reduced lipid accumulation in WAT and BAT. This change in energy balance was primarily due to a marked increase in sympathetic nervous system activity, energy expenditure, and activity levels. In addition E2+/p− mice had severe hypolipidemia and increased triglyceride clearance with markedly improved glucose tolerance and insulin sensitivity in liver, muscle and adipose tissue 66, 93. A similar overall phenotype was also observed in mice with the V159E Oed mutation on the paternal allele (referred to as the Sml mutation when on the paternal allele) 68. These mutations disrupt the coding sequences of both Gsα and the paternally-expressed Gsα isoform XLαs. In contrast to E2+/p− and Sml mice, E1+/p− mice in which only Gsα expression from the paternal allele is disrupted developed a marginal increase in adiposity and very mild insulin resistance 26. It therefore appeared likely that the lean, hypermetabolic phenotype observed in E2+/p− and Sml mice were the direct consequence of XLαs deficiency.
An important role for XLαs in metabolic regulation in mice was confirmed in mice with specific deficiency of XLαs due to a deletion in the upstream XLαs first exon (Gnasxl−) 42. The Gnasxl− deletion had no effect on Gnas imprinting and only produced an abnormal phenotype when it was present on the paternal allele (Gnasxl+/p−) due to fact that XLαs is only expressed from this parental allele. Overall Gnasxl+/p− mice had a phenotype very similar to that of E2+/p− and Sml mice. The initial prominent feature was a failure to suckle which was associated with hypoglycemia and significant neonatal lethality. Reduced suckling in Gnasxl+/p− mice may be a direct effect on orofacial muscle activity as XLαs is expressed in CNS nuclei involved in orofacial motor activity, including the facial, hypoglossal, and trigeminal nuclei 42.
Gnasxl+/p− mice mice, like E2+/p− mice 65, 66, 93, had markedly reduced lipid accumulation in BAT and WAT 28, 42. Studies of adult Gnasxl+/p− mice 28 showed them to continue to maintain a markedly lean phenotype associated with increased energy expenditure, hypolipidemia with increased triglyceride clearance, and markedly improved glucose tolerance and insulin sensitivity. Based upon gene expression studies, it appears that the Gnasxl+/p− phenotype is due to increased lipid oxidation in adipose tissue, particularly in BAT. However these changes were not due to cell-autonomous effects on BAT function as Gnasxl+/p− mice did not have increased metabolic responsiveness to a β3 adrenergic agonist which specifically activates adipose tissue. Moreover XLαs is not expressed in adipose tissue after two weeks of age 28. Rather BAT was overstimulated due to the mice having markedly increased sympathetic nervous system activity as they had increased urinary norepinephrine excretion 28, similar to E2+/p− mice 65.
Based upon the similar findings in Gnasxl+/p−, E2+/p−, and Sml mice, it is clear that the severe metabolic phenotype found in all of these mice is due to XLαs deficiency, which appears to have a dominant effect. It appears that XLαs is involved in regulation of sympathetic output from the central nervous system in a manner opposite to that of Gsα. Whether this occurs by acting at CNS sites that are distinct from Gsα or whether it directly inhibits Gsα signaling remains to be determined. It is unlikely that XLN1 deficiency plays a significant role in producing the Gnasxl+/p− phenotype as the Sml exon 6 mutation, which produces a similar phenotype, dies not alter the coding sequence XLN1 68. Paternal deletion of 20q13 including GNAS has also been associated with feeding difficulties, poor growth, and abnormal adipose tissue distribution in humans 94. However the significant role that XLαs plays in metabolic regulation may be species-specific, as PPHP patients with paternal GNAS mutations that disrupt XLαs expression or function do not produce a similar phenotype.
Tissue-Specific Gsα Knockout Models
Important insights into the effects of Gsα signaling in specific tissues on metabolic regulation have been derived from various mouse models with tissue-specific Gsα deficiency. In these models both Gsα alleles were disrupted and therefore the effects of imprinting are not relevant. Mice with liver-specific Gsα deficiency (LGsKO) showed effects consistent with loss of glucagon action in the liver, with reduced gluconeogenesis and increased hepatic glycogen synthesis, along with increased insulin sensitivity in other tissues 72. LGsKO mice were hypoglycemic and hypoinsulinemic in the fed state but maintained normal glucose and insulin levels after prolonged fasting, probably due to prolonged breakdown of increased hepatic glycogen stores and increased extrahepatic gluconeogenesis. LGsKO mice also developed pancreatic islet cell hyperplasia with markedly increased circulating glucagon and glucagon-like peptide 1 (GLP1) levels and inappropriately high levels of insulin release in response to administered glucose. In addition LGsKO mice had significantly reduced adiposity although the mechanisms underlying this effect on energy balance were not apparent from food intake and energy expenditure studies.
Muscle-specific Gsα knockout mice (MGsKO) showed no differences in body weight or composition but did show evidence of glucose intolerance despite the lack of an apparent defect in insulin sensitivity or glucose-stimulated insulin secretion 73. This may be due to the fact that these mice had a low muscle mass, confirming an important role for β-adrenergic signaling in maintanence of muscle mass. Interestingly the skeletal muscle fibers in MGsKO mice appeared to shift towards type 1 slow twitch (aerobic) fibers based upon myosin heavy chain subtype and kinetic properties but had lower expression of peroxisomal proliferator-activated receptor γ coactivator 1α (PGC1α) and lower oxidative metabolic capacity, which is characteristic of type 2 fast twitch (glycolytic) fibers. Therefore characteristics of muscle fiber type based upon kinetic and other properties can be dissociated from their metabolic characteristics. Finally mice with loss of Gsα expression in pancreatic β cells (βGsKO) developed early postnatal insulin-deficient diabetes with a severe defect in β cell proliferation 70 Although it has been proposed that Gsα signaling promotes β cell growth through activation of the Irs2-Pdx1 pathway 95, expression of both of these signaling proteins was unaffected in β cells in βGsKO mice. These mice also had a severe defect in linear growth and reduced adiposity.
Prader-Willi and Angelman’s Syndromes
Prader-Willi syndrome (PWS) is a common congenital disorder that in addition to severe hyperphagia and obesity, is also associated with short stature, muscular hypotonia, neurobehavior abnormalities and dysmorphic features 96. Severe hyperphagia is presumed to be secondary to a primary hypothalamic defect. PWS is also associated with abnormally high levels of the orexigenic peptide ghrelin, although in one study reducing ghrelin levels in PWS patients with somatostatin did not correct the hyperphagia 97. PWS patients also have increased lipoprotein lipase activity levels suggesting that in addition to hyperphagia, there may be abnormalities in lipid uptake in adipose tissue 98. PWS is associated with uniparental disomies, deletions, or abnormal imprinting of 15q11-13 leading to loss of expression of a series of contiguous paternally-expressed imprinted genes in this region, including NDN, MAGEL2, MKRN3, SNURF-SNRPN, and several sno-RNAs 99. As no specific gene mutation has been found to cause PWS, this disorder is believed to be a contiguous gene syndrome which results from the combined effects of loss of several genes. However there is some evidence that two of these genes (MAGEL2, NDN [Necdin]) may be involved in abnormal hypothalamic development underlying the hyperphagia and obesity in PWS 100.
Angelman’s syndrome (AS) is characterized by severe mental retardation, ataxia, seizures, a happy disposition, and obesity. AS is associated with reciprocal uniparental disomies or deletions of the same 15q11-13 region as that affected in PWS. AS is believed to be primarily due to loss of expression of the ubiquitin protein ligase E3A, the product of the UBE3A gene which is located just telomeric to the PWS region, as mutations affecting this gene have been associated with AS 101. UBE3A undergoes tissue-specific imprinting in the CNS. However Ube3a knockout mice did not show clear evidence of obesity102, 103. More recent studies have identified another gene ATP10C (encoding an ATPase believed to be phospholipid translocase) located nearby UBE3A which also undergoes brain-specific imprinting and which leads to obesity, glucose intolerance, and insulin resistance when mutated in mice 104, 105.
Other Imprinted Genes Associated with Body Weight Dysregulation
Initial evidence for an important role for imprinted genes, particularly those that are paternally-expressed, in hypothalamic development came from the observation in chimeric mice that androgenetic cells (those disomic for the paternal genome) are prevalent in hypothalamus while parthenogenetic cells (those disomic for the maternal genome) are excluded from the hypothalamus 106. In addition to the potential role of the PWS-associated paternally-expressed genes NDN and MAGEL2 in hypothalamic function discussed above, the paternally-expressed imprinted gene Peg3 (paternally-expressed gene 3) which encodes a zinc-finger protein is expressed in the hypothalamus and Peg3 knockout mice develop obesity in adulthood primarily due to reduced sympathetic nervous system activity and metabolic rate 107. While Igf2 (insulin-like growth factor) is a paternally-expressed imprinted gene which promotes prenatal growth, one mouse model in which Igf2 expression was reduced in brain developed increased adiposity, suggesting that it may have an opposite effect on postnatal growth 108.
Two imprinted genes have been linked to obesity primarily via effects on adipocyte differentiation and function. Pref1/Dlk1 (preadipocyte factor 1/Delta, Drosophila Homolog-like 1) is a paternally-expressed gene that is expressed in preadipocytes and which encodes a transmembrane protein which regulates adipocyte differentiation. Pref1/Dlk1 inhibits the differentiation of preadipocytes into mature adipocytes 109 and Pref1/Dlk1-null mice develop adult-onset obesity 110 while mice overexpressing Pref1/Dlk1 in adipose tissue develop a lean phenotype with glucose intolerance 111. Another imprinted gene which affects adiposity through a primary effect on adipocytes is Peg1/Mest (paternally-expressed gene 1/mesoderm-specific transcript) which promotes adipogenesis and obesity when overexpressed and which is induced in WAT in the setting of diet-induced or genetic obesity due to loss of its imprinting 112–115.
Conclusion
The GNAS locus is a unique gene with oppositely imprinted gene products (Gsα and XLαs) which have opposite effects on energy and glucose metabolism. These reciprocal affects occur primarily through opposite CNS regulation of sympathetic nervous system activity and energy expenditure. The effects of maternal and paternal mutations of the GNAS locus on postnatal growth are consistent with pattern predicted by that predicted by the ‘kinship’ theory of imprinting, in that paternal Gnas mutation leads to XLαs deficiency and a severely lean phenotype in mice while maternal GNAS/Gnas mutations in mice and humans lead to Gsα deficiency in specific-tissues results and obesity. However when one examines the effects of other imprinted genes on adult energy balance, this same pattern does not hold up. For example, obesity is a feature of both PWS and AS even though these two syndromes result from loss of paternally- and maternally-expressed imprinted genes from the same chromosomal region, respectively. In addition, paternally-expressed imprinted genes can have opposite effects on sympathetic nervous system activity and energy expenditure (e.g. Gnasxl and Peg3) as well as opposite effects on adipocyte differentiation and function (e.g. Peg1/Mest and Pref1/Dlk1). In addition, both a paternally-expressed (Peg3) and maternally-expressed imprinted gene (Gnas) have similar effects on hypothalamic regulation of sympathetic nervous system activity and energy expenditure.
Acknowledgments
Support: This work was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive, and Kidney Diseases, National Institutes of Health, U.S. Department of Health and Human Services.
References
- 1.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 3.Haig D. Genomic imprinting and kinship: how good is the evidence? Annu Rev Genet. 2004;38:553–85. doi: 10.1146/annurev.genet.37.110801.142741. [DOI] [PubMed] [Google Scholar]
- 4.Lindsay RS, Kobes S, Knowler WC, Bennett PH, Hanson RL. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of type 2 diabetes and BMI in Pima Indians. Diabetes. 2001;50:2850–2857. doi: 10.2337/diabetes.50.12.2850. [DOI] [PubMed] [Google Scholar]
- 5.Gorlova OY, Amos CI, Wang NW, Shete S, Turner ST, Boerwinkle E. Genetic linkage and imprinting effects on body mass index in children and young adults. Eur J Hum Genet. 2003;11:425–432. doi: 10.1038/sj.ejhg.5200979. [DOI] [PubMed] [Google Scholar]
- 6.Dong C, Li WD, Geller F, Lei L, Li D, Gorlova OY, et al. Possible genomic imprinting of three human obesity-related genetic loci. Am J Hum Genet. 2005;76:427–437. doi: 10.1086/428438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rance KA, Fustin JM, Dalgleish G, Hambly C, Bunger L, Speakman JR. A paternally imprinted QTL for mature body mass on mouse chromosome 8. Mamm Genome. 2005;16:567–577. doi: 10.1007/s00335-005-0012-4. [DOI] [PubMed] [Google Scholar]
- 8.Weinstein LS, Xie T, Zhang QH, Chen M. Studies of the regulation and function of the Gsα gene Gnas using gene targeting technology. Pharmacol Ther. 2007;115:271–291. doi: 10.1016/j.pharmthera.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Isolation and characterization of the human Gsα gene. Proc Natl Acad Sci USA. 1988;85:2081–2085. doi: 10.1073/pnas.85.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 11.Bray P, Carter A, Simons C, Guo V, Puckett C, Kamholz J, et al. Human cDNA clones for four species of Gαs signal transduction protein. Proc Natl Acad Sci USA. 1986;83:8893–8897. doi: 10.1073/pnas.83.23.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayward BE, Kamiya M, Strain L, Moran V, Campbell R, Hayashizaki Y, et al. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA. 1998;95:10038–10043. doi: 10.1073/pnas.95.17.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies SJ, Hughes HE. Imprinting in Albright’s hereditary osteodystrophy. J Med Genet. 1993;30:101–103. doi: 10.1136/jmg.30.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell R, Gosden CM, Bonthron DT. Parental origin of transcription from the human GNAS1 gene. J Med Genet. 1994;31:607–614. doi: 10.1136/jmg.31.8.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu S, Yu D, Lee E, Eckhaus ME, Lee R, Corria Z, et al. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein α-subunit (Gsα) knockout mice is due to tissue-specific imprinting of the Gsα gene. Proc Natl Acad Sci USA. 1998;95:8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward BE, Barlier A, Korbonits M, Grossman AB, Jacquet P, Enjalbert A, et al. Imprinting of the Gsα gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest. 001(107):R31–R36. doi: 10.1172/JCI11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstein LS, Yu S, Warner DR, Liu J. Endocrine manifestations of stimulatory G protein α-subunit mutations and the role of genomic imprinting. Endocr Rev. 2001;22:675–705. doi: 10.1210/edrv.22.5.0439. [DOI] [PubMed] [Google Scholar]
- 20.Germain-Lee EL, Ding C, Deng Z, Crane JL, Saji M, Ringel MD, et al. Paternal imprinting of Gαs in the human thyroid as the basis of TSH resistance in pseudohypoparathyroidism type 1a. Biochem Biophys Res Commun. 2002;296:67–72. doi: 10.1016/s0006-291x(02)00833-1. [DOI] [PubMed] [Google Scholar]
- 21.Mantovani G, Ballare E, Giammona E, Beck-Peccoz P, Spada A. The Gsα gene: predominant maternal origin of transcription in human thyroid gland and gonads. J Clin Endocrinol Metab. 002(87):4736–4740. doi: 10.1210/jc.2002-020183. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Erlichman B, Weinstein LS. The stimulatory G protein α-subunit Gsα is imprinted in human thyroid glands: implications for thyroid function in pseudohypoparathyroidism types 1A and 1B. J Clin Endocrinol Metab. 2003;88:4336–4341. doi: 10.1210/jc.2003-030393. [DOI] [PubMed] [Google Scholar]
- 23.Chen M, Wang J, Dickerson KE, Kelleher J, Xie T, Gupta D, et al. Central nervous system imprinting of the G protein Gsα and its role in metabolic regulation. Cell Metab. 2009;9:548–555. doi: 10.1016/j.cmet.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 25.Germain-Lee EL, Schwindinger W, Crane JL, Zewdu R, Zweifel LS, Wand G, et al. A mouse model of Albright hereditary osteodystrophy generated by targeted disruption of exon 1 of the Gnas gene. Endocrinology. 2005;146:4697–4709. doi: 10.1210/en.2005-0681. [DOI] [PubMed] [Google Scholar]
- 26.Chen M, Gavrilova O, Liu J, Xie T, Deng C, Nguyen AT, et al. Alternative Gnas gene products have opposite effects on glucose and lipid metabolism. Proc Natl Acad Sci USA. 2005;102:7386–7391. doi: 10.1073/pnas.0408268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantovani G, Bondioni S, Locatelli M, Pedroni C, Lania AG, Ferrante E, et al. Biallelic expression of the Gsα gene in human bone and adipose tissue. J Clin Endocrinol Metab. 2004;89:6316–6319. doi: 10.1210/jc.2004-0558. [DOI] [PubMed] [Google Scholar]
- 28.Xie T, Plagge A, Gavrilova O, Pack S, Jou W, Lai EW, et al. The alternative stimulatory G protein α-subunit XLαs is a critical regulator of energy and glucose metabolism and sympathetic nerve activity in adult mice. J Biol Chem. 2006;281:18989–18999. doi: 10.1074/jbc.M511752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford JA, Mutchler KJ, Sullivan BE, Lanigan TM, Clark MS, Russo AF. Neural expression of a novel alternatively spliced and polyadenylated Gsα transcript. J Biol Chem. 1993;268:9879–9885. [PubMed] [Google Scholar]
- 30.Hayward BE, Moran V, Strain L, Bonthron DT. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA. 1998;95:15475–15480. doi: 10.1073/pnas.95.26.15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelsey G, Bodle D, Miller HJ, Beechey CV, Coombes C, Peters J, et al. Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics. 1999;62:129–138. doi: 10.1006/geno.1999.6022. [DOI] [PubMed] [Google Scholar]
- 32.Ischia R, Lovisetti-Scamihorn P, Hogue-Angeletti R, Wolkersdorfer M, Winkler H, Fischer-Colbrie R. Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5-HT1B receptor antagonist activity. J Biol Chem. 1997;272:11657–11662. doi: 10.1074/jbc.272.17.11657. [DOI] [PubMed] [Google Scholar]
- 33.Bauer R, Weiss C, Marksteiner J, Doblinger A, Fischer-Colbrie R, Laslop A. The new chromogranin-like protein NESP55 is preferentially localized in adrenaline-synthesizing cells of the bovine and rat adrenal medulla. Neurosci Lett. 1999;263:13–16. doi: 10.1016/s0304-3940(99)00091-9. [DOI] [PubMed] [Google Scholar]
- 34.Bauer R, Ischia R, Marksteiner J, Kapeller I, Fischer-Colbrie R. Localization of neuroendocrine secretory protein 55 messenger RNA in the rat brain. Neuroscience. 1999;91:685–694. doi: 10.1016/s0306-4522(98)00668-x. [DOI] [PubMed] [Google Scholar]
- 35.Lovisetti-Scamiform P, Fischer-Colbrie R, Leitner B, Scherzer G, Winkler H. Relative amounts and molecular forms of NESP55 in various bovine tissues. Brain Res. 1999;829:99–106. doi: 10.1016/s0006-8993(99)01345-1. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Vu TH, Zeng ZL, Nguyen BT, Hayward BE, Bonthron DT, et al. Tissue-specific expression of antisense and sense transcripts at the imprinted Gnas locus. Genomics. 2000;69:295–304. doi: 10.1006/geno.2000.6337. [DOI] [PubMed] [Google Scholar]
- 37.Kehlenbach RH, Matthey J, Huttner WB. XLαs is a new type of G protein. Nature. 1994;372:804–809. doi: 10.1038/372804a0. [DOI] [PubMed] [Google Scholar]
- 38.Abramowitz J, Grenet D, Birnbaumer M, Torres HN, Birnbaumer L. XLαs, the extra-long form of the α-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex. Proc Natl Acad Sci USA. 2004;101:8366–8371. doi: 10.1073/pnas.0308758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastepe M, Gunes Y, Perez-Villamil B, Hunzelman J, Weinstein LS, Jüppner H. Receptor-mediated adenylyl cyclase activation through XLαs, the extra-large variant of the stimulatory G protein alpha subunit. Mol Endocrinol. 2002;16:1912–1919. doi: 10.1210/me.2002-0054. [DOI] [PubMed] [Google Scholar]
- 40.Pasolli HA, Klemke M, Kehlenbach RH, Wang Y, Huttner WB. Characterization of the extra-large G protein α-subunit XLαs. I. Tissue distribution and subcellular localization. J Biol Chem. 2000;275:33622–33632. doi: 10.1074/jbc.M001335200. [DOI] [PubMed] [Google Scholar]
- 41.Pasolli HA, Huttner WB. Expression of the extra-large G protein α-subunit XLαs in neuroepithelial cells and young neurons during development of the rat nervous system. Neurosci Lett. 2001;301:119–122. doi: 10.1016/s0304-3940(01)01620-2. [DOI] [PubMed] [Google Scholar]
- 42.Plagge A, Gordon E, Dean W, Boiani R, Cinti S, Peters J, et al. The imprinted signaling protein XLαs is required for postnatal adaptation to feeding. Nat Genet. 2004;36:818–826. doi: 10.1038/ng1397. [DOI] [PubMed] [Google Scholar]
- 43.Klemke M, Kehlenbach RH, Huttner WB. Two overlapping reading frames in a single exon encode interacting proteins-a novel way of gene usage. EMBO J. 2001;20:3849–3860. doi: 10.1093/emboj/20.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freson K, Jaeken J, Van Helvoirt M, de Zegher F, Wittevrongel C, Thys C, et al. Functional polymorphisms in the paternally expressed XLαs and its cofactor ALEX decrease their mutual interaction and enhance receptor-mediated cAMP formation. Hum Mol Genet. 2003;12:1121–1130. doi: 10.1093/hmg/ddg130. [DOI] [PubMed] [Google Scholar]
- 45.Hayward BE, Bonthron DT. An imprinted antisense transcript at the human GNAS1 locus. Hum Mol Genet. 2000;9:835–841. doi: 10.1093/hmg/9.5.835. [DOI] [PubMed] [Google Scholar]
- 46.Wroe SF, Kelsey G, Skinner JA, Bodle D, Ball ST, Beechey CV, et al. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc Natl Acad Sci USA. 2000;97:3342–3346. doi: 10.1073/pnas.050015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williamson CM, Skinner JA, Kelsey G, Peters J. Alternative non-coding splice variants of Nespas, an imprinted gene antisense to Nesp in the Gnas imprinting cluster. Mamm Genome. 2002;13:74–79. doi: 10.1007/s00335-001-2102-2. [DOI] [PubMed] [Google Scholar]
- 48.Coombes C, Arnaud P, Gordon E, Dean W, Coar EA, Williamson CM, et al. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol. 2003;23:5475–5488. doi: 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa Y, Bianchi C, Nadal-Ginard B, Homcy CJ. Alternative promoter and 5′ exon generate a novel Gsα mRNA. J Biol Chem. 1990;265:8458–8462. [PubMed] [Google Scholar]
- 50.Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, Weinstein LS. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106:1167–1174. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Nealon JG, Weinstein LS. Distinct patterns of abnormal GNAS imprinting in familial and sporadic pseudohypoparathyroidism type IB. Hum Mol Genet. 2005;14:95–102. doi: 10.1093/hmg/ddi009. [DOI] [PubMed] [Google Scholar]
- 52.Bastepe M, Pincus JE, Sugimoto T, Tojo K, Kanatani M, Azuma Y, et al. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet. 2001;10:1231–1241. doi: 10.1093/hmg/10.12.1231. [DOI] [PubMed] [Google Scholar]
- 53.Jan de Beur S, Ding C, Germain-Lee EL, Cho J, Maret A, Levine MA. Discordance between genetic and epigenetic defects in pseudohypoparathyroidism type 1b revealed by inconsistent loss of maternal imprinting at GNAS1. Am J Hum Genet. 2003;73:314–322. doi: 10.1086/377136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williamson CM, Ball ST, Nottingham WT, Skinner JA, Plagge A, Turner MD, et al. A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet. 2004;36:894–899. doi: 10.1038/ng1398. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Chen M, Deng C, Bourc’his D, Nealon JG, Erlichman B, et al. Identification of the control region for tissue-specific imprinting of the stimulatory G protein α-subunit. Proc Natl Acad Sci USA. 2005;102:5513–5518. doi: 10.1073/pnas.0408262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinstein LS, Chen M, Xie T, Liu J. Genetic diseases associated with heterotrimeric G proteins. Trends Pharmacol Sci. 2006;27:260–266. doi: 10.1016/j.tips.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Long DN, McGuire S, Levine MA, Weinstein LS, Germain-Lee EL. Body mass index differences in pseudohypoparathyroidism type 1a versus pseudopseudohypoparathyroidism implicate paternal imprinting of Gαs in the development of human obesity. J Clin Endocrinol Metab. 2007;92:1073–1079. doi: 10.1210/jc.2006-1497. [DOI] [PubMed] [Google Scholar]
- 58.Carel JC, Le Stunff C, Condamine L, Mallet E, Chaussain JL, Adnot P, et al. Resistance to the lipolytic action of epinephrine: a new feature of protein Gs deficiency. J Clin Endocrinol Metab. 1999;84:4127–4131. doi: 10.1210/jcem.84.11.6145. [DOI] [PubMed] [Google Scholar]
- 59.Dekelbab BH, Aughton DJ, Levine MA. Pseudohypoparathyroidism type 1A and morbid obesity in infancy. Endocr Pract. 2009;15:249–253. doi: 10.4158/EP.15.3.249. [DOI] [PubMed] [Google Scholar]
- 60.Germain-Lee EL, Groman J, Crane JL, Jn de Beur SM, Levine MA. Growth hormone deficiency in pseudohypoparathyroidism type 1a: another manifestation of multihormone resistance. J Clin Endocrinol Metab. 2003;88:4059–4069. doi: 10.1210/jc.2003-030028. [DOI] [PubMed] [Google Scholar]
- 61.Nwosu BU, Lee MM. Pseudohypoparathyroidism type 1a and insulin resistance in a child. Nat Rev Endocrinol. 2009;5:345–350. doi: 10.1038/nrendo.2009.81. [DOI] [PubMed] [Google Scholar]
- 62.Hahn S, Frey UH, Siffert W, Tan S, Mann K, Janssen OE. The CC genotype of the GNAS T393C polymorphism is associated with obesity and insulin resistance in women with polycystic ovary syndrome. Eur J Endocrinol. 2006;155:763–770. doi: 10.1530/eje.1.02275. [DOI] [PubMed] [Google Scholar]
- 63.Frey UH, Michalsen A, Merse S, Dobos GJ, Siffert W. A functional GNAS promoter polymorphism is associated with altered weight loss during short-term fasting. Eur J Med Res. 2008;13:576–578. [PubMed] [Google Scholar]
- 64.Frey UH, Hauner H, Jockel KH, Manthey I, Brockmeyer N, Siffert W. A novel promoter polymorphism in the human gene GNAS affects binding of transcription factor upstream stimulatory factor 1, Gαs protein expression and body weight regulation. Pharmacogenet Genomics. 2008;18:141–151. doi: 10.1097/FPC.0b013e3282f49964. [DOI] [PubMed] [Google Scholar]
- 65.Yu S, Gavrilova O, Chen H, Lee R, Liu J, Pacak K, et al. Paternal versus maternal transmission of a stimulatory G protein α subunit knockout produces opposite effects on energy metabolism. J Clin Invest. 2000;105:615–623. doi: 10.1172/JCI8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu S, Castle A, Chen M, Lee R, Takeda K, Weinstein LS. Increased insulin sensitivity in Gsα knockout mice. J Biol Chem. 2001;276:19994–19998. doi: 10.1074/jbc.M010313200. [DOI] [PubMed] [Google Scholar]
- 67.Plagge A, Isles AR, Gordon E, Humby T, Dean W, Gritsch S, et al. Imprinted Nesp55 influences behavioral reactivity to novel environments. Mol Cell Biol. 2005;25:3019–3026. doi: 10.1128/MCB.25.8.3019-3026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelly ML, Moir L, Jones L, Whitehill E, Anstee QM, Goldin RD, et al. A missense mutation in the non-neural G-protein α-subunit isoforms modulates susceptibility to obesity. Int J Obes (Lond) 2009;33:507–518. doi: 10.1038/ijo.2009.30. [DOI] [PubMed] [Google Scholar]
- 69.Xie T, Chen M, Gavrilova O, Lai EW, Liu J, Weinstein LS. Severe obesity and insulin resistance due to deletion of the maternal Gsα allele is reversed by paternal deletion of the Gsα imprint control region. Endocrinology. 2008;149:2443–2450. doi: 10.1210/en.2007-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie T, Chen M, Zhang QH, Ma Z, Weinstein LS. β cell-specific deficiency of the stimulatory G protein α-subunit Gsα leads to reduced β cell mass and insulin-deficient diabetes. Proc Natl Acad Sci USA. 2007;104:19601–19606. doi: 10.1073/pnas.0704796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen A, Gupta D, Gavrilova O, Chen M, Weinstein LS. Mice with adipose-tissue specific deficiency of the stimulatory G protein α-subunit Gsα are resistant to diet-induced obesity and maintain diet-induced, but not cold-induced, thermogenesis. Diabetes. 2006;(Supp) (Abstract) [Google Scholar]
- 72.Chen M, Gavrilova O, Zhao W-Q, Nguyen A, Lorenzo J, Shen L, et al. Increased glucose tolerance and reduced adiposity in the absence of fasting hypoglycemia in mice with liver-specific Gsα deficiency. J Clin Invest. 2005;115:3217–3227. doi: 10.1172/JCI24196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen M, Feng HZ, Gupta D, Kelleher J, Dickerson KE, Wang J, et al. Gsα deficiency in skeletal muscle leads to reduced muscle mass, fiber-type switching, and glucose intolerance without insulin resistance or deficiency. Am J Physiol Cell Physiol. 2009;296:C930–C940. doi: 10.1152/ajpcell.00443.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nogueiras R, Wiedmer P, Perez-Tilve D, Veyrat-Durebex C, Keogh JM, Sutton GM, et al. The central melanocortin system directly controls peripheral lipid metabolism. J Clin Invest. 2007;117:3475–3488. doi: 10.1172/JCI31743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butler AA, Cone RD. The melanocortin receptors: lessons from knockout models. Neuropeptides. 2002;36:77–84. doi: 10.1054/npep.2002.0890. [DOI] [PubMed] [Google Scholar]
- 76.Brito MN, Brito NA, Baro DJ, Song CK, Bartness TJ. Differential activation of the sympathetic innervation of adipose tissues by melanocortin receptor stimulation. Endocrinology. 2007;148:5339–5347. doi: 10.1210/en.2007-0621. [DOI] [PubMed] [Google Scholar]
- 77.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 78.Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O’Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 79.Marsh DJ, Hollopeter G, Huszar D, Laufer R, Yagaloff KA, Fisher SL, et al. Response of melanocortin-4 receptor-deficient mice to anorectic and orexigenic peptides. Nat Genet. 1999;21:119–122. doi: 10.1038/5070. [DOI] [PubMed] [Google Scholar]
- 80.Chen AS, Metzger JM, Trumbauer ME, Guan XM, Yu H, Frazier EG, et al. Role of the melanocortin-4 receptor in metabolic rate and food intake in mice. Transgenic Res. 2000;9:145–154. doi: 10.1023/a:1008983615045. [DOI] [PubMed] [Google Scholar]
- 81.Fan W, Dinulescu DM, Butler AA, Zhou J, Marks DL, Cone RD. The central melanocortin system can directly regulate serum insulin levels. Endocrinology. 2000;141:3072–3079. doi: 10.1210/endo.141.9.7665. [DOI] [PubMed] [Google Scholar]
- 82.Obici S, Feng Z, Tan J, Liu L, Karkanias G, Rossetti L. Central melanocortin receptors regulate insulin action. J Clin Invest. 2001;108:1079–1085. doi: 10.1172/JCI12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Butler AA, Marks DL, Fan W, Kuhn CM, Bartolome M, Cone RD. Melanocortin-4 receptor is required for acute homeostatic responses to increased dietary fat. Nat Neurosci. 2001;4:605–11. doi: 10.1038/88423. [DOI] [PubMed] [Google Scholar]
- 84.Voss-Andreae A, Murphy JG, Ellacott KLJ, Stuart RC, Nillni EA, Cone RD, et al. Role of the central melanocortin circuitry in adaptive thermogenesis of brown adipose tissue. Endocrinology. 2007;148:1550–1560. doi: 10.1210/en.2006-1389. [DOI] [PubMed] [Google Scholar]
- 85.Greenfield JR, Miller JW, Keogh JM, Henning E, Satterwhite JH, Cameron GS, et al. Modulation of blood pressure by central melanocortinergic pathways. N Engl J Med. 2009;360:44–52. doi: 10.1056/NEJMoa0803085. [DOI] [PubMed] [Google Scholar]
- 86.Tallam LS, Stec DE, Willis MA, da Silva AA, Hall JE. Melanocortin-4 receptor-deficient mice Are not hypertensive or salt-sensitive despite obesity, hyperinsulinemia, and hyperleptinemia. Hypertension. 2005;46:326–332. doi: 10.1161/01.HYP.0000175474.99326.bf. [DOI] [PubMed] [Google Scholar]
- 87.Kublaoui BM, Holder JL, Jr, Tolson KP, Gemelli T, Zinn AR. SIM1 overexpression partially rescues agouti yellow and diet-induced obesity by normalizing food intake. Endocrinology. 2006;147:4542–4549. doi: 10.1210/en.2006-0453. [DOI] [PubMed] [Google Scholar]
- 88.Kublaoui BM, Holder JL, Jr, Gemelli T, Zinn AR. Sim1 haploinsufficiency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons. Mol Endocrinol. 2006;20:2483–2492. doi: 10.1210/me.2005-0483. [DOI] [PubMed] [Google Scholar]
- 89.Holder JL, Jr, Butte NF, Zinn AR. Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum Mol Genet. 2000;9:101–108. doi: 10.1093/hmg/9.1.101. [DOI] [PubMed] [Google Scholar]
- 90.Kublaoui BM, Gemelli T, Tolson KP, Wang Y, Zinn AR. Oxytocin deficiency mediates hyperphagic obesity of Sim1 haploinsufficient mice. Mol Endocrinol. 2008;22:1723–1734. doi: 10.1210/me.2008-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holder JL, Jr, Zhang L, Kublaoui BM, DiLeone RJ, Oz OK, Bair CH, et al. Sim1 gene dosage modulates the homeostatic feeding response to increased dietary fat in mice. Am J Physiol Endocrinol Metab. 2004;287:E105–E113. doi: 10.1152/ajpendo.00446.2003. [DOI] [PubMed] [Google Scholar]
- 92.Faivre L, Cormier-Daire V, Lapierre JM, Colleaux L, Jacquemont S, Genevieve D, et al. Deletion of the SIM1 gene (6q16.2) in a patient with a Prader-Willi-like phenotype. J Med Genet. 2002;39:594–596. doi: 10.1136/jmg.39.8.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen M, Haluzik M, Wolf NJ, Lorenzo J, Dietz KR, Reitman ML, et al. Increased insulin sensitivity in paternal Gnas knockout mice Is associated with increased lipid clearance. Endocrinology. 2004;145:4094–4102. doi: 10.1210/en.2004-0038. [DOI] [PubMed] [Google Scholar]
- 94.Genevieve D, Sanlaville D, Faivre L, Kottler ML, Jambou M, Gosset P, et al. Paternal deletion of the GNAS imprinted locus (including Gnasxl) in two girls presenting with severe pre- and post-natal growth retardation and intractable feeding difficulties. Eur J Hum Genet. 2005;13:1033–1039. doi: 10.1038/sj.ejhg.5201448. [DOI] [PubMed] [Google Scholar]
- 95.Jhala US, Canettieri G, Screaton RA, Kulkarni RN, Krajewski S, Reed J, et al. cAMP promotes pancreatic β-cell survival via CREB-mediated induction of IRS2. Genes Dev. 2003;17:1575–1580. doi: 10.1101/gad.1097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldstone AP. Prader-Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol Metab. 2004;15:12–20. doi: 10.1016/j.tem.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 97.Tan TM, Vanderpump M, Khoo B, Patterson M, Ghatei MA, Goldstone AP. Somatostatin infusion lowers plasma ghrelin without reducing appetite in adults with Prader-Willi syndrome. J Clin Endocrinol Metab. 2004;89:4162–4165. doi: 10.1210/jc.2004-0835. [DOI] [PubMed] [Google Scholar]
- 98.Schwartz RS, Brunzell JD, Bierman EL. Elevated adipose tissue lipoprotein lipase in the pathogenesis of obesity in the Prader-Willi syndrome. Trans Assoc Am Physicians. 1979;92:89–95. [PubMed] [Google Scholar]
- 99.Bernhard Horsthemke JW. Mechanisms of imprinting of the Prader-Willi/Angelman region. American Journal of Medical Genetics Part A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- 100.Caqueret A, Yang C, Duplan S, Boucher F, Michaud JL. Looking for trouble: a search for developmental defects of the hypothalamus. Horm Res. 2005;64:222–230. doi: 10.1159/000088977. [DOI] [PubMed] [Google Scholar]
- 101.Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 103.Miura K, Kishino T, Li E, Webber H, Dikkes P, Holmes GL, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- 104.Dhar MS, Sommardahl CS, Kirkland T, Nelson S, Donnell R, Johnson DK, et al. Mice Heterozygous for Atp10c, a Putative Amphipath, Represent a Novel Model of Obesity and Type 2 Diabetes. J Nutr. 2004;134:799–805. doi: 10.1093/jn/134.4.799. [DOI] [PubMed] [Google Scholar]
- 105.Dhar MS, Yuan JS, Elliott SB, Sommardahl C. A type IV P-type ATPase affects insulin-mediated glucose uptake in adipose tissue and skeletal muscle in mice. J Nutr Biochem. 2006;17:811–820. doi: 10.1016/j.jnutbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 106.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res. 1996;92:91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 107.Curley JP, Pinnock SB, Dickson SL, Thresher R, Miyoshi N, Surani MA, et al. Increased body fat in mice with a targeted mutation of the paternally expressed imprinted gene Peg3. FASEB J. 2005;19:1302–1304. doi: 10.1096/fj.04-3216fje. [DOI] [PubMed] [Google Scholar]
- 108.Jones BK, Levorse J, Tilghman SM. Deletion of a nuclease-sensitive region between the Igf2 and H19 genes leads to Igf2 misregulation and increased adiposity. Hum Mol Genet. 2001;10:807–814. doi: 10.1093/hmg/10.8.807. [DOI] [PubMed] [Google Scholar]
- 109.Smas CM, Sul HS. Characterization of Pref-1 and its inhibitory role in adipocyte differentiation. Int J Obes Relat Metab Disord. 1996;20 (Suppl 3):S65–72. [PubMed] [Google Scholar]
- 110.Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee K, Villena JA, Moon YS, Kim KH, Lee S, Kang C, et al. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1) J Clin Invest. 2003;111:453–461. doi: 10.1172/JCI15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takahashi M, Kamei Y, Ezaki O. Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. Am J Physiol Endocrinol Metab. 2005;288:E117–E124. doi: 10.1152/ajpendo.00244.2004. [DOI] [PubMed] [Google Scholar]
- 113.Shi W, Lefebvre L, Yu Y, Otto S, Krella A, Orth A, et al. Loss-of-imprinting of Peg1 in mouse interspecies hybrids is correlated with altered growth. Genesis. 2004;39:65–72. doi: 10.1002/gene.20027. [DOI] [PubMed] [Google Scholar]
- 114.Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2:e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nikonova L, Koza RA, Mendoza T, Chao PM, Curley JP, Kozak LP. Mesoderm-specific transcript is associated with fat mass expansion in response to a positive energy balance. FASEB J. 2008;22:3925–3937. doi: 10.1096/fj.08-108266. [DOI] [PMC free article] [PubMed] [Google Scholar]