Abstract
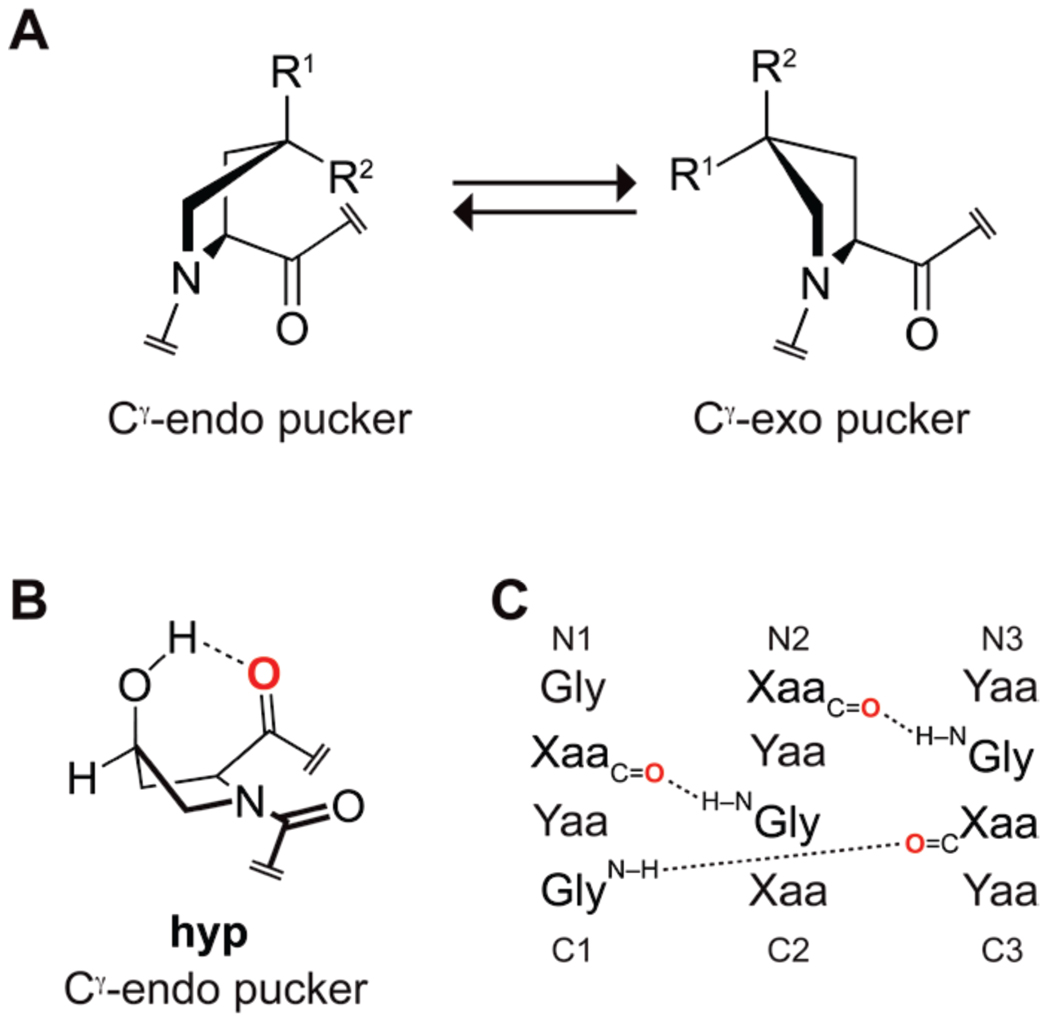
Prolyl 4-hydroxylases install a hydroxyl group in the 4R configuration on the γ-carbon atom of certain (2S)-proline (Pro) residues in tropocollagen, elastin, and other proteins and other proteins to form (2S,4R)-4-hydroxyproline (Hyp). The gauche effect arising from this prevalent post-translational modification enforces a Cγ-exo ring pucker and stabilizes the collagen triple helix. The Hyp diastereomer (2S,4S)-4-hydroxyproline (hyp) has not been observed in a protein, despite the ability of electronegative 4S substituents to enforce the more common Cγ-endo ring pucker of Pro. Here, we use density functional theory, spectroscopy, crystallography, and calorimetry to explore the consequences of hyp incorporation on protein stability using a collagen model system. We find that the 4S-hydroxylation of Pro to form hyp does indeed enforce a Cγ-endo ring pucker, but a transannular hydrogen bond between the hydroxyl moiety and the carbonyl of hyp distorts the main-chain torsion angles that typically accompany a Cγ-endo ring pucker. This same transannular hydrogen bond enhances an n→π* interaction that stabilizes the trans conformation of the peptide bond preceding hyp, endowing hyp with the unusual combination of a Cγ-endo ring pucker and high trans:cis ratio. O-Methylation of hyp to form (2S,4S)-4-methoxyproline (mop) eliminates the transannular hydrogen bond and restores a prototypical Cγ-endo pucker. mop residues endow the collagen triple helix with much more conformational stability than do hyp residues. These findings highlight the critical importance of the configuration of the hydroxyl group installed on Cγ of proline residues.
Introduction
The hydroxylation of proline residues by prolyl 4-hydroxylase to form (2S,4R)-4-hydroxyproline (Hyp) is the most common post-translational modification in animals.1 All known prolyl 4-hydroxylases install the hydroxyl group in the R configuration, that is, on the face of the pyrrolidine ring opposite from the carboxyl group.2 The ensuing gauche effect stabilizes the Cγ-exo ring pucker, which enables an n→π* interaction that stabilizes the trans peptide bond (Figure 1A).3 The preorganization that arises from these stereoelectronic effects has important consequences for the conformational stability of collagen and other proteins.4
Figure 1.
Attributes of Pro derivatives. (A) The pucker of the pyrrolidine ring can be enforced by the gauche effect arising from an electronegative Cγ substituent (R1 or R2), which adopts the pseudo-axial position. Hyp: R1=OH, R2=H; hyp: R1=H, R2=OH; Mop: R1=OMe, R2=H; mop: R1=H, R2=OMe. (B) Intramolecular hydrogen bond in hyp. (C) Interstrand hydrogen bonds in a collagen triple helix. Oi of a hyp residue in the Xaa position is red in panels B and C.
In marked contrast to Hyp, its 4S diasteromer—(2S,4S)-4-hydroxyproline (hyp)—has not been observed in a natural protein. This absence is intriguing given the recent discovery that nonnatural analogs of hyp can enhance the conformational stability of proteins. For example, (2S,4S)-4-fluoroproline (flp) has a demonstrated ability to modulate the stability of structural motifs (e.g., polyproline I- and II-type helices5 and β-turns6) and whole proteins (e.g., triple-helical collagen,7 barstar,8 and green fluorescent protein9). The flp residue prefers a Cγ-endo ring pucker, as expected from a gauche effect, and has a low trans:cis ratio.3d
Although Hyp is especially abundant in collagen, its use is judicious. Structures of collagen triple helices reveal that proline and its derivatives in the Yaa position of the Xaa–Yaa–Gly repeat have a Cγ-exo ring pucker, whereas those in the Xaa position have a Cγ-endo ring pucker.4,10 Hyp is prevalent in the Yaa position,11 consistent with its preferred pucker. Seminal studies of synthetic collagen triple helices indicate that hyp in the Yaa position severely destabilizes the triple helix.12 Surprisingly, hyp in the Xaa position is also severely destabilizing,12–13 despite its preference for the Cγ-endo ring pucker that is desirable in the Xaa position.14
Here, we explore the consequence of incorporating hyp into a protein using collagen as a model system. We were driven by the results of a recent gas-phase microwave spectroscopic analysis of the free amino acid, hypOH, which revealed a transannular hydrogen bond between the hydroxyl and carboxyl groups.14 We use O-methylation to form (2S,4S)-4-methoxyproline (mop) as a probe, because a methoxy group retains the electron-withdrawing15 and hyperconjugative16 ability of a hydroxyl group, but loses the ability to donate a hydrogen bond.17 Our theoretical, spectroscopic, crystallographic, and calorimetric analyses of hyp and mop in a small molecule model and in a collagen triple helix reveal that hydrogen bonding can compromise the potential benefits of an underlying stereoelectronic effect to the conformational stability of a protein.
Results and Discussion
Computational Analyses of Ac-hyp-OMe
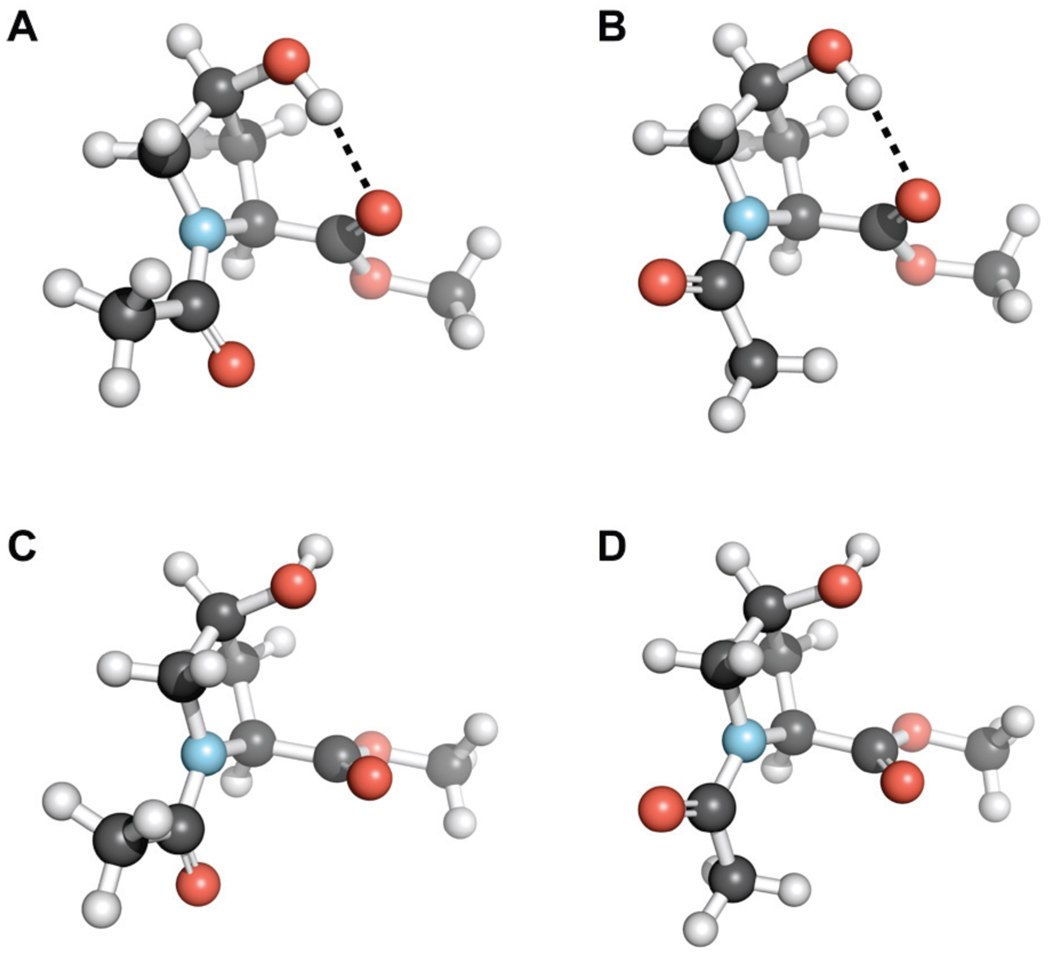
We began by exploring the conformational preferences of Ac-hyp-OMe with density functional theory (DFT) calculations at the B3LYP/6-311+G(2d,p) level. We chose Ac-hyp-OMe as a model to avoid the γ-turn formation that has been observed in Ac-Xaa-NHMe.18 First, gas-phase geometry optimization and frequency calculations were performed on six conformations of Ac-hyp-OMe. We found that a transannular hydrogen bond, formed between the 4S hydroxyl group and the ester carbonyl group of Ac-hyp-OMe (Figure 1B), was an important conformational determinant. The lowest energy conformations of Ac-hyp-OMe have this hydrogen bond in both cis and trans conformers (Figure 2). The conformation of Ac-hyp-OMe with a hydrogen bond, Cγ-endo ring pucker, and trans amide bond (Figure 2A) is >3 kcal/mol more stable than any non-hydrogen bonded conformation (Figures 2C and 2D). Gas-phase calculations can overestimate the stabilization conferred on a particular conformation by hydrogen bonding, and we were interested in understanding how hyp would behave both in a protein environment and in aqueous solution. Therefore, we also performed complete geometry optimizations of six conformations of Ac-hyp-OMe using the conductor-like polarizable continuum (CPCM) model at the B3LYP/6-311+G*(2d,p) level of theory.19 Analysis of the resulting self-consistent field (SCF) energies showed that the most stable conformation in chloroform is the internally hydrogen bonded conformation depicted in Figure 2A, whereas in water the non-hydrogen bonded trans Cγ-endo conformation in Figure 2C is preferred. These results are consistent with the hypothesis that in nonpolar or aprotic solvents (or in a protein environment) hyp prefers a Cγ-endo ring pucker with an intramolecular transannular hydrogen bond; that preference is obviated by aqueous solvation. The transannular hydrogen bond within Ac-hyp-OMe alters the main-chain torsion angles ϕ (C′i−1–Ni–Cαi-C′i) and ψ (Ni–Cαi–C′i–Ni+1) from those observed in standard Cγ-endo ring puckers (ϕ ≈ −70°, ψ ≈ 152°).3b,3d The value of ψ = 140° in Ac-hyp-OMe (Figure 2A) is closer to that of a typical Cγ-exo ring pucker (ψ ≈ 143°).3b,3d Similarly, the value of ϕ = −65° in Ac-hyp-OMe is intermediate between that of a typical Cγ-exo ring pucker (which has ϕ ≈ −59°) and a typical Cγ-endo ring pucker (ϕ ≈ −70°).3b,3d
Figure 2.
Ac-hyp-OMe conformations optimized at the B3LYP/6-311+G(2d,p) level of theory and depicted with the program PyMOL (Delano Scientific, Palo Alto, CA). (A) Cγ-endo ring pucker, trans peptide bond, and intramolecular hydrogen bond. (B) Cγ-endo ring pucker, cis peptide bond, and intramolecular hydrogen bond. (C) Cγ-endo ring pucker, trans peptide bond. (D) Cγ-endo ring pucker, cis peptide bond.
Spectroscopic Analyses of Ac-hyp-OMe and Ac-mop-OMe
Next, we synthesized Ac-hyp-OMe and Ac-mop-OMe and explored their solution conformations by 1H NMR spectroscopy. In aqueous solution, both Ac-hyp-OMe3c and Ac-mop-OMe have particularly low values of Ktrans/cis (Table 1), suggesting that they adopt a Cγ-endo ring pucker. In CDCl3, however, Ac-hyp-OMe but not Ac-mop-OMe has a high value of Ktrans/cis. The chemical shift of the hydroxyl proton of Ac-hyp-OMe decreases with temperature (δ = 3.16 ppm, T = 17.0 °C; 3.12, 22.8; 3.06, 29.6; 3.01, 36.4), as expected from its participation in a hydrogen bond (Figure 1B).20 Because the stability of that hydrogen bond should be enhanced in an aprotic solvent (as predicted by the CPCM calculations), we propose that the high Ktrans/cis value of Ac-hyp-OMe in CDCl3 occurs because the transannular hydrogen bond distorts the ϕ and ψ angles of Ac-hyp-OMe in CDCl3 to values similar to those typically observed in a Cγ-exo ring pucker, as observed in our computational analysis. These ϕ and ψ angles enable an n→π* interaction,3d,3h which increases the value of Ktrans/cis. Indeed, second-order perturbation theory using natural bond orbital analysis21 indicates that the n→π* interaction in the lowest energy hydrogen-bonded, Cγ-endo conformation of Ac-hyp-OMe (Figure 2A) could be worth as much as 1.0 kcal/mol. This same interaction is worth <0.05 kcal/mol in the lowest energy non-hydrogen bonded conformation, Cγ-endo conformation of Ac-hyp-OMe (Figure 2C).
Table 1.
Values of Ktrans/cis and υester for Ac-Pro-OMe, Ac-hyp-OMe, and Ac-mop-OMe
We also used infrared spectroscopy to examine the proline derivatives. An ester carbonyl stretching vibration (υester) decreases with decreasing C=O bond order. In CHCl3, Ac-hyp-OMe has a much lower υester than does either Ac-Pro-OMe or Ac-mop-OMe (Table 1; Figure S1 in the Supporting Information). The n→π* interaction in Ac-hyp-OMe could contribute up to 6 cm−1 to this difference.3c The unusually large magnitude of the shift in υester for Ac-hyp-OMe provides additional support for the existence of a transannular hydrogen bond, which decreases the C=O bond order substantially.
In addition to extracting Ktrans/cis values from 1H NMR spectroscopy, we discerned the dominant ring pucker of Ac-hyp-OMe in solution from the coupling constants between hydrogen atoms on its pyrrolidine ring.22 We could not evaluate its conformation in CDCl3 because of overlapping peaks in its 1H NMR spectrum. In D2O, however, we observed small coupling constants (~2.0 Hz) for H%–Hα, Hβ′–Hγ, and Hβ′–Hδ′ (see: Table S1 in the Supporting Information), which are indicative of a predominantly Cγ-endo conformation.
Crystallographic Analyses of hyp and mop
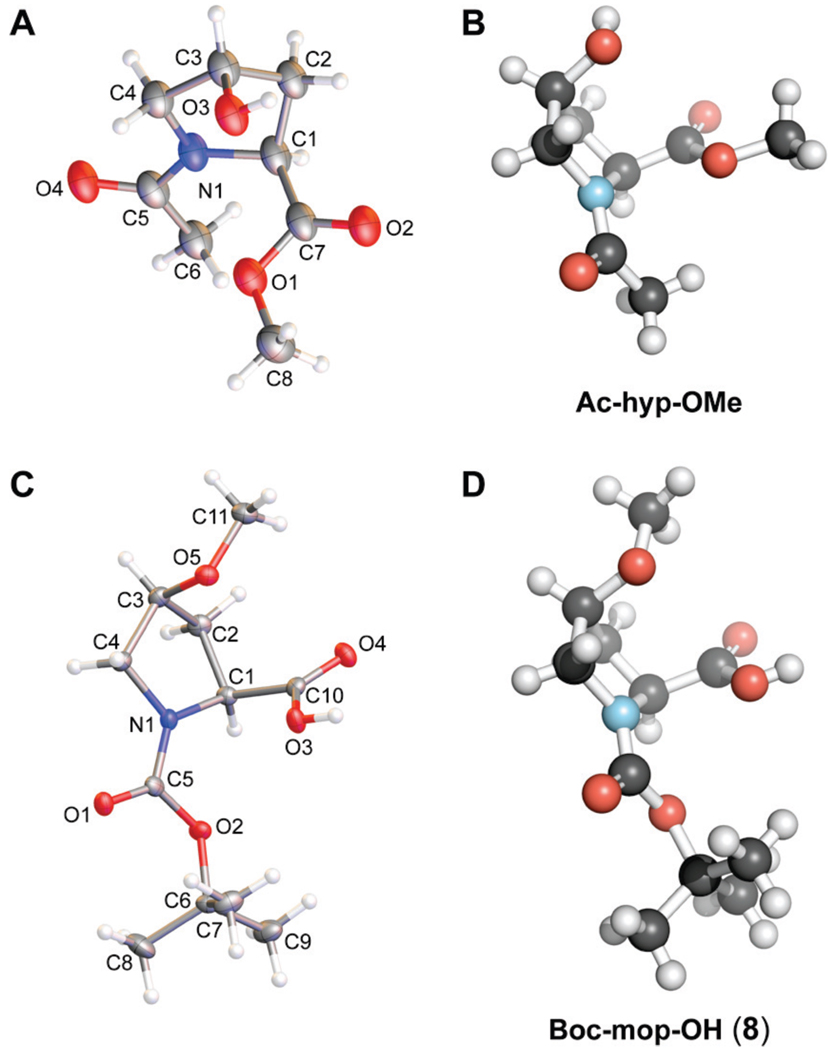
Next, we crystallized Ac-hyp-OMe to explore its conformation in the solid state. The crystal structure of Ac-hyp-OMe (Figures 3A and 3B) displayed a cis peptide bond and the expected Cγ-endo ring pucker. In the solid state, the hydroxyl group formed a hydrogen bond with the amide oxygen of an adjacent molecule in the crystal lattice. A crystal structure of Ac[S]-hyp-OMe (which is the thioamide analog of Ac-hyp-OMe) was similar to that of Ac-hyp-OMe, displaying a Cγ-endo ring pucker but with a trans peptide bond.23
Figure 3.
Crystal structures of hyp and mop derivatives. (A) Molecular drawing of Ac-hyp-OMe with 50% probability ellipsoids. (B) Structure of Ac-hyp-OMe depicted with the program PyMOL. (C) Molecular drawing of Boc-mop-OH (8) with 50% probability ellipsoids. (D) Structure of Boc-mop-OH (8) depicted with the program PyMOL.
Despite an extensive effort, we were unable to obtain a crystal structure of Ac-mop-OMe. Nonetheless, we did obtain a structure of a related molecule, Boc-mop-OH (8) (Figures 3C and 3D). As expected, Boc-mop-OH (8) displayed the Cγ-endo ring pucker in the crystalline state. The methoxy group was oriented away from the ring, perhaps to avoid unfavorable steric interactions with the carboxylate group. A crystal structure of Ac[S]-mop-OMe (which is the thioamide analog of Ac-mop-OMe) reiterates these results, displaying a Cγ-endo ring pucker and main-chain torsion angles typical for that pucker.23
Summary of Data on Ac-hyp-OMe and Ac-mop-OMe
Taken together, our computational, spectroscopic, and crystallographic data suggest that a transannular hydrogen bond in hyp has a strong impact on its conformation. Distortion of the main-chain torsion angles typically adopted by a Pro derivative with Cγ-endo ring pucker results in an anomalously high Ktrans/cis value due to a strengthened n→π* interaction. In contrast, elimination of the transannular hydrogen bond by O-methylation to form mop results in a strong preference for a prototypical Cγ-endo ring pucker with standard main-chain torsion angles. Hence, the O-methylation of hyp residues in proteins could revert the unusual consequences of the transannular hydrogen bond in hyp.
Effects of hyp and mop Residues on the Conformational Stability of a Protein
Collagen provides an appealing system to explore the effects of hyp and mop on protein structure. As described above, collagen triple helices are usually stabilized by Cγ-endo puckered Pro derivatives in the Xaa position, though hyp is known to destabilize the triple helix when in that position.12–13 Collagen-related peptides (CRPs) are widely employed to study the effects of specific amino-acid residues on triple-helix structure and stability.24 Hence, we expected that a set of CRPs with hyp, mop, and Pro in the Xaa position could provide valuable insight on the ability of these residues to modulate protein stability.
Synthesis of (hypProGly)15, (mopProGly)15, and (ProProGly)15
Collagen triple helices with hyp in the Xaa position are notoriously unstable, and have not been characterized to date. Because triple-helix stability is length-dependent, we believed that the 45-residue peptide (hypProGly)15 might form a triple helix of stability sufficient to allow for its detailed characterization.13 Hence, we synthesized (hypProGly)15, (mopProGly)15, and (ProProGly)15.
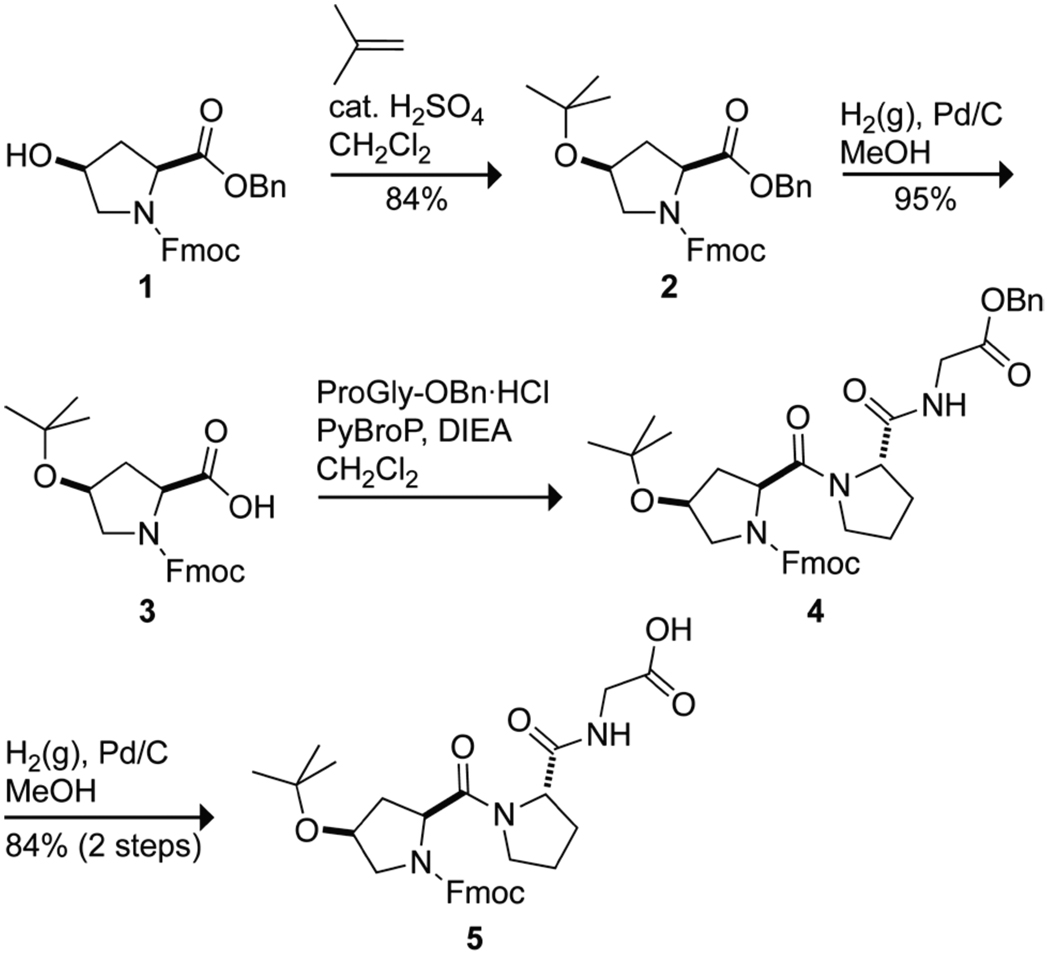
The peptide (hypProGly)15 was synthesized by the condensation of Fmoc-hyp(tBu)ProGly-OH (5) segments on a solid-phase. Tripeptide 5 was synthesized by the route shown in Scheme 1. Briefly, Fmoc-hyp-OBn (1)25 was converted into its O-t-butoxy derivative Fmoc-hyp(tBu)-OBn (2) by treatment with isobutylene and then into the free carboxylic acid Fmoc-hyp(tBu)-OH (3) by hydrogenolysis. PyBroP-mediated coupling with the hydrochloride salt of H-ProGly-OBn26 and hydrogenolysis yielded Fmoc-hyp(tBu)ProGly-OH (5) in 67% overall yield.
Scheme 1.
Synthesis of Fmoc-hyp(tBu)ProGly-OH (5)
The peptide (mopProGly)15 was prepared by segment condensation of Fmoc-mopProGly-OH (11) on a solid-phase. Tripeptide 11 was synthesized by the route shown in Scheme 2. Briefly, Boc-hyp-OBn (6)27 was converted to Boc-mop-OBn (7) by treatment with methyl iodide in the presence of Ag(I) oxide.28 Hydrogenolysis yielded Boc-mop-OH (8). Boc deprotection under acidic conditions followed by treatment with Fmoc-OSu under basic conditions provided Fmoc-mop-OH (9). PyBroP-mediated coupling of 9 and the hydrochloride salt of H-ProGly-OBn and hydrogenolysis of the benzyl ester yielded Fmoc-mopProGly-OH (11) in 25% overall yield.
Scheme 2.
Synthesis of Fmoc-mopProGly-OH (11).
The peptide (ProProGly)15 was synthesized by the segment condensation of Fmoc-ProProGly-OH26 on a solid phase.
Conformational Analyses of (hypProGly)15, (mopProGly)15, and (ProProGly)15
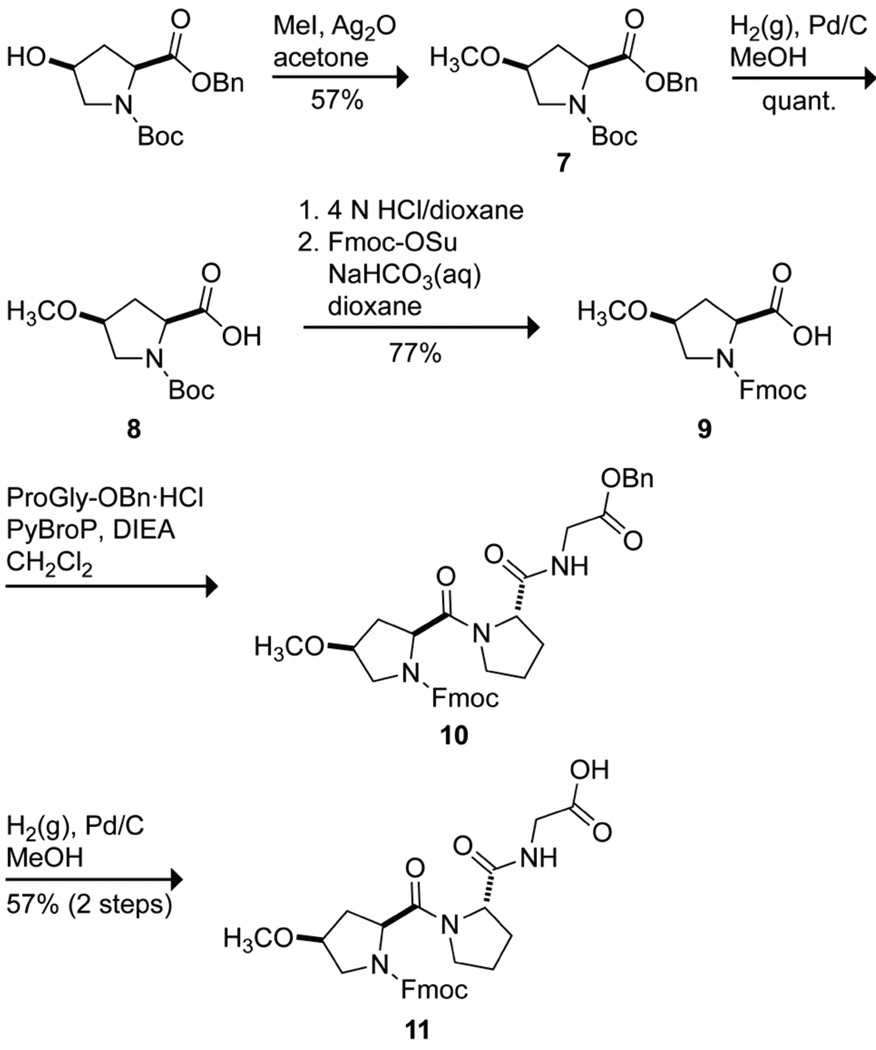
We used circular dichroism (CD) spectroscopy to analyze the conformations of (hypProGly)15, (mopProGly)15, and (ProProGly)15 in solution. The signature CD spectrum of the collagen triple helix has a maximum near 225 nm and a minimum near 210 nm. By this criterion, (hypProGly)15, (mopProGly)15, and (ProProGly)15 form a triple helix at 4 °C (Figure 4A). Upon heating, a cooperative transition of the molar ellipticity at 226 nm, another hallmark of the collagen triple helix, was observed for all three CRPs (Figure 4B). Values of Tm, which is the temperature at the midpoint of the thermal transition between folded and unfolded states, were determined by fitting the data to a two-state model (Table 2).29 We observed that (ProProGly)15 forms a highly stable triple helix with Tm = 65 °C. In contrast, (hypProGly)15 forms a weakly stable triple helix, with Tm = 25 °C. O-Methylation of hyp rescues much of the instability of (hypProGly)15, yielding a triple helix with Tm = 47 °C. Sedimentation equilibrium experiments confirmed the self-assembly of (hypProGly)15, (mopProGly)15, and (ProProGly)15 at 4 °C and the complete disassembly of (hypProGly)15 at 40 °C (see: Figure S2 in the Supporting Information).
Figure 4.
Conformational analysis of (hypProGly)15, (mopProGly)15, and (ProProGly)15 by CD spectroscopy. (A) Spectra of peptide solutions (0.2 mM in 50 mM acetic acid) incubated at ≤4 °C for ≥24 h. (B) Effect of temperature on the molar ellipticity at 226 nm. Data were recorded at intervals of 3 °C after equilibration for ≥5 min.
Table 2.
Thermodynamic Parameters for Folding of Triple-Helical Collagen-Related Peptides
| peptide | Tma | ΔH (kcal/mol)b | –TΔS (kcal/mol)b | ΔG (kcal/mol)b |
|---|---|---|---|---|
| (hypProGly)15 | 25 | −29.9 | 20.5 | −9.4 |
| (mopProGly)15 | 47 | −24.7 | 13.1 | −11.6 |
| (ProProGly)15 | 65 | −32.8 | 19.0 | −13.8 |
Values (±1 °C) were determined in triplicate by CD spectroscopy.
Values were estimated by DSC and reported here at 46 °C. The largest source of error (±5%) was in the determination of [peptide].
The increased stability of (mopProGly)15 triple helices relative to (hypProGly)15 triple helices is likely due to two factors: First, the carbonyl moiety of hyp required to form the essential interstrand hydrogen bond in the collagen triple helix (Figure 1C), is distracted by an intramolecular transannular hydrogen bond with the hydroxyl moiety of hyp (Figure 1B). This hydrogen bond is less accessible to solvent than is the transannular hydrogen bond in Ac-hyp-OMe. O-Methylation removes the transannular hydrogen bond, thereby strengthening the interstrand hydrogen bond. Second, our computational analysis of Ac-hyp-OMe suggests that its transannular hydrogen bond results in peptide main-chain torsion angles inconsistent with the Xaa position of the collagen triple helix. This deleterious preorganization substantially reduces the stability of a (hypProGly)15 triple helix. In contrast, O-methylation results in proper preorganization.
Thermodynamic Analyses of (hypProGly)15, (mopProGly)15, and (ProProGly)15
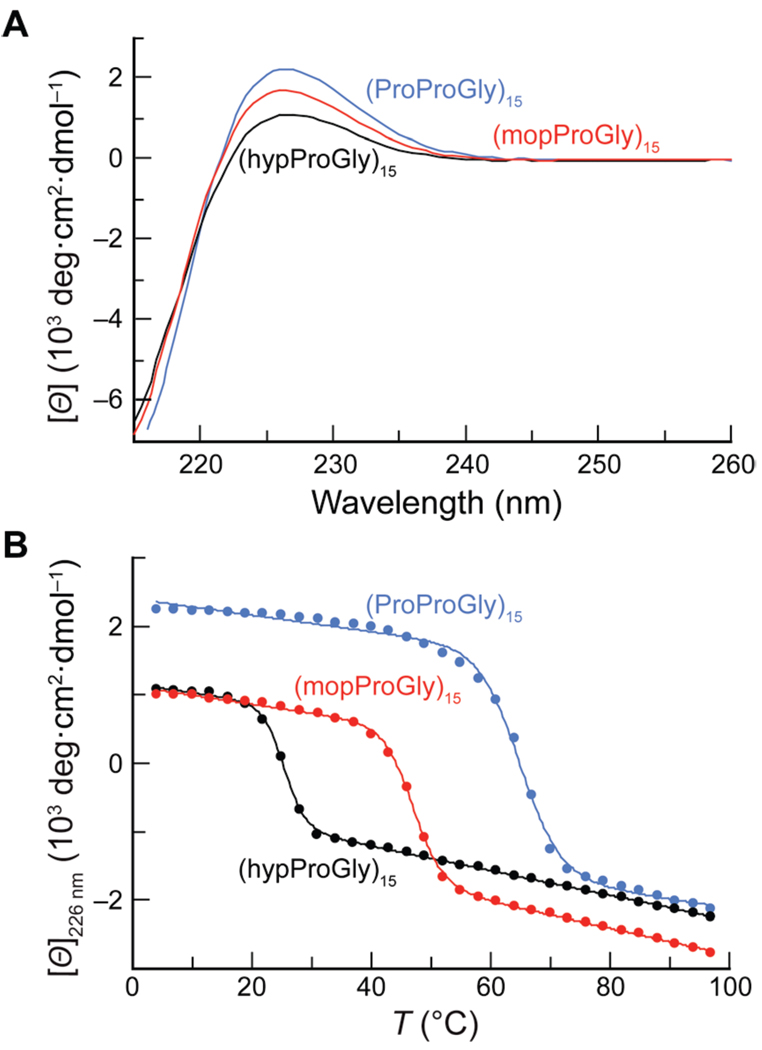
Given the seemingly favorable consequences of hyp O-methylation for triple-helix stabilization, we were surprised to observe that a (mopProGly)15 triple helix is less stable than a (ProProGly)15 triple helix. To determine the thermodynamic basis for the relative stabilities of (hypProGly)15, (mopProGly)15, and (ProProGly)15 triple helices, we analyzed their thermal denaturation by differential scanning calorimetry (DSC) (Figure 5). Thermodynamic parameters were obtained as described in the Experimental section, and are listed in Table 2.
Figure 5.
DSC scans of (hypProGly)15 (122 µM), (mopProGly)15 (122 µM), and (ProProGly)15 (96 µM) in 50 mM HOAc (pH 3.0; scan rate = 6 °C/h).
Triple-helix formation is made difficult by the need to assemble three strands and the absence of a substantial hydrophobic core to stabilize the resulting assembly.22c These factors account for the strongly unfavorable ΔS value observed for all three triple helices (Table 2). It is noteworthy that the most stable triple helix, that formed by (ProProGly)15, also has the most favorable ΔH value. The entropic cost for the folding of (ProProGly)15 is between that for (hypProGly)15 and (mopProGly)15.
The striking instability of (hypProGly)15 relative to (ProProGly)15 derives from both enthalpic and entropic effects. The less favorable ΔH value of (hypProGly)15 could result from the transannular hydrogen bond in hyp (Figure 1B) reducing the favorable enthalpy from the interstrand hydrogen bond in the triple helix (Figure 1C). This reduction in the value of ΔH upon 4S-hydroxylation of Pro in the Xaa position of (ProProGly)n is in stark contrast to the highly enthalpically favorable effect of 4R-hydroxylation of Pro in the Yaa position of (ProProGly)n.4,17,30 The latter result is due to the enthalpically favorable (but entropically disfavorable) hydration of the hydroxyl moiety of Hyp in the Yaa position of folded triple helices. We might anticipate an even greater reduction in the value of ΔH for (hypProGly)15 relative to (ProProGly)15 due to disruption of the interstrand hydrogen bond, but similar hydration of the hydroxyl moiety in hyp could compensate for a reduced value of ΔH (but, once again, at the expense of entropy). The instability of (hypProGly)15 triple helices is also due, in part, to another increase in the entropic cost for their formation. The hyp residues are preorganized poorly for the Xaa position of the triple helix, as the transannular hydrogen bond disrupts the main-chain torsion angles that accompany a normal Cγ-endo ring pucker.
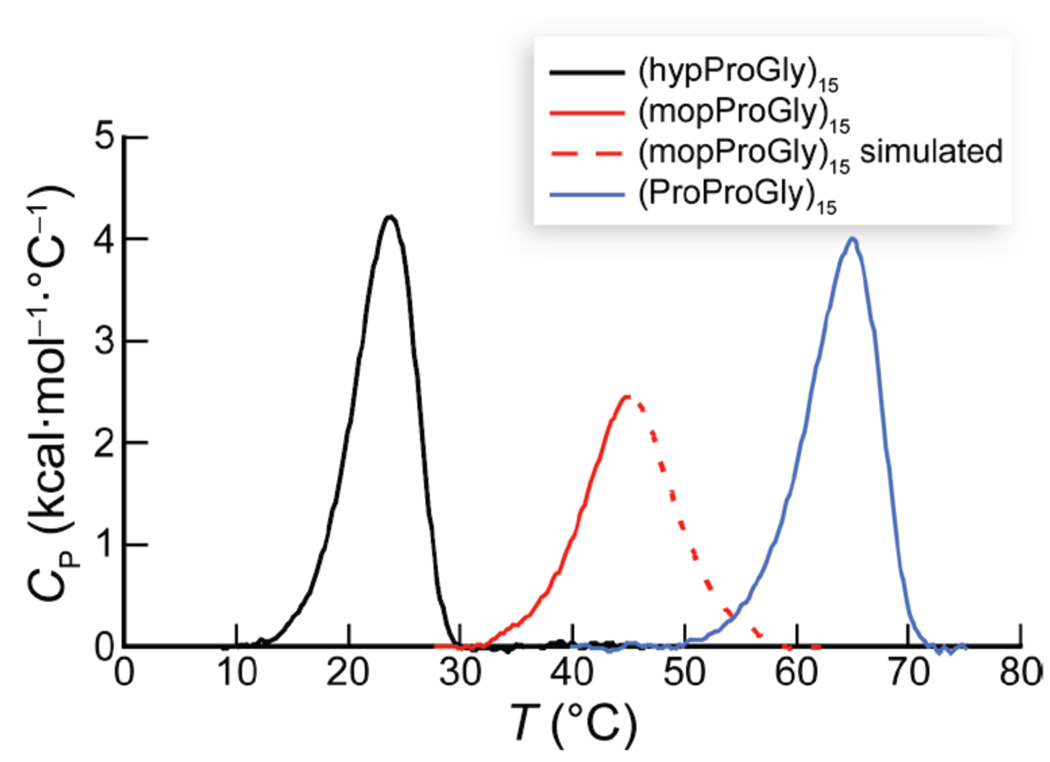
Previously, we showed that (ProMopGly)n strands form much more stable triple helices than do (ProProGly)n strands.17 This hyperstability is the result of an entropic advantage that likely arises from preorganization. Like (ProMopGly)n triple helices, (mopProGly)n triple helices have a more favorable entropy of formation than do (ProProGly)n triple helices (Table 2). Yet, (mopProGly)n triple helices are less stable than (ProProGly)n triple helices due to a less favorable enthalpy of formation. To understand the dichotomy between the relative stabilities of (mopProGly)n and (ProMopGly)n triple helices, we resorted to molecular modeling. Our models show that the 4R-methoxy group of a Mop residue in the Yaa position does not interact unfavorably with any neighboring atoms in a folded triple helix (Figure 6A). In contrast, the 4S-methoxy group of a mop residue in the Xaa position is sterically hindered (Figure 6B). In a triple helix, the carbonyl group of the residue in the Xaa position engages the N–H of a Gly residue in another strand (Figure 1C). The unfavorable steric interaction of the methoxy residue in a (mopProGly)n triple helix with the neighboring strand (Figure 6B) could weaken this essential interstrand hydrogen bond, leading to the reduced enthalpy of triple-helix formation for (mopProGly)15 relative to (hypProGly)15 and (ProProGly)15. This competing enthalpic effect is likely to be the reason that triple helices formed from (mopProGly)15 are less stable than (ProProGly)15 triple helices, despite their apparently high degree of preorganization.
Figure 6.
Space-filling models of a segment of a (ProMopGly)n and (mopProGly)n triple helix. Models were constructed from the three-dimensional structure of a (ProProGly)n triple helix (PDB entry 1k6f10) by replacing the appropriate hydrogen with a methoxy group in its least hindered conformation using the program SYBYL (Tripos, St. Louis, MO). Models were depicted with the program PyMOL.
Further evidence that sterics underlie the reduced conformational stability of (mopProGly)15 triple helices relative to (ProProGly)15 triple helices derives from earlier findings for triple helices with (2S,4S)-4-chloroproline (clp) in the Xaa position of the triple helix.31 Despite the tendency of clp to adopt the Cγ-endo ring pucker, a (clpProGly)10 triple helix is slightly less stable than a (ProProGly)10 triple helix. The destabilization arising from the bulky 4S-chloro group is consistent with a steric basis for the slightly reduced stability of (mopProGly)15 triple helices relative to (ProProGly)15.
Implications for Other Proteins
Our findings have important implications for the burgeoning application of Pro derivatives. hyp is the only known Pro derivative that can adopt a Cγ-endo ring pucker concomitant with a high trans:cis ratio. As such, hyp or related Pro derivatives that can engage in a transannular hydrogen bond could be the only Cγ-endo puckered Pro derivatives that form stable polyproline II-type helices, which usually require Pro derivatives with a Cγ-exo ring pucker. In contrast, mop, like flp and clp, adopts a Cγ-endo ring pucker with prototypical main-chain torsion angles and a low trans:cis ratio.
4R-Hydroxylation inverts the inherent preference of Pro for a Cγ-endo ring pucker3d and preorganizes the main-chain torsion angles that accompany a Cγ-exo ring pucker. In contrast, 4S-hydroxylation results in a derivative with the main-chain torsion angles mandated by 4R-hydroxylation but a diminished capacity to accept a hydrogen bond (Figure 1B). It is interesting to speculate that the evolution of prolyl 3S-hydroxylases was favored by their ability to enforce the Cγ-endo ring pucker in proline residues.32 These enzymes contain the key active-site residues of prolyl 4R-hydroxylases,33 and likewise install a hydroxyl group on the face of the pyrrolidine ring opposite from the carboxyl group, averting transannular hydrogen-bond formation. Collagen prolyl 3R-hydroxylases, like collagen prolyl 4S-hydroxylases, have not been identified in natural proteomes.
Finally, we note a general implication of our data. In the Yaa position, a Mop residue is more beneficial than a Hyp residue to triple-helix stability.17 Likewise, in the Xaa position, mop is more beneficial than hyp (Figures 4 and 5; Table 2). These parallel findings have a common origin with important ramifications—hydrogen bonding can compromise the benefits of an underlying stereoelectronic effect to the conformational stability of a protein.
Experimental Section
General
Commercial chemicals were of reagent grade or better, and were used without further purification. Anhydrous CH2Cl2 was obtained from a CYCLE-TAINER® solvent delivery system (J. T. Baker, Phillipsburg, NJ). In all reactions involving anhydrous solvents, glassware was either oven- or flame-dried. NaHCO3(aq) and brine (NaCl) refer to saturated aqueous solutions. Flash chromatography was performed with columns of silica gel 60, 230–400 mesh (Silicycle, Québec City, Canada). Semi-preparative HPLC was performed at 60 °C with a Macherey–Nagel C-8 reversed-phase column after preheating peptides to 70 °C for ≥30 min to disassemble any oligomers. Analytical HPLC was performed with a Varian C-18 reversed-phase column. HPLC purifications and analyses employed linear gradients of solvent A (H2O containing 0.1% v/v TFA) and solvent B (CH3CN containing 0.1% v/v TFA).
The term “concentrated under reduced pressure” refers to the removal of solvents and other volatile materials using a rotary evaporator at water aspirator pressure (<20 torr) while maintaining the water-bath temperature below 40 °C. Residual solvent was removed from samples at high vacuum (<0.1 torr). The term “high vacuum” refers to vacuum achieved by a mechanical belt-drive oil pump.
NMR spectra were acquired with a Bruker DMX-400 Avance spectrometer (1H, 400 MHz; 13C, 100.6 MHz) at the National Magnetic Resonance Facility at Madison (NMRFAM). NMR spectra were obtained at ambient temperatures on samples dissolved in CDCl3, unless noted otherwise. Mass spectrometry was performed with either a Micromass LCT (electrospray ionization, ESI) in the Mass Spectrometry Facility in the Department of Chemistry or an Applied Biosystems Voyager DE-Pro (matrix-assisted laser desorption/ionization, MALDI) mass spectrometer in the University of Wisconsin Biophysics Instrumentation Facility (BIF). The infrared spectrum of Ac-hyp-OMe was acquired with a Bruker Equinox 55 FTIR.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-tert-butoxyproline Benzyl Ester (2)
Fmoc-hyp-OBn (1) (1.25 g, 2.8 mmol), described previously,25 was dissolved in anhydrous CH2Cl2 (20 mL) and cooled to −78 °C. Concentrated H2SO4 (30 µL) was added followed by isobutylene (~20 mL, ~220 mmol, condensed at −78 °C). The resulting solution was capped with a rubber septum, sealed with copper wire around the neck of the round-bottom flask, and allowed to warm slowly to room temperature with stirring. After 5 d, the solution was cooled to −78 °C, and the septum was removed to allow slow evaporation of isobutylene. Upon warming to room temperature, the solution was concentrated under reduced pressure. The crude product was purified by flash chromatography over silica gel (40% v/v EtOAc in hexanes) to afford Fmoc-hyp(tBu)-OBn (2) (1.18 g, 84%) as a colorless oil. 1H NMR δ: 1.14 and 1.17 (s, 9H), 2.03–2.13 (m, 1H), 2.33–2.48 (m, 1H), 3.33–3.44 (m, 1H), 3.71–3.82 (m, 1H), 3.98–4.55 (m, 5H), 4.97–5.29 (m, 2H), 7.19–7.44 (m, 9H), 7.47–7.64 (m, 2H), 7.70–7.79 (m, 2H); 13C NMR δ: 28.3, 37.9, 38.9, 47.3, 53.5, 53.9, 57.7, 57.9, 66.9, 66.9, 67.6, 68.7, 69.6, 74.2, 120.0, 120.1, 125.1, 125.3, 125.5, 127.2, 127.8, 128.2, 128.3, 128.4, 128.6, 135.8, 135.9, 141.4, 141.4, 141.5, 143.8, 144.4, 154.6, 155.0, 171.8, 172.0; ESI–EMM (m/z): [M + Na]+ calcd for C31H33NO5Na 522.2256; found 522.2231.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-tert-butoxyproline (3)
MeOH (100 mL) was added carefully to a mixture of Fmoc-hyp(tBu)-OBn (2) (1.79 g, 3.6 mmol) and Pd/C (10% w/w, 0.38 g) under Ar(g), and the resulting black suspension was stirred under H2(g) for 45 min. Careful monitoring by thin-layer chromatography (TLC) was necessary to prevent hydrogenolysis of the Fmoc group. The suspension was filtered through a pad of Celite and concentrated under reduced pressure. The crude product was purified by flash chromatography over silica gel (95% v/v CH2Cl2 in MeOH) to afford Fmoc-hyp(tBu)-OH (3) (1.39 g, 95%) as a white solid. 1H NMR (MeOH-d4) δ: 1.18 (s, 9H), 1.90–2.08 (m, 1H), 2.35–2.50 (m, 1H), 3.17 and 3.25 (dd, J = 4.5, 11.0, 1H), 4.14–4.46 (m, 5H), 7.26–7.43 (m, 4H), 7.57–7.68 (m, 2H), 7.75–7.83 (m, 2H); 13C NMR (MeOH-d4) δ: 28.5, 38.8, 39.7, 48.4, 54.5, 55.0, 58.8, 68.6, 69.0, 69.9, 70.6, 75.2, 120.9, 126.1, 126.2, 128.1, 128.8, 142.5, 142.6, 142.7, 145.1, 145.4, 145.4, 156.5, 156.7, 175.5, 175.7; ESI–EMM (m/z): [M + Na]+ calcd for C24H27NO5Na 432.1787; found 432.1800.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-tert-butoxyprolyl-(2S)-prolylglycine Benzyl Ester (4)
A solution of Fmoc-hyp(tBu)-OH (3) (1.31 g, 3.2 mmol) and the hydrochloride salt of H-ProGly-OBn (3.60 g, 12.0 mmol, prepared as described previously26) in anhydrous CH2Cl2 (160 mL) was cooled to 0 °C. PyBroP (1.49 g, 3.2 mmol) and DIEA (3.31 g, 25.6 mmol) were added, and the resulting solution was allowed to warm slowly to room temperature and then stirred for 15 h. The reaction mixture was washed with 10% w/v aqueous citric acid (60 mL), NaHCO3(aq) (60 mL), and brine (60 mL). The organic layer was dried over anhydrous MgSO4(s), and concentrated under reduced pressure. The crude residue was purified by flash chromatography over silica gel (gradient: 30% v/v EtOAc in hexanes to 100% v/v EtOAc) to afford Fmoc-hyp(tBu)ProGly-OBn (4) (1.74 g) as a white solid containing a slight impurity that was removed after the succeeding step. ESI–EMM (m/z): [M + Na]+ calcd for C38H43N3O7Na 676.2999; found 676.2982.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-tert-butoxyprolyl-(2S)-prolylglycine (5)
MeOH (100 mL) was added carefully to a mixture of Fmoc-hyp(tBu)ProGly-OBn (4) (1.74 g, 2.7 mmol) and Pd/C (10% w/w, 0.32 g) under Ar(g), and the resulting black suspension was stirred under H2(g) for 2 h. Careful monitoring by TLC was necessary to prevent hydrogenolysis of the Fmoc group. The suspension was filtered through a pad of Celite and concentrated under reduced pressure. The crude product was purified by flash chromatography over silica gel (CH2Cl2 to elute byproducts, then 3% v/v MeOH in CH2Cl2 containing 0.1% v/v formic acid). The fractions containing the reaction product were concentrated under reduced pressure and the formic acid was removed by dissolving the residue in 10% v/v MeOH in toluene and concentrating under reduced pressure to afford Fmoc-hyp(tBu)ProGly-OH (5) (1.27 g, 84%, two steps) as a white solid. 1H NMR (MeOH-d4) δ: 1.16 and 1.19 (s, 9H), 1.61–2.23 (m, 5H), 2.33–2.58 (m, 1H), 2.93–3.10 (m, 1.4H), 3.33–3.42 (m, 0.5H) 3.47–3.67 (m, 1.3H), 3.68–3.86 (m, 1.7H), 3.90–4.03 (m, 1H), 4.04–4.53 (m, 5.5H), 4.66–4.74 (m, 0.5H), 7.25–7.45 (m, 4H), 7.53–7.66 (m, 2H), 7.75–7.85 (m, 2H); 13C NMR (MeOH-d4) δ: 22.9, 23.1, 25.7, 25.8, 28.5, 28.6, 30.1, 30.2, 32.5, 33.2, 37.5, 37.7, 38.3, 41.7, 42.1, 47.8, 53.4, 53.7, 47.3, 57.8, 58.1, 67.5, 68.5, 68.7, 69.7, 70.3, 75.0, 75.1, 75.3, 120.9, 125.7, 126.1, 128.2, 128.2, 128.3, 128.8., 142.6, 142.7, 144.9, 145.1, 145.5, 145.7, 156.0, 156.5, 172.6, 172.9, 173.0, 173.3, 174.0, 174.5, 174.7; ESI–EMM (m/z): [M – H]− calcd for C31H36N3O7 562.2558; found 562.2556.
N-tert-Butoxycarbonyl-(2S,4S)-4-methoxyproline benzyl ester (7)
Boc-hyp-OBn (6) (6.94 g, 21.6 mmol), prepared as described previously,27 was dissolved in anhydrous acetone (150 mL) under Ar(g). MeI (10.73 g, 75.6 mmol) was added, followed by Ag2O (16.01 g, 69.1 mmol). The resulting suspension was stirred at room temperature for 24 h. The suspension was filtered and concentrated under reduced pressure. The residue was dissolved again in anhydrous acetone (150 mL), MeI (10.73 g, 75.6 mmol) and Ag2O (16.01 g, 69.1 mmol) were added, and the resulting suspension was stirred at room temperature for 24 h. The suspension was filtered, and concentrated under reduced pressure. Flash chromatography over silica gel (50% v/v EtOAc in hexanes) afforded Boc-mop-OBn (7) (4.13 g, 57%) as a colorless oil. Unreacted starting material Boc-hyp-OBn (6) was also recovered. 1H NMR δ: 1.35 and 1.46 (s, 9H), 1.57–1.72 (m, 0.3H), 2.11–2.40 (m, 1.7H), 3.16 and 3.20 (s, 3H), 3.26–3.74 (m, 2H), 3.85–3.94 (m, 1H), 4.31–4.52 (m, 1H), 5.03–5.32 (m, 2H), 7.28–7.39 (m, 5H); 13C NMR δ: 28.4, 28.5, 34.7, 35.9, 51.2, 52.0, 56.5, 56.6, 57.5, 57.9, 66.8, 78.1, 79.1, 80.1, 80.2, 128.2, 128.3, 128.4, 128.5, 128.7, 135.9, 136.1, 154.0, 172.0, 172.3; ESI–EMM (m/z): [M + H]+ calcd for C18H26NO5 336.1806; found 336.1800.
N-9-tert-Butoxycarbonyl-(2S,4S)-4-methoxyproline (8)
MeOH (100 mL) was added carefully to a mixture of Boc-mop-OBn (7) (1.80 g, 5.4 mmol) and Pd/C (10% w/w, 1.00 g) under Ar(g), and the resulting black suspension was stirred under H2(g) for 14 h. The suspension was filtered through a pad of Celite and concentrated under reduced pressure to afford Boc-mop-OH (8) (1.32 g, quant.) as a white solid. 1H NMR (DMSO-d6) δ: 1.34 and 1.40 (s, 9H), 1.93–2.03 (m, 1H), 2.21–2.42 (m, 1H), 3.17 (s, 3H), 3.17–3.24 (m, 1H), 3.47–3.59 (m, 1H), 3.85–3.96 (m, 1H), 4.08–4.20 (m, 1H); 13C NMR δ: 28.4, 32.3, 35.4, 51.5, 53.2, 56.5, 56.8, 78.7, 81.1, 82.1, 154.2, 157.0, 173.1, 175.1; ESI–EMM (m/z): [M + Na]+ calcd for C11H19NO5Na 268.1161; found 268.1161.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-methoxyproline (9)
Boc-mop-OH (8) (2.15 g, 8.8 mmol) was dissolved in 4 N HCl in dioxane (150 mL), and the resulting solution was stirred at room temperature under Ar(g) for 2.5 h. The reaction mixture was concentrated under reduced pressure and dried briefly under high vacuum. The resultant white solid was dissolved in saturated NaHCO3(aq) (75 mL). A solution of Fmoc-OSu (2.96 g, 8.8 mmol) in dioxane (75 mL) was added, and the resulting white suspension was stirred for 20 h. The reaction mixture was concentrated under reduced pressure, and the residue was diluted with water (150 mL) and washed with ether (3 × 150 mL). The aqueous layer was acidified to pH 1.5 with 2 N HCl, extracted with ether (3 × 150 mL), dried over anhydrous MgSO4(s), and concentrated under reduced pressure to afford Fmoc-mop-OH (9) (2.50 g, 77%) as a white solid. 1H NMR (DMSOd6) δ: 2.05–2.43 (m, 2H), 3.30–3.37 (m, 1H), 3.55–3.62 (m, 1H), 3.92–4.01 (m, 1H), 4.14–4.41 (m, 4H), 7.28–7.47 (m, 4H), 7.61–7.71 (m, 2H), 7.86–7.94 (m, 2H); 13C NMR (DMSO-d6) δ: 34.00, 35.1, 46.6, 46.7, 50.9, 51.3, 55.9, 57.2, 57.4, 66.6, 66.9, 77.5, 78.4, 120.1, 125.2, 125.3, 127.2, 127.7, 140.7, 140.8, 143.8, 154.0, 172.6, 173.0; ESI–EMM (m/z): [M + Na]+ calcd for C21H21NO5Na 390.1317; found 390.1134.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-methoxyprolyl-(2S)-prolylglycine Benzyl Ester (10)
A solution of Fmoc-mop-OH (9) (2.45 g, 6.7 mmol) and the hydrochloride salt of H-ProGly-OBn (3.59 g, 12.0 mmol, prepared as described previously26) in anhydrous CH2Cl2 (160 mL) was cooled to 0 °C. PyBroP (3.12 g, 6.7 mmol) and DIEA (3.46 g, 26.8 mmol) were added. The resulting solution was allowed to warm slowly to room temperature and then stirred for 15 h. The reaction mixture was washed with 10% w/v aqueous citric acid (60 mL), NaHCO3(aq) (60 mL), and brine (60 mL). The organic layer was dried over anhydrous MgSO4(s), and concentrated under reduced pressure. The crude residue was purified by flash chromatography over silica gel (gradient: 50% v/v EtOAc in hexanes to 100% v/v EtOAc) to afford Fmoc-mopProGly-OBn (10) (3.23 g) as a white solid containing a slight impurity that was removed after the succeeding step. ESI–EMM (m/z): [M + Na]+ calcd for C35H37N3O7Na 634.2529; found 634.2527.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-methoxyprolyl-(2S)-prolylglycine (11)
MeOH (100 mL) was added carefully to a mixture of Fmoc-mopProGly-OBn (10) (3.23 g, 5.3 mmol) and Pd/C (10% w/w, 0.63 g) under Ar(g), and the resulting black suspension was stirred under H2(g) for 45 min. Careful monitoring by TLC was necessary to prevent hydrogenolysis of the Fmoc group. The suspension was filtered through a pad of Celite and concentrated under reduced pressure. The crude product was purified by flash chromatography over silica gel (CH2Cl2 to elute byproducts, then 5% v/v MeOH in CH2Cl2 containing 0.1% v/v formic acid). The fractions containing the reaction product were concentrated under reduced pressure, and the formic acid was removed by dissolving the residue in 10% v/v MeOH in toluene and concentrating under reduced pressure to afford Fmoc-mopProGly-OH (11) (1.98 g, 57%, two steps) as a white solid. 1H NMR (DMSO-d6) δ: 1.58–2.04 (m, 4H), 2.55–2.70 (m, 1H), 2.99–3.15 (m, 1H), 3.20 and 3.23 (s, 3H), 3.23–3.26 (m, 1H), 3.31–4.02 (m, 7.7H), 4.09–4.17 (m, 0.3H), 4.19–4.66 (m, 4H), 7.26–7.47 (m, 4H), 7.51–7.70 (m, 2H), 7.84–8.07 (m, 2H), 12.53 (bs, 1H); 13C NMR (MeOH-d4) δ: 25.8, 25.9, 30.0, 30.1, 35.2, 35.8, 41.8, 47.9, 49.9, 52.5, 52.8, 57.3, 57.4, 58.1, 58.5, 61.3, 61.6, 67.6, 68.8, 78.9, 79.6, 120.9, 125.7, 125.9, 126.2, 128.2, 128.9, 142.6, 142.7, 145.0, 145.1, 145.4, 145.7, 156.2, 156.7, 172.6, 172.7, 172.8, 174.5, 174.7; ESI–EMM (m/z): [M + Na]+ calcd for C28H31N3O7Na 544.2060; found 544.2065.
N-Acetyl-(2S,4S)-4-methoxyproline Methyl Ester
Ac-hyp-OMe (0.34 g, 1.8 mmol), described previously,3c was dissolved in anhydrous acetone (50 mL) under Ar(g). MeI (1.14 g, 8.0 mmol) was added, followed by Ag2O (0.94 g, 7.4 mmol). The resulting suspension was stirred at room temperature for 24 h. The suspension was filtered and evaporated under reduced pressure. The resulting residue was dissolved in EtOAc (100 mL), washed with water (2 × 100 mL), dried over anhydrous MgSO4(s), and concentrated under reduced pressure. Flash chromatography over silica gel (gradient: 0% v/v MeOH in CH2Cl2 to 10% v/v MeOH in CH2Cl2) afforded Ac-mop-OH (0.25 g, 69%) as a colorless oil. 1H NMR δ: 2.04 and 2.10 (s, 3H), 2.19–2.28 (m, 1H), 2.30–2.36 (m, 0.65H), 2.56–2.63 (m, 0.35H), 3.25 and 3.30 (s, 3H), 3.55–3.71 (m, 2H), 3.72 and 3.76 (s, 3H), 3.91–4.05 (m, 1H), 4.37–4.41 (m, 0.35H), 4.65 (dd, J = 3.5, 8.7 Hz, 0.65H); 13C NMR δ: 22.2, 22.4, 34.2, 36.3, 51.7, 52.5, 52.8, 53.1, 56.3, 56.8, 57.0, 59.0, 77.8, 79.2, 105.2, 169.7, 170.3, 172.0; ESI–EMM (m/z): [M + H]+ calcd for C9H16NO4 202.1074; found 202.1070.
Attachment of Fmoc-hyp(tBu)ProGly-OH (5) to 2-Chlorotrityl Resin
Under Ar(g), 80 mg of 2-chlorotrityl chloride resin (loading: 1.7 mmol/g) was swelled in anhydrous CH2Cl2 (0.7 mL) for 5 min. A solution of compound 5 (75 mg, 0.13 mmol) and DIEA (32 mg, 0.25 mmol) in anhydrous CH2Cl2 (1.3 mL) was added by syringe. Additional anhydrous CH2Cl2 (1.3 mL) was used to ensure complete transfer of 5. After 2 h, anhydrous MeOH (0.6 mL) was added to cap any remaining active sites on the resin. The resin-bound peptide was isolated by gravity filtration, washed with several portions of anhydrous CH2Cl2 (~25 mL), and dried under high vacuum. The mass of the resin-bound peptide was 90 mg. Loading was measured by ultraviolet spectroscopy34 to be 0.36 mmol/g.
Attachment of Fmoc-mopProGly-OH (11) and Fmoc-ProProGly-OH to 2-Chlorotrityl Resin
Fmoc-tripeptide 11 and Fmoc-ProProGly-OH were loaded onto 2-chlorotrityl resin in similar fashion to that described for 5. Loadings were measured by ultraviolet spectroscopy to be 0.32 mmol/g for 11 and 0.35 mmol/g for Fmoc-ProProGly-OH.34
Synthesis of (hypProGly)15, (mopProGly)15, and (ProProGly)15
The three 45-mer peptides were synthesized by the segment condensation of their corresponding Fmoc-tripeptides (5, 11, and Fmoc-ProProGly-OH) on a solid phase using an Applied Biosystems Synergy 432A Peptide Synthesizer at the University of Wisonsin–Madison Biotechnology Center. The first trimer was loaded onto the resin as described above. Fmoc-deprotection was achieved by treatment with 20% (v/v) piperidine in DMF. The trimers (3 equivalents) were converted to active esters by treatment with HBTU, DIEA, and HOBt. Double extended couplings (60–90 min) were employed at room temperature for all couplings except the first three couplings in the synthesis of (hypProGly)15, in which single extended couplings were employed. Peptides were cleaved from the resin in 95:3:2 TFA:triisopropylsilane:H2O (1.5 mL), precipitated from methyl t-butyl ether at 0 °C, and isolated by centrifugation. Semi-preparative HPLC was used to purify the peptides (hypProGly)15 (10% B for 5 min, increasing to 35% B over 45 min), (mopProGly)15 (gradient: 10% B to 70% B over 50 min), and (ProProGly)15 (gradient: 10% B to 70% B over 50 min). All three peptides were >90% pure by analytical HPLC and MALDI–TOF mass spectrometry (m/z) [M + Na]+ calcd for C180H257N45O61Na 4048.8, found 4048.6 for (hypProGly)15, calcd for C195H287N45O61Na 4260.1, found 4258.0 for (mopProGly)15; calcd for C180H257N45O46Na 3809.9, found 3809.5 for (ProProGly)15.
Circular Dichroism Spectroscopy of (hypProGly)15, (mopProGly)15, and (ProProGly)15
Peptides were dried under high vacuum for at least 24 h before being weighed and dissolved to 0.2 mM in 50 mM HOAc(aq) (pH 2.9). The solutions were incubated at ≤4 °C for ≥24 h before CD spectra were acquired with an Aviv Associates (Lakewood, NJ) 202SF CD spectrophotometer. Spectra were measured with a 1-nm band pass in cuvettes with a 0.1-cm pathlength. The signal was averaged for 5 s during wavelength scans and denaturation experiments using a 0.6 °C temperature deadband. During denaturation experiments, CD spectra were acquired at intervals of 3 °C. At each temperature, solutions were equilibrated for 5 min before data acquisition. Values of Tm were determined in triplicate by fitting the molar ellipticity at 225 nm to a two-state model.29
Differential Scanning Calorimetry and Thermodynamic Analyses of (hypProGly)15, (mopProGly)15, and (ProProGly)15
DSC measurements were made with a VP-DSC instrument (MicroCal, LLC, Northampton, MA). Instrument baselines were established by filling both the sample and the reference cells with degassed 50 mM HOAc(aq) and scanning from 5–98 °C at a scan rate of 6 °C/h with a 10 h pre-equilibration period, until the baseline was reproducible. The final buffer–buffer scan was used as the baseline for subsequent peptide scans. Peptide solutions (~100 µM in 50 mM HOAc(aq)) were incubated at 4 °C for ≥24 h prior to degassing and loading in the sample cell (the reference cell solution was not replaced) during the cooldown from the previous buffer-buffer scan (at ~10 °C). Samples were scanned from 5–98 °C at a scan rate of 6 °C/h with a 10-h pre-equilibration period; the first scan of each sample was used in the analysis. Subsequent scans of (mopProGly)15 and (ProProGly)15 showed high reversibility with >90% recovery. (hypProGly)15 folds significantly more slowly than (mopProGly)15 and (ProProGly)15, but data could be reproduced reliably by re-incubating the sample at 4 °C for ≥24 h, loading the solution in the sample cell, and re-scanning from 5–98 °C. After DSC measurements, peptide concentrations were determined by quantitative amino acid analysis (Protein Chemistry Core, Biomolecular Resource Facility, University of Texas Medical Branch). Peptide concentrations of 122, 122, and 96 µM were obtained for (hypProGly)15, (mopProGly)15, and (ProProGly)15, respectively.
Data were processed using the MicroCal software in the Origin 7 program (OriginLab, Northampton, MA). The appropriate reference scan was subtracted from the first sample scan and the data were normalized to monomer concentrations. For (hypProGly)15 and (ProProGly)15, a progress baseline was subtracted from the data, yielding the traces shown in Figure 5. Although its transition was fully reversible, (mopProGly)15 exhibited a strong exotherm likely due to peptide aggregation beginning at ~50 °C, which prevented determination of a high-temperature baseline (see: Figure S3 in the Supporting Information). A linear baseline based on the low temperature data was therefore subtracted from the data and only the first half of the endotherm was used to extract thermodynamic parameters. We also evaluated the transition of (mopProGly)15 at a second concentration, which produced equivalent results and confirmed the validity of our approach.
Values of ΔH (per mole of monomer) were obtained by direct integration of the DSC endotherms for (hypProGly)15 and (ProProGly)15, and by calculating twice the integral of the first half of the DSC endotherm for (mopProGly)15 (under the reasonable assumption based on the endotherms for the other two CRPs that the endotherm was symmetric—see simulated data in Figure 6). The value of ΔS at the Tm was calculated as Tm = ΔH/(ΔS + R · ln(0.75c2)), where c is the concentration of monomeric peptide determined from amino-acid analysis and Tm is the maximum of the DSC endotherm.35 Because our model assumes ΔCP = 0 for triple-helix unfolding (an approximation commonly employed for triple helices formed from collagen-related peptides36,17), ΔH and ΔS are independent of temperature. Values of ΔG at T = 46 °C were calculated as ΔG = ΔH – TΔS, where T = 46 °C (Table 2), which is the average of the Tm values for the three peptides determined by DSC. The thermodynamic parameters are most accurate at temperatures near the Tm.
Measurement of Ktrans/cisValues for Ac-hyp-OMe and Ac-mop-OMe
Ac-hyp-OMe and Ac-mop-OMe (5–10 mg) were dissolved in D2O or CDCl3 (~0.8 mL). 1H NMR spectra were acquired and worked up using NUTS software.37 Values of Ktrans/cis were determined from the relative areas of the trans and cis peaks. NOE difference experiments were performed to confirm the peak assignments.
Computational Methodology
The conformational preferences of Ac-hyp-OMe were examined by hybrid DFT as implemented in Gaussian 03.38 Geometry optimizations and frequency calculations at the B3LYP/6-311+G(2d,p) level of theory were performed to obtain intramolecularly hydrogen-bonded conformers of Ac-hyp-OMe in both the trans and cis geometries, as well as on cis and trans Cγ-endo Ac-hyp-OMe conformers lacking the intramolecular hydrogen bond (Figure 2). Frequency calculations on the optimized structures yielded no imaginary frequencies, indicating true stationary points on the potential energy surface. The resulting SCF energies were corrected by the zero-point vibrational energy (ZPVE) determined in the frequency calculations (see: Table S2 in the Supporting Information). CPCM calculations were performed at B3LYP/6-311+G(2d,p) level of theory using Gaussian 0338 with OFac = 0.8 and RMin = 0.5, and dielectric constants of ε = 4.9 for chloroform and ε = 80 for water. The error on total polarization charges was <0.05 for all cases. Optimized geometries were analyzed by NBO 5.0 at the B3LYP/6-311+G(2d,p) level of theory to estimate the energetic stabilization attributable to the intramolecular hydrogen bond and the n→π* interaction (see: Table S2 in the Supporting Information).21
Supplementary Material
Acknowledgements
We are grateful to D. R. McCaslin for contributive discussions, and to C. D. Brown for technical contributions during the early stages of this work. M. D. S. was supported by graduate fellowships from the US Department of Homeland Security and the ACS Division of Medicinal Chemistry. F. W. K. was supported by NIH postdoctoral fellowship AR50881. The BIF was established with grants BIR-9512577 (NSF) and S10 RR13790 (NIH). NMRFAM is supported by Grant P41RR02301 (NIH). This work was supported by Grant R01 AR044276 (NIH).
Footnotes
Supporting Information Available: Detailed procedures and data for AUC experiments, computational analyses, and X-ray diffraction analyses of Ac-hyp-OMe and Boc-mop-OH (8), IR spectrum of Ac-mop-OMe, and 1H and 13C NMR spectra for novel compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gorres KL, Raines RT. Crit. Rev. Biochem. Mol. Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Fischer E. Chem. Ber. 1902;35:2660–2665. [Google Scholar]; (b) Baldwin JE, Field RA, Lawrence CC, Merritt KD, Schofield CJ. Tetrahedron Lett. 1993;34:7489–7492. [Google Scholar]
- 3.(a) Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. J. Phys. Chem. 1992;96:6472–6484. [Google Scholar]; (b) Vitagliano L, Berisio R, Mazzarella L, Zagari A. Biopolymers. 2001;58:459–464. doi: 10.1002/1097-0282(20010415)58:5<459::AID-BIP1021>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]; (c) Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. J. Am. Chem. Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]; (d) DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. J. Am. Chem. Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]; (e) Hinderaker MP, Raines RT. Protein Sci. 2003;12:1188–1194. doi: 10.1110/ps.0241903. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Raines RT. Protein Sci. 2006;15:1219–1225. doi: 10.1110/ps.062139406. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Hodges JA, Raines RT. Org. Lett. 2006;8:4695–4697. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Choudhary A, Gandla D, Krow GR, Raines RT. J. Am. Chem. Soc. 2009;131:7244–7246. doi: 10.1021/ja901188y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Jakobsche CE, Choudhary A, Raines RT, Miller SJ. J. Am. Chem. Soc. 2010;132:6651–6653. doi: 10.1021/ja100931y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Choudhary A, Pua KH, Raines RT. Amino Acids. 2010;38 doi: 10.1007/s00726-010-0504-8. In Press. DOI: 10.1007/s00726-010-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoulders MD, Satyshur KA, Forest KT, Raines RT. Proc. Natl. Acad. Sci. USA. 2010;107:559–564. doi: 10.1073/pnas.0909592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Horng J-C, Raines RT. Protein Sci. 2006;15:74–83. doi: 10.1110/ps.051779806. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ruzza P, Siligardi G, Donella-Deana A, Calderan A, Hussain R, Rubini C, Cesaro L, Osler A, Guiotto A, Pinna LA, Borin G. J. Pept. Sci. 2006;12:462–471. doi: 10.1002/psc.750. [DOI] [PubMed] [Google Scholar]; (c) Kümin M, Sonntag L-S, Wennemers H. J. Am. Chem. Soc. 2007;129:466–467. doi: 10.1021/ja067148o. [DOI] [PubMed] [Google Scholar]; (d) Chiang Y-C, Lin Y-J, Horng J-C. Protein Sci. 2009;18:1967–1977. doi: 10.1002/pro.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Kim W, McMillan RA, Snyder JP, Conticello VP. J. Am. Chem. Soc. 2005;127:18121–18132. doi: 10.1021/ja054105j. [DOI] [PubMed] [Google Scholar]; (b) Kim W, Hardcastle KI, Conticello VP. Angew. Chem. Int. Ed. 2006;45:8141–8145. doi: 10.1002/anie.200603227. [DOI] [PubMed] [Google Scholar]
- 7.(a) Hodges JA, Raines RT. J. Am. Chem. Soc. 2003;125:9262–9263. doi: 10.1021/ja035881z. [DOI] [PubMed] [Google Scholar]; (b) Doi M, Nishi Y, Uchiyama S, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. J. Am. Chem. Soc. 2003;125:9922–9923. doi: 10.1021/ja035997v. [DOI] [PubMed] [Google Scholar]
- 8.Renner C, Alefelder S, Bae JH, Budisa N, Huber R, Moroder L. Angew. Chem. Int. Ed. 2001;40:923–925. [PubMed] [Google Scholar]
- 9.Steiner T, Hess P, Bae JH, Wiltschi B, Moroder L, Budisa N. PLoS One. 2008;3:e1680. doi: 10.1371/journal.pone.0001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berisio R, Vitagliano L, Mazzarella L, Zagari A. Protein Sci. 2002;11:262–270. doi: 10.1110/ps.32602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Biochemistry. 2000;39:14960–14967. doi: 10.1021/bi001560d. [DOI] [PubMed] [Google Scholar]
- 12.Inouye K, Sakakibara S, Prockop DJ. Biochim. Biophys. Acta. 1976;420:133–141. doi: 10.1016/0005-2795(76)90352-4. [DOI] [PubMed] [Google Scholar]
- 13.Nishi Y, Kawahara K, Uchiyama S, Doi M, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. Peptide Sci.; Proc. Jap. Pept. Symp. 2006;43:169–170. [Google Scholar]
- 14.Lesarri A, Cocinero EJ, López JC, Alonso JL. J. Am. Chem. Soc. 2005;127:2572–2579. doi: 10.1021/ja045955m. [DOI] [PubMed] [Google Scholar]
- 15.Janesko BG, Gallek CJ, Yaron DJ. Phys. Chem. A. 2003;107:1655–1663. [Google Scholar]
- 16.Alabugin IV, Zeidan TA. J. Am. Chem. Soc. 2002;124:3175–3185. doi: 10.1021/ja012633z. [DOI] [PubMed] [Google Scholar]
- 17.Kotch FW, Guzei IA, Raines RT. J. Am. Chem. Soc. 2008;130:2952–2953. doi: 10.1021/ja800225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang G-B, Rito CJ, Gellman SH. Biopolymers. 1992;32:293–301. doi: 10.1002/bip.360320309. [DOI] [PubMed] [Google Scholar]
- 19.Barone V, Cossi M. J. Phys. Chem. A. 1998;102:1995–2001. [Google Scholar]
- 20.Ohnishi M, Urry DW. Biochem. Biophys. Res. Commun. 1969;36:194–202. doi: 10.1016/0006-291x(69)90314-3. [DOI] [PubMed] [Google Scholar]
- 21.Glendening ED, Badenhoop JK, Reed AE, Carpenter JE, Bohmann JA, Morales CM, Weinhold F. NBO 5.0. 2001 [Google Scholar]
- 22.(a) Gerig JT, McLeod RS. J. Am. Chem. Soc. 1973;95:5725–5729. doi: 10.1021/ja00798a046. [DOI] [PubMed] [Google Scholar]; (b) Sonntag L-S, Schweizer S, Ochsenfeld C, Wennemers H. J. Am. Chem. Soc. 2006;128:14697–14703. doi: 10.1021/ja0654938. [DOI] [PubMed] [Google Scholar]; (c) Shoulders MD, Kamer KJ, Raines RT. Bioorg. Med. Chem. Lett. 2009;19:3859–3862. doi: 10.1016/j.bmcl.2009.03.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhary A, Raines RT. Unpublished results [Google Scholar]
- 24.(a) Shoulders MD, Raines RT. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fields GB. Org. Biomol. Chem. 2010;8:1237–1258. doi: 10.1039/b920670a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Malkar NB, Lauer-Fields JL, Borgia JA, Fields GB. Biochemistry. 2002;41:6054–6064. doi: 10.1021/bi012071w. [DOI] [PubMed] [Google Scholar]; (b) Agarkov A, Greenfield SJ, Ohishi T, Collibee SE, Gilbertson SR. J. Org. Chem. 2004;69:8077–8085. doi: 10.1021/jo049103g. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins CL, Vasbinder MM, Miller SJ, Raines RT. Org. Lett. 2005;7:2619–2622. doi: 10.1021/ol050780m. [DOI] [PubMed] [Google Scholar]
- 27.Hodges JA, Raines RT. J. Am. Chem. Soc. 2005;127:15923–15932. doi: 10.1021/ja054674r. [DOI] [PubMed] [Google Scholar]
- 28.Krapcho J, Turk C, Cushman DW, Powell JR, DeForrest JM, Spitzmiller ER, Karanewsky DS, Duggan M, Rovnvak G, Schwartz J, Natarajan S, Godfrey JD, Ryono DE, Neubeck R, Atwa KS, Petrillo EWJ. J. Med. Chem. 1988;31:1148–1160. doi: 10.1021/jm00401a014. [DOI] [PubMed] [Google Scholar]
- 29.Becktel WJ, Schellman JA. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 30.Nishi Y, Uchiyama S, Doi M, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. Biochemistry. 2005;44:6034–6042. doi: 10.1021/bi047887m. [DOI] [PubMed] [Google Scholar]
- 31.Shoulders MD, Guzei IA, Raines RT. Biopolymers. 2008;89:443–454. doi: 10.1002/bip.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins CL, Bretscher LE, Guzei IA, Raines RT. J. Am. Chem. Soc. 2003;125:6422–6427. doi: 10.1021/ja034015j. [DOI] [PubMed] [Google Scholar]
- 33.Vranka JA, Sakai LY, Bächinger HP. J. Biol. Chem. 2004;279:23615–23621. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- 34. [Accessed April 12, 2010];AppliedBiosystems. http://www3.appliedbiosystems.com/cms/groups/psm_marketing/documents/generaldocum ents/cms_040640.pdf.
- 35.Engel J, Bächinger HP. Top. Curr. Chem. 2005;247:7–33. [Google Scholar]
- 36.(a) Engel J, Chen H-T, Prockop DJ, Klump H. Biopolymers. 1977;16:601–622. doi: 10.1002/bip.1977.360160310. [DOI] [PubMed] [Google Scholar]; (b) Mizuno K, Hayashi T, Peyton DH, Bächinger HP. J. Biol. Chem. 2004;279:38072–38078. doi: 10.1074/jbc.M402953200. [DOI] [PubMed] [Google Scholar]
- 37.NUTS–NMR Utility Transform Software. Livermore, California: Acorn NMR; [Google Scholar]
- 38.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JAJ, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Revision C.02 ed. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.