Abstract
The liver contains a population of small bipotential facultative progenitor cells that reconstitute liver function when mature hepatocytes and/or cholangiocytes are unable to proliferate. Mesenchymal markers, including members of the forkhead transcription factor gene family, have been detected in hepatic progenitor cells. The winged helix transcription factor Foxl1 localizes to mesenchymal cells in the intestine; however its expression in the liver has not been reported. We found that Foxl1 is expressed in rare cells in the normal liver but is dramatically induced in the livers of mice that have undergone bile duct ligation or were fed a DDC-containing or CDE diet. In addition, we employed genetic lineage tracing using a Foxl1-Cre transgenic mouse crossed with the Rosa26R lacZ reporter line to demonstrate that Foxl1-Cre-expressing cells are present within the periportal region shortly following injury. These cells give rise to both hepatocytes (marked by HNF-4α expression) and cholangiocytes (marked by CK19 expression), indicating that these cells were derived from Foxl1-Cre expressing cells. Foxl1-Cre expressing cells are distinct from hepatic stellate cells, portal fibroblasts and myofibroblasts, although they are located in close proximity to portal fibroblasts. These results demonstrate that the early Foxl1-Cre lineage cell gives rise to both cholangiocytes and hepatocytes following liver injury and suggest the potential for progenitor-portal fibroblast cell interactions. Conclusion: We propose that Foxl1 is a bona fide marker of the facultative progenitor cell in the mouse liver.
Keywords: cholangiocyte, oval cell, progenitor, stem cell, portal fibroblast
In the normal liver, the majority of hepatocytes and biliary epithelial cells (cholangiocytes) are quiescent. In response to liver injury or loss of liver mass, proliferation of mature liver cells represents the first-line defense to restore homeostasis. However, in patients or animal models with chronic liver disease, hepatocytes and cholangiocytes are blocked from proliferating and small bipotential progenitors are activated (1–5). These facultative stem cells, described in rodent models as “oval cells” or “intermediate hepatobiliary cells”, have the appearance of small hepatocytes with scant cytoplasm and are postulated to arise from a niche close to the terminal bile ducts, called the canal of Hering. Oval cells are frequently found in “ductular reactions” present in both experimental models of liver injury and in patients with chronic liver disease (6, 7). Their presence is strongly associated with impaired proliferation of mature hepatocytes, suggesting that they represent a reserve hepatic progenitor cell population. Repopulation studies in animal models suggest that these progenitors possess the potential to restore function in patients with both acute and chronic liver disease (8).
A better understanding of the pathways involved in the differentiation of hepatic stem cells and the role these cells play during liver recovery requires molecular tools for their isolation and characterization. Likewise, methods for the isolation and ex vivo expansion of human hepatic stem cells are necessary if this approach is to be used in regenerative medicine. Expression profiling studies have identified a subpopulation of hepatic progenitors that express markers consistent with a mixed epithelial/mesenchymal phenotype and that include members of the forkhead winged helix transcription factor family (9, 10). The forkhead winged helix factor Foxl1 had previously been identified as a mesenchymal factor in the intestine with undetectable expression in the developing and adult liver (11). We therefore investigated whether Foxl1 is activated in hepatic progenitor cells during liver injury using genetic lineage tracing to identify the Foxl1 expressing lineage in vivo. We demonstrate that Foxl1 (Forkhead Box l1, formerly Fkh6; 11–14) indeed marks facultative stem cells and their descendants in the liver and represents a population distinct from both hepatic stellate cells and portal fibroblasts.
Experimental Procedures
Mice and experimental protocols
For lineage tracing studies, Foxl1-Cre mice were crossed to Rosa26R lacZ reporter mice (15) and subjected to bile duct ligation (BDL). Foxl1-Cre-negative or sham-operated mice were used as controls. Animals (10 to 12 weeks old) were anesthetized with (2.5% V/V) vaporized isofluorane. A midline laparotomy was performed and the common bile duct ligated twice with 4.0 silk suture. Sham animals underwent a similar laparotomy, after which the common bile duct was exposed and manipulated without placement of ligatures. Mice received a subcutaneous injection of buprenorphine at 0.5mg/kg immediately after surgery, were placed on a warming pad and allowed to recover. All protocols were approved by the IACUC of the University of Pennsylvania. At times of sacrifice, livers were rinsed in PBS and placed in 4% PFA for 45 minutes, rinsed in PBS and cryoprotected in 30% sucrose/PBS overnight at 4°C.
Special diets
Mice were fed a diet containing 3, 5-diethoxycarbonyl-1, 4- dihydrocollidine (DDC) (Sigma-Aldrich, St. Louis, MO) (0.1% wt/wt) in # 5015, PMI Mouse diet (Harlan Teklad, Madison, WI) for 3, 7, 14 or 21 days. Other mice were fed a choline deficient diet for five weeks (Harlan-Teklad, Diet TD88052) with drinking water supplemented with 0.165% ethionine (Sigma, St. Louis, MO). Livers were harvested and processed as described above.
RNA isolation and Quantitative real-time PCR
Total RNA was extracted from liver samples using the Totally RNA Kit (Ambion, Applied Biosystems, Foster, City, CA). Liver RNA was reverse transcribed using oligo dT priming and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). PCR reactions were performed using SyBr Green QPCR Master Mix (Invitrogen, Carlsbad, CA) on an Mx3000 PCR cycler (Stratagene, Agilent Technologies, Santa Clara, CA). Reactions were performed in triplicate with reference dye normalization, and median CT values were used in the analyses. Primers sequences were as follows: Foxl1 (F): TGCCGCATTCCACAGCATAGTC; Foxl1(R): CAAAGTGAGTTCCAGGACAGCCAG. CK19: (F): CCAGGAAGCCCACTACAACAAT; CK19 (R): TCGAGGGAGGGGTTAGAGTAAA. TATA binding protein (TBP) (F): CCCCTTGTACCCTTCACCAAT; TBP (R): GAAGCTGCGGTACAATTCCAG.
Whole Mount Embryo β-galactosidase staining
Embryos (12.5 dpc) were dissected in ice-cold PBS and fixed in 0.2% glutaraldehyde, 1% formaldehyde, 2mM MgCl2, 5mM EGTA and 0.2% NP-40 for 1h. Embryos were rinsed in PBS and incubated in X-gal staining solution overnight (17). Embryos were rinsed in PBS and post-fixed in 4% PFA overnight at 4°C. Embryos were transferred to PBS, dehydrated through a graded PBS/methanol series, cleared in a 1:1 solution of benzyl alcohol:benzyl benzoate for 1h, and imaged using a LeicaM212 microscope. For sectioning, embryos were re-hydrated using the reverse graded PBS/methanol series, rinsed in PBS, and incubated in 30% sucrose in PBS overnight at 4°C. Embryos were equilibrated in OCT for 60 minutes, oriented in fresh OCT and placed on dry ice to freeze. Cryo-sections were washed for 5 minutes each in a sequence of PBS, 4% PFA and then water. The slides were stained with Kernechtrot nuclear fast red (Poly Scientific, Bay Shore, NY) 0.1% for 30 seconds and then sealed with crystal mount (Electron Microscopy Sciences, Hatfield, PA) to dry overnight. Cover slips were mounted on the slides using Histomount (Zymed, Invitrogen, Carlesbad, CA). Sections were imaged on a LeicaDMRE microscope.
Histology and Immunohistochemistry
10 μm cryosections were cut, warmed to room temperature and air dried for 5 minutes. Sections were incubated with X-gal (β-gal) staining solution (17) at 37°C for 3–5 hours. Sections were post-fixed for 5 minutes in 4% PFA, washed briefly in PBS, and mounted in aqueous mounting media (Kirkegaard, Gaithersburg, MD).
For antibody co-localization studies for CK19, HNF-4α and Ki-67, β-gal stained sections were fixed for 5 minutes in 4% PFA. Antigen retrieval was performed by microwaving slides in 10 mM citric acid monohydrate buffer pH 6.0 for 6 minutes. The Vector ABC Elite kit detection method was used according to manufacturer’s instructions (Vector Laboratories, Burlingame, CA). The primary antibodies were diluted in PBT (1X PBS, 0.1% BSA, 0.2% Triton-X) as follows: CK19 (Hybridoma Dev) 1:20; HNF-4α (Santa Cruz Biotechnology, Santa Cruz, CA, sc-8947) 1:1000; Ki-67 (Vector Laboratories, Burlingame, CA, VP-RM04) 1:1500 and incubated with sections overnight at 4°C. For sequential staining of β-gal and CK19, slides were stained as described for β-gal, mounted in PBS and imaged. Slides were then post-fixed in 4% PFA and incubated with α-CK19 as described above with second image capture performed of the same area. β-gal+, CK19+, and HNF-4α+ cells were quantified by counting positive cells in ten 20X microscope fields and calculating the percent β-gal+/CK19+ and β-gal+/HNF-4α+ cells. The percent β-gal+/CK19−/HNF-4α− cells was calculated as a percentage of the sum of all CK19+, HNF-4α+ and β-gal single+ cells. Two or three slides were examined for each BDL and DDC time point.
For co-localization of β-gal/desmin and β-gal/α-SMA, frozen sections were warmed to room temperature for 10 minutes, stained overnight in X-gal substrate solution and post-fixed with 4% PFA at room temperature for 10 minutes. Sections were rinsed in PBS and blocked with PBS/1% BSA at room temperature for 30 minutes. Sections were incubated with primary antibody at the following dilutions overnight at 4°C (α-SMA 1:1600, Sigma, St. Louis, MO, Clone 1A4, Cat #A2457; desmin 1:1000, Sigma, Clone DE-U-10 Cat # D1033). Sections were rinsed in PBS and incubated with Cy5-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, Westgrove, PA, Cat # 715-175-500, 1:600 dilution). Sections were washed twice in PBS for 5 minutes, ddH20 twice for 5 minutes and mounted with fluorescent mounting media (KPL, Cat # 71-00-16). Co-localization of β-gal/elastin was performed as above with the following modifications: After post-fixation with 4% PFA for 10 minutes at room temperature, sections were washed with TBS three times for 5 minutes followed by antigen retrieval with 0.5% hyaluronidase for 1 hour at room temperature and three 5 minute washes in TBS. Blocking was performed with TBS/3% BSA/0.9% NaCl in TBS at room temperature for 30 minutes. Sections were incubated with α-elastin antibody (Cedarlane Laboratories Limited, Hornby, Ont, Cat # CL55041AP, 1:200 dilution) in TBS/3% BSA/1% normal goat serum, rinsed in TBS followed by a 30 minute room temperature incubation with Cy5-conjugated donkey anti-rabbit IgG 1:600 dilution (Jackson ImmunoResearch Laboratories, Cat # 715-175-500), and mounted with fluorescent mounting media after rinses in PBS and water. Immunofluorescent and β-gal images were merged using digital “pseudocolorization” using the IP Lab imaging software application (BD Biosciences, Rockville, MD).
Laser Capture Microdissection, RNA isolation and QPCR
Harvested livers were washed in PBS, embedded in OCT, and frozen in a dry ice/ethanol bath. Four 8μm serial sections were obtained using a Microm HM 505E (Richard Allen Scientific, Kalamazoo, MI) cryostat and placed on RNase free, membrane mounted metal-framed slides (Molecular Machines & Industries, Haslett, MI). Sections were brought to room temperature, placed in 75% EtOH for 30 sec, DEPC-treated water for 30 sec, Hematoxylin for 10 sec, DEPC-treated water for 15 sec, DEPC-treated water for 20 sec, twice in 95% EtOH for 30 secs, 100% EtOH for 30 sec, and xylene for 2 min and air-dried for ten seconds. An uncharged glass slide was placed over the polymer film and immediately processed for microdissection. Liver portal tracts and parenchymal tissues were collected using the SLμCut instrument (Molecular Machines & Industries) fitted to a Nikon Eclipse TE2000-S inverted microscope and collected on caps with adhesive lids (Molecular Machines & Industries, Haslett, MI). RNA was extracted using the PicoPure RNA Isolation Kit (Molecular Devices, Sunnyvale, CA) and samples (30 ng each) reverse transcribed using the WT-Ovation RNA Amplification System (NuGEN, San Carlos, CA). Quantitative PCR reactions were performed using SyBr Green QPCR Master Mix (Invitrogen, Carlsbad, CA) as described above.
Statistical analysis
Students t-tests with equal variance and two-tailed distribution were used to determine the significance of differences between two groups (Excel statistical analysis software, Microsoft, Redmond, VA). A p value of 0.05 was considered statistically significant. Results where indicated are expressed as mean ± SE.
Results
Foxl1 is activated in the liver following injury
The mammalian forkhead box transcription factor Foxl1 is expressed in the gastrointestinal mesenchyme, but unlike its relatives of the Foxa and Foxo classes, its mRNA is not detected in the quiescent liver (11). Because Foxl1 is a marker of the mesenchyme in the intestine, and because hepatic progenitor cells have been reported to express mesenchymal markers, we examined Foxl1 mRNA expression in the bile duct ligation (BDL) model, in which cholestatic injury induces a ductular reaction by qRT-PCR. Expression of Foxl1 was dramatically increased as compared to quiescent liver (Figure 1A). To determine whether the expression of Foxl1 was confined to the portal tract region where the ductular reaction occurs, we used laser capture microdissection (LCM) to isolate portal tracts and adjacent parenchyma for gene expression analysis. Foxl1 expression was enriched in the portal tracts and absent from the surrounding parenchyma (Figure 1B, C). As expected, Foxl1 expression was absent from portal tracts isolated from sham-operated animals or Foxl1 null mice (Figure 1C). All samples were analyzed for cytokeratin 19 (CK19) expression to confirm minimal contamination of the parenchymal samples (Figure 1D). These data clearly show that the Foxl1 gene is activated in cells within or near the portal triad following cholestatic liver injury.
Figure 1. Foxl1 expression is induced following cholestatic liver injury.
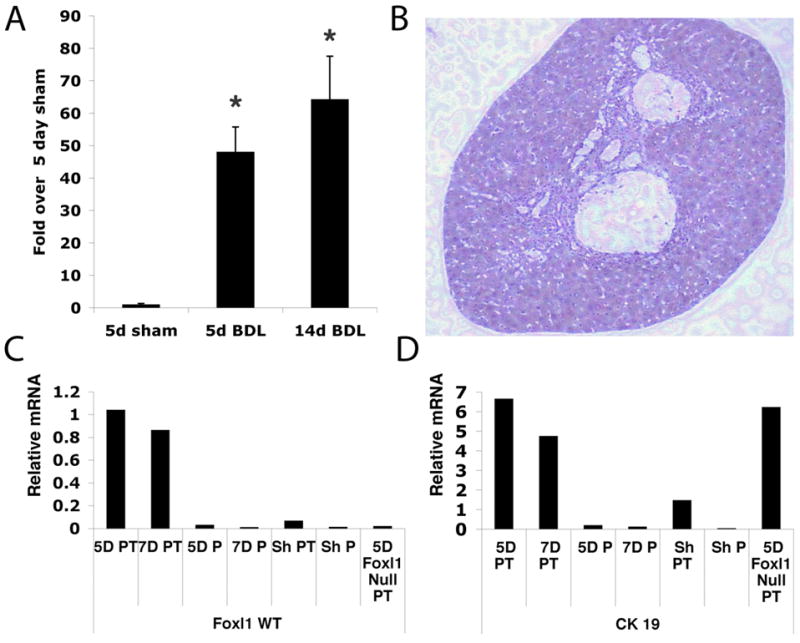
A) Expression of Foxl1 mRNA was significantly increased in livers 5 and 14 days following bile duct ligation (BDL). 5 day (n=4), 2 week (n=4), WT sham n=2. *P-value < 0.02. (B–D) Laser capture microdissection of liver tissue following BDL detects Foxl1 mRNA in portal tracts but not the liver parenchyma. Portal tracts and adjacent parenchyma of Foxl1 WT livers following 3, 5 and 7 day BDL, Foxl1 null 5 day BDL and 7 day sham WT BDL were microdissected and RNA isolated for gene expression analysis. (B) Representative portal tract isolated by laser capture microdissection for gene expression analysis. (C) qRT-PCR for Foxl1 establishes that Foxl1 is expressed in the portal tract but not liver parenchyma of BDL mice. Portal tracts of Foxl1 null mice or sham-operated animals did not express Foxl1. (D) qRT-PCR confirmed expression of the bile duct marker CK19 in portal tracts but not in surrounding parenchyma of livers after BDL. Abbreviations, WT (Wild type), PT (Portal Tract), P (Parenchyma), Sh (Sham).
Lineage tracing of Foxl1-positive cells following bile duct ligation
In order to precisely localize Foxl1 expressing cells and their descendants in the injured liver, we employed genetic lineage tracing with the Cre/loxP technology. A BAC transgenic line in which expression of Cre recombinase is under the control of 170 kB of cis-regulatory elements of the murine Foxl1 gene recapitulates the expression pattern of Foxl1 in the gastrointestinal tract (13). We crossed the Foxl1-Cre line to Rosa26-lacZ reporter mice, in which beta-galactosidase (β-gal) is expressed only in those cells in which Cre has been active. At present the ontogeny of facultative bipotential progenitors in the liver is unknown. One possibility is that these cells are remnants of the fetal hepatoblast lineage, which differentiates into both hepatocytes and cholangiocytes in mid- to late gestation. Therefore, we investigated whether the Foxl1-Cre transgene is expressed in fetal hepatoblasts. We collected Foxl1-Cre; Rosa26R embryos on day 12.5 of gestation, a time when the hepatic primordium is clearly visible but has not yet differentiated into mature cell types, and performed β-gal staining. As shown in Figure 2, hepatoblasts were negative for β-gal, and thus for Foxl1-Cre expression, suggesting that the Foxl1-positive progenitor in the adult liver is either not a remnant of the fetal hepatoblast population, or that this gene is activated in this lineage later in life.
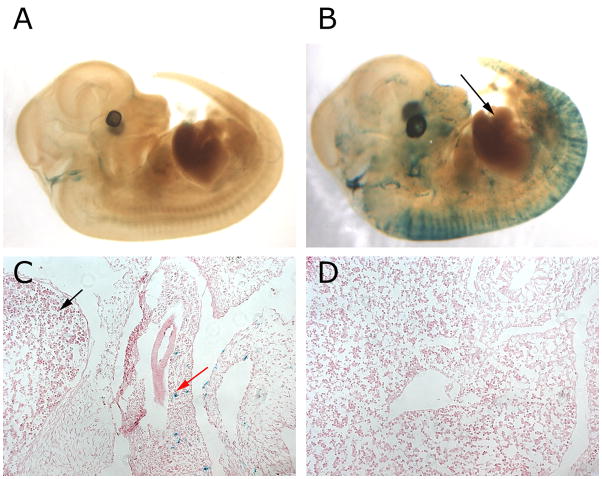
Figure 2. Foxl1-Cre does not mark fetal hepatoblasts.
β-gal staining of Rosa26R control embryo (A) and Foxl1-Cre; Rosa26R embryos (B–D) at day 12.5 of gestation. A) Control embryo with only some blue staining near the otic vesicle. B) Foxl1-Cre; Rosa26R embryo with β-gal positive cells in the developing spinal cord, but none in the liver (marked by black arrow). C) Section of a 12.5 dpc Foxl1-Cre; Rosa26R embryo, showing β-gal positive cells in the gastrointestinal mesenchyme (red arrow) but not in the liver (black arrow). D) Another view of the embryo sectioned in C, showing that there are no blue cells present in the liver, indicating that the Foxl1-Cre transgene is not active in fetal hepatoblasts.
In adult mice, a limited number of cholangiocytes were β-gal positive in the livers of sham-operated animals, consistent with the mRNA data shown in Figure 1A (Figure 3A). However, following BDL, β-gal activity was strongly induced, appearing first in isolated cells within ductular reactions adjacent to the portal vein (Figure 3B), and increasing and extending away from the portal triad over time (Figure 3C–H). There are no reliable antibodies for either Foxl1 or β-gal. We therefore performed sequential staining and image capture to confirm the colocalization of β-gal and CK19 (Figure 3I and J). This expression pattern is consistent with that of a facultative hepatic progenitor cell that gives rise to both hepatocytes and cholangiocytes.
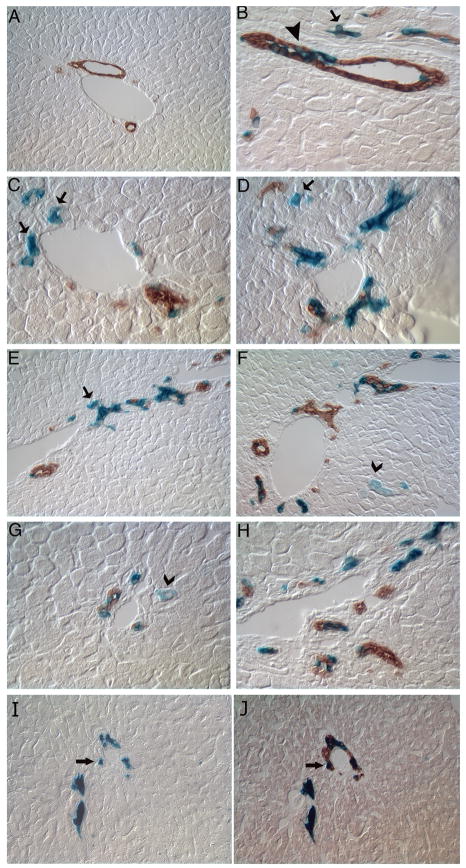
Figure 3. Lineage tracing of Foxl1-positive cells following bile duct ligation.
Foxl1Cre; Rosa26RlacZ mice underwent BDL and livers were harvested 3, 5, 7 and 14 days later. Livers were analyzed for β-gal activity reflecting Foxl1-Cre activation (blue) and co-stained with the cholangiocyte marker CK19 (brown). We observed a steady increase in the number of β-gal positive cells over time, which were seen as early as 3 days in β-gal+/CK19+ co-labeled bile ductular cells (arrowhead) and β-gal+/CK19− cells in periportal regions (arrow). By 5 days (C, D) and 7 days (E, F) we found increasing numbers of β-gal+/CK19− cells (arrow) in periportal regions and the emergence of β-gal stained cells with the morphologic appearance of hepatocytes (F, G, chevron arrow). Figure (A) 5 day sham, 20× magnification, (B) 3 day BDL 40× magnification, (C, D) 5 day BDL 40× magnification, (E, F) 7 day BDL 20× magnification, (G,H) 14 day BDL, 40× magnification. Sequential stain of 14 day BDL at 20× magnification with β-gal (I) followed by CK19 (J). Representative β-gal+/CK19+ cells are indicated by arrows.
In order to confirm that Foxl1 marks bipotential progenitors, we performed triple label experiments with the livers of bile duct ligated Foxl1-Cre; Rosa26R mice for β-gal, CK19 and HNF-4α (hepatocyte nuclear factor 4 alpha), an orphan nuclear receptor specific for hepatocytes. The Foxl1-Cre positive lineage gives rise to both hepatocytes and cholangiocytes, consistent with the proposed role of Foxl1 as a marker of hepatic progenitor cells (Figure 4). We performed quantitative analysis of β-gal+ cells that coexpressed either CK19+, HNF-4α+ or were positive only for β-gal. As shown in Figure 4G, the percentage of β-gal positive cells in bile ducts that co-stained with CK19 increased three-fold beginning 3 days after BDL surgery, consistent with an enrichment for Foxl1-Cre expressing cholangiocytes in this model. Five and seven days post-BDL, cells that were β-gal+/CK19−/HNF-4α− increased sixteen and thirteen-fold over sham levels, respectively (Figure 4D, H). The number of cells that were β-gal+/HNF-4α+ increased six-fold 14 days after BDL, following the peak of β-gal+/CK19−/HNF-4α− cells (Figure 4F, H). These data suggest that Foxl1-Cre lineage positive cells gave rise to mature hepatocytes through an intermediate cell stage in which neither CK19 nor HNF-4α are expressed (Figure 4D). We never observed cells co-staining for CK19 and either HNF-4α, or C/EBPα, another established marker of hepatoblasts/hepatocytes (18) (data not shown). While our inability to detect triple positive cells does not exclude the possibility that they are present in low numbers in the Foxl1-Cre; Rosa26R mice, our data suggests that the cholangiocyte and hepatocyte fates are mutually exclusive in this model.
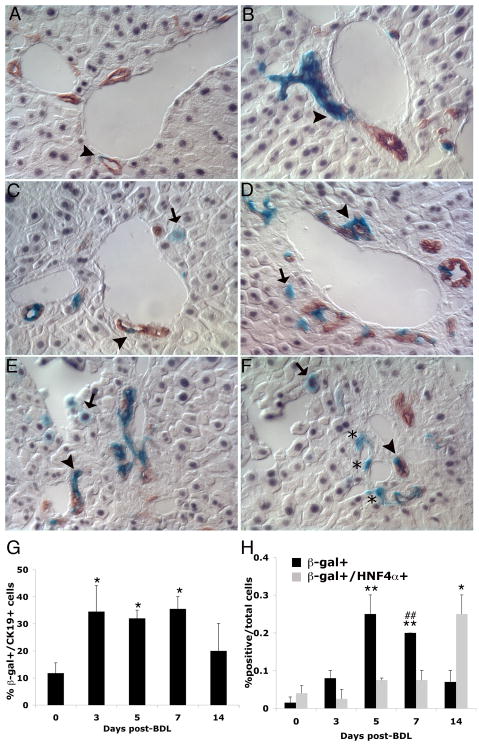
Figure 4. Lineage tracing of Foxl1-Cre-positive cells following bile duct ligation.
Triple labeling for β-gal, CK19 and HNF4α allowed the identification of Foxl1-Cre-lineage positive cholangiocytes (β-gal/CK19 double positive cells) and hepatocytes (β-gal/HNF-4α double positive cells). Liver cells were identified with the hepatocyte marker HNF4α (grey) and cholangiocyte marker CK19 (brown), and Foxl1-Cre-positive cells and their descendants by β-gal staining (blue). Rare double-labeled β-gal/CK19 cells appeared in sham operated livers (A) and were frequently observed in ductular reactions beginning at 3 days post-BDL and throughout the time course examined (B–F, arrowhead). Single-labeled β-gal positive cells in the 14 day BDL liver (Panel F) are identified by asterisk. Double labeled β-gal/HNF-4α hepatocytes are shown in day 5 post-BDL livers (arrows in C, D, and E). (A) Sham-operated, (B) 3 day BDL, (C) 5 day BDL, (D) 7 day BDL, (E, F) 14 day BDL, 40× magnification. Quantification of percent β-gal+/CK19+ cells (G) and percent β-gal+ and β-gal+HNF-4α+/total cells at indicated time points post-BDL (H). *p-value < 0.05 relative to control, **p-value <0.001 relative to control. ##p-value < 0.05 percent β-gal+/total cells relative to β-gal+/HNF-4α+/total cells 7 day BDL.
The Foxl1-positive cell lineage is enriched for proliferating cells
Oval cells are proliferating cells that appear in the liver under conditions of injury when hepatocyte proliferation is limited. To determine if the Foxl1-Cre lineage is enriched for proliferating cells, we performed dual-labeling studies for β-gal and the proliferation marker Ki-67. As expected, we detected very few proliferating cells in the livers of sham-operated animals (Figure 5A). However, three days following BDL, we observed cells singly positive for Ki-67 or β-gal in bile ducts and periportal areas, and some double-positive cells in bile ducts (Figure 5B). The frequency of double positive cells in the portal triad increased five and seven days following surgery; in addition, a few hepatocytes were found to be β-gal and Ki-67 positive in 5-day BDL animals (Figure 5C, D). By 14 days post-surgery, there was an overall decrease in the number of proliferating cells (Figure 5E–H). These data show that the Foxl1-Cre marked lineage proliferates following BDL, consistent with a facultative progenitor phenotype.
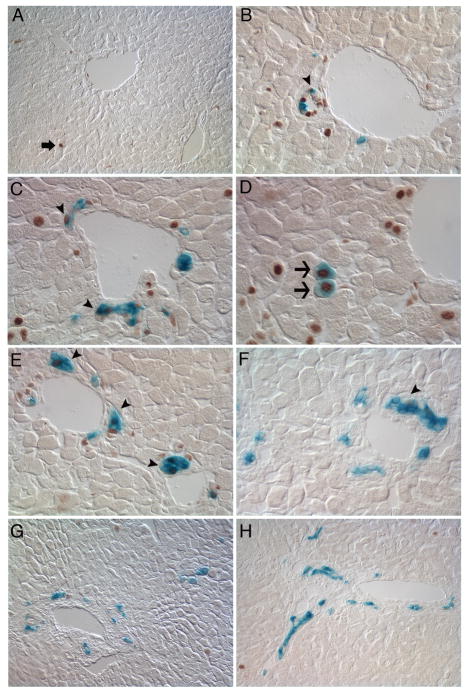
Figure 5. The Foxl1-Cre-positive cell lineage is enriched for proliferating cells.
β-gal and Ki-67 double-labeling reveals co-localization of β-gal and Ki-67 (brown labeled nuclei), indicating that Foxl1-Cre positive cells are actively proliferating following BDL injury. Only rare Ki-67 positive hepatocytes are detected in sham-operated livers (A, arrow). In the 3-day BDL liver (B, arrowhead)), Ki-67 and β-gal co-localized in cells within bile ductules. Co-localization of Ki-67 and β-gal in 5 (C) and 7-day BDL livers (E) was detected in ductular reactions (arrowheads). Dual stained β-gal/Ki-67 cells with hepatocyte morphology were seen in 5 day BDL (D, arrows). By 14 days post-BDL, the majority of β-gal cells were Ki-67 negative (F–H). (A) sham, 20×, (B) 3 d BDL, 40× (C, D) 5 d BDL 40×, (E) 7 d BDL (40×) and, (F) 14 d BDL, 40×, (G–H), 14 d BDL, 20× magnification.
Foxl-1Cre activation following DDC diet
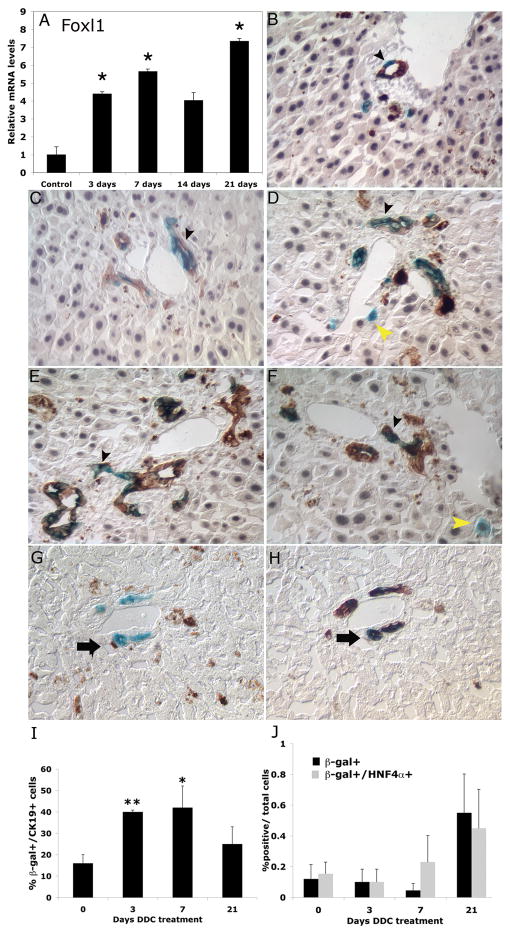
In order to demonstrate that activation of the Foxl1 locus is not unique to bile duct ligated livers, we also tested the DDC model of liver injury, which has been shown to induce a massive oval cell response in rodents (19–21). As demonstrated in Figure 6A, Foxl1 mRNA levels were significantly increased in mice fed a DDC diet, although the fold-change was less than that seen in the BDL model. Next we investigated whether the Foxl1-Cre transgene is also activated after DDC treatment, and whether the Foxl1 lineage contributes to mature epithelial cells in this model. β-gal-positive cells were significantly increased in the DDC injured liver, again within ductular reactions (Figure 6B–H). Furthermore, subsets of the β-gal positive cells also expressed CK19 or HNF-4α (Figure 6F), consistent with our notion that the Foxl1-Cre transgene marks bipotential progenitors in the injured liver. Similar to the BDL model, we detected a 2.6 fold increase in the percentage of β-gal positive cells in bile ducts that co-stained with cytokeratin 19 after seven days of DDC-induced injury (Figure 6I). However, in contrast to the BDL model, the number of single β-gal positive and β-gal, HNF-4α double positive cells did not increase significantly during DDC injury (Figure 6J), which may reflect the inter-animal variability in response to this dietary genotoxin. β-gal and CK19 were also coexpressed in a subset of CK19 cells in mice that received a choline deficient, ethionine supplemented (CDE) diet, another well established model associated with hepatic progenitor cell proliferation (Supplementary Figure S1). Thus, we have shown that the Foxl1 gene is activated in three models of liver injury, and marks cells that ultimately express genes specific to the mature epithelial cells of the liver.
Figure 6. The Foxl1-Cre-positive lineage in DDC-treated mice.
(A) Expression of Foxl1 mRNA was significantly increased in livers of mice fed a diet containing 0.01% DDC. Triple label staining of liver sections of Foxl1-Cre; Rosa26RlacZ mice: β-gal (blue), CK19 (brown) and HNF-4α (gray) 3 days (B), 7 days (C), 14 days (D), or 21 days (E, F) after treatment with a DDC-containing diet. Cells within ductular reactions were co-labeled with β-gal and CK19 beginning 3 days after initiation of DDC diet (B, black arrowhead) and detected throughout the 21-day timecourse (C–F). Co-localization of β-gal and HNF4-α is detected in 14 day and 21 day DDC diet livers (yellow arrowhead, D, F). Brown background staining represents heme-containing breakdown products of DDC. Quantitation of percent β-gal+/CK19+ cells (G) and percent β-gal+ and β-gal+HNF-4α+/total cells (H). *p-value < 0.05 relative to control, **p-value <0.01 relative to control. Sequential stain of 21 day DDC 20× magnification with β-gal (I) followed by CK19 (J). Representative β-gal+/CK19+ cells are indicated by arrows.
Foxl1-expressing cells are closely apposed to portal fibroblasts
Foxl1 expression in the intestinal tract is restricted to the mesenchyme (11, 22, 23). However, the expression of Foxl1 in hepatic myofibroblast-like cells, including portal fibroblasts and stellate cells, has not been examined previously. Because portal fibroblasts and hepatic stellate cells are located in close proximity to progenitor cells and can signal in a paracrine fashion to hepatic progenitor cells (24–26), we investigated the localization of Foxl1 expression in relationship to portal fibroblasts and hepatic stellate cells in two injury models. We detected significant expression of elastin, a specific marker of portal fibroblasts in DDC-injured livers (Figure 7B) (27, 28). There was minimal expression of desmin (a marker of hepatic stellate cells) and α-SMA (a marker of both portal myofibroblasts and hepatic stellate cells which have undergone myofibroblastic differentiation) in the DDC injured livers, suggesting that this paradigm does not stimulate significant activation of hepatic stellate cells or other myofibroblasts (7D, F). Notably, elastin positive cells were increased in number and encircled the β-gal positive cells, but did not co-stain the same cell population. In BDL injured livers (Figure S2), we detected significant expression of elastin, desmin and α-SMA (Figure S2, B, D, F). Similar to the DDC model, there was no overlap between Foxl1-Cre (β-gal) expression and the markers of either portal fibroblasts or hepatic stellate cells. The close proximity of portal fibroblasts to Foxl1-Cre positive progenitor cells in both DDC and BDL paradigms suggests that portal fibroblasts rather than hepatic stellate cells participate in paracrine signaling with Foxl1-Cre positive progenitors.
Figure 7. Foxl1-Cre does not co-localize with myofibroblastic cell markers in the DDC injury paradigm.

β-gal/desmin, β-gal/elastin and β-gal/α-SMA co-staining in control (A, C, E) and 21 day DDC diet treated mice (B, D,F). β-gal staining appears aqua blue, while desmin, elastin and SMA are shown as royal blue pseudocolorization of the original immunofluorescence images. Elastin, α-SMA and desmin staining in control livers were confined to perivascular cells (A, C, E). Elastin positive cells were observed in close proximity to, but not overlapping with the β-gal positive cells in DDC fed mice (B, arrow). There was minimal expression of desmin and α-SMA in the DDC injured livers (D, F). 20× magnification.
Discussion
Identification of unique oval cell markers that can be used to isolate resident hepatic progenitors has been impeded by their heterogeneity and by the fact that the majority of markers identified to date are shared by other cell types within the liver (29). Foxl1 is expressed in a limited number of cells in the normal adult liver, but is dramatically induced in two established models of liver injury associated with hepatic progenitor cell proliferation. Although Foxl1-specific antibodies do not yet exist, our genetic lineage tracing experiments strongly suggests that the Foxl1-Cre lineage gives rise to both cholangiocytes and hepatocytes following cholestatic or genotoxic liver injury. Thus, Foxl1 appears to be a bona fide marker of the facultative progenitor cell in the mouse liver.
Progenitor cells proliferate in close proximity to hepatic stellate cells and portal fibroblasts in chronic liver injury, suggesting that they constitute a progenitor cell niche that influences differentiation and proliferation (25, 26). In contrast to its intestinal expression in the mesenchyme, Foxl1 is not expressed in either hepatic stellate cells or portal fibroblasts. These results are consistent with those of Yovchev and colleagues whose characterization of oval cell lineages indicate that a subpopulation of hepatic progenitor cells expressing markers of a mixed epithelial-mesenchymal lineage were able to repopulate injured rat liver and were distinct from thy-1 positive portal fibroblasts and hepatic stellate cells (9).
A better understanding of the cellular organization of the hepatic stem cell niche is essential for elucidating hepatic progenitor cell behavior in response to liver injury. Kuwahara and colleagues have used label retention assays in combination with acetaminophen-induced liver injury to identify four predicted sources of hepatic progenitors (30). Although we employed different liver injury paradigms, the locations of Foxl1-Cre positive cells in the BDL, DDC and CDE livers were similar to a subset of the label-retaining cell populations identified in Kuwahara’s analysis, specifically, the peribiliary hepatocytes and intraductular cells. Our quantitative analysis demonstrating an increase in the number of Foxl1-Cre+/CK19−/HNF-4α− cells preceding the increase in Foxl1-Cre+/HNF-4α+ positive cells is consistent with a model in which Foxl1-Cre progenitor cells progress through an intermediate stage in their differentiation to mature cholangiocytes and hepatocytes. While our findings strongly suggest that Foxl1 positive cells represent bipotential progenitors, we cannot exclude the possibility that Foxl1-Cre+/HNF-4α+ and Foxl1-Cre+/CK19+ cells arise from separate progenitor niches that may be activated independently during the course of BDL and DDC induced liver injury. We are currently developing an inducible Foxl1-Cre transgenic mouse that will allow us to perform more definitive fate mapping analyses.
Our co-staining studies indicate that portal fibroblasts are located in close proximity to Foxl1-Cre progenitor cells in both injury paradigms investigated, and in many instances these cells appear to encircle the Foxl1-Cre positive cells. The juxtaposition of Foxl1-Cre-expressing progenitor cells and portal fibroblasts suggests the potential for paracrine signaling between these two cell populations. At the present time, the signals that direct hepatic bipotential progenitor differentiation to either hepatocyte or cholangiocyte lineages are unknown. The potential for crosstalk between hepatic progenitor cells and portal fibroblasts has not been investigated in vivo, due in part to the lack of suitable genetic models. However, recent studies have demonstrated that hedgehog mediated paracrine interactions between hepatic stellate cells and hepatic progenitors are required for hepatic progenitor cell viability and proliferation (31, 32). Preliminary studies indicate that Foxl1 is a downstream target of hedgehog signaling in the intestine (KHK, unpublished observations). These findings suggest that Foxl1 may also be a critical mediator of hedgehog signaling cross talk with portal fibroblasts in the liver, and could be involved in the maintenance of liver progenitor cell viability, proliferative response to injury and/or differentiation. To investigate the possibility that Foxl1 itself participates in hepatic progenitor lineage commitment decisions, we will investigate in the future whether Foxl1−/−; Foxl1-Cre; Rosa26R mice exhibit differences in cell lineage allocation during recovery from liver injury.
While hepatic progenitors possess some characteristics of both epithelial and mesenchymal lineages (9, 33), the absence of co-localization of β-gal with myofibroblast markers argues strongly against the possibility that Foxl1-Cre progenitor cells have undergone a lineage switch to a myofibroblastic cell in either BDL or DDC injury paradigms within the time frame examined. Nevertheless, the activation of Foxl1, a mesenchymally expressed gene in the gastrointestinal tract, in hepatic progenitors could be important for the acquisition of mesenchymal characteristics associated with epithelial-mesenchymal transition (EMT), such as increased motility and invasiveness, cellular characteristics that facilitate migration of progenitors to sites of liver injury.
Finally, we would like to suggest that the Foxl1-Cre mouse represents a unique tool that will allow investigators to isolate progenitor cells that can be used in co-culture experiments to investigate mechanisms of progenitor cell differentiation and self-renewal, and to specifically ablate any gene of interest in the progenitor cell lineage.
Supplementary Material
Acknowledgments
Financial Support: University of Pennsylvania Institute for Regenerative Medicine to L.E.G. and K.H.K. Support from NIH R01-DK58123 to R.G.W and NIH R01-DK53839 to K.H.K... Z.L. is a postdoctoral fellow of the Fred and Suzanne Biesecker Pediatric Liver Center. NIH Center Grant P-30-DK050306
We thank Beth Helmbrecht for the care of the animal colony, Yonah Esterson for technical assistance and Gary Swain and members of the University of Pennsylvania Center for Digestive Disease Morphology Core for their histology service.
List of Abbreviations
- β-gal
Beta-galactosidase
- α-SMA
alpha smooth muscle actin
- DDC
3,5-diethoxycarbonyl-1,4-dihydrocollidine
- CK19
cytokeratin 19
- HNF-4α
Hepatocyte nuclear factor-4alpha
- BDL
Bile duct ligation
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
References
- 1.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 3.Ponfick V. Ueber leberresection und leberreaction. Verhandl Deutsch Gesellesch Chir. 1890:19. [Google Scholar]
- 4.Shiojiri N. Analysis of differentiation of hepatocytes and bile duct cells in developing mouse liver by albumin immunofluorescence. Dev Growth Differ. 1984;26:555–561. doi: 10.1111/j.1440-169X.1984.00555.x. [DOI] [PubMed] [Google Scholar]
- 5.Shiojiri N, Inujima S, Ishikawa K, Terada K, Mori M. Cell lineage analysis during liver development using the spf(ash)-heterozygous mouse. Lab Invest. 2001;81:17–25. doi: 10.1038/labinvest.3780208. [DOI] [PubMed] [Google Scholar]
- 6.Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 7.Popper H, Kent G, Stein R. Ductular cell reaction in the liver in hepatic injury. J Mt Sinai Hosp N Y. 1957;24:551–556. [PubMed] [Google Scholar]
- 8.Sell S. Comparison of liver progenitor cells in human atypical ductular reactions with those seen in experimental models of liver injury. Hepatology. 1998;27:317–331. doi: 10.1002/hep.510270202. [DOI] [PubMed] [Google Scholar]
- 9.Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- 10.Dan YY, Riehle KJ, Lazaro C, Teoh N, Haque J, Campbell JS, Fausto N. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc Natl Acad Sci U S A. 2006;103:9912–9917. doi: 10.1073/pnas.0603824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaestner KH, Silberg DG, Traber PG, Schutz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 12.Kaestner KH, Bleckmann SC, Monaghan AP, Schlondorff J, Mincheva A, Lichter P, Schutz G. Clustered arrangement of winged helix genes fkh-6 and MFH-1: possible implications for mesoderm development. Development. 1996;122:1751–1758. doi: 10.1242/dev.122.6.1751. [DOI] [PubMed] [Google Scholar]
- 13.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 14.Kaestner KH, Lee KH, Schlondorff J, Hiemisch H, Monaghan AP, Schutz G. Six members of the mouse forkhead gene family are developmentally regulated. Proc Natl Acad Sci U S A. 1993;90:7628–7631. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biological programmers. Methods in Molecular biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 17.Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. Embo J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamasaki H, Sada A, Iwata T, Niwa T, Tomizawa M, Xanthopoulos KG, Koike T, et al. Suppression of C/EBPalpha expression in periportal hepatoblasts may stimulate biliary cell differentiation through increased Hnf6 and Hnf1b expression. Development. 2006;133:4233–4243. doi: 10.1242/dev.02591. [DOI] [PubMed] [Google Scholar]
- 19.Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- 20.Swenson ES, Kuwahara R, Krause DS, Theise ND. Physiological variations of stem cell factor and stromal-derived factor-1 in murine models of liver injury and regeneration. Liver Int. 2008;28:308–318. doi: 10.1111/j.1478-3231.2007.01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen BE, Grossbard B, Hatch H, Pi L, Deng J, Scott EW. Mouse A6-positive hepatic oval cells also express several hematopoietic stem cell markers. Hepatology. 2003;37:632–640. doi: 10.1053/jhep.2003.50104. [DOI] [PubMed] [Google Scholar]
- 22.Perreault N, Katz JP, Sackett SD, Kaestner KH. Foxl1 controls the Wnt/beta-catenin pathway by modulating the expression of proteoglycans in the gut. J Biol Chem. 2001;276:43328–43333. doi: 10.1074/jbc.M104366200. [DOI] [PubMed] [Google Scholar]
- 23.Perreault N, Sackett SD, Katz JP, Furth EE, Kaestner KH. Foxl1 is a mesenchymal Modifier of Min in carcinogenesis of stomach and colon. Genes Dev. 2005;19:311–315. doi: 10.1101/gad.1260605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramadori G, Neubauer K, Odenthal M, Nakamura T, Knittel T, Schwogler S, Meyer zum Buschenfelde KH. The gene of hepatocyte growth factor is expressed in fat-storing cells of rat liver and is downregulated during cell growth and by transforming growth factor-beta. Biochem Biophys Res Commun. 1992;183:739–742. doi: 10.1016/0006-291x(92)90545-v. [DOI] [PubMed] [Google Scholar]
- 25.Cassiman D, Libbrecht L, Desmet V, Denef C, Roskams T. Hepatic stellate cell/myofibroblast subpopulations in fibrotic human and rat livers. J Hepatol. 2002;36:200–209. doi: 10.1016/s0168-8278(01)00260-4. [DOI] [PubMed] [Google Scholar]
- 26.Cassiman D, Denef C, Desmet VJ, Roskams T. Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology. 2001;33:148–158. doi: 10.1053/jhep.2001.20793. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Dranoff JA, Chan EP, Uemura M, Sevigny J, Wells RG. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- 28.Lorena D, Darby IA, Reinhardt DP, Sapin V, Rosenbaum J, Desmouliere A. Fibrillin-1 expression in normal and fibrotic rat liver and in cultured hepatic fibroblastic cells: modulation by mechanical stress and role in cell adhesion. Lab Invest. 2004;84:203–212. doi: 10.1038/labinvest.3700023. [DOI] [PubMed] [Google Scholar]
- 29.Jelnes P, Santoni-Rugiu E, Rasmussen M, Friis SL, Nielsen JH, Tygstrup N, Bisgaard HC. Remarkable heterogeneity displayed by oval cells in rat and mouse models of stem cell-mediated liver regeneration. Hepatology. 2007;45:1462–1470. doi: 10.1002/hep.21569. [DOI] [PubMed] [Google Scholar]
- 30.Kuwahara R, Kofman AV, Landis CS, Swenson ES, Barendswaard E, Theise ND. The hepatic stem cell niche: identification by label-retaining cell assay. Hepatology. 2008;47:1994–2002. doi: 10.1002/hep.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung Y, Brown KD, Witek RP, Omenetti A, Yang L, Vandongen M, Milton RJ, et al. Accumulation of hedgehog-responsive progenitors parallels alcoholic liver disease severity in mice and humans. Gastroenterology. 2008;134:1532–1543. doi: 10.1053/j.gastro.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleig SV, Choi SS, Yang L, Jung Y, Omenetti A, VanDongen HM, Huang J, et al. Hepatic accumulation of Hedgehog-reactive progenitors increases with severity of fatty liver damage in mice. Lab Invest. 2007;87:1227–1239. doi: 10.1038/labinvest.3700689. [DOI] [PubMed] [Google Scholar]
- 33.Inada M, Follenzi A, Cheng K, Surana M, Joseph B, Benten D, Bandi S, et al. Phenotype reversion in fetal human liver epithelial cells identifies the role of an intermediate meso-endodermal stage before hepatic maturation. J Cell Sci. 2008;121:1002–1013. doi: 10.1242/jcs.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sicklick JK, Choi SS, Bustamante M, McCall SJ, Perez EH, Huang J, Li YX, et al. Evidence for epithelial-mesenchymal transitions in adult liver cells. Am J Physiol Gastrointest Liver Physiol. 2006;291:G575–583. doi: 10.1152/ajpgi.00102.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.