Abstract

In an effort to develop a rational approach to identify anticancer agents with selective polypharmacology, we mined millions of docked protein−ligand complexes involving more than a thousand cancer targets from multiple signaling pathways to identify new structural templates for proven pharmacophores. Our method combines support vector machine-based scoring to enrich the initial library of 1592 molecules, with a fingerprint-based search for molecules that have the same binding profile as the EGFR kinase inhibitor erlotinib. Twelve new compounds were identified. In vitro activity assays revealed three inhibited EGFR with IC50 values ranging from 250 nM to 200 μM. Additional in vitro studies with hERG, CYP450, DNA, and cell culture-based assays further compared their properties to erlotinib. One compound combined suitable pharmacokinetic properties while closely mimicking the binding profile of erlotinib. The compound also inhibited H1299 and H460 tumor cell proliferation. The other two compounds shared some of the binding profile of erlotinib, and one gave the most potent inhibition of tumor cell growth. Interestingly, among the compounds that had not shown inhibition of EGFR, four blocked H1299 and H460 proliferation, one potently with IC50 values near 1 μM. This compound was from the menogaril family, which reached phase II clinical trials for the treatment of lymphomas. This suggests that our computational approach comparing binding profiles may have favored molecules with anticancer properties like erlotinib.
Keywords: Docking, proteome, support vector machine, scoring, lung cancer, systems biology
The large number of mutations in cancer cells supports the description of cancer as a systems biology disease.1 This is especially true for lung cancer,2 often the result of years of chemical insults that lead to multiple mutations. Effective therapy for such cancers may require the design of small molecules that target multiple proteins across several signaling pathways.3 It has been argued that the effectiveness of anticancer drugs such as Gleevec is likely enhanced by off-target effects. In fact, clinically used anticancer drugs have been found to possess a greater degree of promiscuity than other FDA-approved drugs. Approaches to search for molecules that mimic the properties of existing anticancer drugs are highly desirable. It is reasonable to assume that molecules that bind to the same proteins across the human proteome as existing cancer drugs are likely going to possess a similar efficacy and pharmacokinetic (PK) profile.
Toward this end, we have followed a unique approach whereby we define a binding profile of molecules and drugs that are docked to a large number of structures within the human proteome.4 In a previous effort, thousands of compounds originating from the National Cancer Institute (NCI) diversity set were docked to structures collected from the human proteome, resulting in a large database of protein−ligand complexes known as the Docked Protein Interaction Network (DOPIN).4 Here, we extend this effort to dock the more than 1000 FDA-approved drug molecules to the human proteome. The resulting millions of protein−small molecule complexes can be used to identify compounds that specifically target proteins within DOPIN or to find small molecules that simultaneously bind and inhibit multiple targets across multiple signaling pathways. Such multitargeted agents may result in more effective cancer agents for the treatment of lung cancer.
Here, we attempt to shift the standard paradigm for screening of compounds by searching for molecules that mimic the “binding profile” of an FDA-approved drug, erlotinib. The binding profile is defined by predicted off-targets of the drug, which span multiple signaling pathways. We postulate that capturing the binding profile across a large set of targets will lead to molecules that are more likely to possess more drug-like rather than hit-like properties. The search for compounds is conducted by mining the DOPIN database that consists of millions of protein−small molecule complexes that were obtained from docking thousands of compounds and FDA-approved drugs to thousands of proteins from the human proteome. The database can be found at the following Web site: http://www.biodrugscreen.org.4
Because epidermal growth factor receptor (EGFR) is known to be the principal target of erlotinib,6 our search began by training a support vector machine (SVM) algorithm to enrich our initial library toward molecules likely to target EGFR. We report a mechanism for generating a SVM model tailored to a specific target using pair potentials from knowledge-based scoring functions. The training of a machine learning algorithm is performed using a set of positive and negative descriptors. In our approach, the positive descriptors consisted of pair potentials obtained from three-dimensional structures of protein−ligand complexes from the PDB. The negative descriptors were pair potentials computed from protein−ligand complexes that were obtained by docking a randomly selected set of 10000 molecules to EGFR. Once the training of the SVM model was completed, it was applied to identify active molecules from among the 1592 compounds of the NCI diversity set capable of binding to EGFR. Unlike existing scoring functions, the SVM algorithm classifies molecules as either “active” or “inactive”. From the NCI diversity set, 168 compounds were identified as “active”.
The next step in the search for compounds that mimic erlotinib was to compare the binding profiles of these 168 compounds to that of erlotinib. The binding profile is defined by the collection of computed binding affinities of the compounds across a large number of cancer targets from the HCPIN database known as “pathway” proteins; these targets originate from multiple signaling pathways.7 To compare the binding profile of compounds to that of erlotinib, a fingerprint is defined for each molecule as a string of bits with length equal to the total number of targets for each molecule. These strings are compared to each other using a Tanimoto coefficient as described in the Supporting Information.8,9 Because there are more than a thousand structures in our collection of the human proteome, it was felt that fingerprints containing a large number of bits would dilute the effects of the off-targets of each molecule. Instead, a select number of targets were used to define a fingerprint using predicted off-targets of erlotinib. These targets were identified by searching for those complexes that possess a computed score that was better than that of erlotinib bound to EGFR. The score used in this effort was a consensus score consisting of the average value of CHEMSCORE and GOLD. It was found that for erlotinib, 10 targets met that criterion. Including EGFR, the resulting 11 targets are listed in Table S1 of the Supporting Information. Six of the targets are kinases. This is very interesting since erlotinib is a kinase inhibitor. These findings add confidence in the simple consensus approach that was used to identify the off-targets. The kinases span several signaling pathways including MAPK, cell cycle, apoptosis, JAK, and TLR. Among the other targets, there were two GTPases, namely, Rap1A and Rac1. Analysis of the docked structure of erlotinib to these proteins revealed that the drug was occupying the substrate binding site. These GTPases affect the MAPK and TLR signaling pathways. Finally, Mad2 protein, p300, and activin receptor type IIB were also identified as potential off-targets. These proteins span multiple signaling pathways including cell cycle, JAK, and TGF.
For each compound within DOPIN, a fingerprint consisting of 11 bits was defined. A bit of 1 is assigned if the computed affinity of the compound to one of the 11 targets is greater than that of erlotinib to EGFR. The fingerprint of all compounds within DOPIN was compared to that of erlotinib. The 12 molecules that shared the greatest binding profile similarity to erlotinib were selected and are shown in Table S1 of the Supporting Information. These compounds were acquired from the NCI.
Compounds 1−12 were tested for inhibition of EGFR kinase activity at a concentration of 50 μM, using a fluorescence-based coupled-enzyme assay. Three of the twelve compounds were active (25, 30, and 90% inhibition). A concentration-dependent study of these three compounds is shown in Figure 1A. Compounds 3, 7, and 12 inhibited EGFR with IC50 values of 115, 200, and 0.25 μM, respectively. The structures of the three compounds are shown in Figure 1B. As expected, erlotinib inhibited potently, with an estimated IC50 of 2 nM.
Figure 1.

(A) Concentration-dependent inhibition of EGFR kinase activity. (B) Chemical structure of erlotinib and EGFR inhibitors.
An attempt to understand the structural basis for inhibition of EGFR by compounds 7 and 12 was carried out. The binding mode of erlotinib and these compounds to the EGFR active site is shown in Figure 2. Like erlotinib, both compounds exploit a deep cavity within the ATP binding pocket of EGFR. Erlotinib binds to this pocket through its acetylene and anilino ring. Compounds 7 and 12 bind to this pocket through a bromoaniline and six-membered pyran substituent, respectively. Erlotinib also occupies a long tunnel-like solvent-exposed region of the cavity through its quinazoline and ether linkages. This cavity is not exploited by compounds 7 and 12.
Figure 2.

Stereoviews of the three-dimensional structures of (A) erlotinib, (B) compound 7, and (C) compound 12 in complex with EGFR. The structure of the EGFR−erlotinib complex was obtained from the PDB databank (accession code: 1M17). The protein is depicted in ribbon representation colored in cyan. The binding pocket was rendered as a Connolly surface representation color-coded by its electrostatic potential. Compounds are shown in capped stick representation (yellow for carbon, red for oxygen, and blue for nitrogen).
To test whether there exist any alternative binding sites for compounds 7 and 12, we docked these compounds to cavities on the structure of EGFR that were located using the program SiteMap from the Schrodinger Inc. package. Among the five sites that were identified, the score for compound 12 bound to the ATP binding site was the most favorable by nearly 2 kcal/mol when compared to the next most favorable site. For compound 7, however, it appears that the score for binding to the ATP site was slightly less favorable in energy than the most favorable binding site by a negligible 0.2 kcal/mol. We resorted to another approach to establish the binding site for this compound by using a blind docking procedure such that the molecule was docked onto the entire protein surface. Out of the 10 docking runs, eight resulted in compound 7 binding to the ATP site, strongly suggesting that the ATP site is favored over the other site.
Finally, it is worth mentioning that EGFR may not be the only kinase target for compounds 3, 7, and 12. To provide insight into other potential kinase off-targets for compounds 7 and 12, we searched the DOPIN database (http://www.biodrugscreen.org). We found a number of kinases that exhibited scores that were better than those of 7 and 12 bound to EGFR, providing additional evidence that these compounds target multiple signaling pathways. The kinase potential off-targets are listed in Table S2 of the Supporting Information.
It is encouraging that the combined SVM scoring and multi-target approach led to molecules that inhibit the known target of erlotinib EGFR. However, our main objective in this work is to find molecules that share some of the efficacy and suitable PK properties of erlotinib. To further assess the similarities between these compounds and erlotinib, in vitro evaluation of three PK properties, namely, human ether-a-go-go related gene (hERG) K+ channel binding, cytochrome p450 CYP2C9 inhibition, and DNA binding, was performed. The hERG K+ channel is a cardiac ion channel whose inhibition is a major risk factor for arrhythmia.10 CYPs detoxify harmful compounds and catalyze key reactions in the biosynthesis of endogenous hormones,11 thus making their inhibition highly undesirable. Finally, DNA binding serves as an indicator of specificity and may suggest the potential for toxicity.
Data in Figure 3 reveal that at a 6 μM concentration, erlotinib does not block the hERG K+ channel, as should be expected for an approved drug. E-4031, a known hERG K+ channel blocker, shows complete inhibition. At this same concentration, compounds 3 and 12 exhibited significant blockage, while compound 7 showed little effect. Among the compounds that did not inhibit EGFR activity but showed inhibition of tumor cell proliferation (see below), a range of properties was observed. Compound 2 and 10 significantly inhibited CYP2C9, while 8 and 11 showed milder inhibition at 30 and 40%, respectively. hERG blockage was observed for 2 and 8, while 10 and 11 showed a less significant effect (see Figure S1 for chemical structures). Erlotinib also shows little inhibition of the CYP2C9 isozyme (Figure 3), while compound 12 showed significant inhibition of this enzyme, suggesting its potential for unfavorable effects with respect to metabolism. DNA binding assays revealed that compounds did not have a high affinity to DNA, suggesting that their effect is likely due to binding and inhibition of a protein target (Figure 3B).
Figure 3.

Percent inhibition of hERG K+ channels and CYP2C9 isozyme, with E-4031 and sulfaphenazole used as controls, respectively. All experiments were performed at 6 μM concentration.
Cell culture-based proliferation assays compared the anticancer properties of compounds 1−12 to erlotinib. In two non-small cell lung cancer cell lines (H1299 and H460), erlotinib inhibited the growth of both cell lines with IC50 values in the 10 μM range (Figure 4 and Figure S3 of the Supporting Information). Among the three compounds that inhibited EGFR activity, only 7 and 12 showed tumor cell inhibition. Compound 7 inhibited H460 and H1299 proliferation with IC50 values of 20 and 40 μM, respectively. This 6-(halogen-substituted aniline)pyrimidine compound was synthesized several decades ago.12 It was found not to possess any biological activity against leukemia and Walker carcinosarcoma tumors. Compound 12, which inhibited EGFR at a nanomolar level, exhibited potent inhibition with an IC50 nearing 1 μM in the H1299 cell line. The antitumor activities of compound 12, a natural product also known as chaetochromin A, have been known for several decades.13 Chaetochromin A is a mycotoxin that was first isolated from Chaetochromin thielavioideum.14 It has shown efficacy in breast, ovarian, and lymphatic leukemia. To the best of our knowledge, this is the first study that documents its effectiveness blocking the growth of lung tumor cells.
Figure 4.
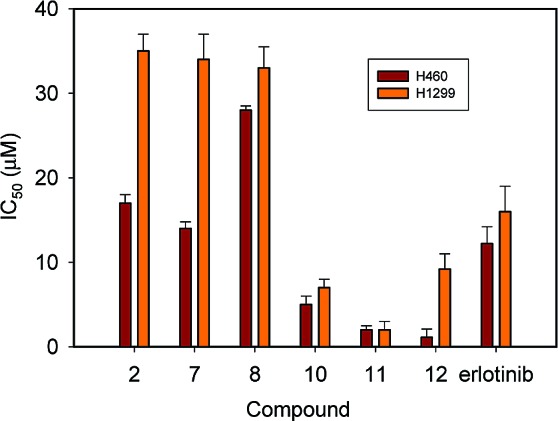
Concentration at 50% inhibition (IC50) of H460 and H1299 lung tumor cell growth by compounds and erlotinib.
It was interesting that among the compounds that did not inhibit EGFR kinase activity, four blocked cell proliferation of H1299 and H460 tumor cells, namely, compounds 2, 8, 10, and 11 (Figure 4). Compound 2, which inhibited cell growth with an IC50 of 20 and 40 μM for H460 and H1299 tumor cells, is a disubstituted pvrazolopyrimidine compound that was found to have anticancer properties several decades ago.15 It is believed that its anticancer properties arise from the isomeric relationship to purines. Compound 8, an N-substituted indole, showed inhibition at the 30 μM level. A search at the PubChem Web site did not reveal any bioassays for these compounds, although derivatives have been shown to possess antiparasitic effects. The coumarin-based compound 10 exhibited more significant inhibition of tumor cell proliferation (IC50 about 10 μM for both cell lines), exceeding the potency of erlotinib. The Pubchem Web site does not report that this compound was heretofore tested in any type of bioassay. To the best of our knowledge, this is the first study that documents anticancer properties of compounds 2 and 10. Finally, compound 11 belongs to the menogaril family of compounds, which are well-known to be potent anticancer agents.16 This is reflected by the nearly 1 order of magnitude greater potency of this compound over erlotinib. The high level of efficacy and favorable PK properties of the menogaril compounds has led to studies in humans for the treatment of stomach and breast cancer.16 A phase II clinical study for the treatment of non-Hodgkin's lymphomas was also carried out.17
The discovery that the compounds with anticancer properties have been previously known to possess anticancer properties is highly encouraging, particularly considering the fact that one molecule has made it to clinical trials. Two molecules, namely, 2 and 10, have never been previously known to possess anticancer properties. These results suggest that we have achieved our main objective toward identifying anticancer agents by exploiting a large number of targets from the human proteome across multiple signaling pathways. No less important was the fact that we arrived at these molecules by targeting multiple proteins that were identified by using the FDA-approved drug erlotinib. This drug was particularly helpful in limiting the number of targets that were considered in the search. Future studies will have to identify other mechanisms to select these targets for the design of molecules with limited polypharmacology. Another significant aspect of this work was the fact that some of these molecules shared similar PK properties to that of erlotinib, suggesting that by targeting multiple proteins that are predicted to bind to the drug, we may have captured some of the favorable PK properties of the drug, such as compound 7. While compound 12 was a potent inhibitor of EGFR, the high level of inhibition of CYP2C9 and hERG suggests that this molecule may possess excessive toxicity in vivo and may not be worth pursuing. Both compounds 7 and 12 could serve as chemical probes to study the role of EGFR and other kinases in cancer, although future efforts will have to improve the potency of compound 7 toward EGFR. Ultimately, animal studies will establish whether these molecules are viable leads for the development of cancer therapeutics.
It is also worth mentioning the innovations in the computational search. To our knowledge, machine learning has never been to date used to successfully score receptor−ligand complexes in structure-based virtual screening. We expect that the success of this approach, as evidenced by the discovery of three molecules that inhibited EGFR, will spawn additional efforts toward using this method for the ranking of receptor−ligand complexes in virtual screening. We anticipate a series of studies from our own laboratory probing this method in more detail and comparing its performance to other existing scoring functions.
We performed a computational search that combines machine learning scoring with a fingerprint analysis to search for molecules that mimic the binding profile of erlotinib, an FDA-approved oncology drug used in the clinic to treat lung cancer. It was encouraging that among 12 molecules that were selected from an original set of 1592, three showed inhibition of EGFR kinase activity. Further comparison of in vitro binding PK properties as well as tumor cell proliferation assays reveals that the compounds shared similar binding profiles to erlotinib. Compound 7, in particular, exhibited a nearly identical profile to erlotinib except for its weaker potency in inhibiting EGFR kinase activity. Compound 12, on the other hand, was a potent inhibitor of EGFR and, like erlotinib, inhibited tumor cell growth and did not bind DNA. However, the compound displayed greater promiscuity by strongly binding to hERG and CYP2C9. Interestingly, four compounds (2, 8, 10, and 11) showed little inhibition of EGFR and exhibited in some cases strong inhibition of H1299 and H460 tumor cells growth, suggesting that these compounds may potentially share off-targets with erlotinib and may also provide a basis for the development of cancer therapeutics. These results suggest that a search based on a binding profile could be a viable approach to identify molecules that mimic the properties of existing drugs.
Acknowledgments
We are grateful to Jed F. Fisher for reading the manuscript and his valuable comments.
Supporting Information Available
Supporting Information contains experimental and computational procedures as well as chemical structures of all compounds discussed in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
This research was supported by the NIH (S.O.M.) and the INGEN grant from the Lilly Endowment, Inc. (S.O.M.). Computer time on the Big Red supercomputer at Indiana University was funded by the National Science Foundation and by Shared University Research grants from IBM, Inc. to Indiana University. We thank the Cancer Free Lungs for a fellowship to L.L.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hornberg J. J.; Bruggeman F. J.; Westerhoff H. V.; Lankelma J. Cancer: A systems biology disease. Biosystems 2006, 83, 81–90. [DOI] [PubMed] [Google Scholar]
- Pleasance E. D.; Stephens P. J.; O'Meara S.; McBride D. J.; Meynert A.; Jones D.; Lin M. L.; Beare D.; Lau K. W.; Greenman C.; Varela I.; Nik-Zainal S.; Davies H. R.; Ordonez G. R.; Mudie L. J.; Latimer C.; Edkins S.; Stebbings L.; Chen L.; Jia M.; Leroy C.; Marshall J.; Menzies A.; Butler A.; Teague J. W.; Mangion J.; Sun Y. A.; McLaughlin S. F.; Peckham H. E.; Tsung E. F.; Costa G. L.; Lee C. C.; Minna J. D.; Gazdar A.; Birney E.; Rhodes M. D.; McKernan K. J.; Stratton M. R.; Futreal P. A.; Campbell P. J. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010, 463, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenwitheesuk E.; Horst J. A.; Rivas K. L.; Van Voorhis W. C.; Samudrala R. Novel paradigms for drug discovery: Computational multitarget screening. Trends Pharmacol. Sci. 2008, 29, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Bum-Erdene K.; Baenziger P. H.; Rosen J. J.; Hemmert J. R.; Nellis J. A.; Pierce M. E.; Meroueh S. O. BioDrugScreen: A computational drug design resource for ranking molecules docked to the human proteome. Nucleic Acids Res. 2010, 38, D765–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd F. A.; Rodrigues Pereira J.; Ciuleanu T.; Tan E. H.; Hirsh V.; Thongprasert S.; Campos D.; Maoleekoonpiroj S.; Smylie M.; Martins R.; van Kooten M.; Dediu M.; Findlay B.; Tu D.; Johnston D.; Bezjak A.; Clark G.; Santabarbara P.; Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 2005, 353, 123–132. [DOI] [PubMed] [Google Scholar]
- Huang Y. J.; Hang D.; Lu L. J.; Tong L.; Gerstein M. B.; Montelione G. T. Targeting the human cancer pathway protein interaction network by structural genomics. Mol. Cell. Proteomics 2008, 7, 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bull. Soc. Vaudoisedes Sci. Naturelles 1901, 37, 241–272. [Google Scholar]
- Butina D. Unsupervised data base clustering based on Daylight's fingerprint and Tanimoto similarity: A fast and automated way to cluster small and large data sets. J. Chem. Inf. Comput. Sci. 1999, 39, 747–750. [Google Scholar]
- Keating M. T.; Sanguinetti M. C. Molecular and cellular mechanisms of cardiac arrhythmias. Cell 2001, 104, 569–580. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 2001, 14, 611–650. [DOI] [PubMed] [Google Scholar]
- O'Brien D. E.; Weinstock L. T.; Cheng C. C. Pyrimidines. XXII. 2,4-Diamino-6-aryl-amino-5-pyrimidinecarboxaldehydes and related compounds. J. Med. Chem. 1968, 11, 387–388. [DOI] [PubMed] [Google Scholar]
- Koyama K.; Ominato K.; Natori S.; Tashiro T.; Tsuruo T. Cytotoxicity and antitumor activities of fungal bis(naphtho-gamma-pyrone) derivatives. J. Pharmacobiodyn. 1988, 11, 630–635. [DOI] [PubMed] [Google Scholar]
- Sekita S.; Yoshihira K.; Natori S.; Udagawa S.; Sakabe F.; Kurata H.; Umeda M. Chaetoglobosins, cytotoxic 10-(indol-3-yl)-[13]cytochalasans from Chaetomium spp. I. Production, isolation and some cytological effects of chaetoglobosins A-J. Chem. Pharm. Bull. (Tokyo) 1982, 30, 1609–1617. [DOI] [PubMed] [Google Scholar]
- Skipper H. E.; Robins R. K.; Thomson J. R.; Cheng C. C.; Brockman R. W.; Schabel F. M. Jr. Structure-activity relationships observed on screening a series of pyrazolopyrimidines against experimental neoplasms. Cancer Res. 1957, 17, 579–596. [PubMed] [Google Scholar]
- Inaba M.; Sato S.; Yamori T.; Tashiro T.; Ohnishi Y.; Maruo K.; Ueyama Y.; Tsuruo T. Anticancer activities of orally administered menogaril against human stomach and breast cancers implanted in nude mice. Anticancer Res. 1992, 12, 1953–1956. [PubMed] [Google Scholar]
- Moore D. F. Jr.; Brown T. D.; LeBlanc M.; Dahlberg S.; Miller T. P.; McClure S.; Fisher R. I. Phase II trial of menogaril in non-Hodgkin's lymphomas: A Southwest Oncology Group trial. Invest. New Drugs 1999, 17, 169–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


