Abstract
Eight novel families of miniature inverted repeat transposable elements (MITEs) were discovered in the African malaria mosquito, Anopheles gambiae, by using new software designed to rapidly identify MITE-like sequences based on their structural characteristics. Divergent subfamilies have been found in two families. Past mobility was demonstrated by evidence of MITE insertions that resulted in the duplication of specific TA, TAA, or 8-bp targets. Some of these MITEs share the same target duplications and similar terminal sequences with MITEs and other DNA transposons in human and other organisms. MITEs in A. gambiae range from 40 to 1340 copies per genome, much less abundant than MITEs in the yellow fever mosquito, Aedes aegypti. Statistical analyses suggest that most A. gambiae MITEs are in highly AT-rich regions, many of which are closely associated with each other. The analyses of these novel MITEs underscored interesting questions regarding their diversity, origin, evolution, and relationships to the host genomes. The discovery of diverse families of MITEs in A. gambiae has important practical implications in light of current efforts to control malaria by replacing vector mosquitoes with genetically modified refractory mosquitoes. Finally, the systematic approach to rapidly identify novel MITEs should have broad applications for the analysis of the ever-growing sequence databases of a wide range of organisms.
Keywords: transgenic insects, interspersed repeats, genome, evolution, bioinformatics
Mosquitoes transmit a number of diseases that are among the deadliest in human history. Malaria, the most devastating mosquito-borne disease, is responsible for more than a million deaths every year in tropical and subtropical countries. The impact of malaria and other mosquito-borne diseases is on the rise because of increasing insecticide resistance by mosquitoes and drug resistance by the pathogens. Novel strategies to control the transmission of these diseases are clearly urgently needed. One approach is to create a disease-resistant mosquito by genetic manipulation and to replace vector mosquitoes in wild populations with the genetically modified refractory mosquitoes. The success of this strategy hinges on three major steps: the identification of genes that confer refractory traits, the development of efficient and stable transformation systems, and a clear understanding of the mechanisms of the spread of genetic elements in mosquito populations. Major efforts are underway and significant progress has been made in these research areas (1, 2). In addition to these specific steps, a better understanding of the basic genetics of the vector mosquitoes is essential to ensure a sustained success of such a sophisticated genetic approach and to minimize potential risks. Genetic information on endogenous DNA transposable elements in mosquitoes is specially relevant considering that most of the transformation tools tested in mosquitoes are derived from exogenous DNA transposable elements (1, 2), some of which have been shown to interact with endogenous elements (3, 4). In addition, analysis of endogenous DNA transposable elements will provide useful information regarding their spread, evolution, and interactions with the mosquito genomes.
DNA transposable elements such as P, hobo, and mariner are characterized by terminal inverted repeats (TIRs) flanking a gene encoding a transposase. Recently, several families of short interspersed elements with TIRs have been found in a wide range of organisms, including plants, vertebrates, insects, and a nematode (e.g., refs. 5–16). These elements, named miniature inverted repeat transposable elements (MITEs), share common structural characteristics such as TIRs, small size, no coding potential, AT richness, and the potential to form stable secondary structures (17). MITEs may have been using the transposition machinery of autonomous DNA transposable elements, taking advantage of shared TIRs (7, 9, 18, 19). However, MITEs are a distinct group of elements that are not simply deletion derivatives of the autonomous elements. MITEs are generally homogeneous in size. The sequence similarity between most MITEs and their corresponding autonomous elements is limited to the TIRs (7, 9, 19). It has been shown that MITEs are significant components in several eukaryotic genomes (6–12, 17). Many MITEs have been found near genes where they could potentially be involved in gene regulation and/or defining chromatin domains (17, 20).
Several DNA transposable elements have been documented in Anopheles gambiae, the primary vector of human malaria (refs. 21 and 22; http://bioweb.pasteur.fr/BBMI). Here I report the discovery of eight novel families of MITEs in a newly released A. gambiae sequence tagged site (STS) database (Genoscope and Institut Pasteur, Paris), by using a computer program specifically designed to rapidly search for MITEs according to their common characteristics. This study represents a systematic analysis of a large group of endogenous transposable elements in A. gambiae, which revealed tremendous diversity. The characteristics, abundance, genomic distribution, and evolution of these elements have been analyzed. The relationship between these MITEs and DNA transposable elements with coding capacities have been explored. These discoveries have important implications to the current efforts to control malaria by genetic modification of mosquitoes.
Materials and Methods
Database Searches Using findmite.
findmite is a C program designed to rapidly search a database for sequences that have the characteristics of MITEs. The program searches the database for inverted repeats flanked by user-defined direct repeats within a specified distance range. The program uses the idea of the Knuth–Morris–Pratt string matching algorithm (23) to speed up the pattern match shifts. Two major modifications include replacing A, T, G, and C with integers and allowing mismatches. The program was tested with simulated data as well as small databases containing known MITEs. A copy of the software will be provided upon request and it will also be posted on the internet for download (http://www.biochem.vt.edu/aedes). The database used in this study contains 17,509 STSs with an average size of 829 bp, generated by Genoscope and the Institut Pasteur (http://www.genoscope.cns.fr, February 2000 release). Potential MITEs were searched for that satisfy the following specifications: direct repeat, NNNN, NNNNNNNN, TAA, TAT, TTA, or TA, respectively; length of the TIR, 11 bp; allowed mismatch, 1; distance between the inverted repeats, 30–650 bp. TIRs solely composed of A/T strings, C/G strings, or simple repeats were filtered out. These parameters were selected according to the common features of known MITEs. Each search was completed within a few minutes on a SGI Unix server. To identify incomplete or degenerate copies, and to confirm their repetitive nature, potential MITEs identified in the above analysis were used to search the same STS database with blast (24) and fasta of GCG (Genetics Computer Group, Madison, WI, Version 10, 1999) after removing unlikely candidates by visual inspection.
Analysis of MITEs and Flanking Sequences.
GCG programs were used for sequence analysis. These include gap and bestfit for pairwise comparison, pileup for multiple sequence alignment, and pretty for consensus construction. Both mfold of GCG and genequest of Lasergene (DNASTAR, Madison, WI) were used to predict secondary structures, which gave consistent results. The following formula was used to estimate the copy number of MITEs: copy no. = (no. in database × genome size)/database size. The A. gambiae haploid genome is 270 Mbp (25). The number of elements in the database was determined for each family by the number of entries that matched the consensus at a P value below 0.001 during a blast search. There is a 3% redundancy in the search results, as noted in Table 1. However, it does not affect the estimation of the copy number as similar redundancy rate would likely apply to the entire database. The flanking sequences of confirmed MITEs were used to search the A. gambiae genome database to identify evidence of MITE insertions that resulted in target duplications. AT contents were calculated using a C program named atcontent, which implements the following formula: AT content = (number of A + T + W)/(number of A + T + W + G + C + S). Ambiguous nucleotides other than W (A or T) or S (G or C) were not counted. Poly(A) tails of expressed sequence tags (ESTs) were ignored.
Table 1.
Characteristics of A. gambiae MITEs
| Element | Target | TIR | Length | No. in database | No. in genome | No. full-length* | AT†, % | Variation‡, % | −ΔG§, kcal/mol |
|---|---|---|---|---|---|---|---|---|---|
| TA-Iα-Ag | TA | CAGGCGGTCCCCGAGATACACGGT | 365 | 72 | 1340 | 10 | 62.2 | 3 to 28 | 98 |
| TA-Iβ-Ag | TA | CAGTCTKTYCCCGAGTTACGCGGWT | 346 | 27 | 500 | 8 | 65.6 | 7 to 38 | 55 (91) |
| TA-IIα-Ag | TA | CAGTGGAGCGCCGTTTATCCGGG | 358 | 34 | 630 | 9 | 61.9 | 10 to 23 | 91 |
| TA-IIβ-Ag | TA | CAGTAGAACGTCGATTATCCGGG | 379 | 24 | 450 | 6 | 60.2 | 3 to 23 | 101 |
| TA-III-Ag | TA | CAGGGTTTCCCACGATTTATTGGT (54 bp) | 245 | 52 | 970 | 24 | 62 | 0.4 to 26 | 121 |
| TA-IV-Ag | TA | CAGTAGGTGACCGCTAACTG | 363 | 7 | 130 | 3 | 63.7 | 5 to 9 | 86 |
| TA-V-Ag | TA | CAGTgAACcCTCTCTTATTTGA | 348 | 16 | 300 | 5 | 62.8 | 3 to 20 | 45 (70) |
| TAA-I-Ag | TAA | CGGCCAAGCTACACGTACCGGACGACATCGGACRATGC | 184 | 2 | 40 | 2 | 46.7 | 7 | 53 (95) |
| TAA-II-Ag | TAA | TACGGACGTCACACGAGGCGTAAACT | 142 | 17 | 320 | 9 | 56.8 | 2 to 25 | 59 |
| Joey¶ | TAA | AGGCCGGGGTACAYTGTCCGTACTCGCTAGT (69 bp) | 351 | 60 | 1120 | 10 | 56.5 | 2 to 24 | 146 |
| 8bp-I-Ag | NTTTANAN | CAGGGGTCTCCAAACT | 320 | 39 | 725 | 14 | 61.8 | 2 to 39 | 40 (75) |
| Pegasus‖ | NNNNNNNN | CAGTGTTG | 534 | 5 | 90 | 0 | 64.5 | 1 to 5 | 117 |
The new MITEs are named according to their target sequences, which are followed by Roman numerals. Ag represents the first letters of the genus and species names. α and β indicate subfamilies.
A total of 103 full-length MITEs were identified, three of which were redundant copies. Thus the rate of redundancy is approximately 3%.
Average AT content of the full-length sequences. The sample sizes of these MITEs are listed in the 7th column of the table, except for Pegasus. Note that TAA-I-Ag is the only GC-rich element.
The variations were calculated by using paup (see Materials and Methods) based on pairwise differences of the full-length copies.
These are negative ΔG value of the consensus sequence of each family. Some families have smaller negative ΔG values because of a large number of degenerate bases in their consensus sequences. Shown in each bracket is the lowest ΔG value of an individual element of the family. ΔG values were calculated using mfold of GCG (Genetics Computer Group, Madison, WI, Version 10, 1999).
Joey was first discovered as an insertion in a Pegasus element by Besansky et al. (22). Current analysis identified multiple copies of Joey elements that provided the basis for the characterization and the estimation of its copy number.
Six full-length Pegasus elements were discovered by Besansky et al. (22). The structural information provided here is based on analysis of these elements. None of the five Pegasus elements identified in the STS database is full-length. The copy number of Pegasus estimated here is higher than the 34 copies estimated with use of hybridization methods (22), which may not be able to detect highly degenerate copies.
Statistical Analysis.
The two-sample Mann–Whitney test was used for the nonparametric comparison between medians of different datasets. For parametric analyses of the means, either a pooled-variance t test or a “Welch's approximate t test” was used based on the result of an F-test which estimates the probability of equal variance between two data populations (26). An one-tailed binomial test was used to estimate the probability of finding N or more sequences that contain at least two MITEs, assuming random distribution (26). All statistical tests and calculations were performed by using minitab 10.5 (MINITAB, State College, PA). Unless otherwise noted, statistical tests were performed at α = 0.05.
Results
Discovery and Characterization of Eight Novel Families of MITEs in A. gambiae.
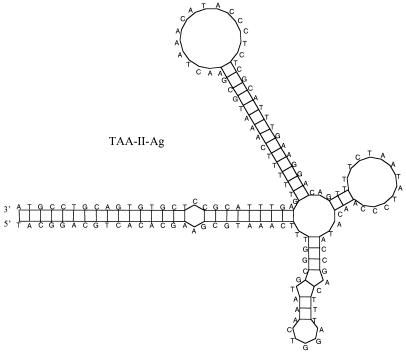
As shown in Table 1, eight novel families of MITEs were discovered in the A. gambiae STS database by using findmite as described in Materials and Methods. Two of these consist of divergent subfamilies as described below. In addition, multiple copies of a previously identified single insertion sequence named Joey (22) were also found during the search, establishing them as an independent family of MITEs. The boundaries of each family/subfamily and the putative target site duplications were determined based on multiple sequence alignments, which have been deposited in the EMBL alignment database (accession nos. DS43373–DS43385). Insertion events were identified for six of the eight new families, which demonstrated their previous mobility (Fig. 1). The alignments in Fig. 1 also confirmed the actual boundaries between the TIRs and the target duplications. The names and classifications of these MITEs are described in Table 1, which are based mainly on their target sequences. Consensus sequences were constructed by using alignments of the full-length elements within each family. Analyses described in Table 1 confirmed that these families are novel MITEs as they share all or most of the characteristics of MITEs including TIRs, no coding potential, AT richness, small size, and the potential to form stable secondary structures. In addition, most complete copies in a family are homogeneous in size. Shown in Fig. 2 is the predicted secondary structure of TAA-II-Ag, which is a good representation of the nonhairpin structures of most of the A. gambiae MITEs. One exception is TAA-I-Ag, which has the potential to form a simple hairpin structure (data not shown).
Figure 1.
Evidence of past mobility of some of the newly discovered MITEs in A. gambiae. The names of these MITEs are described in Table 1. The sequences were aligned by using gap of GCG (Genetics Computer Group, Madison, WI, Version 10, 1999) with gap weight = 40 and gap length weight = 0. The top sequences in the alignments contain MITE insertions that are not present in the bottom sequences. The bottom sequences were identified in the A. gambiae sequence tagged site (STS) database during blast searches using sequences flanking MITEs as queries. Two elements, one from the TA-IIα-Ag family (AL151950) and the other from the TA-III-Ag family (AL155989), are inserted in a middle repetitive sequence (37). The putative target duplications are underlined. Note that the target duplication flanking the TAA-II-Ag in AL141968 is different from the target consensus TAA.
Figure 2.
Predicted secondary structure of the consensus sequence of TAA-II-Ag. Multiple sequence alignment of the full-length elements used to create the consensus sequence has been deposited in the EMBL database (accession no. DS43382). The structure was plotted by using genequest of Lasergene (DNASTAR, Madison, WI), which is identical to the structure predicted by using mfold of GCG (Genetics Computer Group, Madison, WI, Version 10, 1999).
As shown in Table 1, five of the MITEs are flanked by TA target duplications (TA-I-Ag to TA-V-Ag). Three additional MITEs, TAA-I-Ag, TAA-II-Ag, and Joey are flanked by TAA duplications, whereas one other MITE, 8bp-I-Ag, and a previously characterized element Pegasus (22), are flanked by 8-bp repeats. Although there are no overall sequence similarities between different families of MITEs, the first 3 bases of the TIRs of all TA-specific and 8-bp MITEs are invariably CAG. However, the TIRs of the three TAA-specific MITEs in A. gambiae do not share any similarities. Analysis of the sequence alignments (accession nos. DS43375 and DS43383) and phylogenetic inference (data not shown) suggests that TA-I-Ag and TA-II-Ag consist of divergent subfamilies, namely TA-Iα-Ag (accession no. DS43384), TA-Iβ-Ag (accession no. DS43385); and TA-IIα-Ag (accession no. DS43376), TA-IIβ-Ag (accession no. DS43377). Subgroupings were also found in these subfamilies. As shown in Fig. 3, the consensus sequences of TA-Iα-Ag and TA-Iβ-Ag are 66.2% similar, whereas the consensus sequences of TA-IIα-Ag and TA-IIβ-Ag are 61.7% similar. In addition to the TIRs, the subterminal repeats are also conserved between the subfamilies, which is consistent with the hypothesis that subterminal repeats may play important structural or functional roles in some MITEs (12). These subfamilies are analyzed independently in subsequent analyses. The copy number of these MITEs ranges from 40 to 1340. Together they constitute up to 0.8% of the entire genome.
Figure 3.
(A) Pairwise comparison between the consensus sequences of the two subfamilies of TA-I-Ag: TA-Iα-Ag and TA-Iβ-Ag. Multiple sequence alignments of the full-length elements used to create the two consensus sequences were deposited in the EMBL database (accession nos. DS43384 and DS43385). The two consensus sequences were aligned by using gap of GCG (Genetics Computer Group, Madison, WI, Version 10, 1999) with gap weight = 30 and gap length weight = 1. Thick arrows mark the TIRs, and thin arrows mark the subterminal repeats. Flanking TA target duplications are not shown. D = A, G, T; H = A, C, T; K = G, T; M = A, C; N = A, C, G, T; R = A, G; S = G, C; V = G, A, C; W = A, T; Y = C, T. (B) Pairwise comparison between the consensus sequences of the two subfamilies of TA-II-Ag: TA-IIα-Ag and TA-IIβ-Ag. Multiple sequence alignments used to create the consensus sequences were deposited in the EMBL database (accession nos. DS43376 and DS43377). The two consensus sequences were aligned by using gap as described in A. All symbols are as in A.
High AT Content of MITEs and Flanking Sequences.
The average AT content of the A. gambiae genome is 54.7 ± 0.05% (mean ± SEM), based on the content of the 17,509 STSs. As described in the Fig. 4 legend, the average AT contents of the forward and reverse ESTs in the A. gambiae EST database (27) are 49.6 ± 0.72% and 47.9 ± 0.63%, respectively, significantly lower than the genome average (P < 0.0001). All TA-specific and 8-bp MITEs have significantly higher AT contents (60.2–65.6%) than the genome average, the forward and reverse ESTs, and the three TAA-specific MITEs. Although TAA-II-Ag and Joey contain significantly more AT (56.5–56.8%) than the ESTs, they are not significantly different from the genome average. Although TAA-I-Ag contains significantly less AT (46.7%) than the genome average, it is not significantly different from the ESTs. The flanking sequences of all MITEs but TAA-I-Ag contain quite high levels of AT (61.2–65.5%), significantly higher than both the genome average and the ESTs. The AT contents of sequences flanking TAA-I-Ag are not significantly different from either the genome average or the ESTs. It should be noted that the statistical tests involving TAA-I-Ag were not very powerful because only two copies of TAA-I-Ag were found in the database.
Figure 4.
Average AT contents of MITEs and their flanking sequences compared with STS and EST sequences in the A. gambiae database. AT contents of all full-length MITEs (see Table 1 for sample sizes) and their flanking sequences (STS minus MITE, indicated by the suffix “F”), and all of the 17,509 STS sequences in the A. gambiae genome database were calculated. Calculations of the Pegasus elements and their flanking regions were based on sequences reported by Besansky et al. (22). The forward and reverse sequences of the A. gambiae ESTs were analyzed separately because many of them represent pairs of sequences covering different regions of the same clone. Two hundred ESTs were randomly selected from each of the 2,990 forward ESTs and the 2,936 reverse ESTs (27). They were analyzed by using blast to remove redundancy that resulted from multiple copies of cDNAs. AT contents of 186 nonredundant forward ESTs (EST-For) and 181 nonredundant reverse ESTs (EST-Rev) were calculated and analyzed. Data points represent the mean AT contents. The error bar represents the SEM. Note that the standard errors for several data points are too small to be shown at the current scale. Mann–Whitney tests were used to compare the medians at α = 0.05. In most cases, t-tests were also used to compare the means, which gave the same conclusions. Samples in tier I have significantly higher AT contents than samples in tier II and III, whereas most samples in tier II have significantly higher AT contents than samples in tier III. One exception is TAA-I-AgF of tier II, which has a small sample size. Its AT content is neither significantly higher than samples in tier III nor significantly lower than TA-Iα-AgF, TA-IIα-AgF, TA-IV-AgF, and TA-IV-Ag of tier I. The other exception is the comparison between TAA-I-Ag and TA-IV-Ag, which is not significantly different. Samples in tier II are not significantly different from each other while EST-For is slightly more AT-rich than EST-Rev in tier III (P = 0.045). A few samples in tier I are slightly more AT-rich than others.
Distribution of MITEs.
Seventeen of the 340 MITE-containing STSs contain two MITEs, whereas one STS contains three. Under the assumption of random distribution, the probability of finding 18 or more sequences that contain at least two MITEs, P(X ≥ 18), can be calculated using an one-tailed binomial test. X is the number of sequences that contain at least two MITEs. The probability that a given STS contains at least two MITEs is P′ = P2 = (340/17509)2 =0.0003764, where P is the probability that a given STS contains at least one MITE. P(X ≥ 18) = 1− P(X ≤ 17). P(X ≤ 17) is the cumulative binomial probability of 17 or fewer successes in 17,509 trials given the probability of P′. P(X ≤ 17) = 0.9998 and P(X ≥ 18) = 0.0002. Therefore, it is significantly more likely to find a second MITE in a MITE-containing sequence than by random chance.
Discussion
The study presented here represents a systematic analysis of a large group of endogenous transposable elements in the primary malaria vector, A. gambiae. As discussed below, such analyses have important implications to the current efforts to control malaria by genetic modifications of mosquitoes. In addition, results described in this study demonstrated a homology-independent approach to rapidly identify novel MITEs through a systematic analysis of a relatively large database. This is especially important for the study of MITEs because database analysis based on homology to known MITEs has limited applications because of the lack of significant overall sequence conservation between MITEs in divergent species. Furthermore, in contrast to the previously described method for identifying inverted repeats, which is computationally intensive (6), the current method is able to handle large databases because of the speed afforded by fully incorporating the common characteristics of MITEs such as flanking direct repeats, TIRs, and small size. Because a whole-genome database is not required, this systematic approach could have broad applications for the analysis of the model genomes as well as the vast majority of the less sequenced genomes.
Diverse Families of MITEs in A. gambiae.
There is a tremendous diversity in the A. gambiae MITEs. These MITEs, including the eight families discovered in this study and two previously identified MITE-like families Joey and Pegasus (22), can be grouped into three categories based on their insertion target sequences. There are five families of TA-specific MITEs, three TAA-specific MITEs, and two 8-bp MITEs. In addition, highly divergent subfamilies with distinct subgroups were present in two of the TA-specific MITEs (Fig. 3, and EMBL alignments DS43375 and DS43383), which is consistent with the hypothesis that more than one source gene was amplified during the evolution of some MITEs (12). Unlike the yellow fever mosquito, Aedes aegypti, and a few vertebrate species, no 4-bp MITEs were found in A. gambiae. Although the search parameters were selected to encompass the common features of known MITEs, as described in Materials and Methods, there may exist other MITEs in A. gambiae that do not fit these parameters, which will of course not be identified in this survey.
As shown in Table 1, the three TAA-specific MITEs in A. gambiae do not share any sequence similarities even at the TIRs. They have no sequence similarities to the MITEs in plants that are flanked predominantly by TAA (or TTA) repeats. On the other hand, the first three bases of the TIRs of the TA-specific and the 8-bp MITEs in A. gambiae are invariably CAG. As shown in Table 2, more than half of the TA-specific MITEs in A. gambiae share similar terminal repeats with a TA-specific MITE in man and a Mimo element in a Culex mosquito (10, 16). These MITEs also share similar TIRs with a few Tc1-pogo DNA transposons including Tsessebe I of A. gambiae (21). The phrase “DNA transposon” here refers to a DNA element that has the coding capacity for its transposase, which may or may not be still active. A number of TA-specific MITEs have been found in a nematode Caenorhabditis elegans and the yellow fever mosquito, A. aegypti (7, 11, 12). However, MITEs containing the Tc1-pogo-type TIRs are not the major elements in these species. In addition, 8bp-I-Ag in A. gambiae shares the same AT-rich 8-bp target and very similar TIRs with the human MER30 (10). Both elements have TIRs similar to the autonomous Ac element, a member of the hAT superfamily (10). All of the sequence similarities described above are limited to the target site and the TIRs only. Nevertheless, it is intriguing that the TA-specific Tc1-pogo type MITEs and the 8-bp MITEs are the predominant MITEs in both human and the A. gambiae genomes. It is also interesting to note that Tc1-Pogo and hAT DNA transposons, which share similar TIRs with diverse families of MITEs in divergent organisms, are two groups of the most widely distributed DNA transposons in eukaryotes.
Table 2.
Conservation in target sequences and terminal inverted repeats between MITEs and autonomous DNA transposons in diverse organisms
| Element | Target | TIR* | Size |
|---|---|---|---|
| Ac† | 8-bp | CAGGGaTGaaaA | 4560 |
| 8bp-I-Ag‡ | NTTTANAN | CAGGGGTcTCCAAaCt | 320 |
| MER30§ | NTYTANAN | CAGGGGTGTCCAAtC | 230 |
| pogo¶ | TA | CAGTA-TaattCGcTTAgCTGctcga | 2121 |
| Tsessebe I‡ | TA | CAGTA-TcgaCaGaaWgataG | 2055 |
| TA-IV-Ag‡ | TA | CAGTAGgtgaCCGcTaA-CTGGt | 363 |
| Mimo‖ | TA | CAGTAGTtgttCGgTaA-CTGGGc | 324 |
| TA-IIα-Ag‡ | TA | CAGTgGagCgCCGtTTATCcaGGt | 358 |
| TA-IIβ-Ag‡ | TA | CAGTAGaaCgtCGaTTATCcGGG | 379 |
| MER44A§ | TA | CAGTAGTcCcCCc-TTATCcGcGg | 333 |
| TA-V-Ag‡ | TA | CAGTgaacCctCtcTTATtTGa | 348 |
Consensus of each family is used in the comparison. Uppercase letters indicate nucleotides that are the same as the majority in the group. Lowercase letters indicate nucleotides that are different from the majority.
Ac is a maize autonomous DNA transposon of the hAT superfamily (19).
8bp-I-Ag, TA-IIα-Ag, TA-IIβ-Ag, TA-IV-Ag, and TA-V-Ag are A. gambiae MITEs reported in this paper. Tsessebe I is a Tcl-pogo DNA transposon in A. gambiae (21).
MER30 and MER44A are MITEs found in man (10).
pogo is a DNA transposon in the fruit fly, Drosophila melanogaster (38).
Mimo is a MITE in a mosquito Culex pipiens (16).
MITEs and DNA Transposons in A. gambiae.
The similarities to different DNA transposons at the insertion target and the TIRs support the hypothesis that MITEs may have been borrowing the transposition machinery from autonomous DNA transposons. As discussed above, Tsessebe I, a Tc1-pogo type DNA transposon in A. gambiae (21), share similar TIRs with some of the A. gambiae MITEs. However, it is not clear whether Tsessebe I had been involved in mobilizing these MITEs because the similarities between their TIRs are limited. The discovery of diverse families of MITEs in A. gambiae suggests that its genome once harbored, or may still harbor a variety of DNA transposons that may be responsible for the mobilization of these MITEs. In addition to Tsessebe I, a few families of DNA transposons including mariner and hobo-like elements, have been documented in the analysis accompanying the release of the A. gambiae STS database (http://bioweb.pasteur.fr/BBMI). Because of the short length of the STS, the sequences of these DNA transposons are not complete. It will be interesting to see whether every A. gambiae MITE shares similar TIRs with an endogenous DNA transposon in the genome. With a few possible exceptions (18), most MITEs that share similar terminal sequences and insertion targets have no internal homology to each other or to the “related” DNA transposons. Consistent with the above observation, no A. gambiae MITEs were found to have internal sequence similarities with any known DNA transposons. Therefore, it is reasonable to hypothesize that many families of MITEs could have originated from chance events that generated a pair of inverted repeat sequences that can be mobilized by endogenous DNA transposons (12, 19). The above hypothesis and the hypothesis of MITEs being derived from internal deleted autonomous DNA transposons (18) are not mutually exclusive.
MITEs and Mosquito Genomes.
The availability of a large number of A. gambiae STS sequences provided an opportunity to analyze the distribution of MITEs in the context of the host genome. Statistical analyses indicated that the distribution of A. gambiae MITEs is highly biased toward AT-rich regions and there is a nonrandom association between different families of MITEs. Biased distribution of some MITEs has also been shown in a recent survey of the C. elegans genome (28). A blast search of the consensus sequences of A. gambiae MITEs against an A. gambiae EST database representing 2,380 independent genes (27) indicated that only three of the ESTs contain a MITE, which is consistent with the observation that MITEs are seldom found in gene exons (5, 8, 11, 29). However, MITEs in many plants and in the yellow fever mosquito, A. aegypti, are frequently found in the flanking regions and introns of genes (5, 8, 11, 29). There is evidence that some MITEs in the flanking regions of plant genes may be involved in gene regulation, either by providing regulatory sequences, or by serving as matrix attachment regions that help define chromatin domains (17, 20). It is not yet clear whether the A. gambiae MITEs have similar distributions relative to genes because the short length of STS sequences may severely reduce the chance for identifying nearby genes. There are high degrees of variation in genome size and organization between different mosquitoes. The genome of A. gambiae is 270 Mbp, organized in a pattern of “long period interspersion” in which single copy DNA is less interrupted by repetitive elements (30). In contrast, the A. aegypti genome is 800 Mbp, organized in a pattern of “short period interspersion” in which single copy DNA is partitioned into small blocks by repetitive elements (31). The copy number of A. gambiae MITEs ranges from 40 to 1,340, which is much lower compared with 2,100 to 10,000 copies in A. aegypti (11, 12). This is consistent with the hypothesis that there may be a correlation between the copy number of MITEs and the genome size of the hosts (11). The differences in the relative abundance of MITEs may have also contributed to the different organizations of the mosquito genomes and reflect different types of interactions between the hosts and these widespread transposable elements.
Implications for the Genetic Approach to Control Malaria and Other Mosquito-Borne Diseases.
As described in the Introduction, major efforts are underway to develop a strategy to control malaria and other mosquito-borne diseases by replacing vector mosquitoes in wild populations with genetically modified refractory mosquitoes. A few exogenous DNA transposons including Tc1-mariner-like elements and the hAT-like elements are being developed as transformation tools for mosquitoes (e.g., refs. 32 and 33). Interactions with endogenous transposable elements that have TIRs similar to the introduced transposon have been shown to be a potential problem (3, 4). Because MITEs are likely mobilized by autonomous DNA transposons sharing similar TIRs, the diverse families of MITEs discovered in this study could act as potential substrates if the introduced transposon uses similar TIRs. Analyses of endogenous MITEs and DNA transposons may provide information that could help better devise transposon-based transformation tools to reduce possible inactivation by endogenous elements and cross-mobilization of endogenous elements that may cause high rates of mutation. Because MITEs are significant components in a wide range of eukaryotes, these considerations may be broadly relevant as transposon-based transgenic technology is being applied to manipulate the genomes of insects, plants, and more recently mammals (34, 35). On the other hand, further analyses of MITEs may lead to the identification of active DNA transposons that may be mobilizing MITEs in mosquitoes. It is not yet clear how effective it will be to use endogenous DNA transposons as transformation tools in the same species. However, active DNA transposons found in one species may at least have the potential to serve as transformation tools in related vector mosquitoes. Moreover, some of the MITEs may be used to develop markers for genetic mapping and population studies, if insertion polymorphism is demonstrated. Markers derived from a MITE family have been used to construct a relatively detailed genetic map for maize, taking advantage of a newly developed assay to rapidly screen a large number of transposon insertion sites (36). Such markers, when developed in mosquitoes, could also be powerful tools to investigate the spread of genetic elements in mosquito populations. Therefore, a better understanding of the characteristics, behavior, and evolution of endogenous transposable elements and their interactions with the mosquito genomes is of great importance to the long-term success of the current genetic approach to control malaria and other mosquito-borne diseases.
Acknowledgments
findmite was developed in collaboration with Kun Li in the Department of Computer Science at the University of Arizona, who was responsible for the implementation of the program. Min Liang in the Department of Computer Science at Virginia Tech and Chunhong Mao at Virginia Tech Library Systems provided modifications to findmite. Chunhong Mao implemented AT content. Keying Ye in the Department of Statistics at Virginia Tech helped in the design of statistical analysis. I thank David Bevan, James Biedler, and Kathy Chen for critical comments on the manuscript. This work was supported by National Institutes of Health Grant AI42121 (to Z.T.) and by the Agricultural Experimental Station at Virginia Tech.
Abbreviations
- EST
expressed sequence tag
- MITEs
miniature inverted repeat transposable elements
- STS
sequence tagged site
- TIR
terminal inverted repeat
Footnotes
Data deposition: The sequence alignments reported in this paper have been deposited in EMBL alignment database (accession nos. DS43373–DS43385).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041593198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041593198
References
- 1.Beaty B. Proc Natl Acad Sci USA. 2000;97:10295–10297. doi: 10.1073/pnas.97.19.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enserink M. Science. 2000;290:440–441. doi: 10.1126/science.290.5491.440. [DOI] [PubMed] [Google Scholar]
- 3.Sundararajan P, Atkinson P, O'Brochta D A. Insect Mol Biol. 1999;8:359–368. doi: 10.1046/j.1365-2583.1999.83128.x. [DOI] [PubMed] [Google Scholar]
- 4.Jasinskiene N, Coates C J, James A A. Insect Mol Biol. 2000;9:11–18. doi: 10.1046/j.1365-2583.2000.00153.x. [DOI] [PubMed] [Google Scholar]
- 5.Bureau T E, Wessler S R. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oosumi T, Garlick B, Belknap W R. Proc Natl Acad Sci USA. 1995;92:8886–8890. doi: 10.1073/pnas.92.19.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oosumi T, Garlick B, Belknap W R. J Mol Evol. 1996;43:11–18. doi: 10.1007/BF02352294. [DOI] [PubMed] [Google Scholar]
- 8.Bureau T E, Ronald P C, Wessler S R. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan G T. J Mol Biol. 1995;254:1–5. doi: 10.1006/jmbi.1995.0593. [DOI] [PubMed] [Google Scholar]
- 10.Smit A F A, Riggs A D. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu Z. Proc Natl Acad Sci USA. 1997;94:7475–7480. doi: 10.1073/pnas.94.14.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu Z. Mol Biol Evol. 2000;17:1313–1325. doi: 10.1093/oxfordjournals.molbev.a026415. [DOI] [PubMed] [Google Scholar]
- 13.Izsvák Z, Ivics Z, Shimoda N, Mohn D, Okamoto H, Hacket P B. J Mol Evol. 1999;48:13–21. doi: 10.1007/pl00006440. [DOI] [PubMed] [Google Scholar]
- 14.Surzycki S A, Belknap W R. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Arbuckle J, Wessler S R. Proc Natl Acad Sci USA. 2000;97:1160–1165. doi: 10.1073/pnas.97.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feschotte C, Mouches C. Gene. 2000;250:109–116. doi: 10.1016/s0378-1119(00)00187-6. [DOI] [PubMed] [Google Scholar]
- 17.Wessler S R, Bureau T E, White S E. Curr Opin Gene Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 18.Feschotte C, Mouches C. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 19.MacRae A F, Clegg M T. Genetica. 1992;86:55–66. doi: 10.1007/BF00133711. [DOI] [PubMed] [Google Scholar]
- 20.Tikhonov A P, Bennetzen J L, Avramova Z V. Plant Cell. 2000;12:249–264. doi: 10.1105/tpc.12.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossman G L, Cornel A J, Rafferty C S, Robertson H M, Collins F H. Genetica. 1999;105:69–80. doi: 10.1023/a:1003690102610. [DOI] [PubMed] [Google Scholar]
- 22.Besansky N J, Mukabayire O, Bedell J A, Lusz H. Genetica. 1996;98:119–129. doi: 10.1007/BF00121360. [DOI] [PubMed] [Google Scholar]
- 23.Cormen T H, Leiserson C E, Rivest R L. Introdution to Algorithms. Cambridge, MA: MIT Press; 1990. pp. 869–875. [Google Scholar]
- 24.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besansky N J, Powell J R. J Med Entomol. 1992;29:125–128. doi: 10.1093/jmedent/29.1.125. [DOI] [PubMed] [Google Scholar]
- 26.Zar J H. Biostatistical Analysis. Upper Saddle River, NJ: Prentice Hall; 1996. [Google Scholar]
- 27.Dimopoulos G, Casavant T L, Chang S, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, et al. Proc Natl Acad Sci USA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surzycki S A, Belknap W R. Proc Natl Acad Sci USA. 2000;97:245–249. doi: 10.1073/pnas.97.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bureau T E, Wessler S R. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rai K S, Black W C, IV. Adv Genet. 1999;41:1–33. doi: 10.1016/s0065-2660(08)60149-2. [DOI] [PubMed] [Google Scholar]
- 31.Warren A M, Crampton J M. Genet Res. 1991;58:225–232. doi: 10.1017/s0016672300029979. [DOI] [PubMed] [Google Scholar]
- 32.Jasinskiene N, Coates C J, Benedict M Q, Cornel A J, Salazar Rafferty C, James A A, Collins F H. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catteruccia F, Nolan T, Loukeris T G, Blass C, Savakis C, Kafatos F C, Crisanti A. Nature (London) 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 34.O'Brochta D A, Atkinson P. Insect Biochem Mol Biol. 1996;26:739–753. doi: 10.1016/s0965-1748(96)00022-7. [DOI] [PubMed] [Google Scholar]
- 35.Yant S R, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay M A. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 36.Casa A M, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler S. Proc Natl Acad Sci USA. 2000;97:10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biessmann H, Kobeski F, Walter M F, Kasravi A, Roth C W. Insect Mol Biol. 1998;7:83–93. doi: 10.1046/j.1365-2583.1998.71054.x. [DOI] [PubMed] [Google Scholar]
- 38.Tudor M, Lobocka M, Goodell M, Pettitt J, O'Hare K. Mol Gen Genet. 1992;232:126–134. doi: 10.1007/BF00299145. [DOI] [PubMed] [Google Scholar]