Abstract
Dipeptidyl (acyloxy)methyl ketones (AOMKs) have been identified as mechanism-based inhibitors of certain cysteine proteases. These compounds are also inhibitors of the integral membrane proteins Rce1p and Ste24p, which are proteases that independently mediate a cleavage step associated with the maturation of certain isoprenylated proteins. The enzymatic mechanism of Rce1p is ill-defined, whereas Ste24p is a zinc metalloprotease. Rce1p is required for the proper processing of the oncoprotein Ras and is viewed as a potential target for cancer therapy. In this study, we synthesized a small library of dipeptidyl AOMKs to investigate the structural elements that contribute to the inhibitor properties of this class of molecules toward Rce1p and Ste24p. The compounds were evaluated using a fluorescence-based in vitro proteolysis assay. The most potent dipeptidyl AOMKs contained an arginine residue and the identity of the benzoate group strongly influenced potency. A “warhead” free AOMK inhibited Rce1p and Ste24p. The data suggest that the dipeptidyl AOMKs are not mechanism-based inhibitors of Rce1p and Ste24p and corroborate the hypothesis that Rce1p is not a cysteine protease.
Keywords: Ras converting enzyme (Rce1p), sterile mutant 24 (Ste24p), (acyloxy)methyl ketone, protease, Ras, CaaX protein, post-translational modification
1. Introduction
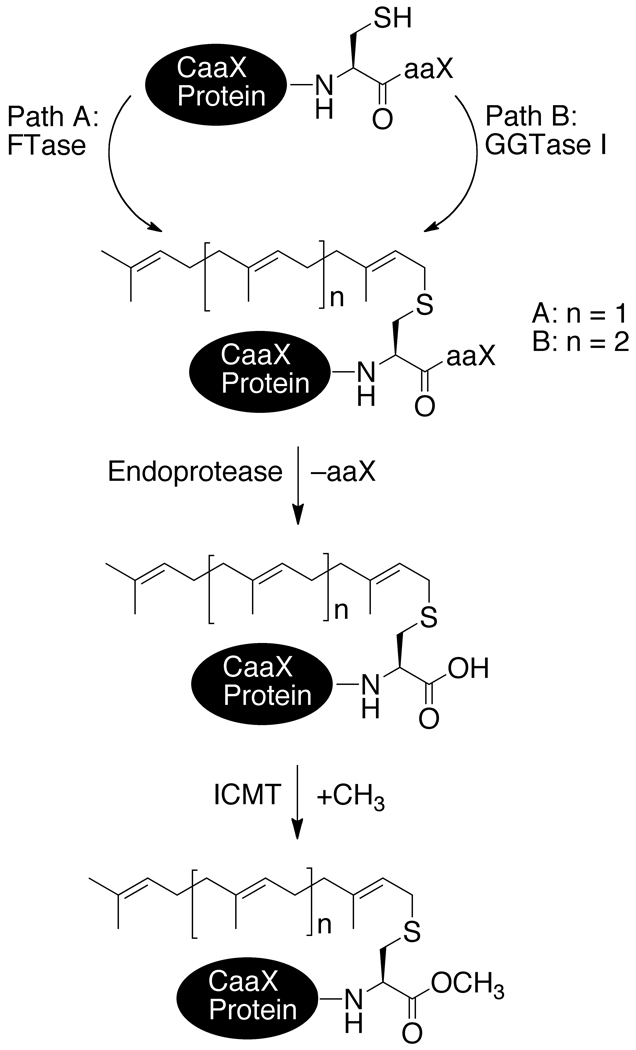
Many eukaryotic proteins bear a C-terminal CaaX tetrapeptide motif, where C is cysteine, a is typically an aliphatic amino acid, and X is one of several amino acids, that directs an ordered series of post-translational modifications (Figure 1).1–3 These include the covalent addition of an isoprenoid lipid to the cysteine by either farnesyl or geranylgeranyl transferase (FTase or GGTase),4 a proteolytic step that trims away the aaX portion,5, 6 and methyl esterification of the resultant new carboxyl terminus by isoprenylcysteine methyltransferase (ICMT).7, 8 These modifications are critical to the activity of many CaaX proteins; defects in the processing pathway can result in non-functional or mislocalized protein, or enhanced turnover of the unprocessed intermediate.5, 9, 10 The Ras subfamily of small GTP-binding proteins11 are CaaX proteins having a prominent role in carcinogenesis.2, 3 Hence, Ras proteins and Ras-regulatory proteins are considered targets for anticancer therapeutics.2, 12
Figure 1.
Post-translational modifications associated with CaaX proteins.
The CaaX endoproteases Ras converting enzyme 1 (Rce1p) and sterile mutant 24 (Ste24p), both first identified in S. cerevisiae, mediate the carboxyl terminal proteolysis step of CaaX protein maturation.5 Despite functional similarity, Rce1p and Ste24p lack primary sequence similarity.5 Orthologs of both proteases are present in humans,13, 14 mice,15 C. elegans,16 plants, and other eukaryotic organisms.17, 18 Both are integral membrane proteins with multiple predicted transmembrane spans19, 20 that localize to the endoplasmic reticulum (ER) with the catalytic sites presumably facing the cytosol where their substrates are located.19, 21 Ste24p is a zinc-dependent metalloprotease,5, 19, 22 possessing a consensus HEXXH motif (H is histidine, E is glutamate, and X is any amino acid) that is required for proteolytic activity. The zinc chelator 1,10-phenanthroline inhibits the proteolytic activity, but the addition of zinc reactivates it.19, 22
Several reports have implicated Rce1p as a cysteine protease,23–25 but mutation20 and bioinformatic26 studies provide a stronger case that it is a metalloprotease. The only conserved cysteine among eukaryotic Rce1p orthologs (Cys251 in yeast Rce1p) is not required for enzyme function,20 seriously weakening the case for a cysteine-based mechanism. Seven residues appear important for optimum enzymatic activity of yeast Rce1p: Glu156, His194, His248, Glu157, Tyr160, Asn252.20 The first three are critical for the proteolytic activity of yeast Rce1p and probably define the catalytic site, while the latter four optimize activity and substrate specificity. Glu156, His194, and His248 are not in close proximity in the primary sequence. These presumed catalytic residues are conserved among Rce1p orthologs, and in the case of trypanosomal Rce1p, are also required for activity.27 These residues must be in close three-dimensional proximity if they indeed are part of the active site and are hypothesized to reside at or within the membrane.20 Such a membrane-embedded active site is atypical for proteases, but has been proposed for presenilins, site-2 protease, rhomboid, and the signal peptide peptidase.28
Rce1p and Ste24p have distinct, but partially overlapping, substrate specificities, which suggests that they have different cellular functions. Rce1p cleaves Ras and Ras-related proteins, whereas Ste24p does not cleave yeast Ras2p5 or human farnesylated K-Ras,21 which have CaaX motifs consisting of CIIS and CVIM residues, respectively. Mammalian Ste24p cleaves pre-lamin A (CSIM).29, 30 Both proteases act on the precursor to the yeast a-factor mating pheromone (CVIA).31 Knockout studies in mice have shown that Rce1p is required for embryonic and cardiac development,15, 32 and Ste24p is required for proper skeletal and muscular development.29, 30 Because of its involvement in pre-lamin A processing, human Ste24p deficiency is also connected to human progeroid disorders.33–35
Inhibition of Rce1p is an attractive anticancer strategy because it would be expected to impede Ras-induced oncogenic transformation, while not affecting the maturation of Ste24p-dependent substrates.2, 3 Furthermore, mouse embryonic fibroblasts deficient in Rce1p are more sensitive to an FTase inhibitor than wild type cells,36 indicating the potential for combination therapies. Inhibitors of Rce1p fall into four categories: non-specific protease inhibitors (N-tosyl-l-phenylalanine chloromethyl ketone (TPCK),24, 25, 37 organomercurials,25 Zn2+ and Cu2+ 25), substrate mimetics (non-hydrolyzable isoprenylated peptides and isoprenoids),23, 24, 37, 38 natural products (barangcadoic and rhopaloic acids39 and scalaranes40), and small-molecules discovered in biochemical screens of chemical libraries.41, 42
We previously reported that peptidyl (acyloxy)methyl ketones (AOMKs) inhibit both Rce1p and Ste24p.43 This compound class is best known for potently and irreversibly modifying certain cysteine proteases, presumably through covalent modification of the active site cysteine.44, 45 Curiously though, only one cysteine residue is conserved among Rce1p orthologs and none is conserved among Ste24p enzymes. Furthermore, the conserved cysteine in the Rce1p family (Cys251 in yeast Rce1p sequence) is not required for activity, and AOMKs inhibit the C251A mutant.20, 43 The inhibition of the CaaX proteases by AOMKs indicates that this compound class represents an important new tool for the study of the CaaX proteases. By comparison, AOMKs perform more consistently than TPCK, a widely described chloromethyl ketone Rce1p inhibitor.16, 37, 43, 46 Moreover, AOMKs are the first agents described that inhibit both Rce1p and Ste24p. Thus, these compounds have potential for leading to a better understanding of CaaX protease enzymology.
In this study, we investigated the structural elements of AOMKs (Figures 2 and 3) as they contribute to the inhibitory properties of this compound class against yeast Rce1p and Ste24p in a fluorescence-based in vitro proteolysis assay. In particular, we have determined how the structural profile of the benzoate moiety and amino acid substitutions of the peptidyl group modulate the inhibitory properties of AOMKs.
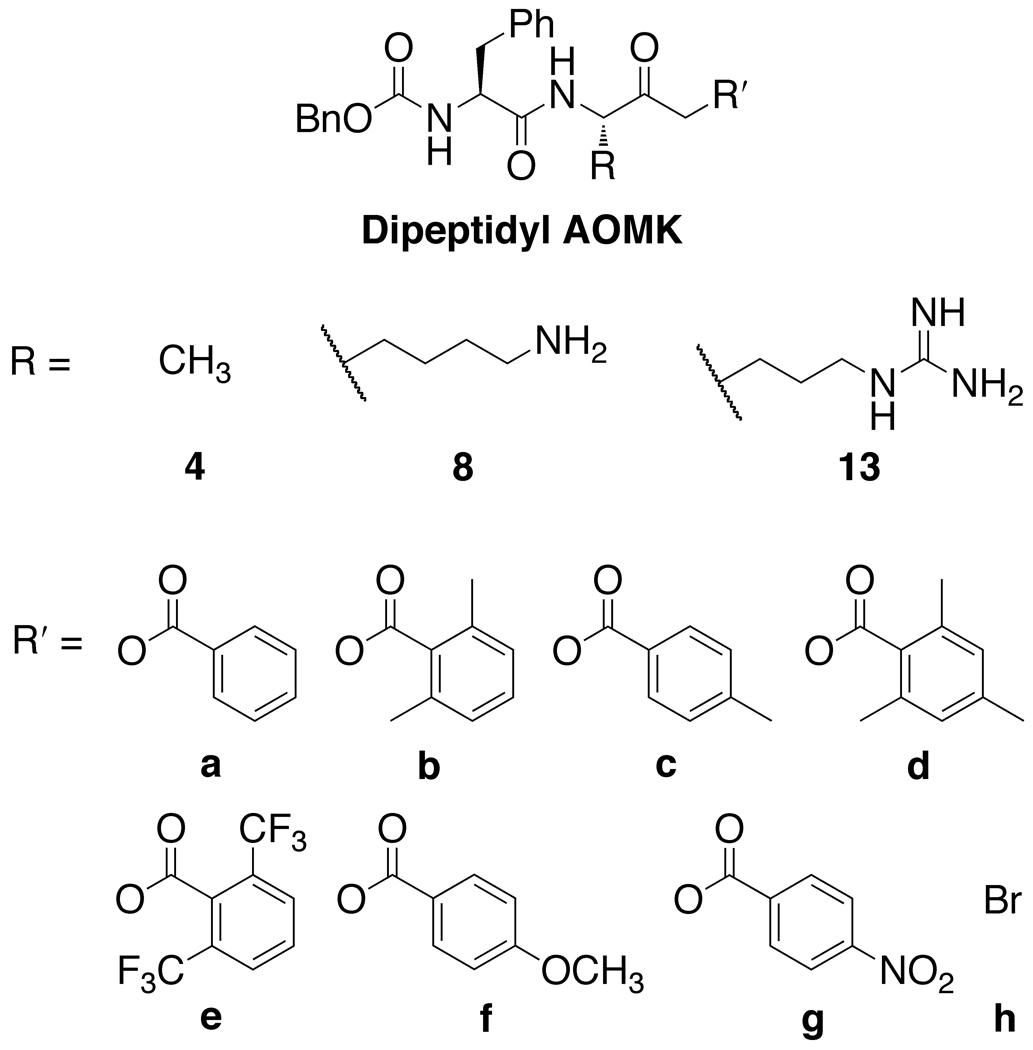
Figure 2.
Dipeptidyl AOMK compounds synthesized for this study.
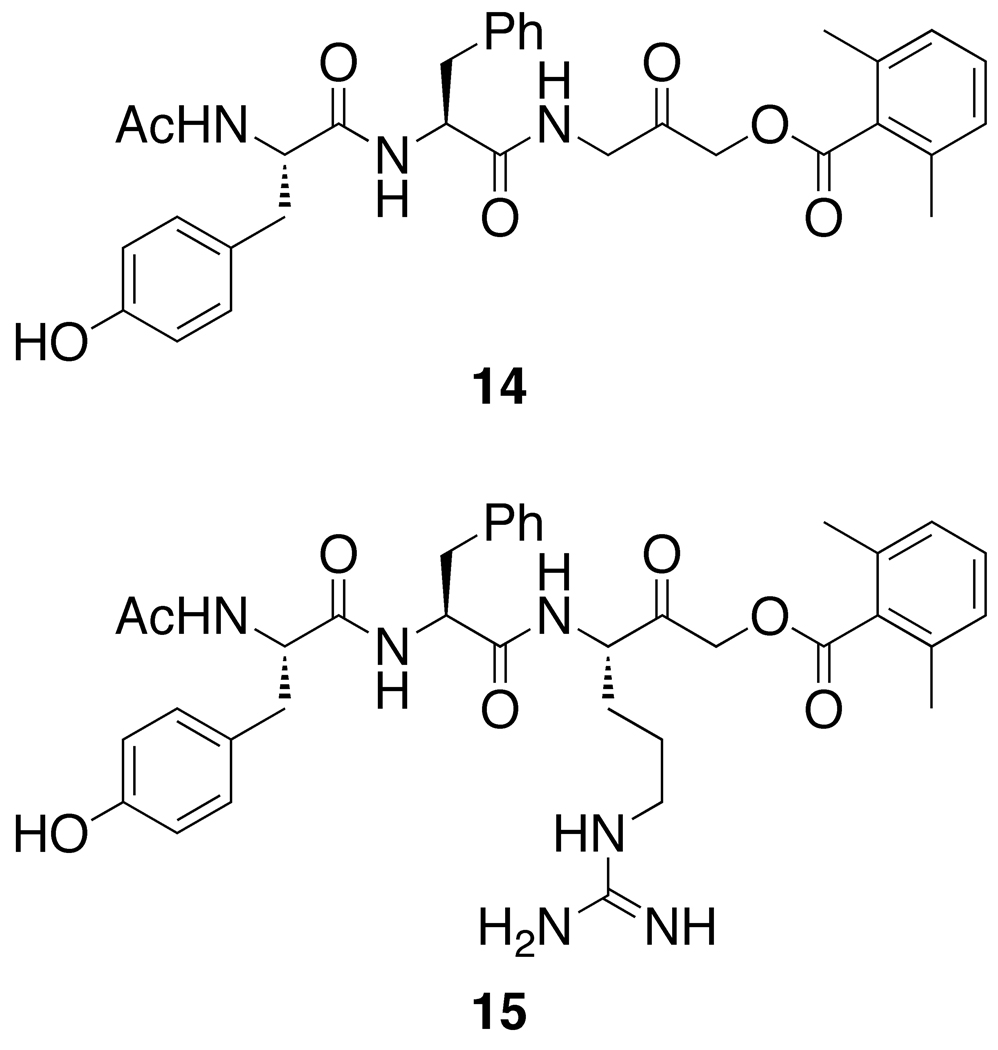
Figure 3.
Other AOMKs.
2. Results
2.1. Dipeptidyl AOMK Synthesis
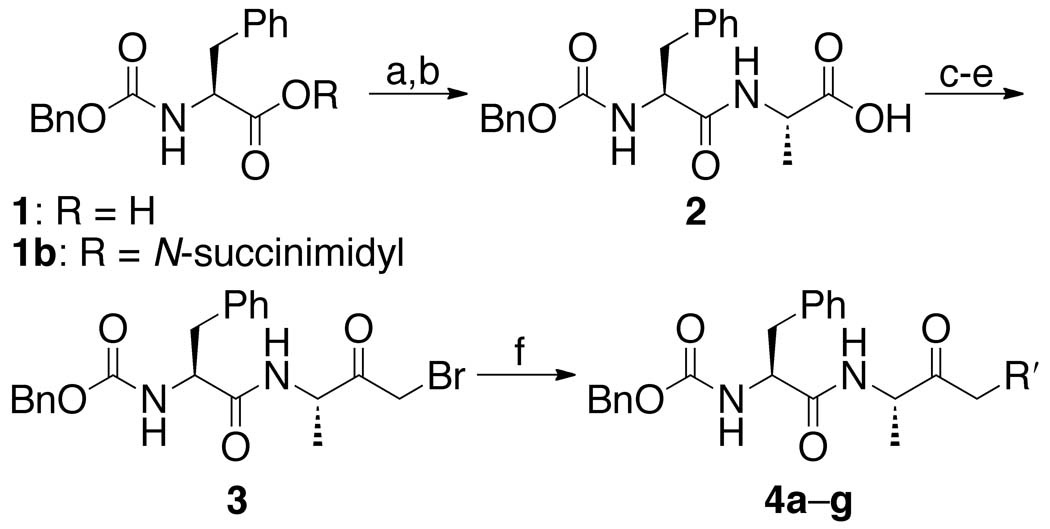
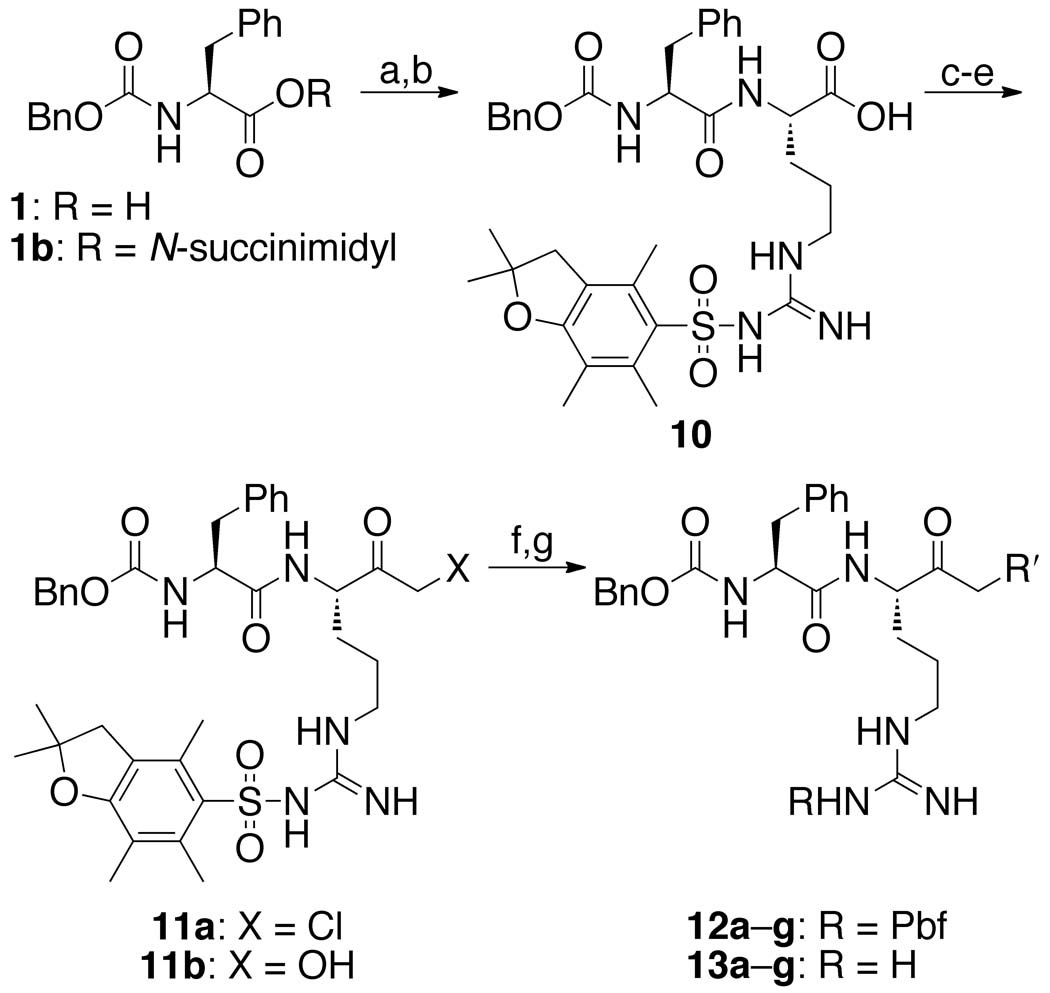
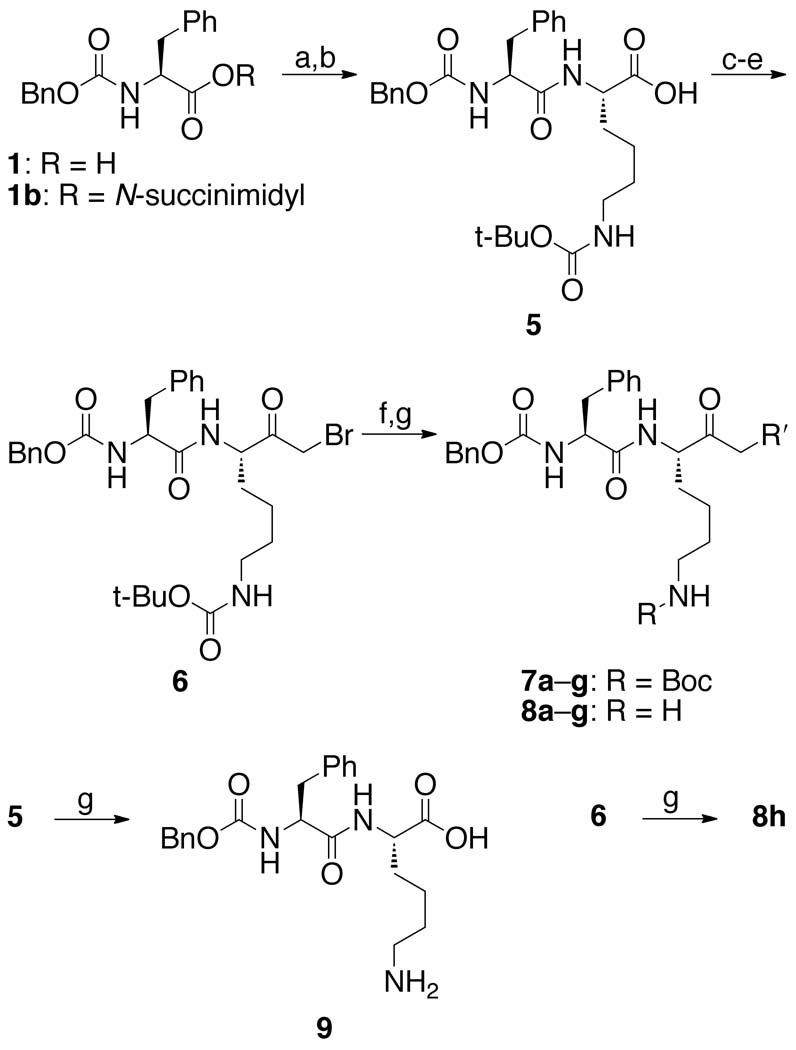
A library of three series of dipeptidyl AOMKs, each with different benzoyloxy groups (Figure 2), were synthesized using the method described by Krantz 44 and outlined in Schemes 1–3. Benzyloxycarbonyl-protected phenylalanine (1, Scheme 1) was reacted with N-hydroxysuccinimide and DCC to generate the succinimide ester 1b, which was coupled to alanine to provide dipeptide 2. The isobutyl formate of 2 was treated with diazomethane to generate the α-diazo ketone, which was converted to the bromomethyl ketone 3 by brief exposure to HBr in acetic acid. Displacement of the bromide by the benzoates a–g resulted in the AOMKs 4a–g in yields of 8–75%. Similarly, DCC coupling of ε-Boc-protected lysine to the succinimide 1b provided dipeptide 5 (Scheme 2), which was converted to the bromomethyl ketone 6 using the Krantz procedure in 73% yield. The bromomethyl ketone 6 served as the starting point for the syntheses of the benzoyl esters 7a–g, which proceeded smoothly, albeit in low yields (~35%). Deprotection of 7a–g with trifluoroacetic acid (TFA) provided AOMKs 8a–g after purification by HPLC. Removal of the Boc group from 5 with TFA in dichloromethane gave dipeptide 9 for control experiments. Deprotection of 6 under similar conditions provided bromide 8h in 54% yield.
Scheme 1.
See Figure 2 for definitions of R′. Reagents: (a) N-hydroxysuccinimide, DCC, THF, 0 °C, 18 h, 95%; (b) l-alanine, TEA, CO2, dioxane, H2O, pH 8, rt, 20 h, 51%; (c) isobutyl chloroformate, N-methylmorpholine, THF, −20 °C, 2 h; (d) CH2N2, Et2O, 0 °C; (e) HBr, AcOH, CH2Cl2, 0 °C, 1 min, 15% for three steps; (f) benzoic acids a–g, KF, DMF, rt, 4 h, 8–75% yield.
Scheme 3.
See Figure 2 for definitions of R′. Reagents: (a) N-hydroxysuccinimide, DCC, THF, 0 °C, 15 h, 93%; (b) l-arginine(Pbf), TEA, CO2, dioxane, H2O, pH 8, rt, 20 h, 48%; (c) isobutyl chloroformate, N-methylmorpholine, THF, −20 °C, 2 h; (d) CH2N2, Et2O, 0 °C; (e) HCl, AcOH, 0 °C, 1 min, 22% for three steps; (f) benzoic acids a–g, KF, DMF, rt, 4 h, 22–45%; (g) TFA/H2O (95:5), 0 °C, 4 h.
Scheme 2.
See Figure 2 for definitions of R′. Reagents: (a) N-hydroxysuccinimide, DCC, THF, 0 °C, 15 h, 93%; (b) l-lysine(ε-Boc), TEA, CO2, dioxane, H2O, pH 8, rt, 20 h, 55%; (c) isobutyl chloroformate, N-methylmorpholine, THF, −20 °C, 2 h; (d) CH2N2, Et2O, 0 °C; (e) HBr, AcOH, CH2Cl2, 0 °C, 1 min, 73% for three steps; (f) benzoic acids a–g, KF, DMF, rt, 4 h, ~35%; (g) TFA, CH2Cl2, 0 °C, 53–93%.
The synthesis of the arginine series of AOMKs proved more difficult (Scheme 3). Coupling of Pbf-protected l-arginine to the benzyloxycarbonyl-protected phenylalanine produced dipeptide 10 in 93% yield, but the subsequent reactions to convert 10 to a halomethyl ketone behaved inconsistently, despite the report of a related Pbf-protected arginine substrate undergoing the transformation in 65% yield.47 Quenching the diazo intermediate with HBr gave unsatisfactory results; using HCl generated the chloro ketone 11a. Regardless the acid, reactions typically yielded hydroxymethyl ketone 11b, and attempts to convert it to the desired AOMK with the appropriate benzoyl chloride failed. When the reaction sequence was successful, we obtained 11a in 22% yield. Displacement of the chloride with benzoic acids a–g provided Pbf-protected AOMKs 12a–g in 22–45% yield. Pbf-protected dipeptides 12a–g were successfully deprotected to give the arginine series of AOMKs 13a–g after purification by HPLC. The deprotection step to convert bis(trifluoromethyl)benzoate 12e to 13e often resulted in decomposition, but we were able to obtain a sufficient quantity of 13e for biological studies.
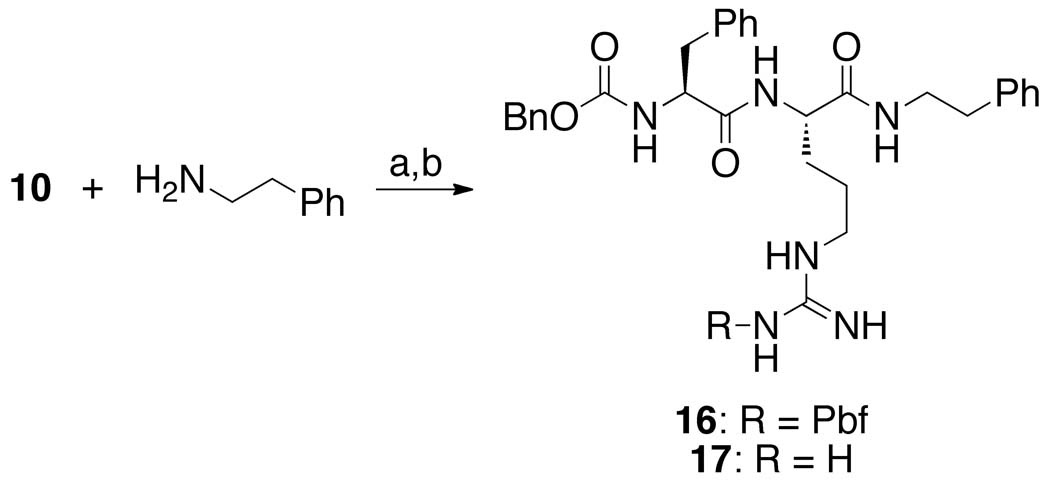
To explore the role of the benzoate group, we prepared a dipeptidyl AOMK possessing an N-(β-phenethylamide) (17, Scheme 4) in place of the benzoyloxymethyl group. This “warhead-free” dipeptide was synthesized by coupling β-phenethylamine to protected dipeptide 10, followed by removal of the Pbf group with TFA.
Scheme 4.
Reagents: (a) HOBt, HBTU, DIPEA, CH2Cl2, 0 °C, then rt overnight, 39%; (b) TFA/H2O (95:5), 0 °C, 2 h, 33%.
2.2. Fluorescence-Based Proteolysis Assay
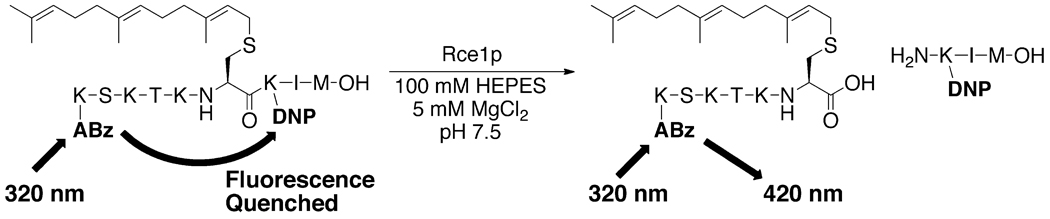
The inhibition by the dipeptidyl AOMK compounds was evaluated using a fluorescence-based in vitro CaaX proteolysis assay (Figure 4).42, 43, 48 ER membranes enriched for either yeast Rce1p or Ste24p were used as the source of enzyme activity. Two different fluorogenic substrates based on K-Ras4b were used to monitor the proteolytic activity. For Rce1p, ABz-KSKTKC(farnesyl)QLIM was used, where ABz is ortho-aminobenzoate and QL is ε-dinitrophenyl lysine. For Ste24p, ABz-KSKTKC(farnesyl)VIQL was used since the Rce1p substrate is not effectively recognized by Ste24p.43 In both substrates, fluorescence resulting from excitation of the ABz group is quenched by QL until CaaX protease-mediated proteolysis cleaves the peptide to liberate the quenching group. The assay was carried out in 96-well plates with fluorescence output measured using a fluorescence microplate reader. Decreased fluorescence output compared to a DMSO control indicated inhibition of proteolysis.
Figure 4.
Fluorescence-based assay for Rce1p endoprotease activity. Assay for Ste24p is identical except substrate is ABz-KSKTKC(farnesyl)VIK-DNP. ABz is ortho-aminobenzoate and DNP is ε-dinitrophenyl.
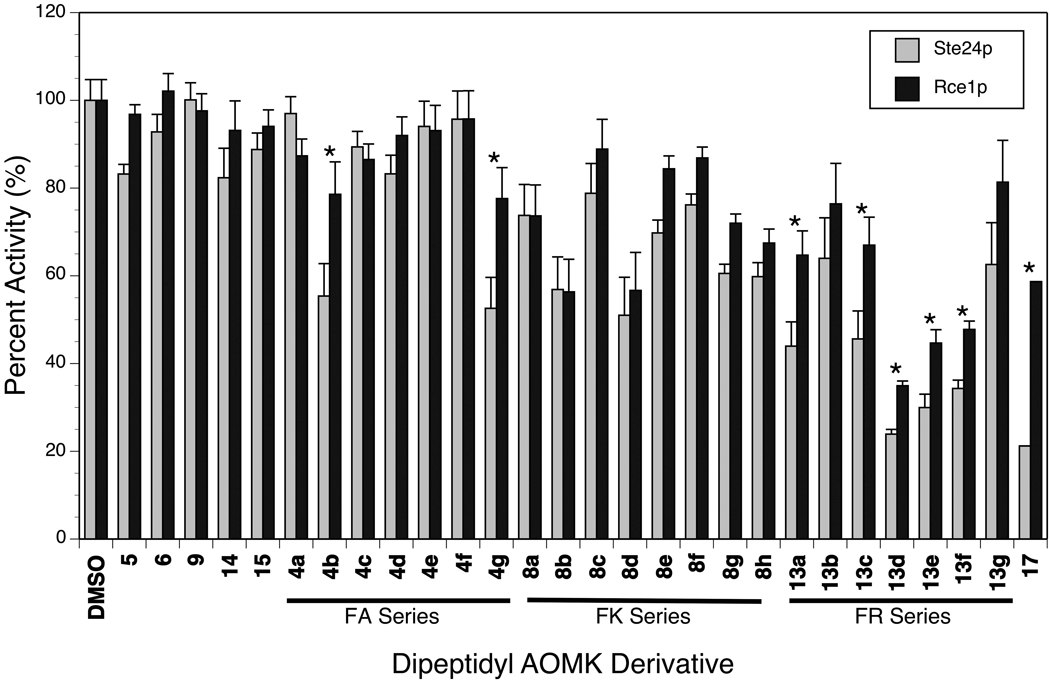
The proteolytic activities of Rce1p and Ste24p relative to a DMSO control in the presence of 100 µM AOMK-based inhibitors are summarized in Figure 5. Control peptides lacking the benzoyl ester group (5, 6, 9) and tripeptidyl AOMKs (14, 15) were not significant inhibitors of either Rce1p or Ste24p. Overall, AOMKs containing alanine (4a–g) were the least inhibitory, while those with arginine (13a–g) were the most potent. The most inhibitory compound in the assay was Z-FR-CH2OBz(2,4,6-trimethyl) (13d). The dipeptidyl AOMKs demonstrated similar inhibitory strength against Ste24p and Rce1p, though certain compounds demonstrated slightly preferential inhibition of Ste24p over Rce1p (4b, 4g, 13a, 13c–f, 17), with “warhead-free” dipeptide 17 having the most dramatic preference.
Figure 5.
Effect of dipeptidyl AOMK derivatives (100 µM) on the endoprotease activity of Ste24p and Rce1p relative to a DMSO control in the fluorescence-based assay. Values are the average of at least 4 replicates. The error bars represent the standard deviation of the measurement. Statistically significant (p < 0.05) differences between Ste24p and Rce1p inhibition are marked with an asterisk.
3. Discussion
Peptidyl AOMKs are mechanism-based inhibitors of cysteine proteases.44 They covalently and irreversibly attach to the active-site cysteine. The potency and reactivity of the AOMK can be tuned by modifying the benzoate leaving group moiety. The better the leaving group capability of the benzoate, the more potent the inhibitor when there is rapid, reversible binding of the inhibitor. Since Ste24p is a zinc-dependent metalloprotease, it is surprising that the AOMKs inhibit its activity. Earlier studies suggest that Rce1p is a cysteine protease, in part because it responds to classic cysteine protease inhibitors, such as chloromethyl ketones and organomercurials, but not to classic serine, metallo-, and aspartyl protease inhibitors.23–25 A mechanistic-based inhibition of Rce1p by AOMKs is thus plausible if it were indeed a cysteine protease. This conclusion, however, should be viewed cautiously. One concern is that mechanism-based protease inhibitors often have cross-reactivity with side chains other than the intended targets, which obscures the identification of the mechanism of the catalytic pathway. Second, mutational analyses on the only cysteine conserved among Rce1p eukaryotic orthologs, specifically Cys251 of yeast Rce1p, have presented conflicting results. In one study, the C251A mutant is reported to be inactive,25 but another found that it retains wild type activity in assays for protease activity.20 Third, bioinformatic studies across a broader range of potential orthologs do not reveal a conserved cysteine.26 It is considerably more likely that Rce1p is not a cysteine protease and that, as with Ste24p, AOMKs act in a non-mechanistic fashion to inhibit Rce1p.
In a previous study,43 we investigated the inhibition of Rce1p and Ste24p by compounds 4d, 8d, 13b, 14, and 15 and found that dipeptidyl AOMKs 8d and 13b inhibited yeast Rce1p and Ste24p. In the current study, the in vitro activity of these five compounds was reassessed, and we found good agreement with the previous data, except for the inhibition of Rce1p by 13b and of Ste24p by 8d. In the current work, we found that these AOMKs were weaker inhibitors than previously reported. Possible explanations for the difference include the following: (1) the membranes containing Rce1p and Ste24p had different specific activities (i.e., the amount of active enzyme per milligram of membrane was different), so the AOMKs could have partitioned into the membrane differently, altering the concentration of free AOMK present,43 and (2) the AOMKs and substrate peptides used in the previous study were from different sources, so there might be batch to batch variations in the samples.
Our previous work43 showed that inhibition of Rce1p by 8d was not readily reversible through washing with salt or urea buffers, but 8d did not inhibit Rce1p in a time-dependent manner, suggesting that binding was reversible. Compound 8d reduced the Vmax of Rce1p in a dose-dependent manner without significantly changing the Km value from that observed in the absence of inhibitor. Analysis of the kinetic data suggested that 8d was a tightly bound, reversible, noncompetitive inhibitor of Rce1p.
Our current studies indicate that the leaving group ability of the benzoyl ester does not correlate well with the inhibitory properties of the dipeptidyl AOMKs. For example, the conjugate acids of the benzoates in AOMKs 13b, d, and g have similar pKa values (3.21, 3.43, and 3.44, respectively), but 13b and g are far less potent than 13d in affecting the activity of Rce1p and Ste24p. This finding is inconsistent with a cysteine protease mechanism, because a better leaving group would be expected to better inhibit the enzyme,44 assuming that the AOMKs undergo rapid and reversible binding and have similar Km values. The kinetic data previously reported for 8d43 suggest that this is a reasonable assumption. The observation that the bromide 8h, which possesses an excellent leaving group, is not much better at inhibiting Rce1p than other AOMKs also does not support mechanism-based inhibition. Furthermore, if the dipeptidyl AOMKs were acting as mechanism-based inhibitors, then dipeptide 17, where the α-acyloxy group has been replaced with an amide, should not inhibit the proteases. Dipeptide 17 is characterized as “warhead-free” because it lacks a benzoate leaving group, which renders it incapable of covalently attaching to a putative sulfur nucleophile in the active site. As an alternative, we propose that dipeptidyl AOMKs are likely reversible, non-competitive inhibitors of the CaaX proteases.
4. Conclusion
Based upon the discovery that peptidyl (acyloxy)methyl ketones are inhibitors of Rce1p and Ste24p,43 we investigated the structural elements of AOMKs that contribute to the inhibitory properties of this compound class. In particular, we explored how the structural profile of the benzoate moiety and amino acid substitutions of the second amino acid modify the inhibitory properties of AOMKs. We generated a 21-member library of AOMKs by coupling three benzyloxycarbonyl-protected dipeptides to seven benzoic acids, and evaluated their ability to inhibit Rce1p and Ste24p in a fluorescence-based proteolysis assay. The data reveal that the inhibitory activity of the peptidyl AOMKs is strongly influenced by the identity of the amino acid in the second position (Z-FA < Z-FK < Z-FR) and the structure of the benzoate. Activity by a “warhead free” AOMK suggests that the inhibition of both Rce1p and Ste24p is not mechanism based, as it is with cysteine proteases, but rather is a result of non-covalent binding of the AOMK to the active site of the protease. This corroborates other work that suggests that Rce1p is not a cysteine protease.20, 26
5. Material and Methods
5.1. Reagents and general methods
All reagents were purchased from commercial sources and used without further purification with the following exceptions: THF was dried on an activated alumina column immediately prior to use and DMF was distilled under vacuum and stored over molecular sieves. Thin layer and column chromatography were performed on precoated silica gel 60 F254 plates and 230–400 mesh silica gel 60, respectively. Flash chromatography was performed on a Flash+ system (Biotage, Charlottesville, VA). HPLC analysis (analytical and preparative) was performed on a Varian ProStar HPLC system using Microsorb C-18 reverse phase columns and a diode array UV detector. 1H NMR spectra were recorded on Varian MercuryPlus 400 MHz spectrometer. Mass spectrometry was performed on a Sciex API-1 Plus quadruple mass spectrometer with an electrospray ionization source or a Bruker Autoflex MALDI-TOF. ESI-HRMS was performed on a Bruker 4.7 T FT-MS. CaaX protease substrates ABz-KSKTKC(farnesyl)QLIM and ABz-KSKTKC(farnesyl)VIQL were commercially synthesized by Anaspec.
5.2. Synthesis of dipeptidyl (acyloxy)methylketones
Compounds 1b, 2, 3, 4a–g, 7a, 7d–f, 8a, 8d–f,44 5, 6,49 9,50 and 13b45 are known in the literature. N-Hydroxysuccinimide ester 1b is also available commercially. All of the dipeptidyl (acyloxy)methylketones were synthesized by the method described by Krantz et al.44 except compounds 13b, 14, and 15, which were obtained from Matthew Bogyo.45 All compounds were purified by HPLC prior to the protease assay, then stored at −20 °C in neat form or at −80 °C as a stock solution in DMSO. HPLC analysis of several dipeptidyl AOMKs (8c and g and 13a, c, d, and f) did not reveal decomposition of the compounds during prolonged storage (>6 months) under these conditions. 1H NMR analysis of 8h after prolonged storage (>3 years) as a solid at −20 °C did not indicate that it had degraded.
Z-Phe-Lys(Boc)-CH2-O-(2,6-dimethyl)benzoate (7b)
2,6-Dimethylbenzoic acid (50.7 mg, 0.338 mmol) was added to a solution of bromide 6 (100.5 mg, 0.166 mmol) in dry DMF (3 mL) and KF (70.1 mg, 1.21 mmol). After the reaction was stirred at room temperature overnight, it was diluted with ether (20 mL) and washed with water (3 × 5 mL). With each wash, EtOAc was added to help separate the layers. The organic portion was washed with saturated NaHCO3 (2 × 5 mL) and brine (2 × 5 mL), dried over MgSO4, and concentrated under vacuum to a white solid (98.0 mg). The product was purified by recrystallization from EtOAc and hexanes to give a white powder (7b, 39.1 mg, 0.0580 mmol, 35%), mp 160–161 °C: 1H NMR (400 MHz, CDCl3) δ 7.40-7.15 (m, 11H), 7.05 (d, J = 7.8 Hz, 2H), 6.48 (m, 1H), 5.35 (m, 1H), 5.09 (s, 2H), 4.92 (d, J = 17 Hz, 1H), 4.79 (d, J = 16.6 Hz, 1H), 4.68 (m, 1H), 4.62 (m, 1H), 4.45 (m, 1H), 3.08 (m, 4H), 2.39 (s, 6H), 1.91 (m, 1H), 1.63 (m, 1H), 1.58 (m, 2H), 1.56 (s, 9H), 1.26 (m, 2H); MS (ESI) m/z calcd for (C38H47N3O8) 673.3, found 674.3 [M + H]+, 696.3 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 674.3441, found 674.3451.
Z-Phe-Lys(Boc)-CH2-O-(4-methyl)benzoate (7c)
mp 149–151 °C; 1H NMR (400 MHz, CDCl3) δ 7.97 (d, J = 8.3 Hz, 2H) 7.40-7.18 (m, 12H), 6.52 (m, 1H), 5.35 (m, 1H), 5.08 (s, 2H), 4.92 (d, J = 17.1 Hz, 1H), 4.81 (d, J = 17.1 Hz, 1H), 4.79 (m, 1H), 4.67 (m, 1H), 4.45 (m, 1H), 3.08 (m, 4H), 2.43 (s, 3H), 1.88 (m, 1H), 1.60, (m, 1H), 1.46 (m, 2H), 1.43 (s, 9H), 1.24 (m, 2H); MS (ESI) m/z calcd for (C37H45N3O8) 659.3, found 660.3 [M + H]+, 682.3 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 660.3285, found 660.3322.
Z-Phe-Lys(Boc)-CH2-O-(4-nitro)benzoate (7g)
mp 159–160 °C; 1H NMR (400 MHz, CDCl3) δ 8.33 (d, J = 8.8 Hz, 2H), 8.25 (d, J = 8.8 Hz, 2H) 7.40-7.18 (m, 10H), 6.48 (m, 1H), 5.32 (m, 1H), 5.09 (s, 2H), 5.00 (d, J = 17.1 Hz, 1H), 4.91 (d, J = 16.6 Hz, 1H), 4.72 (m, 1H), 4.60 (m, 1H), 4.46 (m, 1H), 3.10 (m, 4H), 1.88 (m, 1H), 1.60, (m, 1H), 1.48 (m, 2H), 1.43 (s, 9H), 1.25 (m, 2H); MS (ESI) m/z calcd for (C36H42N4O10) 690.3, found 691.3 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 691.2979, found 691.2998.
Z-Phe-Lys-CH2-O-(2,6-dimethyl)benzoate (8b)
TFA (1 mL) was added dropwise over 5 min to a solution of 7b (53.0 mg, 0.0787 mmol) in CH2Cl2 (3 mL) at 0 °C. The solution was warmed to room temperature over 2.5 h. The solvent was evaporated, and the resulting sticky residue was reprecipitated with EtOAc and ether, filtered, and dried under vacuum to give a brownish solid, which was purified by HPLC, using a gradient of 25% CH3CN in water/TFA (0.1%) to 100% CH3CN, to provide a white solid (8b, 24.1 mg, 0.0420 mmol, 53%), mp 136–139 °C: MS (ESI) m/z calcd for (C33H39N3O6) 573.3, found 574.2 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 574.2917, found 574.2909.
Z-Phe-Lys-CH2-O-(4-methyl)benzoate (8c)
mp 112–114 °C; MS (ESI) m/z calcd for (C32H37N3O6) 559.3, found 560.3 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 560.2760, found 560.2781.
Z-Phe-Lys-CH2-O-(4-nitro)benzoate (8g)
mp 134–145 °C; MS (ESI) m/z calcd for (C31H34N4O8) 590.2, found 591.2 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 591.2455, found 591.2449.
Z-Phe-Lys-CH2-Br (8h)
TFA (1.5 mL) was added dropwise over 5 min to a solution of 6 (50 mg, 0.083 mmol) in CH2Cl2 at 0 °C. The solution was warmed to room temperature over 4 h. The solvent was evaporated, and the resulting sticky residue was purified by HPLC with a gradient of 20% CH3CN in water/TFA (0.1%) to 100% CH3CN, providing a white solid (8h, 22.7 mg, 0.0450 mmol, 54%), mp 141–143 °C: 1H NMR (400 MHz, CD3OD) δ 7.28 (m, 10H), 5.05 (s, 2H), 4.53 (m, 1H), 4.38 (m, 1H), 3.9 (s, 2H), 3.07 (dd, J = 13.7, 6.8 Hz,1H), 2.94 (dd, J = 13.7, 8.3 Hz, 1H), 2.86 (t, J = 7.3 Hz, 2H), 1.87 (m, 1H), 1.60 (m, 3H), 1.36 (m, 2H); MS (ESI) m/z calcd for (C24H30BrN3O4) 503.1 (79Br), 505.1 (81Br), found 504, 506 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 504.1498 (79Br), 506.1477 (81Br), found 504.1519, 506.1497.
Z-Phe-Arg(Pbf)-OH (10)
l-Arginine(Pbf) (250 mg, 0.586 mmol) was dissolved in a mixture of dioxane (10 mL) and water (10 mL). Et3N (0.163 mL) was added followed by solid carbon dioxide pieces until pH 8 was achieved. Z-l-Phe-N-hydroxysuccinimide ester (1b, 230 mg, 0.586 mmol) was added and the solution was stirred for 20 h. The reaction was quenched with 1N HCl (50 mL) and the product was extracted with EtOAc. The EtOAc extracts were washed successively with 1N HCl (50 mL), water (50 mL), and brine (25 mL); dried over MgSO4; and concentrated under vacuum. The crude product was repreciptated from EtOAc and hexanes to produce Z-Phe-Arg(Pbf)-OH (10) as a white solid (230 mg, 0.325 mmol, 55%), mp 95–100 °C: 1H NMR (400 MHz, CDCl3) δ 7.60 (br, 1H), 7.35-7.10 (m, 10H), 5.90 (m, 1H), 5.10-4.80 (m, 3H), 4.65-4.38 (m, 3H), 3.11 (m, 3H), 2.95 (m, 1H), 2.90 (s, 2H), 2.52 (d, J = 6.8 Hz, 3H), 2.45 (d, J = 5.9 Hz, 3H), 2.05 (s, 3H), 1.95-1.20 (m, 6H), 1.43 (s, 6H); MS (MALDI) m/z calcd for (C36H45N5O8S) 707.3, found 708.3 [M + H]+, 730.3 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 708.3067, found 708.3046.
Z-Phe-Arg(Pbf)-CH2-Cl (11a)
Under a nitrogen atmosphere, isobutyl chloroformate (0.109 mL, 0.830 mmol) was added dropwise to a solution of Z-Phe-Arg(Pbf)-OH (10, 509.6 mg, 0.722 mmol) and N-methylmorpholine (0.099 mL, 0.902 mmol) in anhydrous THF (5 mL) at −20 °C. The solution was allowed to stir for 15 min. Diazomethane in ether (prepared from Diazald®) was added to the reaction flask dropwise. The solution was allowed to warm to room temperature over 3 h, then cooled to 0 °C with an ice bath. A 1:1 mixture of HCl and acetic acid (5 mL) was added dropwise. Once the evolution of N2 subsided, the reaction was diluted with EtOAc and washed with water (2 × 25 mL), brine (2 × 25 mL), and saturated NaHCO3 (2 × 25 mL). The solvent was evaporated to yield a yellow solid, which upon purification by flash chromatography with 80% EtOAc in hexanes provided Z-Phe-Arg(Pbf)-CH2-Cl (11a, 117.7 mg, 0.1590 mmol, 22%): 1H NMR (400 MHz, CDCl3) δ 7.76 (m, 1H), 7.27-7.13 (m, 10H), 6.36 (s, 2H), 6.10 (br, 1H), 5.93 (m, 1H), 4.93 (m, 1H), 4.94 (d, J = 12.2 Hz, 1H), 4.86 (d, J = 12.2 Hz, 1H), 4.58 (m, 1H), 4.51 (m, 1H), 4.09 (m, 1H), 4.00 (m, 1H), 3.08 (m, 2H), 2.95 (m, 1H), 2.91 (s, 2H), 2.55 (s, 3H), 2.47 (s, 3H), 2.07 (s, 3H), 1.81 (m, 2H), 1.61, (m, 2H), 1.44 (s, 6H); MS (ESI) m/z calcd for (C37H46ClN5O7S) 739.3, found 740.2 [M + H]+, 762.2 [M + Na]+, 778.2 [M + K]+; HRMS (ESI) m/z calcd for [M + H]+ 740.2884 (35Cl) and 742.2854 (37Cl), found 740.2907, 742.2895.
Z-Phe-Arg(Pbf)-CH2-O-benzoate (12a)
Under a nitrogen atmosphere, Z-Phe-Arg(Pbf)-CH2-Cl (11a, 50 mg, 0.068 mmol) was dissolved in dry DMF (2 mL). Benzoic acid (25 mg, 0.20 mmol) and KF (40 mg, 0.69 mmol) were added, and the reaction was stirred at room temperature overnight. The reaction was diluted with EtOAc, then successively washed with water (5 mL), saturated NaHCO3 (5 mL), and brine (5 mL). The solvent was dried over MgSO4 and evaporated to yield a residue, which upon purification by flash chromatography with 70% EtOAc in hexanes provided Z-Phe-Arg(Pbf)-CH2-O-benzoate (12a, 24.8 mg, 0.0300 mmol, 44%.), mp 94–96 °C: 1H NMR (400 MHz, CDCl3) δ 8.02 (m, 2H), 7.72 (br, 1H), 7.55 (m, 1H), 7.41 (m, 2H), 7.28-7.13 (m, 10H), 6.31 (br, 2H), 6.10 (br, 1H), 5.86 (br, 1H), 4.92 (m, 4H), 4.56 (m, 2H), 3.12 (m, 3H), 2.95 (m, 1H), 2.91 (s, 2H), 2.57 (s, 3H), 2.50 (s, 3H), 2.07 (s, 3H), 1.89 (m, 2H), 1.70 (m, 2H), 1.44 (s, 6H); MS (ESI) m/z calcd for (C44H51N5O9S) 825.3, found 826.2 [M + H]+, 848.4 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 740.2884, found 740.2907.
Z-Phe-Arg(Pbf)-CH2-O-(2,6-dimethyl)benzoate (12b)
mp 99–102 °C; 1H NMR (400 MHz, CDCl3) δ 7.35-7.10 (m, 11H), 7.04 (d, J = 7.3 Hz, 2H), 6.08 (br, 2 H), 5.55 (br, 1H), 4.98 (s, 2H), 4.80 (m, 2H), 4.61 (m, 1H), 4.51 (m, 1H), 3.11 (m, 3H), 3.03 (m, 1H), 2.92 (s, 2H), 2.57 (s, 3H), 2.51 (s, 3H), 2.38 (s, 6H), 2.07 (s, 3H), 1.96 (m, 2H), 1.68 (m, 2H), 1.44 (s, 6H); MS (MALDI) m/z calcd for (C46H55N5O9S) 853.37, found 854.44 [M + H]+, 876.51 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 854.3798, found 854.3775.
Z-Phe-Arg(Pbf)-CH2-O-(4-methyl)benzoate (12c)
mp 88–91 °C; 1H NMR (400 MHz, CDCl3) δ 7.92 (t, J = 8 Hz, 2H), 7.30-7.18 (m, 12H), 6.08 (br, 2H), 5.58 (br, 0.67H), 5.40 (br, 0.33H), 5.00 (d, J = 16.6 Hz, 2H), 4.98 (m, 1H), 4.83 (m, 1H), 4.62 (m, 1H), 4.45 (m, 1H), 3.11 (m, 3H), 3.00 (m, 1H), 2.93 (s, 2H), 2.58 (s, 3H), 2.51 (s, 3H), 2.41 (s, 3H), 2.08 (s, 3H), 1.90 (m, 2H), 1.67 (m, 2H), 1.45 (s, 6H); MS (ESI) m/z calcd for (C45H53N5O9S) 839.4, found 840.4 [M + H]+, 862.30 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 840.3642, found 840.3637.
Z-Phe-Arg(Pbf)-CH2-O-(2,4,6-trimethyl)benzoate (12d)
mp 82–84 °C; 1H NMR (400 MHz, CDCl3) δ 7.28-7.08 (m, 10H), 6.86 (s, 2H), 6.13 (br, 2H), 5.60 (br, 0.67H), 5.46 (br, 0.33H), 4.93 (m, 3H), 4.78 (m, 1H), 4.52 (m, 2H), 3.11 (m, 2H), 3.01 (m, 1H), 2.92 (s, 2H), 2.57 (s, 3H), 2.50 (s, 3H), 2.35 (s, 6H), 2.29 (s, 3H), 2.07 (s, 3H), 1.96 (m, 2H), 2.63 (m, 2H), 1.45 (s, 6H); MS (ESI) m/z calcd for (C47H57N5O9S) 867.4, found 868.2 [M + H]+, 890.4 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 868.3955, found 868.3994.
Z-Phe-Arg(Pbf)-CH2-O-(bis-2,6-trifluoromethyl)benzoate (12e)
1H NMR (400 MHz, CDCl3) δ 7.94 (d, J = 8.3 Hz, 2H), 7.74 (t, J = 7.8 Hz, 1H), 7.44 (br, 1H), 7.17 (m, 10H), 6.17 (br, 2H), 6.00 (br, 1H), 5.63 (d, J = 6.8 Hz, 1H), 4.94 (m, 4H), 4.62 (m, 1H), 4.55 (m, 1H), 3.12 (m, 3H), 2.97 (m, 1H), 2.92 (s, 2H), 2.55 (s, 3H), 2.49 (s, 3H), 2.07 (s, 3H), 1.90 (m, 2H), 1.63 (m, 2H), 1.44 (s, 6H); MS (ESI) m/z calcd for (C46H49F6N5O9S) 961.3, found 962.4 [M + H]+, 984.2 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 962.3233, found 962.3281.
Z-Phe-Arg(Pbf)-CH2-O-(4-methoxy)benzoate (12f)
mp 83–85 °C; 1H NMR (400 MHz, CDCl3) δ 8.00 (m, 2H), 7.30-7.15 (m, 10H), 6.91 (m, 2H), 6.11 (br, 2H), 5.60 (br, 0.67H), 5.43 (br, 0.33H), 4.99 (m, 3H), 4.83 (m, 1H), 4.64 (m, 1H), 4.48 (m, 1H), 3.86 (s, 3H), 3.11 (m, 3H), 2.98 (m, 1H), 2.94 (s, 2H), 2.58 (s, 3H), 2.52 (s, 3H), 2.09 (s, 3H), 1.93 (m, 2H), 1.67 (m, 2H), 1.45 (s, 6H); MS (ESI) m/z calcd for (C45H53N5O10S) 855.4, found 856.4 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 856.3591, found 856.3574.
Z-Phe-Arg(Pbf)-CH2-O-(4-nitro)benzoate (12g)
mp 108–111 °C; 1H NMR (400 MHz, CDCl3) δ 8.22 (m, 4H), 7.45 (br, 1H), 7.30-7.14 (m, 10H), 6.18 (br, 2H), 5.63 (br, 0.67H), 5.48 (br, 0.33H), 4.99 (m, 4H), 4.55 (m, 2H), 3.12 (m, 2H), 3.05 (m, 1H), 2.93 (s, 2H), 2.58 (s, 3H), 2.51 (s, 3H), 2.08 (s, 3H), 1.93 (m, 2H), 1.67 (m, 2H), 1.45 (s, 6H); MS (ESI) m/z calcd for (C44H50N6O11S) 870.3, found 871.4 [M + H]+, 893.2 [M + Na]+; HRMS (ESI) m/z calcd for [M + H]+ 871.3336, found 871.3368.
Z-Phe-Arg-CH2-O-benzoate (13a)
Z-Phe-Arg(Pbf)-CH2-O-benzoate (12a, 24.4 mg, 0.295 mmol) was stirred at 0 °C in 95:5 TFA/water (10 mL) until TLC indicated the reaction complete. The solvent was evaporated and the remaining residue purified by HPLC, using 40% CH3CN in water/TFA (0.1 %) as the eluant, to provide Z-Phe-Arg-CH2-O-benzoate (13a, 5.6 mg, 0.00975 mmol, 33%), mp 104–106 °C: MS (ESI) m/z calcd for (C31H35N5O6) 573.3, found 574.3 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 574.2665, found 574.2648.
Z-Phe-Arg-CH2-O-(4-methyl)benzoate (13c)
MS (MALDI) m/z calcd for (C32H37N5O6) 587.3, found 588.2 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 588.2822, found 588.2806.
Z-Phe-Arg-CH2-O-(2,4,6-trimethyl)benzoate (13d)
MS (MALDI) m/z calcd for (C34H41N5O6) 615.3, found 616.2 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 616.3135, found 616.3161.
Z-Phe-Arg-CH2-O-(bis-2,6-trifluoromethyl)benzoate (13e)
MS (ESI) m/z calcd for (C33H33F6N5O6) 709.2, found 710.0 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 710.2413, found 710.2395.
Z-Phe-Arg-CH2-O-(4-methoxy)benzoate (13f)
MS (MALDI) m/z calcd for (C32H37N5O7) 603.3, found 604.3 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 604.2771, found 604.2768.
Z-Phe-Arg-CH2-O-(4-nitro)benzoate (13g)
mp 101–103 °C; MS (ESI) m/z calcd for (C31H34N6O8) 618.2, found 619.0 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 619.2516, found 619.2499.
Z-Phe-Arg(Pbf)-(β-phenethylamine) (16)
Z-Phe-Arg(Pbf)-OH (10, 90.2 mg, 0.127 mmol) was dissolved in CH2Cl2 (4.5 mL) and cooled to 0 °C. HOBt (19.5 mg, 0.147 mmol) was added and the solution was stirred for 10 min at 0 °C. HBTU (48.3 mg, 0.127 mmol) and DIPEA (0.024 mL, 0.127 mmol) were added sequentially, and the solution was allowed to warm to room temperature over 0.5 h. β-Phenethylamine (0.018 mL, 0.127 mmol) was added and the solution was stirred overnight. The reaction was diluted with CH2Cl2 (50 mL) and washed successively with water (25 mL) and brine (25 mL). The solvent was dried over Na2SO3 and evaporated to yield a residue, which upon purification by flash chromatography with 1:1 THF/CHCl3 provided Z-Phe-Arg(Pbf)-(β-phenethylamine) (16, 40.2 mg, 0.0496 mmol, 39%): 1H NMR (400 MHz, CDCl3) δ 7.40-7.10 (m, 15H), 6.18 (br, 2H), 5.50 (br, 0.67H), 5.44 (br, 0.33H) 4.99 (m, 2H), 4.38 (m, 2H), 3.44 (m, 3H), 3.17 (m, 3H), 2.94 (s, 2H), 2.78 (m, 2H), 2.59 (s, 3H), 2.51 (s, 3H), 2.09 (s, 3H), 1.93 (m, 2H), 1.67 (m, 2H), 1.45 (s, 6H); MS (ESI) m/z calcd for (C44H54N6O7S) 810.4, found 811.4 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 811.3853, found 811.3858.
Z-Phe-Arg-(β-phenethylamine) (17)
A solution of Z-Phe-Arg(Pbf)-(β-phenethylamine) (16, 24 mg, 0.029 mmol) in 95:5 TFA/water (10 mL) was stirred for 2 h at 0 °C. The solvent was evaporated and the remaining residue was purified by HPLC using a 20-min linear eluant gradient from 30% acetonitrile CH3CN in water/TFA (0.1 %) to 100% CH3CN to provide Z-Phe-Arg-(β-phenethylamine) (17, 5.4 mg, 0.0097 mmol, 33%) as a clear yellow film: 1H NMR (400 MHz, CD3OD) δ 8.30 (d, J = 7.8 Hz, 1H), 8.13 (d, J = 7.8, 1H), 7.99 (t, J = 4.9, 1H), 7.70 (br, 1H), 7.25 (m, 15H), 5.05 (m, 2H), 4.35 (m, 1H) 4.26 (m, 1H), 4.08 (br, 1H), 3.37 (m, 2H), 3.27 (m, 1H), 3.07 (m, 2H), 2.92 (m, 3H), 2.76 (m, 2H), 1.76 (m, 2H), 1.60 (m, 2H); MS (ESI) m/z calcd for (C31H39N6O4) 558.3, found 559.3 [M + H]+; HRMS (ESI) m/z calcd for [M + H]+ 559.3033, found 559.3021.
5.3. Yeast Strains
The yeast strain used in this study was SM3614 (MATa trp1 leu2 ura3 his4 can1 ste24Δ::LEU2 rce1Δ::TRP1).6, 51, 52 Plasmid-bearing versions of the strain were generated according to published methods.53 Strains were routinely grown at 30 °C using synthetic complete dropout (SC–ura) medium.54 The over-expression plasmids encoding yeast Rce1p (pWS479) and yeast Ste24p (pSM1282) have been reported.19, 20, 42, 55
5.4. In vitro fluorescence-based CaaX proteolysis assay
An established fluorescence-based assay was used to monitor CaaX protease activity in the presence of candidate inhibitors.42, 43, 48 In brief, the assay involved the mixing of a quenched fluorogenic substrate with yeast membranes enriched for the appropriate CaaX protease. Membranes used as the source of activity were isolated according to our reported methods20, 22 and stored at −80 °C as 1 mg/mL stocks in lysis buffer (50 mM Tris, pH 7.5, 0.2 M sorbitol, 1 mM EDTA, 0.2% NaN3) containing a protease inhibitor cocktail (chymostatin, leupeptin, pepstatin, aprotinin, and phenylmethylsulphonyl fluoride) that does not inhibit either of the CaaX proteases. The membranes were typically diluted with assay buffer (100 mM HEPES, pH 7.5, 5 mM MgCl2) to 0.5 mg/mL immediately before use. The substrate was diluted to 40 µM from a 1 mM stock prepared in 4% DMSO. Assay assembly involved the dispensing of 50-µL aliquots of diluted membranes into the wells of a black, clear-bottom, 96-well microplate. This was followed by the addition of each compound to duplicate wells and a 15-min pretreatment incubation at 30 °C. After pretreatment, the assay was initiated by adding 50 µL of diluted substrate, resulting in final concentrations of 0.25 mg/mL membrane, 20 µM substrate, and 100 µM inhibitor. The sample fluorescence was measured every 30 to 60 s over a 60-min time course at 30 °C using a Bio Tek Synergy™ HT microplate fluorometer equipped with a 320/420-nm excitation/emission filter set. The collected data were exported to Microsoft Excel and graphed as a function of fluorescence versus time, and the initial velocities determined. These values were used to calculate the percentage activities relative to a DMSO-treated sample, which was included as a control in each reaction set.
Supplementary Material
Acknowledgments
We are grateful to Dr. Margot Paulick for technical advice and critical discussions and to Dr. Matthew Bogyo for donations of several AOMKs. This work was supported in part through a Georgia Cancer Coalition Distinguished Cancer Clinician/Scientist Scholar Award and National Institutes of Health grant (GM067092) (WKS), an NSF CAREER Award (TMD), a University of Georgia (UGA) BHSI Summer Research Fellowship (SRB), a UGA SURO Summer Research Fellowship (ACR), and a UGA CURO Summer Research Fellowships (NWH and JPM).
Abbreviations
- ABz
ortho-aminobenzoate
- AOMK
(acyloxy)methyl ketone
- DCC
N,N′-dicyclohexylcarbodiimide
- DIPEA
N,N-diisopropylethylamine
- HEPES
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- HOBt
1-hydroxybenzotriazole
- HBTU
O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- ICMT
isoprenylcysteine methyltransferase
- Pbf
pentamethyl-2,3-dihydrobenzofuran-5-sulfonyl
- QL
ε-dinitrophenyl lysine
- TEA
triethylamine
- TFA
trifluoroacetic acid
- TPCK
N-tosyl-l-phenylalanine chloromethyl ketone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data (1H NMR spectra and HPLC chromatograms for new compounds) associated with this article can be found in the online version at http...???
References
- 1.Young SG, Ambroziak P, Kim E, Clarke S. In: The Enzymes. Tamanoi F, Sigman DS, editors. Vol. 22. New York: Academic Press; 2001. p. 155. [Google Scholar]
- 2.Winter-Vann AM, Casey PJ. Nat. Rev. Cancer. 2005;5:405. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Nat. Rev. Drug Disc. 2007;6:541. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 4.Casey PJ, Seabra MC. J. Biol. Chem. 1996;271:5289. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 5.Boyartchuk VL, Ashby MN, Rine J. Science. 1997;275:1796. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 6.Tam A, Nouvet FJ, Fujimura-Kamada K, Slunt H, Sisodia SS, Michaelis S. J. Cell Biol. 1998;142:635. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sapperstein S, Berkower C, Michaelis S. Mol. Cell Biol. 1994;14:1438. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai Q, Choy E, Chiu V, Romano J, Steitz SR, Steitz SA, Michaelis S, Philips MR. J. Biol. Chem. 1998;273:15030. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 9.Backlund PS., Jr J. Biol. Chem. 1997;272:33175. doi: 10.1074/jbc.272.52.33175. [DOI] [PubMed] [Google Scholar]
- 10.Wright LP, Philips MR. J. Lipid Res. 2006;47:883. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Takai Y, Sasaki T, Matozaki T. Physiol. Rev. 2001;81:153. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 12.Gelb MH, Brunsveld L, Hrycyna CA, Michaelis S, Tamanoi F, Van Voorhis WC, Waldmann H. Nat. Chem. Biol. 2006;2:518. doi: 10.1038/nchembio818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freije JMP, Blay P, Pendas AM, Cadinanos J, Crespo P, Lopez-Otin C. Genomics. 1999;58:270. doi: 10.1006/geno.1999.5834. [DOI] [PubMed] [Google Scholar]
- 14.Otto JC, Kim E, Young SG, Casey PJ. J. Biol. Chem. 1999;274:8379. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 15.Kim E, Ambroziak P, Otto JC, Taylor B, Ashby M, Shannon K, Casey PJ, Young SG. J. Biol. Chem. 1999;274:8383. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 16.Cadiñanos J, Schmidt WK, Fueyo A, Varela I, López-Otín C, Freije JMP. Biochem. J. 2003;370:1047. doi: 10.1042/BJ20021514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bracha K, Lavy M, Yalovsky S. J. Biol. Chem. 2002;277:29856. doi: 10.1074/jbc.M202916200. [DOI] [PubMed] [Google Scholar]
- 18.Cadiñanos J, Varela I, Mandel DA, Schmidt WK, Diaz-Perales A, Lopez-Otin C, Freije JMP. J. Biol. Chem. 2003;278:42091. doi: 10.1074/jbc.M306700200. [DOI] [PubMed] [Google Scholar]
- 19.Tam A, Schmidt WK, Michaelis S. J. Biol. Chem. 2001;276:46798. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- 20.Plummer LJ, Hildebrandt ER, Porter SB, Rogers VA, McCracken J, Schmidt WK. J. Biol. Chem. 2006;281:4596. doi: 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11175. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt WK, Tam A, Michaelis S. J. Biol. Chem. 2000;275:6227. doi: 10.1074/jbc.275.9.6227. [DOI] [PubMed] [Google Scholar]
- 23.Ma YT, Gilbert BA, Rando RR. Biochemistry. 1993;32:2386. doi: 10.1021/bi00060a033. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Ma Y-t, Rando RR. Biochemistry. 1996;35:3227. doi: 10.1021/bi952529s. [DOI] [PubMed] [Google Scholar]
- 25.Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. Biochemistry. 2000;39:4096. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 26.Pei J, Grishin NV. Trends Biochem. Sci. 2001;26:275. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- 27.Mokry DZ, Manandhar SP, Chicola KA, Santangelo GM, Schmidt WK. Eukaryotic Cell. 2009;8:1891. doi: 10.1128/EC.00169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper NM, Lendeckel U, editors. Intramembrane-Cleaving Proteases (I-CLiPs) Dordrecht, The Netherlands: Springer; 2007. [Google Scholar]
- 29.Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, Mohr A, Meta M, Genant H, Jiang Y, Wisner ER, Van Bruggen N, Carano RAD, Michaelis S, Griffey SM, Young SG. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13049. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pendas AM, Zhou Z, Cadinanos J, Freije JMP, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, Lopez-Otin C. Nat. Genet. 2002;31:94. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 31.Trueblood CE, Boyartchuk VL, Picologlou EA, Rozema D, Poulter CD, Rine J. Mol. Cell Biol. 2000;20:4381. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergo MO, Lieu HD, Gavino BJ, Ambroziak P, Otto JC, Casey PJ, Walker QM, Young SG. J. Biol. Chem. 2004;279:4729. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- 33.Barrowman J, Michaelis S. Biol. Chem. 2009;390:761. doi: 10.1515/BC.2009.080. [DOI] [PubMed] [Google Scholar]
- 34.Worman HJ, Fong LG, Muchir A, Young SG. J. Clin. Invest. 2009;119:1825. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young SG, Meta M, Yang SH, Fong LG. J. Biol. Chem. 2006;281:39741. doi: 10.1074/jbc.R600033200. [DOI] [PubMed] [Google Scholar]
- 36.Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagase H, Casey PJ, Balmain A, Young SG. Mol. Cell Biol. 2002;22:171. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y. Cancer Lett. 1998;131:191. doi: 10.1016/s0304-3835(98)00146-3. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y. Ann. NY Acad. Sci. 1999;886:103. doi: 10.1111/j.1749-6632.1999.tb09405.x. [DOI] [PubMed] [Google Scholar]
- 39.Craig KS, Williams DE, Hollander I, Frommer E, Mallon R, Collins K, Wojciechowicz D, Tahir A, Van Soest R, Andersen RJ. Tetrahedron Lett. 2002;43:4801. [Google Scholar]
- 40.Williams DE, Hollander I, Feldberg L, Frommer E, Mallon R, Tahir A, van Soest R, Andersen RJ. J. Nat. Prod. 2009;72:1106. doi: 10.1021/np900042r. [DOI] [PubMed] [Google Scholar]
- 41.Schlitzer M, Winter-Vann A, Casey PJ. Bioorg. Med. Chem. Lett. 2001;11:425. doi: 10.1016/s0960-894x(00)00685-5. [DOI] [PubMed] [Google Scholar]
- 42.Manandhar SP, Hildebrandt ER, Schmidt WK. J. Biomol. Screen. 2007;12:983. doi: 10.1177/1087057107307226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter SB, Hildebrandt ER, Breevoort SR, Mokry DZ, Dore TM, Schmidt WK. Biochim. Biophys. Acta. 2007;1773:853. doi: 10.1016/j.bbamcr.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krantz A, Copp LJ, Coles PJ, Smith RA, Heard SB. Biochemistry. 1991;30:4678. doi: 10.1021/bi00233a007. [DOI] [PubMed] [Google Scholar]
- 45.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KAH, Salvesen GS, Bogyo M. Nat. Chem. Biol. 2005;1:33. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 46.Dolence EK, Dolence JM, Poulter CD. J. Comb. Chem. 2000;2:522. doi: 10.1021/cc000026m. [DOI] [PubMed] [Google Scholar]
- 47.Wood WJL, Huang L, Ellman JA. J. Comb. Chem. 2003;5:869. doi: 10.1021/cc034008r. [DOI] [PubMed] [Google Scholar]
- 48.Hollander I, Frommer E, Mallon R. Anal. Biochem. 2000;286:129. doi: 10.1006/abio.2000.4795. [DOI] [PubMed] [Google Scholar]
- 49.Rauber P, Walker B, Stone S, Shaw E. Biochem. J. 1988;250:871. doi: 10.1042/bj2500871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bezas B, Zervas L. J. Am. Chem.Soc. 1961;83:719. [Google Scholar]
- 51.Siliciano PG, Tatchell K. Cell. 1984;37:969. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- 52.Powers S, Michaelis S, Broek D, Santa Anna-A S, Field J, Herskowitz I, Wigler M. Cell. 1986;47:413. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- 53.Elble R. Bio Techniques. 1992;13:18. [Google Scholar]
- 54.Michaelis S, Herskowitz I. Mol. Cell Biol. 1988;8:1309. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romano JD, Schmidt WK, Michaelis S. Mol. Biol. Cell. 1998;9:2231. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.