Abstract
The hormone-secreting cell types of the anterior pituitary differentiate in a specific spatial and temporal manner. The α-subunit of the glycoprotein hormones appears at embryonic d 11.5 in the mouse, followed by steroidogenic factor-1, which distinguishes the gonadotrope progenitor cells, around embryonic d 14. Gonadotrope maturation is marked by the onset of LHβ-gene expression 2 d later. The αT3-1 and LβT2 immortalized mouse pituitary cell lines correspond to these later sequential stages of gonadotrope differentiation. In addition to the early markers of the gonadotrope lineage present in αT3-1 cells, LβT2 cells also express markers of a mature gonadotrope, including LHβ and FSHβ. Using transient transfections to compare expression among gonadotrope and non-gonadotrope-derived cell types, we show that the rat 1.8-kb LHβ promoter directs reporter gene expression specifically to the mature gonadotrope LβT2 cell line. Promoter truncation and mutagenesis analyses indicate that the homeodomain (HD) element located at approximately −100 bp relative to the transcriptional start site is essential for this selectivity to LβT2 cells when compared with αT3-1 cells. In EMSAs, this HD site binds a protein present in LβT2 but not other gonadotrope-derived cells. Antibody supershift and competition experiments indicate that this LβT2 nuclear protein is a K50 HD protein related to the Otx family, though it is not a known pituitary homeobox transcription factor protein. These studies indicate a role for a novel Otx-related HD protein in gonadotrope maturation during development.
The molecular basis of development lies in the specific and differential expression of genes that characterize individual cell types. An important step in understanding cell fate determination is the identification of factors involved in cell-type specific expression of genes. In the neuroendocrine system, the anterior pituitary is an ideal model for studying cell fate decisions involved in specifying individual phenotypes. The mature anterior pituitary consists of five distinct cell populations that develop from a common primordium. These different cell types result from the sequential activation and inactivation of lineage-specific genes during development and are characterized by their ability to produce specific hormones.
The gonadotropins, LH and FSH, are essential reproductive hormones that are specifically produced in the gonadotrope population of the anterior pituitary. LH and FSH are both members of the glycoprotein hormone family that also includes TSH, made in pituitary thyrotropes, and CG, made in human and primate placenta (1). Each of these hormones shares a common α-subunit and contains a unique β-subunit that confers the physiological specificity of the hormone. The β-subunit genes of LH and FSH are expressed exclusively in the pituitary gonadotrope, thus limiting the synthesis and secretion of LH and FSH to these cells.
The molecular mechanism controlling the gonadotrope-specific expression of the LH and FSH β-subunit genes has been difficult to study due to the heterogeneous nature of the anterior pituitary in vivo and the lack of a differentiated gonadotrope cell line that produces endogenous LHβ and FSHβ in which to study their expression. Our laboratory has used targeted oncogenesis in transgenic mice to develop several pituitary-derived cell lines of the gonadotrope and thyrotrope lineages (2). The αT1-1 cell line expresses the common α-glycoprotein hormone subunit (α-GSU) of the glycoprotein hormones, but is not committed to either the gonadotrope or thyrotrope lineage. The αT3-1 cell line corresponds to a determined, but immature gonadotrope, expressing not only α-GSU, but also specific markers of the gonadotrope lineage, such as steroidogenic factor-1 (SF-1) and GnRH receptor (GnRH-R). In addition to these early markers of gonadotrope differentiation, the LβT2 cell line expresses the β-subunits of LH and FSH (3, 4) and therefore corresponds to a more mature gonadotrope.
LH β-subunit gene expression has been studied extensively using transgenic mice, heterologous cell lines (GH3, CV1) (5, 6), and more recently the αT3-1 and LβT2 cell lines (7–9). Several cis-regulatory elements important for both basal and GnRH regulated transcription of the LHβ gene have been identified. Binding sites for the orphan nuclear receptor SF-1 are located at −127 bp and −59 bp relative to the transcriptional start site of the rat LHβ promoter (10). Within the pituitary, SF-1 expression is gonadotrope specific, making it a key identifier of the gonadotrope cell population (11). In transfected cells, SF-1 has been shown to activate transcription of numerous gonadotrope markers, including GnRH-R (12), the glycoprotein α-subunit (13), and LHβ. Supporting the importance of SF-1 to gonadotrope function in vivo, SF-1 null mice are infertile and have markedly reduced levels of α-GSU, LHβ, FSHβ, and GnRH-R (11). Pituitary-specific SF-1 knockout mice indicate that the impaired gonadotropin expression is a primary defect due to the lack of SF-1 in the pituitary and not a secondary defect due to SF-1 deficiency in other tissues (14). Although SF-1 is an important basal regulator of LHβ gene expression, it is not sufficient to confer gonadotrope specificity of the gonadotropin β-subunit genes because it is expressed earlier in development and in steroidogenic tissues outside of the pituitary (15) as well as in αT3-1 cells, which do not produce the LH or FSH β-subunits.
Binding sites for early growth response protein 1 (Egr-1) are adjacent to the SF-1 binding sites, located at −112 bp and−50 bp relative to the transcriptional start site (5). Egr-1 (NGFI-A, Krox-24, zif/268) is an early-response gene that is important for GnRH regulation of the LHβ gene. Egr-1 null animals are infertile and fail to produce LHβ, basally or in response to GnRH (16, 17). In LβT2 and αT3-1 cells, basal expression of Egr-1 is low; however, expression is dramatically increased upon treatment with GnRH (6, 7, 18). Egr-1 can activate transcription of the LHβ gene by direct physical interactions and synergies with SF-1 and the pituitary homeobox 1 transcription factor (Ptx1, also termed Pitx1 or P-OTX) (5, 6).
Ptx1 is a member of the Ptx family of Paired-like homeobox transcription factors that also includes Ptx2 and Ptx3. Whereas both Ptx1 and Ptx2 are expressed in gonadotrope cells and are important for pituitary development (19–21), Ptx3 is absent from the pituitary (22). Ptx1 expression is first detected in the presumptive pituitary around embryonic d 9.5, after invagination of Rathke’s Pouch (23), and continues throughout development of the anterior pituitary. In the adult pituitary, Ptx1 expression is maintained in all pituitary cells, albeit at varying levels (24, 25). Mice null for Ptx1 die at birth and exhibit developmental defects in the anterior pituitary gland, including decreases in the number of gonadotropes and thyrotropes and corresponding decreases in TSHβ, LHβ, and FSHβ gene transcripts (19). Indeed, using transient transfections of heterologous cells (CV1), it has been demonstrated that Ptx1 is capable of transactivating numerous pituitary-specific promoters, including α-GSU, LHβ, FSHβ, GnRH-R, TSHβ, POMC, PRL, and GH (22, 26). In CV1 cells, Ptx1 can activate LHβ gene expression through the homeodomain (HD) binding element located at −100 bp of the rat LHβ promoter (27). Although this HD element is essential for overall LHβ promoter activity in transgenic mice (9), it is not required for Ptx1 regulation in CV1 cells. Even in the presence of a mutated HD-binding site, Ptx1 can activate transcription of the LHβ gene through synergy with SF1 and Egr-1 (6).
Despite the importance of SF-1, Egr-1, and Ptx1 for expression of the LHβ gene, they are not sufficient to confer LHβ gene expression because all of these factors are present in αT3-1 cells (which do not express endogenous LHβ), as well as being present early in gonadotrope development in vivo before the onset of LHβ gene expression (11, 17, 19). The specific expression of endogenous LHβ in LβT2 cells indicates that there are additional factors, not present in the early gonadotrope precursor αT3-1 cells, which are involved in the maturation of the gonadotrope and the onset of gonadotropin β-subunit gene expression and the subsequent production and secretion of LH. These cell lines present an opportunity to identify lineage specific and developmentally regulated transcription factors involved in gonadotrope differentiation and the cell-specific transcriptional regulation of LHβ gene expression in the more differentiated LβT2 cells when compared with the precursor αT3-1 cells.
Using a transient transfection paradigm to directly compare reporter gene expression in a variety of pituitary- and nonpituitary-derived cell lines, we show that an LHβ-luciferase (Luc) reporter plasmid is specifically expressed in the mature LβT2 gonadotrope cell line. Truncation and mutagenesis analyses indicate that the HD element is necessary for the specific expression in LβT2 cells when compared with precursor αT3-1 cells. Using EMSA, we show that this HD element interacts with a protein in LβT2 cell nuclear extract that does not appear in αT3-1 or αT1-1 cell nuclear extracts. Further analyses indicate that this LβT2 nuclear protein complex does not contain Ptx1 or Ptx2 HD transcription factors but does contain a protein related to Otx HD family members.
RESULTS
A Transfected 1.8-kb Rat LH β-Subunit Promoter Targets Reporter Gene Expression Specifically to LβT2 Cells
Because the endogenous mouse LHβ gene is expressed in LβT2 gonadotrope cells but not in the immature αT3-1 cells, we postulate that LβT2 cells can be used as a model to identify factors involved in the induction of LHβ gene expression during gonadotrope maturation. To study the activity of the LHβ promoter, we first measured the level of reporter gene expression from the LHβ promoter in LβT2 cells as compared with a variety of other pituitary and nonpituitary-derived cell lines. To accomplish this, a fragment encompassing 1.8 kb of the rat LHβ promoter was linked to a Luc reporter gene (LHβLuc) and used in transient transfection studies. A Rous Sarcoma Virus (RSV) enhancer and promoter driving β-galactosidase (RSV-β-gal) was cotransfected as an internal control. Additionally, an RSV-Luc plasmid was transfected in parallel with LHβLuc into each cell type to control for differences in transfection efficiency, transcription rates, reporter mRNA half-life, Luc and β-gal protein stability, and metabolic rates between distinct cell types. The ratio of RSV-Luc to RSV-β-gal was set to one hundred for each cell line, and the LHβLuc values in each cell line were normalized to the RSV-Luc/RSV-β-gal value. To validate this approach, we tested the activities of the promoterless pUC18-Luc and a TK-Luc plasmid and found each to be expressed at equivalent levels in αT3-1 and LβT2 cells relative to the RSV controls (data not shown).
Results from these experiments show that the transfected LHβ reporter gene is specifically expressed in the more differentiated LβT2 pituitary gonadotrope cell line (Fig. 1). LHβ promoter activity is approximately 4.5- to 5-fold higher in LβT2 cells than in the gonadotrope precursor αT3-1 cells and up to 20-fold higher than in nongonadotrope-derived cell lines such as AtT20 (corticotrope-derived), GT1-7 (hypothalamic neuroendocrine), JEG-3 (placental), HeLa (cervical fibroblast), CV1 (kidney-derived), and NIH3T3 (fibroblast-derived). These data show that the 1.8-kb rat LHβ promoter is sufficient to direct LβT2 cell-specific expression.
Fig. 1. The Rat LH β-Subunit Promoter Is Specifically Active in LβT2 Cells.
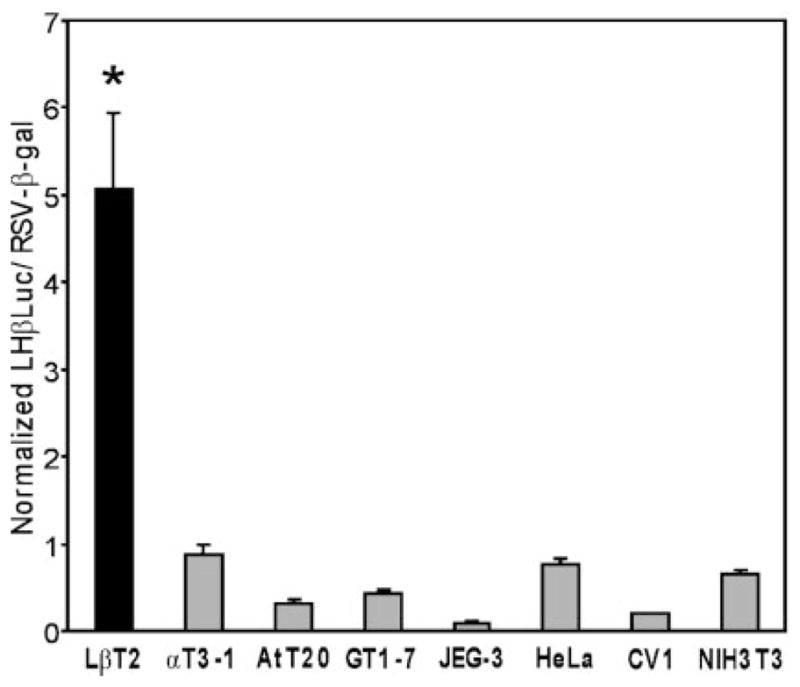
The 1.8-kb rat LHβ promoter was linked to the Luc reporter gene (LHβLuc) and transiently transfected into pituitary-derived LβT2 (differentiated gonadotrope), αT3-1 (precursor gonadotrope), and AtT20 (corticotrope) cells and nonpituitary-derived GT1-7 (hypothalamic), JEG-3 (placental), HeLa (cervical fibroblast), CV1 (kidney), and NIH3T3 (fibroblast) cells. RSV-β-gal was cotransfected as an internal control. To control for differences in transfection efficiencies and transcription rates between distinct cell lines, a parallel control, RSV-Luc was transfected into each cell line with the RSV-β-gal internal control. The ratio of RSV-Luc divided by RSV-β-gal was set at 100 for each cell line (not shown). Results represent the mean ± SEM of at least three independent experiments (n ≥ 3). The bar marked with an asterisk (*) (LβT2 cells) is statistically significant from all other bars (P < 0.05). Significant differences between other cell lines are not shown.
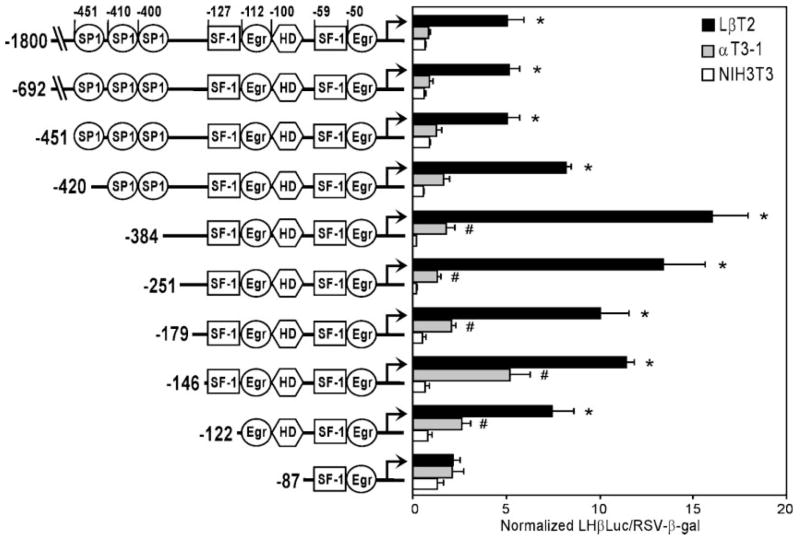
The 1.8-kb rat LHβ promoter contains several previously identified regulatory elements that are involved in both basal and GnRH-regulated expression of the gene. To identify which region(s) of the LHβ promoter are involved in LβT2 cell specificity, 5′ deletions of LHβLuc were transiently transfected into LβT2, αT3-1, and NIH3T3 cells (Fig. 2). Truncation of the LHβ promoter to as little as 179 bp has no effect on specificity between LβT2 and αT3-1 cells. Further deletion to 146 bp reduces the specificity, due to an apparent increase in LHβ promoter activity in αT3-1 cells, but retains an approximate 2-fold difference. The remaining selectivity between LβT2 and αT3-1 cells is eliminated when the promoter is shortened from 122 bp to 87 bp. This results in a decrease of activity in LβT2 cells, indicating that this region is important for LβT2 cell specificity.
Fig. 2. The Region Between −122 and −87 of the Rat LHβ Promoter is Necessary for LβT2 Cell Specificity.
Truncations of the LHβ promoter linked to Luc were transiently transfected into LβT2, αT3-1, and NIH3T3 cells (identified by black, gray, and white bars, respectively). Diagrams at left depict the 5′ deletions used as well as the numerous regulatory elements identified in the rat LHβ promoter, including HD and binding sites for SP1, SF-1, and Egr. Results represent the mean ± SEM of at least three independent experiments (n ≥ 3). Asterisks (*) designate a significant difference from αT3-1 and NIH3T3 cells (P < 0.05); # indicates a significant difference from NIH3T3 cells (P < 0.05).
A Nuclear Protein Complex Present in LβT2 Cells, But Not Other Gonadotrope-Derived Cell Lines, Binds the LHβ Promoter HD Element
The region of the LHβ promoter between −122 bp and −87 bp has been previously shown to include the 5′ Egr-1 binding site that is involved in GnRH regulation of the LHβ gene as well as the HD protein binding sequence that is important for Ptx1 regulation in CV1 cells. To determine whether any LβT2 cell-specific proteins are binding to this region of the LHβ promoter, an EMSA was performed using a 35-bp oligo-nucleotide probe corresponding to this region of the LHβ promoter (LH 121/87) with nuclear extracts from LβT2, αT3-1, αT1-1, and a variety of other pituitary-and nonpituitary-derived cell lines (Fig. 3). Within the gonadotrope-derived cell lines, strong binding of a protein complex is observed using nuclear extract from the differentiated LβT2 cells, but not the precursor αT3-1 or αT1-1 cells (complex 2). A complex with the same migration, but varying levels of intensity, is also observed in AtT20 and to a lesser degree in TαT1 and GT1-7 cells, all of which are endocrine-derived cell lines. Several other complexes were also observed using nuclear extracts from LβT2 cells. Complex 1 is a slowly migrating complex that is present in all cell lines at varying levels. Complex 3, which migrates slightly faster than complex 2, appears equally intense in LβT2, αT3-1, and AtT20 nuclear extracts but less intense in αT1-1, TαT1, and GT1-7 cell nuclear extracts. Two other complexes appear in several cell types but were found to be variable and relatively nonspecific; they are indicated by lines but are not numbered.
Fig. 3. A Complex Present in LβT2 Cells, but Absent from Other Gonadotrope-Derived Cell Lines, Binds the LHβ Promoter.
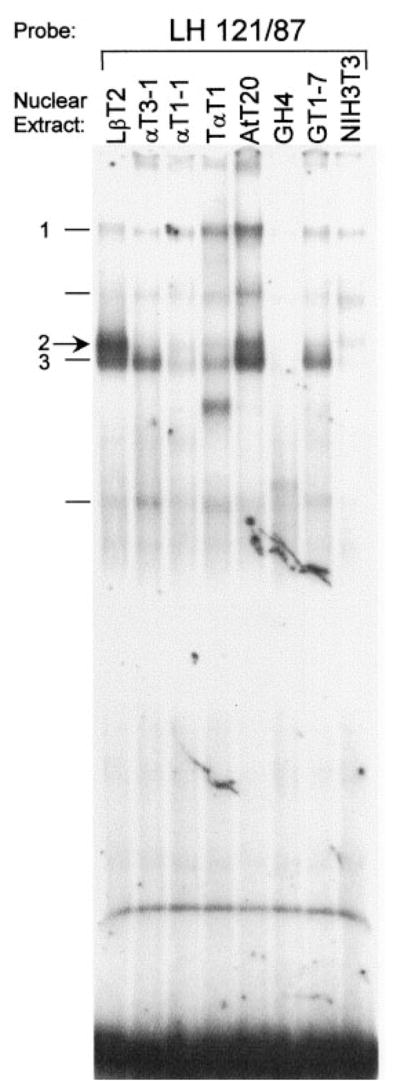
EMSA was conducted using the LH 121/87 probe and nuclear extracts from pituitary and nonpituitary-derived cell lines, indicated above each lane: LβT2 (gonadotrope), αT3-1 (precursor gonadotrope), αT1-1 (uncommitted gonadotrope/thyrotrope), TαT1 (thyrotrope), AtT20 (corticotrope), GH4 (so-matotrope), GT1-7 (hypothalamic neuroendocrine), and NIH3T3 (fibroblast). Complexes in LβT2 cells are indicated at left. Complex 2 exhibits strong binding in the mature gonadotrope LβT2 nuclear extracts, but not the αT3-1 or αT1-1 precursor nuclear extracts.
To further define the sequences within this region of the LHβ promoter that bind to each of the complexes, EMSA analysis was performed using the LH 121/87 probe, LβT2 cell nuclear extract, and shortened oligo-nucleotide competitors consisting of sequences from the LH 121/87 region (Fig. 4A). The sequences of oligonucleotides used in competitions are shown in Table 1. Complex 1 is competed only by the region from −121 bp to −100 bp, which encompasses the previously identified 5′ Egr-1 element (5). Under basal conditions, Egr-1 binding to its 3′ element in the LHβ cells; rather, promoter is not detected in GH3 or αT3-1 antibody supershift experiments indicate that the ubiquitous transcription factor SP1 binds to this GC-rich Egr-1 site (28). Similarly, we detected SP-1 binding to the 5′ Egr-1 element from the LHβ promoter using antibody supershift experiments with LβT2, αT3-1, and NIH3T3 cell extracts (data not shown). Mutation of this Egr-1/SP1 element eliminates competition of complex 1 but has no effect on binding of faster migrating complexes (Fig. 4B, mutant 1, lane 3).
Fig. 4. Complex 2 Binds the HD Element of the LHβ Promoter.
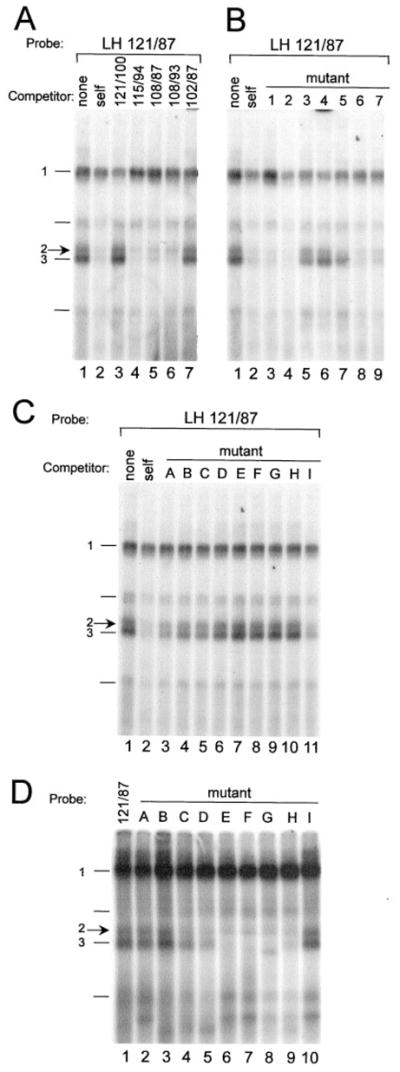
A–C, EMSA was conducted using the LH 121/87 probe and nuclear extract from LβT2 cells. Competitions were performed using 100-fold excess unlabeled oligonucleotide as indicated above each lane: None (no competitor), self (wild-type LH 121/87 competitor). A, Competitions using truncated LH 121/87 oligonucleotide competitors indicate that complex 2 binds in the region from −108 to −94. B and C, Competitions using mutated LH 121/87 oligonucleotides indicate that complex 2 binds the HD element. D, EMSA was conducted using oligonucleotides containing single base-pair mutations as probes and nuclear extract from LβT2 cells. The closed arrow at left indicates complex 2; complexes 1 and 3 are also indicated. The results are summarized and sequences of all oligonucleotide competitors and probes are depicted in Table 1.
Table 1.
Summary of Complex 2, Otx1, and Ptx1 Binding Site Requirements
 |
Wild-type and mutant oligonucleotides are depicted as used for probes and competitions in EMSA in Figs. 4 and 8 and data not shown. The homeodomain (HD) and SP1/Egr binding sequences are indicated. At right, the ability of each oligonucleotide to bind and compete with complex 2, Otx1, or Ptx1 binding to the LH 121/87 probe is depicted (+++, maximum; +, minimum; and −, no binding).
Complexes 2 and 3 are successfully competed by addition of excess unlabeled wild-type LH 121/87 (self competition), as well as the smaller oligonucleotides containing sequences from −108 to −94, indicating that this region is sufficient to bind the proteins comprising these complexes (Fig. 4A, lanes 4–6). Experiments using oligonucleotide competitors containing 3 bp block mutations scanning the sequences from −109 bp to −89 bp (sequences are shown in Table 1) further narrow down the region important for binding of complex 2 to the 6 bases between −103 bp and −98 bp and binding of complex 3 to the 9 bases between −103 and −95 (Fig. 4B, mutants 3–5, lanes 5–7). This difference in binding requirements between complexes 2 and 3 is based on the ability of mutant 5 to compete with complex 2 to a greater extent than it competes with complex 3 (Fig. 4B, mutant 5, lane 7). The region these complexes bind encompasses the HD-binding element of the LHβ promoter. These results were confirmed using the mutant oligonucleotides as probes and examining complex binding directly (data not shown).
To establish that complexes 2 and 3 are actually binding to the HD element, oligonucleotides with single base-pair point mutations spanning the HD binding site and surrounding sequences were used as competitors and probes in EMSA (Fig. 4, C and D, respectively, sequences shown in Table 1). Point mutations within the HD-binding element (mutants D, E, F, G, and H) almost completely block binding and competition of both complexes 2 and 3 to the LH 121/87 probe. In contrast, oligonucleotides containing point mutations outside the core HD-binding element (mutants A, B, and I) bind equally as well as wild-type. Slight differences in binding ability between complexes 2 and 3 were observed using these mutants as probes (5′ end of HD site, mutants C and D, Fig. 4D). These gelshift data indicate that the core bases of the HD-binding element, but not additional bases outside this core, are necessary for binding of complexes 2 and 3 to the LHβ promoter.
The LHβPromoter HD Element Is Necessary for LβT2 Cell-Specific Expression
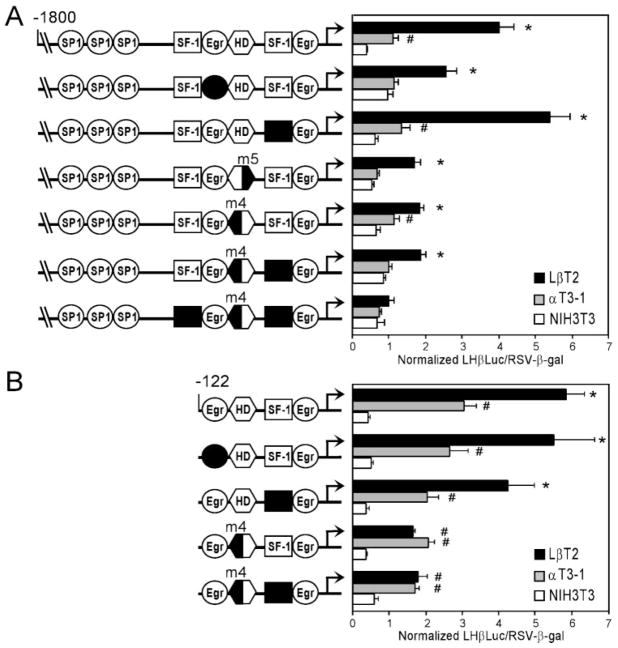
Having examined the proteins binding to the HD and Egr/SP1 elements of the LHβ promoter, mutations were introduced into −1800LHβLuc and −122LHβLuc and transiently transfected into LβT2, αT3-1, and NIH3T3 cells to ascertain whether either of these elements is involved in LβT2 cell-specific expression (Fig. 5). Site-directed mutagenesis was performed to mutate the 5′ Egr-1 binding site, the HD element, and the 3′ SF-1 binding site. The 3′ SF-1 site was mutated because of the importance of SF-1 for LHβ gene expression and the ability of SF-1 to physically interact and synergize with both Ptx1 and Egr-1. Mutation of the element that binds Egr-1 has little or no effect on LβT2 cell-specific expression of the LHβ gene under our basal (non-GnRH stimulated) conditions. Similarly, mutation of the 3′ SF-1 binding site has no effect on LβT2 cell specificity (4-fold for −1800LHβLuc and 2-fold for −122LHβLuc), although it does reduce basal levels in both LβT2 and αT3-1 cells within −122LHβLuc.
Fig. 5. Mutation of the HD Element Abrogates LβT2 Cell Specificity.
Site-directed mutagenesis was performed to mutate the 5′ Egr-1 binding site, the 3′ SF-1 binding site, the 5′ SF-1 binding site, and the HD in both the −1800LHβLuc and −122LHβLuc reporter genes. Two different HD mutations were examined (m4 and m5) as indicated in the diagram at left. HDm4 and HDm5 correspond to mutant 4 and mutant 5 used in Figs. 4 and 8 and shown in Table 1. Transient transfections of mutant LHβLuc plasmids were performed in LβT2, αT3-1, and NIH3T3 cells (identified by black, gray, and white bars, respectively). A, Mutations in the context of −1800LHβLuc. B, Mutations in the context of −122LHβLuc. Results represent the mean ± SEM of at least three independent experiments, each performed in triplicate (n ≥ 9). Asterisks (*) designate a significant difference from αT3-1 and NIH3T3 cells (P < 0.05); # indicates a significant difference from NIH3T3 cells (P < 0.05).
Two different HD mutations were examined in −1800LHβLuc based on protein binding data from Fig. 4. HD mutant 5 (m5), which shows reduced binding of complex 2, reduces basal activity of both LβT2 and αT3-1 cells. The difference between LβT2 and αT3-1 cells for −1800LHβLuc HDm5 is 2.6-fold (decreased from 4-fold). In contrast, HD mutant 4 (m4), which completely blocks binding of complex 2, reduces basal activity only in LβT2 cells. Specificity between LβT2 and αT3-1 cells for HDm4 is reduced to 1.5-fold. Whereas HDm4 only partially reduces the LβT2 cell-specificity of −1800LHβLuc, the same mutation completely abolishes the specificity of −122LHβLuc as compared with αT3-1 cells. Further mutations used HDm4 because it has a greater effect on both binding of complex 2 and specificity between the gonadotrope-derived cell lines. Expression of the HD (m4) and 3′ SF-1 double mutant is similar to that of the HD mutant (m4) alone in both LHβLuc plasmids. The residual difference of the 3′ SF-1 and HD (m4) double mutant within −1800LHβLuc between the gonadotrope-derived cell lines is due to the presence of the 5′ SF-1 site, through which SF-1 and Ptx1 have been shown to synergize when overexpressed in CV1 cells (6). Mutation of this 5′ SF-1 site in combination with the 3′ SF-1 and HD (m4) mutations (i.e. a triple mutant) eliminates all differences between the cell lines. These transfection data indicate that the HD element, which binds complexes 2 and 3, is the key element necessary for expression of LHβ in LβT2 cells. Although both complexes 2 and 3 are present in nongonadotrope-derived cell lines, they may be involved in gonadotrope-specific expression of LHβ by interaction with the gonadotrope-restricted transcription factor, SF-1. We focused further on the identity of complex 2 due to its robust expression in the more mature gonadotrope LβT2 cell line as compared with gonadotrope precursor cell lines, which indicates a likely role in LβT2 cell-specific expression of the LHβ gene.
Complex 2 Contains an Otx-Related Factor that Is Not a Known Member of the Ptx or Otx HD Families
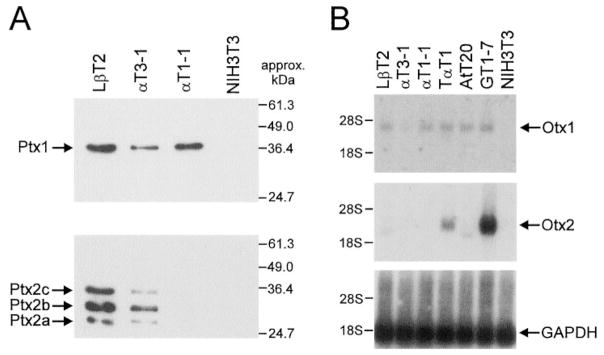
Because of their ability to regulate the LHβ promoter in CV1 cells (26) and their role in pituitary development (19, 21), it was of interest to ascertain whether complex 2 contains Ptx1 or Ptx2 HD transcription factors. Additionally, Otx HD transcription factors were examined because they are related to the Ptx family and Otx1 can also activate transcription of the LHβ promoter in cotransfections (27, 29). Expression of Ptx and Otx family members in gonadotrope-derived cell lines was determined to identify whether any of these factors is restricted to LβT2 cells. Ptx1 RNA expression has previously been shown to be widespread throughout the anterior pituitary (24, 25). Western analysis reveals that Ptx1 protein is present in LβT2, αT3-1, and αT1-1 cells, consistent with its expression throughout pituitary development (Fig. 6A). In contrast, all three isoforms of Ptx2 are expressed in committed gonadotropes (LβT2, αT3-1), but not uncommitted pituitary progenitor cells (αT1-1). Both Ptx1 and Ptx2 levels are somewhat higher in LβT2 cells than αT3-1 cells. Northern analysis of Otx1 and Otx2 expression reveals the presence of Otx1 mRNA in all cell lines tested except NIH3T3 (Fig. 6B). As with the Ptx family members, Otx1 is expressed at higher levels in LβT2 cells than αT3-1 cells. Otx2 is not expressed in any of the gonadotrope-derived cell lines. These expression studies demonstrate the presence of Ptx1, Ptx2, and Otx1 in LβT2 cells but also show that none of these factors are restricted to these mature gonadotrope cells; in particular, they are all expressed in the αT3-1 cells.
Fig. 6. Expression of Otx and Ptx HD Proteins in Pituitary-Derived Cell Lines.
A, Ptx1 and Ptx2 are expressed in both LβT2 and αT3-1 cells. A, Western blot with approximately 15 μg of nuclear extract from LβT2, αT3-1, αT1-1, and NIH3T3 cells was incubated with antibodies specific to Ptx1 or Ptx2. For Ptx1, a single band is observed in LβT2, αT3-1, and αT1-1 cells. The three bands observed with the Ptx2 antibody represent the three isoforms of Ptx2 (21), as indicated. B, Otx1, but not Otx2, is expressed in gonadotrope-derived cell lines. A Northern blot with approximately 2 μg poly(A) mRNA from LβT2, αT3-1, αT1-1, TαT1, AtT20, GT1-7, and NIH3T3 cells was probed with radiolabeled mouse cDNA fragments coding for Otx1 or Otx2. The GAPDH cDNA was used to allow visualization of the quantity of RNA loaded in each lane. Otx1 is detected in all cell types except NIH3T3, although at low levels in αT3-1 cells. Otx2 is detected in TαT1 and GT1-7 cells but not any of the gonadotrope-derived cell lines.
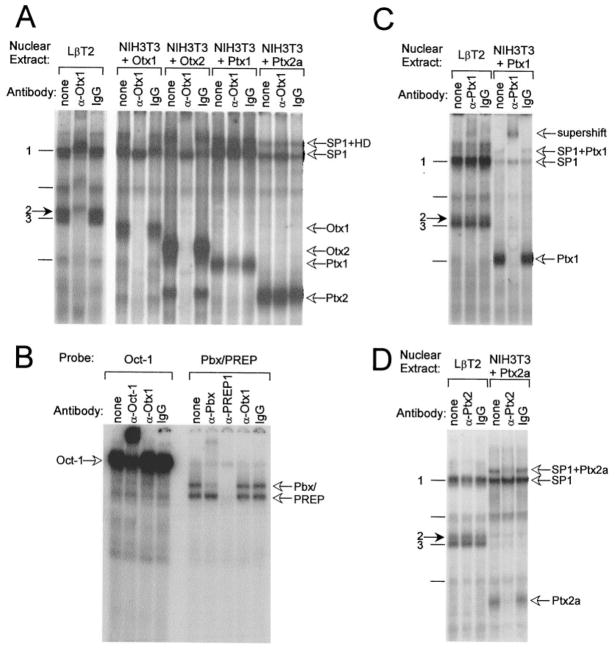
We next determined whether any of these Ptx or Otx proteins bind the LHβ promoter and if their migration corresponds to that of complex 2. Nuclear extracts from NIH3T3 cells transiently transfected with expression vectors coding for Ptx1, Ptx2a, Otx1, and Otx2 were used in EMSA with the LH 121/87 probe (Fig. 7). Although all four of the Ptx and Otx family members tested are capable of binding the LHβ promoter HD element, none of them comigrates with complex 2, nor with complex 3. These data indicate that complex 2 (or 3) is not simply a known member of the Ptx or Otx families.
Fig. 7. Known Members of the Ptx and Otx Families of HD Proteins Do Not Comigrate with Complex 2.
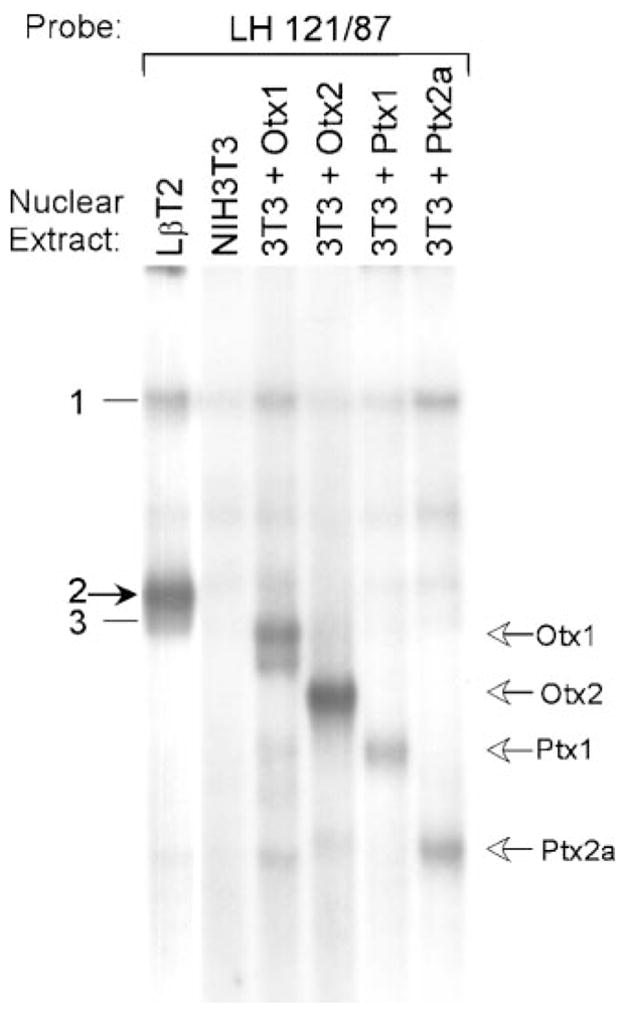
EMSA was performed using the LH 121/87 probe and nuclear extracts from LβT2 cells, NIH3T3 cells, and NIH3T3 cells transiently transfected with an expression vector coding for Otx1 (3T3 + Otx1), Otx2 (3T3 + Otx2), Ptx1 (3T3 + Ptx1), or Ptx2a (3T3 + Ptx2a), as indicated above each lane. Open arrows at right indicate the migrations of the Otx and Ptx family members as labeled. The closed arrow at left indicates complex 2.
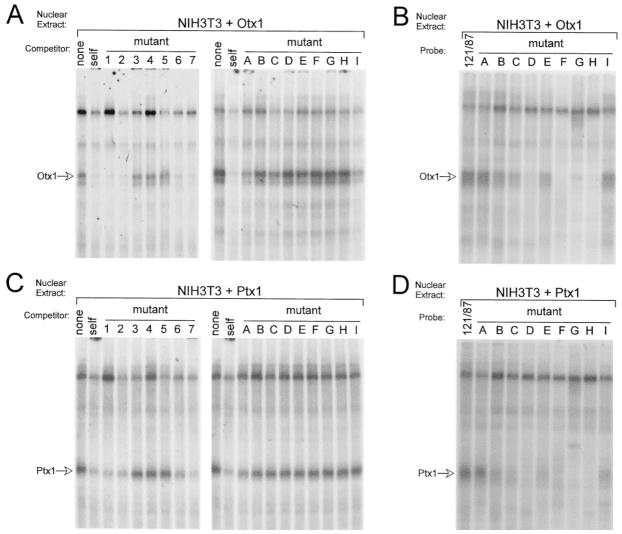
To compare with complex 2, the sequence requirements for Ptx1 and Otx1 binding to the LHβ HD element were examined by EMSA using mutated oligo-nucleotide competitors and probes as shown in Table 1. As with complexes 2 and 3, the oligonucleotides containing 3-bp mutations indicate the HD element is essential for both Otx1 and Ptx1 interaction with the LHβ probe (Fig. 8, A and C, mutants 3–5). Similar results were obtained using probes with these mutations (data not shown). Although the HD element is required for binding both proteins, experiments using competitors and probes containing single base-pair point mutations reveal subtle differences in binding site requirements between Ptx1 and Otx1. Whereas Otx1 binding, similar to complex 2, relies mainly on the core HD recognition sequence (GATTA) (Fig. 8A, mutants D, F, G, and H), Ptx1 binding requires several additional bases outside this core (Fig. 8C, mutants B and I). Although the binding specificities of complex 2 and Otx1 are not identical (summarized in Table 1), these data imply a closer relation of the protein comprising complex 2 to Otx1 than to Ptx1.
Fig. 8. Binding of Otx1 and Ptx1 to the LHβ Promoter HD Element.
A and C, EMSA was conducted using the LH 121/87 probe, nuclear extracts from NIH3T3 cells transiently transfected with an expression vector coding for either Otx1 (NIH3T3 + Otx1) (A) or Ptx1 (NIH3T3 + Ptx1) (C), and oligonucleotide competitors containing either 3 bp or single base-pair point mutations spanning the HD element (Table 1). Competitions were performed using 100-fold excess unlabeled oligonucleotide as indicated above each lane: None (no competitor), self (wild-type LH 121/87 competitor). B and D, EMSA was conducted using oligonucleotides containing single base-pair mutations as probes with NIH3T3 + Otx1 (B) or NIH3T3 + Ptx1 (D) nuclear extracts. The results are summarized and the sequences of all oligonucleotide competitors are depicted in Table 1. A and B, Otx1 binding to the LHβ promoter relies mainly on the core HD recognition sequence (GATTA). Migration of Otx1 is indicated by an open arrow. C and D, Ptx1 binding to the LHβ promoter is more dependent on several nucleotides outside the core HD sequence. Migration of Ptx1 is indicated by an open arrow.
Migration analysis does not rule out the possibility that a Ptx or Otx transcription factor is contained in complex 2 along with an additional protein(s). To further address the possibility that Otx or Ptx is present in complex 2, we performed antibody shift experiments using LβT2 nuclear extracts compared with nuclear extracts prepared from NIH3T3 cells transfected with expression vectors for Otx1, Otx2, Ptx1, or Ptx2a and used antibodies directed against Otx1, Ptx1, and Ptx2 (Fig. 9). The Otx1 antibody disrupts protein-DNA interactions of Otx1 and Otx2 and, to a lesser degree, Ptx1 (Fig. 9A). At the concentration used, the antibody does not cross-react with Ptx2; however, when higher concentrations of antibody are used, the Otx1 antibody can disrupt binding of both Ptx1 and Ptx2 equally as well as it disrupts binding of Otx proteins (data not shown). The Otx1 antibody disrupts binding of several bands in LβT2 cell nuclear extract, including complexes 2 and 3 and a band representing the concurrent binding of proteins to both the SP1 and HD sites (Fig. 9A, uppermost bands, SP1+HD). Although this antibody is not Otx1 specific, its ability to disrupt protein-DNA interactions appears to be limited to Ptx and Otx HD family members as it fails to interact with other HD proteins such as Oct-1, Pbx1/2/3, PREP-1, Msx1, and Dlx2 (Fig. 9B; and Givens, M. L., and P. L. Mellon, personal communication, January 2002). This indicates that a protein present in complex 2 is structurally related to the Ptx and Otx families.
Fig. 9. Complex 2 Contains an Otx-Related HD Protein.
A, Complex 2 contains a factor that is related to Otx HD transcription factors. EMSA was performed using nuclear extracts from LβT2 cells and NIH3T3 cells transiently transfected with an expression vector coding for Otx1 (NIH3T3 + Otx1), Otx2 (NIH3T3 + Otx2), Ptx1 (NIH3T3 + Ptx1), or Ptx2a (NIH3T3 + Ptx2a). Antibodies directed against Otx1 (α-Otx1) or normal rabbit IgG (IgG) were included in the reactions as indicated. The closed arrow at left indicates complex 2; open arrows at right indicate the migrations of the Otx and Ptx family members, SP1, and bands representing the concurrent binding of proteins to both the SP1 and HD sites (SP1 + HD), as labeled. The LβT2 lanes shown were run on a separate gel from the transfected NIH3T3 lanes shown. B, The Otx1 antibody does not cross-react with TALE or POU-domain containing HD proteins. EMSA was performed using nuclear extracts from LβT2 cells and Oct-1 (5′-TGTCGAATGCAAATCACTAGAA-3′ (top strand)] (51) or Pbx/PREP (5′-AGCGCGGGGCGCATCAATCAATTTCG-3′ (top strand)] (52) consensus probes. Antibodies directed against Oct-1, Pbx 1/2/3 (Pbx), PREP-1, or Otx1 were included in the reactions as indicated. C, Ptx1 from LβT2 cells does bind the LH 121/87 probe, but is not present in complex 2. EMSA was conducted using nuclear extracts from LβT2 cells (left) and NIH3T3 cells transiently transfected with an expression vector coding for Ptx1 (NIH3T3 + Ptx1, right). Antibodies directed against Ptx1 (α-Ptx1) or normal rabbit IgG (IgG) were included in the reactions as indicated. The closed arrow at left indicates complex 2; open arrows at right indicate Ptx1, SP1, SP1+Ptx1, and the Ptx1 supershift as labeled. D, Ptx2 is not present in complex 2. EMSA was conducted using nuclear extracts from LβT2 cells (left) and NIH3T3 cells transiently transfected with an expression vector coding for Ptx2a (NIH3T3 + Ptx2a, right). Antibodies directed against Ptx2 (α-Ptx2) or normal rabbit IgG (IgG) were included in the reactions as indicated. The closed arrow at left indicates complex 2; open arrows at right indicate Ptx2a, SP1, and SP1+Ptx2a as labeled.
Ptx1 and Ptx2 antibodies were examined next. Inclusion of a Ptx1 antibody with LβT2 cell nuclear extract results in a faint supershift. As Ptx1 from transfected NIH3T3 cells comigrates with the faster migrating of the two somewhat nonspecific complexes, it is likely that this faint complex represents the binding of endogenous Ptx1 protein from LβT2 cell nuclear extract (Fig. 9C, indicated by open arrow). However, complexes 2 and 3 are not affected. The supershifted band at the top the gel represents a supershift of the band generated when Ptx1 and SP1 bind simultaneously (data not shown). Although this supershift confirms the binding of some endogenous Ptx1 to the LHβ promoter, Ptx1 is not contained within the LβT2 nuclear protein complex of interest. Similarly, whereas the Ptx2 antibody is capable of binding control Ptx2a protein (NIH3T3 + Ptx2a) and disrupting its interaction with DNA (Fig. 9D), this antibody has no effect on any of the complexes observed using LβT2 cell nuclear extract, including complex 2. As shown in the Western blot in Fig. 6, this antibody detects all three splice variants of the Ptx2 protein. This indicates that endogenous Ptx2 protein in LβT2 cell nuclear extract is not binding the LHβ HD element under these conditions and is not contained within complex 2 or 3. The upper band that disappears in the NIH3T3 + Ptx2a lane with Ptx2 antibody represents the concurrent binding of Ptx2a with SP1 (data not shown). Both Ptx1 and Ptx2 antibodies were tested for cross-reactivity against each other as well as Otx1 and Otx2 and were found to be specific for the intended protein (data not shown).
Complex 2 Contains a K50 HD Transcription Factor
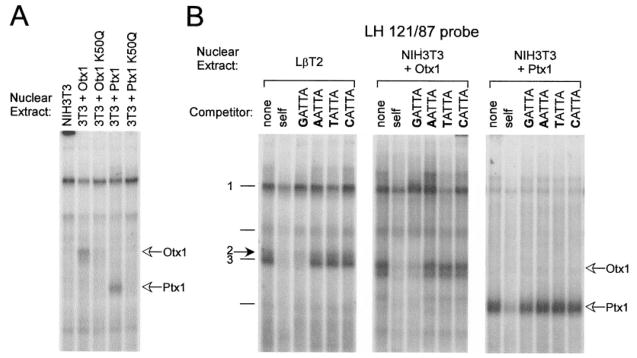
The Ptx and Otx HD proteins are Paired-like HD transcription factors that contain a lysine in the 50th position of the HD and are therefore referred to as K50 HD proteins (30). The 50th amino acid of the HD interacts with the nucleotide bases 5′ to the ATTA HD recognition core and is important in determining DNA-binding specificity of HD transcription factors (31). Whereas K50 HDs exhibit a preference for binding GATTA target sequences, as is found in the LHβ promoter, Q50 HDs (containing a glutamine at the 50th position) show a preferred binding sequence of either AATTA or CATTA (31, 32). To examine the importance of a K50 amino acid for HD protein interactions with the LHβ promoter HD element, we used site-directed mutagenesis to mutate the Ptx1 and Otx1 HDs from K50 to Q50 (AAG → CAG). EMSA was performed using nuclear extracts from NIH3T3 cells transiently transfected with expression vectors coding for either wild-type (NIH3T3 + Otx1 or NIH3T3 + Ptx1) or mutant (NIH3T3 + Otx1 K50Q or NIH3T3 + Ptx1 K50Q) proteins (Fig. 10A). Expression of both wild-type and mutant proteins in the cells was confirmed by Western blotting (data not shown). This single amino acid change from a K50 to Q50 blocks the ability of both Otx1 and Ptx1 to bind to the LHβ promoter, indicating that a lysine at the 50th position of the HD is important for interaction with this HD element.
Fig. 10. Complex 2 Contains a K50 HD Transcription Factor with Binding Specificity Similar to that of Otx1.
A, Lysine at position 50 of the HD is essential for Otx1 and Ptx1 interaction with the LHβ promoter HD element. Site-directed mutagenesis was performed to mutate the coding sequence of both Ptx1 and Otx1 such that the lysine at the 50th position of the HD (K50) was replaced by a glutamine (Q50). EMSA was performed using the LH 121/87 probe and nuclear extracts from NIH3T3 cells transiently transfected with an expression vector coding for wild-type Otx1 (NIH3T3 + Otx1), mutant Otx1 (NIH3T3 + Otx1 K50Q), wild-type Ptx1 (NIH3T3 + Ptx1), or mutant Ptx1 (NIH3T3 + Ptx1 K50Q), as indicated above each lane. Open arrows at right indicate the migrations of Otx1 and Ptx1. B, Complex 2 shares binding specificity with the Otx family of HD transcription factors but not the Ptx family of HD transcription factors. EMSA was performed using LβT2 (left), NIH3T3 + Otx1 (middle), and NIH3T3 + Ptx1 (right) nuclear extracts. Results using NIH3T3 + Otx2 and NIH3T3 + Ptx2a nuclear extracts are not shown, but Otx2 is competed similarly to Otx1 and Ptx2a is competed similarly to Ptx1. Oligonucleotide competitors containing a core HD sequence (5′-TGTACAGTCCTCANATTATTCTCAGG-3′ where N is either G (GATTA), A (AATTA), T (TATTA), or C (CATTA)] (33) were included in the reactions as indicated above each lane. The closed arrow at left indicates complex 2; open arrows at right indicate the migrations of the Otx1 and Ptx1.
To further verify that complex 2 contains a K50 HD protein, oligonucleotide competitors unrelated to the LHβ HD element but containing a consensus HD recognition sequence, NATTA, where N is either G, A, T, or C (33), were used in EMSA. Complexes 2 and 3 are specifically competed by the oligonucleotide containing a K50 HD binding sequence, GATTA, and not the others. Similarly, both Otx1 and Otx2 are also specifically competed by the GATTA containing oligonucleotide (Fig. 10B, and data not shown). Interestingly, when Ptx1 and Ptx2 proteins were tested under the same conditions, none of the oligonucleotides competed, including the one containing GATTA, indicating a requirement for specific flanking sequences that are not required by complex 2, Otx1 or Otx2 (Fig. 10B, and data not shown). The same results were obtained when the consensus oligonucleotides were used as probes and binding of Otx1 and Ptx1 proteins was examined directly (data not shown). These differences in binding specificity between Ptx1 and Otx1 and complex 2 are consistent with the different binding requirements for interaction with the LHβ promoter observed earlier (Table 1). The similarity in binding specificity of complex 2 with Otx1 and Otx2 homeoproteins, but not with the Ptx1 and Ptx2 homeoproteins, suggests that complex 2 contains a protein more closely related to Otx HD transcription factors than Ptx HD transcription factors.
DISCUSSION
During development of the anterior pituitary, distinct endocrine cell types differentiate in defined spatial and temporal patterns (34). In the mouse, a committed gonadotrope population can first be detected by embryonic d 14 with the expression of SF-1 (11). Subsequent maturation of the gonadotrope is marked by the onset of LHβ gene expression around embryonic d 16.5 and FSHβ gene expression around embryonic d 17.5 (35). The factors specifically involved in this induction of gonadotropin β-subunit gene expression during the later stages of gonadotrope differentiation are not well characterized. Whereas αT3-1 cells only express early markers of the gonadotrope lineage, LβT2 cells express markers of a mature gonadotrope as evidenced by the presence of endogenous LHβ and FSHβ. The LβT2 and αT3-1 cell lines thus correspond to these close sequential stages of gonadotrope differentiation in development and are excellent models in which to study the molecular mechanisms governing the maturation of the pituitary gonadotrope.
In the present study, the specificity of expression of LHβ to the more mature gonadotrope was analyzed using transient transfection assays in gonadotrope and nongonadotrope-derived cell lines. Activity of the 1800 bp rat LHβ promoter is 4.5- to 5-fold higher in LβT2 cells as compared with αT3-1 cells. The activity in αT3-1 cells when compared with nongonadotrope-derived cell lines is not significantly different, demonstrating that even these committed gonadotrope precursors lack conditions necessary for gonadotrope-specific LHβ gene expression. This is especially notable because αT3-1 cells do express several factors thought to be important for LHβ gene expression, including SF-1, Egr-1, Ptx1, and Ptx2. Most of the studies concerning LHβ gene regulation have been performed using heterologous cell lines, such as CV1, GH3, and JEG-3 (5–7); however, the use of these cells does not accurately reflect cell-specific responses since distinct sets of transcription factors, kinases, G proteins, receptors, and other classes of molecules are expressed in different cell types. Furthermore, co-transfections with expression vectors for transcription factors not normally expressed in a cell can show activation regardless of the actual role of that factor in regulating LHβ in the gonadotrope. In the current work, the selectivity of LHβ promoter activity to LβT2 cells underscores the importance of studying LHβ gene regulation in a homologous cell system.
As development relies on both the activation and inactivation of lineage specific genes, several models could account for the differential expression of the LHβ gene between LβT2 and αT3-1 cells. One possibility is the presence of inhibitory factors in early gonadotrope cells (αT3-1) preventing LHβ gene expression. Alternatively, there may be activating factors present in mature gonadotrope cells (LβT2) conferring LHβ gene expression. The promoter truncation analysis described herein suggests that both mechanisms of gene regulation are at work within the LHβ promoter. The approximately 4.5-fold difference in LHβ promoter activity between LβT2 and αT3-1 cells is reduced by truncation of two regions of the LHβ promoter. Deletion from −179 bp to −146 bp reduces the difference to 2-fold. This is due to a relative increase in expression of LHβLuc in αT3-1 cells (though notably not in NIH3T3 cells), consistent with the possibility of an αT3-1 specific inhibitor acting on this region of the promoter. Interestingly, down-regulation of the LHβ promoter after 24 h of tonic GnRH treatment also maps to this region (Vasilyev, V. V., M. A. Lawson, and P. L. Mellon, manuscript submitted). Further characterization of this region to identify potential repressors might provide useful insights into both hormonal and basal input into gonadotrope development.
The remaining selectivity in LHβLuc expression between the gonadotrope-derived cell lines is lost upon deletion of the region between −122 bp and −87 bp. This is due to the loss of activity specifically in LβT2 cells, indicating the removal of a promoter element specifically active in the more mature gonadotrope cell line. This region of the promoter contains the previously characterized 5′ Egr-1 binding site and the HD element. Consistent with the negligible levels of Egr-1 in uninduced LβT2 and αT3-1 cells (6, 7, 18), Egr-1 is not detected binding to its promoter elements using nuclear extracts from non GnRH-treated cells; rather, the ubiquitous SP1 transcription factor binds to the GC-rich Egr-1 elements [(28), and data not shown]. Mutation of the 5′ Egr-1 element has little or no effect on either basal (18) or cell-specific activity of the LHβ promoter. This contrasts with the importance of Egr-1 in GnRH regulation of the LHβ gene; responsiveness of the LHβ promoter to GnRH in both αT3-1 (6, 18) and LβT2 (7, 8) cells is attenuated by mutation of the Egr-1 binding sites.
Whereas Egr-1 elements are not involved in basal activity of the LHβ promoter, the HD element is essential for both basal activity as well as cell-specific differences in promoter activity in LβT2 vs. αT3-1 cells. We used two 3-bp HD mutations that exhibited different protein interactions in EMSA to examine the role of the HD element in LHβ gene regulation. The partial decrease in LβT2 cell-specificity observed with HDm5, which reduces basal activity in both LβT2 and αT3-1 cells, may indicate a contribution of complex 3, Otx1, or Ptx1 to LHβ promoter activity; alternatively, the decrease in activity may be due to the reduced binding of complex 2. Unlike HDm5, HDm4 completely blocks binding of complex 2, in addition to the other proteins, and only reduces basal activity in LβT2 cells. The greater effect of the HDm4 mutation (vs. HDm5) on LβT2 cell specificity of −1800LHβLuc correlates with the loss of complex 2 binding, indicating a role for this protein in LβT2 cell-specific expression of LHβ. Whereas HDm4 in the context of the 1800-bp promoter only decreases LβT2 cell-specificity to 1.5-fold, the same mutation in the context of the 122-bp promoter abolishes the difference. One element present in the longer promoter but absent from the shorter version is the 5′ SF-1 binding site. When this element is mutated along with the 3′ SF-1 binding site and the HD element (m4) (a triple mutant), the 1800-bp promoter loses its specific activity in LβT2 cells. The necessity for mutating both SF-1 sites as well as the HD element to eliminate the differences between LβT2 and αT3-1 cells in −1800LHβLuc despite the presence of SF-1 in both cell lines is probably due to interactions of SF-1 with the protein(s) acting through the HD element. SF-1 is capable of interacting with Ptx1, physically and synergistically (26, 27), and may be capable of interacting with additional HD proteins as well. Data from transgenic mice confirms the importance of this HD site for LHβ promoter activity in vivo. A −776/+12 bovine LHβ promoter confers gonadotrope-specific expression and regulation by both GnRH and gonadal steroids to a CAT reporter gene in transgenic mice (36). Recently, it was shown that mutation of the HD element completely abrogates basal activity as well as GnRH responsiveness of this bovine LHβ promoter in transgenic mice (9).
Despite the clear importance of the HD element for LHβ promoter activity, the proteins acting through this element are not well characterized, and no comparative studies have been performed. When we compared the proteins binding to the LHβ promoter HD element in LβT2 and various other cell types, the major difference observed between cell lines of the gonadotrope lineage is the presence of complex 2 in LβT2 but not αT3-1 or αT1-1 cells. This nuclear protein complex has the binding characteristics of a K50 HD protein and is recognized by an Otx1 antibody. Of interest, complex 3, which appears at equal levels in LβT2 and αT3-1 nuclear extracts but to a lesser extent in αT1-1 nuclear extract, has binding specificity almost identical to complex 2 and is also recognized by the Otx1 antibody. This suggests that the proteins comprising complexes 2 and 3 are closely related. These complexes may represent alternatively spliced variants of the same protein or individual members of the same protein family. Although complexes with migration similar to complex 2 appear in some nongonadotrope endocrine-derived cell types, this complex may account for the difference in activity of the LHβ promoter between the mature gonadotrope LβT2 and precursor gonadotrope αT3-1 cells. Tissue-specific gene expression is often conferred by complex control regions in which several cell-restricted, but not necessarily cell-specific, factors interact (37, 38). The spatial and temporal pattern in which regulatory factors are expressed is determining when cell-fate decisions are being made. With regards to the LHβ gene, a factor expressed in mature gonadotropes that is also present elsewhere in the pituitary could still contribute to gonadotrope-specific expression and activation during development by interaction with SF-1, which, within the anterior pituitary, is uniquely expressed in the gonadotrope population.
Because of their known role in early pituitary development and their presence in gonadotrope cells, Ptx1 and Ptx2 were considered as possible candidates for the complex 2 protein. Although both LβT2 and αT3-1 cells express Ptx1 and Ptx2 mRNA and protein (Ref. 22; Fig. 6), previous studies have not demonstrated binding of these proteins to the LHβ promoter HD element. Tremblay et al. (22, 27) established the presence of Ptx1 DNA-binding activity in αT3-1 cells using the POMC gene’s Ptx1 promoter element, yet Ptx1 was not shown binding to the LHβ promoter element. In their work demonstrating the necessity of the HD element for LHβ promoter activity in vivo, Quirk et al. (9) observe several proteins binding to the HD element using LβT2 cell nuclear proteins. They presume one of these bands to be Ptx1 but were unable to detect it using a Ptx1 antibody. Here we demonstrate for the first time that Ptx1 does indeed bind the LHβ promoter HD element, albeit at quite low levels in LβT2 cells. These same experiments show that Ptx1 protein is not contained within complex 2. As with Ptx1, we demonstrate the ability of Ptx2 from transfected NIH3T3 cells to bind the LHβ HD element. Because Ptx2 protein is present in LβT2 cells, our inability to detect Ptx2 binding the LHβ promoter using LβT2 nuclear extracts may indicate that Ptx2 is not interacting with the HD element in these gonadotrope cells. Ptx1 and Ptx2 are capable of transactivating the LHβ promoter in heterologous cell systems (CV1, HeLa) (23, 26) and have been suggested to be involved in cell-specific transcriptional regulation but neither has been proven to be functional in gonadotrope-derived cell lines (LβT2, αT3-1). Ptx1 and Ptx2 HD proteins are necessary for pituitary development and therefore indirectly necessary for gonadotrope function; whether or not they directly regulate LHβ gene expression through the HD element in mature gonadotropes needs further clarification.
Interestingly, an Otx1 antibody that can recognize both Ptx and Otx HD proteins also recognizes complex 2, which is our best candidate for an LβT2 HD protein required for LHβ promoter specificity to mature gonadotropes. The Ptx and Otx families are Paired-like HD proteins, a subclass of Paired-class proteins; they share homology in the HD to Paired-type proteins, but lack a Paired domain (30). The fact that Ptx and Otx proteins lack homology outside the HD (39) suggests that the Otx1 antibody recognizes the Paired-like HD. This is further supported by data demonstrating that the Otx1 antibody does not recognize HD proteins outside the Paired-class, including the TALE-HD proteins, Pbx1/2/3 and PREP-1, the POU domain factor, Oct-1, and the Antennapedia Class proteins, Msx1 and Dlx2 (Givens, M. L., and P. L. Mellon, personal communication, January 2002). Unlike the Antennapedia class, which includes only Q50 HD proteins, the Paired-like subclass includes both K50 and Q50 HD proteins. As we have not tested Q50 Paired-like proteins, we cannot rule out the possibility that the Otx1 antibody also recognizes these HD proteins. However, the DNA-binding data suggest that complex 2 contains a K50 HD protein related to Otx and not a Q50 HD protein.
First of all, the LHβ promoter HD element resembles the preferred target sequence for K50 HD proteins, GATTA, and binds Ptx and Otx family members. Furthermore, the presence of this lysine is essential to the ability of Ptx and Otx to interact with this sequence. When Ptx1 and Otx1 are mutated to convert them from K50 to Q50, they are no longer capable of binding the LHβ promoter HD element. Finally, experiments with consensus oligonucleotides indicate that complex 2 only binds a HD recognition sequence of GATTA. In addition to supporting the presence of a K50 HD protein in complex 2, experiments using these HD recognition sequences reveal differences in binding specificity between the Otx and Ptx families. Whereas Otx proteins bind the GATTA consensus sequence, the Ptx proteins do not bind any of the consensus sequences. Although seemingly unexpected at first, the inability of Ptx to bind the GATTA HD consensus element is not entirely surprising. Ptx1 interacts with the LHβ promoter HD element with binding specificity distinct from both Otx1 and complex 2 (Figs. 4 and 8 and Table 1). Whereas Otx1 and complex 2 binding relies mainly on the core HD recognition sequence in the LHβ probe, Ptx1 binding also requires several bases outside this core. These additional outside bases are not the same in the GATTA HD consensus oligonucleotide used in Fig. 10B, consistent with the inability of Ptx to interact with this site. Ptx and Otx families are grouped together in the K50 Paired-like subclass based on the presence of a lysine at position 50 of the HD. In fact, phylogenetic analysis of Paired-like HD proteins indicates that the Ptx and Otx families evolved from separate ancestral origins (30). Their separate origins are probably the reason for observed differences in binding specificity between the families. This distinction between Ptx and Otx factors leads us to conclude that complex 2 includes a protein more closely related to Otx than Ptx.
Otx HD proteins are related to the orthodenticle (otd) gene of Drosophila and are involved in brain morphogenesis (40). Whereas Otx2 is not expressed in any of the gonadotrope-derived cell lines and is therefore not a candidate for the complex 2 protein, both Otx1 and the Otx-related cone-rod homeobox (Crx) mRNAs are expressed in LβT2 cells (Fig. 6 and data not shown) and were considered as possible candidates. Consistent with a role in the postnatal pituitary, Otx1 knockout mice exhibit transient dwarfism and hypogonadism and a corresponding reduction of LH, FSH, and GH levels; however, these defects are temporary and the mice appear indistinguishable from wild-type by 4 months of age (29). Though Otx1 does not comigrate with complex 2, since the Otx1 antibody cross-reacts with related HD proteins, we cannot completely rule out the possibility that complex 2 contains Otx1 along with an additional protein. Paired-like HD proteins are capable of binding DNA as cooperative heterodimers, but this requires a palindromic repeat of the TAAT recognition sequence (33). The LHβ promoter HD element does not contain this palindromic target element necessary for recognition by such dimers, suggesting that the proteins interacting with this element are monomers and that Otx1 is not interacting with an additional protein to form complex 2. In addition, subtle, but important, differences exist in the binding specificities of Otx1 and complex 2. Although complex 2 could be an unidentified isoform of Otx1, to date no additional isoforms of Otx1 have been reported and only a single band is observed by Northern analysis (Ref. 41 and Fig. 6). Furthermore, we have not observed a protein in LβT2 nuclear extract that comigrates with Otx1 by Western analysis (data not shown) or EMSA.
Crx is an Otx-related HD protein that is expressed in mature and developing photoreceptor cells and the pineal gland (42, 43). As with Otx1 and Otx2, Crx is capable of binding the LHβ promoter HD element, but does not comigrate with complex 2 (data not shown). As an antibody directed against Crx (kindly provided by Dr. Shiming Chen) does not supershift complex 2 (data not shown), it appears that Crx is not the complex 2 protein. Although the binding specificity of complex 2 and its recognition by the Otx1 antibody suggest that it is an Otx-related HD protein, the lack of comigration with Otx1, Otx2, or Crx, the absence of Otx2 from LβT2 cells, and the failure of a Crx antibody to interact with complex 2, indicate that this may be a novel protein. The complete annotation of the mouse and human genomes should assist in the identification of new members of the Otx HD family.
In summary, the studies presented here emphasize the importance of using a homologous cell system to study tissue-specific gene regulation. We have identified the HD binding site as a key element involved in the activation of the LHβ gene in mature LβT2 gonadotrope cells as compared with precursor αT3-1 cells. Complex 2 is an attractive candidate for involvement in this LβT2 cell-specific expression of the LHβ gene: it is present in LβT2 gonadotrope cells but not αT3-1 or αT1-1 precursor gonadotrope cells and binds the HD element of the LHβ promoter that is essential for both basal activity and LβT2 cell-specific expression. Although our data suggest that Ptx1, Ptx2, and Otx1 are not members of complex 2, this same evidence suggests that complex 2 contains a K50 HD protein related to the Otx family. Future work will aim at identifying this protein and examining its role in LHβ gene expression, gonadotrope development, and normal reproductive function. Successful identification of this protein may also lead to a greater understanding of cell-type specification and maturation in the anterior pituitary.
MATERIALS AND METHODS
Plasmid Construction and Preparation
The −1800 LHβ-Luc reporter gene was created by cloning 1800 bp of the rat LHβ 5′ flanking sequence into a modified pUC18 vector containing the Luc reporter gene and SV40 T antigen intron and polyA sequence. The−692,−251, −122, and −87 truncations were made using AccI, NheI, TthIII, and BsteII restriction sites, respectively. The −420 truncation was created by amplifying a fragment of the rat LHβ promoter from −420 bp to −178 bp by PCR, digesting with HindIII and NheI, and subcloning the 170-bp fragment into the −1800LHβLuc plasmid digested with HindIII and NheI. The HindIII site is located at the 5′ cloning end of the promoter in LHβLuc; the NheI site is located at −252 bp of the promoter. The forward primer used in the PCR contained a HindIII restriction site for cloning back into the LHβLuc plasmid: 5′-AGTGCCAAGCTTTCCTGATTAGGGGGCTGGGCGAGG-3′. The reverse primer corresponds to the sequence of the rat LHβ promoter from −200 bp to −178 bp. The −451, −384, −179, and −146 reporter plasmids were provided by V. V. Vasilyev.
The mouse Ptx1 and human Otx1 cDNAs were kindly provided by Dr. Jacques Drouin and Dr. Antonio Simeone, respectively. The Ptx1 and Otx1 cDNAs were amplified using Pfu DNA polymerase (Stratagene, La Jolla, CA) and PCR primers designed to the translational start and stop sites of each cDNA. The cDNAs were ligated into the TopoII vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The sequences of the entire cDNAs were confirmed by automated sequencing performed by the Center for AIDS Research Molecular Biology Core at University of California, San Diego. The cDNAs were then recloned into the pcDNA3 expression vector using the EcoRI restriction sites of both plasmids. The RSV-Ptx2a expression vector was a gift from Dr. Jacques Drouin; the mouse Otx2 expression vector (pSG-mOtx2) has been previously described (44).
Plasmid DNA was prepared from overnight bacterial cultures using a cesium chloride protocol adapted from Sambrook et al. (45).
Cell Culture and Transient Transfections
Cells were grown in 60-mm diameter dishes to 50–60% confluency in DMEM (Cellgro, Mediatech, Inc., Herndon, VA) supplemented with 10% FBS (Omega Scientific Inc., Tarzana, CA) and 1% penicillin-streptomycin (Life Technologies, Inc., Grand Island, NY) at 37 C with 5% CO2. Transient transfections were performed using FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN), after the manufacturer’s protocol. The cells were harvested 48 h after transfection and Luc and β-gal assays performed. Experiments were performed using 3 μg (Figs. 2 and 3) or 2.5 μg (Fig. 4) of the reporter plasmids. One-half microgram of RSV-βgal was used as an internal control. All transfection experiments were performed at least three times.
Normalizing Transfection Data and Statistics
To control for differences in expression between the different cell types, each experiment was normalized. The RSV enhancer fused to the RSV promoter driving Luc (RSV-Luc) was transfected in triplicate in each experiment. The internal control, RSV enhancer and promoter fused to β-galactosidase (RSV-β-gal), was used as an internal control for each transfected plate of cells. The RSV-Luc values were divided by the RSV-β-gal β-gal values, averaged, and set equal to 100. The values for the other plates were normalized to this value in individual cell types; thus, the values from the individual cell types can be directly compared. The mean of at least three experiments is depicted. The error bars represent SEM. Normal or Box Cox Transformed ratios for each promoter construct in each cell type were compared by the ANOVA Factorial test, followed by the Tukey-Kramer HSD post hoc test. In all analyses, P ≤ 0.05 was considered significant.
Luc and β-gal Assays
At harvest, cells were washed twice in 1× PBS and then 1 ml of harvesting buffer (0.15 M NaCl; 1 mM EDTA; 40 mM Tris-HCl pH 7.4) was added to each dish. Cells were scraped from the plate, transferred to 1.5-ml microcentrifuge tubes, and collected by centrifugation at 3000 rpm for 5 min. The harvesting buffer was discarded and the cells were resuspended in 50 μl of lysis solution (100 mM potassium phosphate (pH 7.8), 0.2% Triton X-100). Cells were spun at 14,000 rpm for 5 min and the supernatant was transferred to a new tube and assayed for Luc and β-gal activity. Fifteen microliters of each sample was used per assay.
Luc activity was measured using an EG&G Berthold (Wildbad, Germany) Microplate Luminometer by injecting 100 μl per well of a buffer containing 100 mM Tris-HCl (pH 7.8), 15 mM MgSO4, 10 mM ATP and 65 μM of luciferin. β-gal assays were performed using the Galacto-Light Plus Kit (Tropix, Bedford, MA) after the manufacturer’s protocol. Before each β-gal assay, cell extracts were heat-inactivated at 48 C for 50 min. The Luc and β-gal values for a mock-transfected plate of cells were subtracted from each transfected plate value. Luc values were divided by the β-gal values to control for transfection efficiency and normalized to the ratio of RSV-Luc divided by RSV-β-gal for the appropriate cell type as described earlier.
Mutagenesis
Mutagenesis of LHβLuc reporter genes was performed using the Transformer Site-Directed Mutagenesis Kit (CLONTECH Laboratories, Inc., Palo Alto, CA) according to the manufacturer’s protocol. Oligonucleotides used for mutagenesis were the following: 3′ SF-1 site (5′-GCCTCT-GCTTAGTGGAATTCCCACCCCCACAACCCGC-3′), 5′ SF-1 site (5′-GCTGGTCCCTGGCTTTTCTGAAATTCTCTGTCTC-GCCCCCAAAG-3′), 5′ EGR-1 site (5′-CTGACCTTGTCT-GTCTAGTACTCAAAGAGATTAGTGTCTAGG-3′), HD element mutant 4 (HDm4) (5′-CGCCCCCAAAGATCGTAGTGTCTAG-GTTACCCAAGCC-3′), HD element mutant 5 (HDm5) (5′-CGC-CCCCAAAGAGATGCTTGTCTAGGTTACCCAAGCC-3′). Mutagenesis of Otx1 and Ptx1 expression vectors was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. Oligonucleotides used for mutagenesis were the following: Otx1 K50Q (5′-GAGTCCAGGTCTGGTTCCAGAACCGCCGCGCCAAA-TGCC-3′ (top strand)], Ptx1 K50Q (5′-CGAGTGCGGGTCTG-GTTCCAGAACCGGCGAGCCAAATGG-3′ (top strand). Mutated sequences (underlined) were confirmed by dideoxynucleotide sequencing (46).
Nuclear Extracts and EMSAs
Crude nuclear extracts were prepared by the method of Lee et al. (47). Glycerol was added to a final concentration of 20%. Nuclear extracts from NIH3T3 cells overexpressing Ptx or Otx family members were prepared 2 d after transient transfection of NIH3T3 cells with expression vectors coding for either Ptx1, Ptx2, Otx1, Otx2, Ptx1 K50Q, or Otx1 K50Q. NIH3T3 cells were grown on 100-mm dishes and transiently transfected with 5 μg of the appropriate expression vector using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN), after the manufacturer’s protocol. Extracts were assayed for protein concentration using the Bio-Rad Laboratories, Inc. (Hercules, CA) protein assay reagent, aliquoted, and frozen at −80 C. For the EMSA, annealed oligonucleotides (1 pmol, Operon Technologies, Alameda, CA) were phosphorylated with [γ-32P]ATP (7000 Ci/mmol; ICN Biomedicals, Inc., Aurora, IL) and T4 Polynucleotide Kinase (New England Biolabs, Inc., Beverly, MA). The probes were phenol/chloroform extracted and passed over two Microspin G-25 columns (Amersham Pharmacia Biotech, Piscataway, NJ). Each binding reaction (20 μl) contained 2 fmol probe and 2 μg nuclear extract in 10 mM HEPES (pH 7.9), 50 mM KCl, 5 mM MgCl2, 0.25 mg BSA, 0.1% Nonidet P-40, 0.1 μg poly(deoxyinosine-deoxycytidine), and 5 mM DTT. Reactions were incubated for 5 min at room temperature after addition of probe, loaded onto a 17 cm × 30 cm 7% nondenaturing polyacrylamide gel acrylamide:bis (30:1); 0.25 × 130 mM Tris, 45 mM boric acid, 2.5 mM EDTA; and electrophoresed for 4 h at 250 V. After electrophoresis, gels were dried and subjected to autoradiography. In Figs. 4 and 7–10, the probe was cut off for better viewing of the relevant protein-DNA complexes. For competition and antibody experiments, 100-fold excess unlabeled competitor oligonucleotide (200 fmol) or 0.5 μl antibody (Otx1) or 1 μl antibody (Ptx1, Ptx2, Oct-1, Pbx1/2/3, PREP-1) was added 20 min before addition of probe. Oligonucleotides used as probes and in competitions (Operon Technologies) are described in Table 1 (for Figs. 4 and 8) or legend (Figs. 9 and 10). The Ptx1 (48), Ptx2 (49), and Otx1 (29) antibodies were kindly provided by Dr. Jacques Drouin, Dr. Tord Hjalt, and Dr. Antonio Simeone, respectively. The Oct-1, Pbx 1/2/3, and PREP-1 antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Western Blotting
Preparation of nuclear extracts has been described above. Fifteen micrograms of each nuclear extract were electrophoresed in a 10% SDS-polyacrylamide gel and transferred to Immobilon-P membrane (Millipore Corp., Bedford, MA). Ponceau S staining before immunoblotting was used to ascertain the efficiency of transfer. Membrane was blocked in PBS-T (PBS with 0.2% Tween-20) with 5% nonfat dry milk for 1 h and then incubated with the primary antibody for 1 h. The Ptx1 antibody (48) was diluted 1:2000 and the Ptx2 antibody (49) was diluted 1:100 in blocking buffer (PBS-T/5% milk). The membrane was washed three times for 5 min each with blocking buffer and then incubated with a horseradish peroxidase-labeled anti-rabbit secondary antibody (diluted in blocking buffer 1:2000 for Ptx1 and 1:1000 for Ptx2). The membrane was then washed 3 times for 5 min each before detection using enhanced chemiluminescence (Amersham Pharmacia Biotech). All washes and incubations were done at room temperature.
Northern Analysis
Total RNA was prepared using TRI REAGENT (Sigma, St. Louis, MO) and polyA+ RNA was extracted using PolyATract mRNA Isolation System IV (Promega Corp., Madison, WI) according to the manufacturers’ protocols. Two micrograms of each polyA+ RNA sample were electrophoresed in a denaturing 1% formaldehyde-agarose gel and transferred to Hybond N+ nylon membrane (Amersham Pharmacia Biotech) by capillary blotting. The RNA was fixed by UV cross-linking and then the membrane was hybridized overnight with 32P-labeled cDNA probe at 55 C in a 25% formamide solution. DNA probes were labeled by random priming (50). Excess probe was removed by washing at 65 C (Otx2) or 50 C (Otx1) with 0.2× SSPE and 0.1% SDS. The Otx1 probe consisted of a 396 nucleotide SacI-PstI fragment from the mouse Otx1 cDNA (41) and the Otx2 probe consisted of a 900 nucleotide EcoRI fragment of pSG-mOTX2 (44).
Acknowledgments
We thank Jacques Drouin, Tord Hjalt, and Antonio Simeone for generously providing antibodies and cDNAs. We are grateful to William McGinnis, Sandra Holley, Naama Rave-Harel, Marjory Givens, Mark A. Lawson, Suzie Bailey, Djurdjica Coss, Flavia Pernasetti, Carolyn Kelley, and Shelley Nelson for helpful discussions and critical reading of the manuscript; Vyacheslav V. Vasilyev for providing several of the LHβLuc truncations; and Scott Anderson, Shannon Snyder, and Rachel White for technical support.
This research was supported by NIH Grant R37-HD-20377 (to P.L.M.). This work was also supported by NICHD/NIH through cooperative agreement (U54-HD-12303) as part of the Specialized Cooperative Centers Program in Reproduction Research (P.L.M.). S.B.R. was partially supported by NIH Training Grant T32-DK-07451.
Abbreviations
- Egr-1
Early growth response protein 1
- β-gal
β-galactosidase
- α-GSU
α-glycoprotein hormone subunit
- GnRH-R
GnRH receptor
- HD
homeodomain
- Luc
luciferase
- Ptx1
pituitary homeobox 1 transcription factor
- RSV
Rous sarcoma virus
- SF-1
steroidogenic factor 1
References
- 1.Pierce JG, Parsons TF. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 2.Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- 3.Graham KE, Nusser KD, Low MJ. LβT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to activin A. J Endocrinol. 1999;162:R1–R5. doi: 10.1677/joe.0.162r001. [DOI] [PubMed] [Google Scholar]
- 4.Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang H-J, Miller WL, Mellon PL. Cell-specific transcriptional regulation of FSHβ by activin and GnRH in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- 5.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 6.Tremblay JJ, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem. 1999;274:13870–13876. doi: 10.1074/jbc.274.20.13870. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser UB, Halvorson LM, Chen MT. Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-β gene promoter: an integral role for SF-1. Mol Endocrinol. 2000;14:1235–1245. doi: 10.1210/mend.14.8.0507. [DOI] [PubMed] [Google Scholar]
- 9.Quirk CC, Lozada KL, Keri RA, Nilson JH. A single Pitx1 binding site is essential for activity of the LHβ promoter in transgenic mice. Mol Endocrinol. 2001;15:734–746. doi: 10.1210/mend.15.5.0628. [DOI] [PubMed] [Google Scholar]
- 10.Halvorson LM, Kaiser UB, Chin WW. Stimulation of luteinizing hormone β gene promoter activity by the orphan nuclear receptor, steroidogenic factor-1. J Biol Chem. 1996;271:6645–6650. doi: 10.1074/jbc.271.12.6645. [DOI] [PubMed] [Google Scholar]
- 11.Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 12.Ngan ES, Cheng PK, Leung PC, Chow BK. Steroidogenic factor-1 interacts with a gonadotrope-specific element within the first exon of the human gonadotropin-releasing hormone receptor gene to mediate gonadotrope-specific expression. Endocrinology. 1999;140:2452–2462. doi: 10.1210/endo.140.6.6759. [DOI] [PubMed] [Google Scholar]
- 13.Barnhart KM, Mellon PL. The orphan nuclear receptor, steroidogenic factor-1, regulates the glycoprotein hormone α-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–885. doi: 10.1210/mend.8.7.7527122. [DOI] [PubMed] [Google Scholar]
- 14.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–154. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 15.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 16.Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 17.Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe MW, Call GB. Early growth response protein 1 binds to the luteinizing hormone-promoter and mediates gonadotropin-releasing hormone-stimulated gene expression. Mol Endocrinol. 1999;13:752–763. doi: 10.1210/mend.13.5.0276. [DOI] [PubMed] [Google Scholar]
- 19.Szeto DP, Rodriguez-Esteban C, Ryan AK, O’Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisúa-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisúa-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 21.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 22.Tremblay JJ, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 23.Szeto DP, Ryan AK, O’Connell SM, Rosenfeld MG. P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanctôt C, Gauthier Y, Drouin J. Pituitary homeobox 1 (Ptx1) is differentially expressed during pituitary development. Endocrinology. 1999;140:1416–1422. doi: 10.1210/endo.140.3.6549. [DOI] [PubMed] [Google Scholar]
- 25.Kurotani R, Tahara S, Sanno N, Teramoto A, Mellon PL, Inoue K, Yoshimura S, Osamura RY. Expression of Ptx1 in the adult rat pituitary glands and pituitary cell lines: hormone-secreting cells and folliculo-stellate cells. Cell Tissue Res. 1999;298:55–61. doi: 10.1007/pl00008809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tremblay JJ, Goodyer CG, Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- 27.Tremblay JJ, Marcil A, Gauthier Y, Drouin J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 1999;18:3431–3441. doi: 10.1093/emboj/18.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halvorson LM, Kaiser UB, Chin WW. The protein kinase C system acts through the early growth response protein 1 to increase LHβ gene expression in synergy with steroidogenic factor-1. Mol Endocrinol. 1999;13:106–116. doi: 10.1210/mend.13.1.0216. [DOI] [PubMed] [Google Scholar]
- 29.Acampora D, Mazan S, Tuorto F, Avantaggiato V, Tremblay JJ, Lazzaro D, di Carlo A, Mariano A, Macchia PE, Corte G, Macchia V, Drouin J, Brûlet P, Simeone A. Transient dwarfism and hypogonadism in mice lacking Otx1 reveal prepubescent stage-specific control of pituitary levels of GH, FSH and LH. Development. 1998;125:1229–1239. doi: 10.1242/dev.125.7.1229. [DOI] [PubMed] [Google Scholar]
- 30.Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- 31.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991;30:11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 32.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 33.Tucker SC, Wisdom R. Site-specific heterodimerization by Paired class homeodomain proteins mediates selective transcriptional responses. J Biol Chem. 1999;274:32325–32332. doi: 10.1074/jbc.274.45.32325. [DOI] [PubMed] [Google Scholar]
- 34.Dasen JS, Rosenfeld MG. Combinatorial codes in signaling and synergy: lessons from pituitary development. Curr Opin Genet Dev. 1999;9:566–574. doi: 10.1016/s0959-437x(99)00015-5. [DOI] [PubMed] [Google Scholar]
- 35.Japon MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 36.Keri RA, Wolfe MW, Saunders TL, Anderson I, Kendall SK, Wagner T, Yeung J, Gorski J, Nett TM, Camper SA, Nilson JH. The proximal promoter of the bovine luteinizing hormone β-subunit gene confers gonadotrope-specific expression and regulation by gonadotropin-releasing hormone, testosterone, and 17 β-estradiol in transgenic mice. Mol Endocrinol. 1994;8:1807–1816. doi: 10.1210/mend.8.12.7708066. [DOI] [PubMed] [Google Scholar]
- 37.He X, Rosenfeld MG. Mechanisms of complex transcriptional regulation: Implications for brain development. Neuron. 1991;7:183–196. doi: 10.1016/0896-6273(91)90257-z. [DOI] [PubMed] [Google Scholar]
- 38.Struhl K. Mechanisms for diversity in gene expression patterns. Neuron. 1991;7:177–181. doi: 10.1016/0896-6273(91)90256-y. [DOI] [PubMed] [Google Scholar]
- 39.Drouin J, Lamolet B, Lamonerie T, Lanctôt C, Tremblay JJ. The PTX family of homeodomain transcription factors during pituitary developments. Mol Cell Endocrinol. 1998;140:31–36. doi: 10.1016/s0303-7207(98)00026-4. [DOI] [PubMed] [Google Scholar]
- 40.Acampora D, Simeone A. The TINS Lecture. Understanding the roles of Otx1 and Otx2 in the control of brain morphogenesis. Trends Neurosci. 1999;22:116–122. doi: 10.1016/s0166-2236(98)01387-3. [DOI] [PubMed] [Google Scholar]
- 41.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice R, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm of the gastrulating mouse embryo. EMBO J. 1993;12:2735–2774. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like Paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 43.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 44.Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol. 2000;14:1246–1256. doi: 10.1210/mend.14.8.0509. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee KAW, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 48.Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–859. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- 49.Hjalt TA, Semina EV, Amendt BA, Murray JC. The Pitx2 protein in mouse development. Dev Dyn. 2000;218:195–200. doi: 10.1002/(SICI)1097-0177(200005)218:1<195::AID-DVDY17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 50.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 51.Clark ME, Mellon PL. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol. 1995;15:6169–6177. doi: 10.1128/mcb.15.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Q, Knoepfler PS, Scheele J, Wright DD, Kamps MP. Both Pbx1 and E2A-Pbx1 bind the DNA motif ATCAATCAA cooperatively with the products of multiple murine Hox genes, some of which are themselves oncogenes. Mol Cell Biol. 1995;15:3786–3795. doi: 10.1128/mcb.15.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]