Abstract
Although it has long been established that episodic secretion of gonadotropin-releasing hormone (GnRH) from the hypothalamus is required for normal gonadotropin release, the molecular and cellular mechanisms underlying the synchronous release of GnRH are primarily unknown. We used the GT1-7 mouse hypothalamic cell line as a model for GnRH secretion, because these cells release GnRH in a pulsatile pattern similar to that observed in vivo. To explore possible molecular mechanisms governing secretory timing, we investigated the role of the molecular circadian clock in regulation of GnRH secretion. GT1-7 cells express many known core circadian clock genes, and we demonstrate that oscillations of these components can be induced by stimuli such as serum and the adenylyl cyclase activator forskolin, similar to effects observed in fibroblasts. Strikingly, perturbation of circadian clock function in GT1-7 cells by transient expression of the dominant-negative Clock-Δ19 gene disrupts normal ultradian patterns of GnRH secretion, significantly decreasing mean pulse frequency. Additionally, overexpression of the negative limb clock gene mCry1 in GT1-7 cells substantially increases GnRH pulse amplitude without a commensurate change in pulse frequency, demonstrating that an endogenous biological clock is coupled to the mechanism of neurosecretion in these cells and can regulate multiple secretory parameters. Finally, mice harboring a somatic mutation in the Clock gene are subfertile and exhibit a substantial increase in estrous cycle duration as revealed by examination of vaginal cytology. This effect persists in normal light/dark (LD) cycles, suggesting that a suprachiasmatic nucleus-independent endogenous clock in GnRH neurons is required for eliciting normal pulsatile patterns of GnRH secretion.
Keywords: circadian, GnRH, secretion, pulsatility, Clock, GT1-7
Introduction
Reproductive function hinges on the episodic secretion of gonadotropin-releasing hormone (GnRH) from hypothalamic nerve terminals to stimulate gonadotropin release from the anterior pituitary. This pulsatile GnRH secretion ultimately regulates critical reproductive processes ranging from gametogenesis to ovulation. Although GnRH perikarya and nerve terminals are positioned within the mediobasal hypothalamus to receive multiple neuronal and humoral signals and can respond to exogenous stimuli by modulating GnRH secretion, it has become apparent that species-specific timing of GnRH pulse release is an intrinsic property of GnRH neurons. Studies using immortalized GT1-1 and GT1-7 cell lines reveal that these homogenous clonal cell populations are capable of recapitulating GnRH pulse release patterns observed in vivo from rodents (Martinez de la Escalera et al., 1992; Wetsel et al., 1992; Weiner and Martinez de la Escalera, 1993). In more recent work, pulsatile GnRH release was also observed from tissue explants of premigratory GnRH cells collected from embryonic primate olfactory placode (Terasawa et al., 1999), demonstrating that GnRH neurons do not require the complex array of afferent neuronal connections present in the mature hypothalamus to secrete GnRH in discretely timed pulses. Although the phenomenon of GnRH pulsatility has been well documented, mechanisms mediating coordinated neurosecretion have proven elusive.
Although GnRH pulsatility can be modulated by exogenous signals such as adrenergic (Herbison et al., 1989; Herbison, 1997), glutamatergic (Brann and Mahesh, 1994), and GABAergic (Herbison and Dyer, 1991) inputs, an inherent “pulse generator” synchronizes the activity of populations of GnRH neurons to produce a meaningful secretory output. Because this episodic secretion pattern is species- and cell-specific (Wetsel et al., 1992; Terasawa et al., 1999), a developmentally regulated molecular program is likely responsible for the fundamental machinery of this neuroendocrine pulse generation. In the studies below, we investigated possible molecular mechanisms governing this phenomenon.
Recent discoveries in the field of circadian biology have revealed that molecular feedback loops of gene expression responsible for regulating clock function are comprised of complex interactions among multiple transcription factors, including many PAS-basic helix-loop-helix family members such as Period (mPer1, mPer2, mPer3) (Zylka et al., 1998), Cryptochrome (mCry1 and mCry2) (Kume et al., 1999), Clock (Gekakis et al., 1998), Bmal1 (also termed MOP3) (Honma et al., 1998; Bunger et al., 2000) as well as the orphan nuclear receptor REV-ERBα (Preitner et al., 2002), and the casein kinase isoforms I epsilon (CKIϵ) (Lowrey et al., 2000) and delta (CKIδ) (Lee et al., 2001). Recent studies have demonstrated that functional clocks exist not only in the mammalian central biological clock, the suprachiasmatic nucleus (SCN), but also within many peripheral cell types and cultured cell lines (Balsalobre et al., 1998) in which multiple humoral stimuli are capable of “resetting” molecular clock oscillations. We explored the possibility that expression of intracellular and intercellular components required for ordered GnRH secretion may be orchestrated by a timing mechanism under the control of the molecular clock transcriptional feedback loop.
There is considerable evidence that the circadian biological clock is an important modulator of the reproductive axis, particularly in the generation of gonadotropin surges. Previous experiments demonstrated that ablation or lesioning of the SCN prevents induction of preovulatory luteinizing hormone (LH) surges, resulting in estrous acyclicity and infertility (Gray et al., 1978). In addition, ovariectomized rodents treated with chronically elevated 17β-estradiol (17β-E2) exhibit LH surges on consecutive days that are always temporally confined to the late afternoon and early evening (Legan and Karsch, 1975; Legan et al., 1975), further implicating circadian regulation of ovulation. Thus, many consequent studies concentrated on neuronal connectivity between SCN afferents and GnRH perikarya and have found possible afferent neuromodulators in the form of arginine vasopressin (van der Beek et al., 1997b; Funabashi et al., 2000) and vasoactive intestinal peptide (van der Beek et al., 1993, 1994, 1997a), although the necessity of these peptides for intrinsic pulsatile GnRH secretion has yet to be demonstrated.
In support of the aforementioned studies implicating biological clock function in GnRH surge generation, evidence suggests that the circadian clock may be involved in regulating basal GnRH pulse frequency as well. Hamsters harboring the naturally occurring tau mutation, a mutated CKIϵ unable to fully phosphorylate Per2 (Lowrey et al., 2000), exhibit accelerated LH pulse frequencies (Loudon et al., 1994), suggesting that the underlying GnRH pulsatility may be fundamentally altered by this circadian mutation. In addition, recent studies implicate proper molecular clock function as crucial to normal reproductive patency in Drosophila (Beaver et al., 2002). In the current studies, we provide evidence that not only do functional circadian clocks exist within immortalized GnRH neurons, but also that perturbation of clock function disrupts normal pulse patterns of GnRH secretion as well as estrous cycles in vivo. These results suggest a model in which intrinsic and extrinsic circadian signals could serve to both synchronize and modulate GnRH pulse secretion by influencing endogenous molecular clocks present in these neurons, thus providing tight neuroendocrine control over processes as far-ranging as pubertal progression and estrous cyclicity.
Materials and Methods
Cell culture. GT1-7 and NIH3T3 cells were cultured in DMEM (Mediatech, Herndon, VA) containing 4.5% glucose, supplemented with 10% fetal calf serum (FCS; Omega Scientific, Tarzana, CA), penicillin (100 U/ml), and streptomycin (0.1 mg/ml; Invitrogen, Carlsbad, CA). Cells were incubated in 5% CO2/95% O2 at 37°C. For RNA isolation and nuclear extract collection, multiple plates depending on length of collection of similar passage (passage 6-15) and confluency (∼90%) were all cultured in the same conditions in parallel.
Serum shock and drug treatments. For serum-shock treatment, multiple plates were switched to DMEM supplemented with 50% adult horse serum or 50% fetal bovine serum (FBS; serum shock) at 0 hr. The media containing 50% serum was removed after 2 hr and replaced with serum-free (SF) DMEM, similar to techniques performed previously by Balsalobre et al. (1998). For serum-free treatment, other plates were switched at 0 hr to SF DMEM conditions and remained in the same media for the duration of the collection. Other plates of GT1-7 cells were treated with 1.0 μm forskolin (FSK; Sigma, St. Louis, MO) in SF DMEM for 15-30 min, followed by replacement with SF DMEM for the duration of the experiment. Other plates were treated for 1 hr with 1.0 μm ionomycin (Calbiochem, San Diego, CA), 100 nm phorbol-12-myristate-13-acetate (PMA; Calbiochem), and 100 μm sodium nitroprusside (SNP; Sigma) and were then switched to SF DMEM for the duration of the experiment.
RNA isolation. After serum-shock or switch to serum-free media, cells were harvested every 4 hr by removal of media, followed by an ice-cold PBS wash and the addition of Trizol (Invitrogen) at a concentration of 2 × 106 cells per milliliter. RNA was isolated from cells as described previously (Chomczynski and Sacchi, 1987). Briefly, cell lysates suspended in Trizol were treated with chloroform, followed by isopropanol precipitation of RNA pellets, and were resuspended in autoclaved water treated with diethylpyrocarbonate.
Reverse transcriptase-PCR. Total RNA was isolated from confluent plates of GT1-7 cells using Trizol as described above. Tissues (whole brain and pituitary) were homogenized in Trizol. One microgram of total RNA isolated was reverse transcribed using SuperScript II enzyme (Invitrogen). Twenty microliters of reverse transcriptase (RT) mix were amplified using sequence-specific PCR primers for mPer1 [forward (Fwd), 5′-TCAGAGCAGGACAACCCATCTACCAGT-3′; reverse (Rev) 5′-AAGCGGGCACCCCGAAACAC-3′], mPer2 (Fwd, 5′-CCGACATGAATGGATACG-3′; Rev, 5′-GAGGTAATGCCCTCAACC-3′), mPer3 (Fwd, 5′-AAAAGGCAGCCAGTAACG-3′; Rev, 5′-GTTCATCTGCTGATAGGACG-3′), clock (Fwd, 5′-GCGAGAACTTGGCATTGAAGAG-3′; Rev, 5′-CTGTGTCCACTCATTACACTCTGTTG-3′), Bmal1 (Fwd, 5′-CCACAGGATAAGAGGGTCAT-3′; Rev, 5′-ATGATGAGGAAACACTGGAG-3′), mCry1 (Fwd, 5′-GCGTCTATATCCTCGACC-3′; Rev, 5′-AAGTGCCTCAGTTTCTCC-3′), mCry2 (Fwd, 5′-AGAAGGTGAAGAGGAACAGC-3′; Rev, 5′-ACAGGAAGGGACAGATGC-3′), and CKIϵ (Fwd, 5′-CACTCCAAGAACTTCATCC-3′; Rev, 5′-CAGTCGAAGACGTAGTCG-3′). Reactions without RT served as negative controls.
Western blotting. GT1-7 nuclear extracts were collected every 6 hr for 48 hr. Briefly, cells were rinsed with 1 × PBS and lysed with buffer containing 20 mm Tris-HCl, 140 mm NaCl, 0.5 mm EDTA, protease inhibitors aprotinin, pepstatin, and leupeptin (10 μg/ml each), and PMSF (1.0 mm). Nuclei were pelleted by centrifugation and resuspended in a hypertonic buffer solution containing protease inhibitors. Protein concentrations were determined by Bradford Assay (Bio-Rad, Hercules, CA), and 20 μg of protein was loaded onto an SDS-polyacrylamide gel. After resolution by electrophoresis, proteins were electrically transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore, Bedford, MA) and probed with Bmal- or mPer1-specific antibodies (α Diagnostics International, San Antonio, TX). Immunoreactive bands were detected using anti-rabbit secondary antibodies (Amersham Pharmacia, Piscataway, NJ) linked to horseradish peroxidase and amplified with ECL reagent (Amersham Pharmacia). Quantification of band intensity was performed using a Molecular Dynamics (Sunnyvale, CA) STORM Imager combined with ImageQuant analysis software for the Macintosh computer (Amersham Biosciences, Arlington Heights, IL).
Plasmids and cloning. PCR fragments of mPer1 (289 bp; 59-348), mPer2 (238 bp; 435-673), mCry1 (312 bp; 68-380), and mCry2 (225 bp; 62-287) were cloned into pCR2.1-Topo vectors (Invitrogen) and were sequenced to determine proper orientation. Plasmids were linearized with HindIII, and antisense cRNA probes were transcribed using a Maxiscript kit with either T7 or T3 RNA polymerases (Ambion, Austin, TX). mPer1, mPer2, mCry1, and mCry2 expression plasmids were generously provided by Dr. Steven Reppert (University of Massachussetts School of Medicine, Worcester, MA). The mPer1-luciferase plasmid was kindly provided by Dr. Joe Takahashi (Northwestern University and Howard Hughes Medical Institute, Evanston, IL). pGL3, a promoterless luciferase reporter plasmid (Stratagene, La Jolla, CA) was used as a background control for reporter assays. The expression vector for Clock-Δ19 was provided by Dr. Charles Weitz (Harvard University, Boston, MA). The pcDNA3.1 plasmid (Invitrogen) was used as a control for transfections of mPer1, mCry1, and Clock-Δ19. A thymidine kinase-β-galactosidase (tk-β-gal) plasmid was used as an internal control in all reporter assays to normalize for transfection efficiency.
Ribonuclease protection assays. Antisense RNA probes for mPer1, mPer2, mCry1, mCry2, and cyclophilin were labeled with α-32P UTP at a high specific activity (3-6 × 105 cpm/ng) using a Maxiscript II kit (Ambion) and incubated with 10-15 μg of total RNA from samples collected at 4 hr increments for 60-72 hr. Samples were hybridized with probes for 12-16 hr at 42-55°C and digested using an RNase A/T1 mixture for 30 min at 37°C [ribonuclease protection assays (RPA) III kit; Ambion]. After enzyme inactivation and precipitation, RNA pellets were resuspended in loading buffer and run on a 5% denaturing polyacrylamide gel for 1 hr. Gels were dried and placed on phosphoimager screens for 12-16 hr (Molecular Dynamics). Images were visualized using a STORM phosphoimager, and band intensity was analyzed using ImageQuant software (Molecular Dynamics; Amersham Biosciences).
Transient transfections. For the mPer1-luciferase expression profiles, 1.5 × 105 GT1-7 cells per well were plated onto 24-well plates. Twenty-four hours after plating, cells were transfected with both 0.3 μg of mPer1-luciferase reporter plasmid and 0.1 μg of tk-β-gal plasmid as an internal control for transfection efficiency using FuGene 6 lipofection agent (Roche Molecular Biochemicals, Indianapolis, IN). Replicate wells were cotransfected with equimolar amounts of pcDNA3.1, Clock-Δ19, or mCry1 per plate. Twelve to 16 hr after transfection, cells were washed with 1 × PBS and lysed with 0.1 m K-phosphate buffer, pH 7.8, with 0.2% Triton X-100. Parallel plates were assayed every 4 hr for 72 hr after either a serum shock or media switch to serum-free media. Equal volumes of cell lysate were assayed for luciferase activity using a microplate luminometer (MicroLumat Plus; EG&G, Bertholt, Germany) by injecting 10 μl of a buffer containing 100 mm Tris-HCl, pH 7.8, 15 mm MgSO4, 10 mm ATP, and 65 mm luciferin per well. β-gal activity was measured using the Galacto-light assay (Tropix, Bedford, MA) following the instructions of the manufacturer. All values are presented as the ratio of luciferase activity over β-gal activity to normalize for transfection efficiency. Transfections were done with four to six replicate wells per experiment, and experiments were repeated at least three times. Cells transfected with pGL3, the control vector of mPer1-luc, exhibited nonoscillating luciferase activity (data not shown).
GT1-7 cell perifusion and fraction collection. To measure high-resolution secretion of GnRH from living GT1-7 cells over time, the following perifusion techniques were used. Cells were grown on Cytodex 3 beads (Amersham Pharmacia) for 4-7 d in DMEM containing 10% FCS on nonadherent Petri dishes (Fisher Scientific, Houston, TX). Media was replaced every 48 hr. Twenty-four hours before loading into perifusion columns, GT1-7 cells were transfected as described above with equimolar amounts of pcDNA3.1, Clock-Δ19, or mCry1. Cells on beads were loaded into perifusion columns (1.0 cc volume) to a height of 0.75 cc. A Krebs-Ringers phosphate buffer solution (KRB), aerated with 95% O2/5% CO2 and kept in a 37°C water bath, was perifused through temperature-controlled columns at a flow rate of 0.1 ml/min. After a 6-24 hr equilibration period, 0.5 ml fractions were collected every 5 min for 10.5 hr. The length of the equilibration period was varied to stagger collection start times relative to the time of day. In some experiments, 0.1 ml fractions were collected every minute for 10 hr. At the conclusion of each experiment, buffer was replaced with KRB containing high (30-60 mm) KCl for 0.5-1.0 hr to test for cell viability through observation of depolarization-induced GnRH secretion. Fractions were frozen and stored at -80°C until radioimmunoassay. In addition, some columns were cotransfected with either CMV-enhanced green fluorescent protein (eGFP) or rat GnRH-eGFP plasmids to determine the robustness of expression after prolonged perifusion. After perifusion, GT1-7 cells on beads were removed from columns and placed in slide chambers for FITC excitation and visualization using confocal microscopy in the University of California, San Diego, Imaging Core facility. Cells on beads from additional columns were used to quantify transfection efficiency. Trypsin (1×) was used to remove transfected cells from beads, which were allowed to settle to the bottom of a conical tube in PBS. Ten microliters were removed from the cell layer and placed on a hemocytometer. Transfection efficiency is shown as a percentage of GFP-positive cells divided by total cell number.
Vaginal cytology assessment in mice. All animal procedures were conducted in full accordance with the policies of the University of California's Animal Care and Use Committee. Clock/clock mutant mice (generously provided by Martha Vitaterna and Joe Takahashi, Northwestern University), clock/+ heterozygotes, and wild-type littermates were housed, four to a cage, under either 12 hr LD cycles or constant darkness (DD) cycles and fed lab chow ad libitum. Vaginal cytology was monitored daily for 6 weeks using an infrared camera for visualization in DD. Adult female and male mice were mated in both LD and DD conditions.
Radioimmunoassay. GnRH primary antibody EL-14 was kindly provided by Martha Bosch, Oline Ronnekleiv, and Martin Kelly (University of Oregon Health Sciences Center, Portland, OR). Briefly, 0.1 ml aliquots of each 0.5 ml perifusate sample were incubated for 48 hr with GnRH antibody at 4°C, after which time ∼10,000 cpm sample of radioiodinated GnRH (Amersham Pharmacia) was added. Forty-eight hours later, after ethanol precipitation of bound fractions, radioactivity was detected by a gamma-counter (Micromedic, Huntsville, AL). Intra-assay and inter-assay variability was 4.6 and 6.8%, respectively.
Statistical data analysis. The CLUSTER (Veldhuis; University of Virginia, Charlottesville, VA) statistical pulse analysis program was used to identify significant GnRH pulses. For 5 min sampling, pulse peaks were determined as two points per peak flanked by two nadir points with a 5.0% significance cutoff level as determined by up-stroke and down-stroke t-statistic values of 2.0, and three points per peak flanked by three nadir points with t-statistic values of 3.0 for 1 min sampling (similar to Veldhuis and Johnson, 1986, and Clarke et al., 2002). PeakFit (SPSS, Chicago, IL) was used after cluster analysis to independently confirm pulse peaks using deconvolution analysis as well as to determine individual and cumulative peak area. Mean frequency of release was determined as the mean number of pulses per hour within the 10 hr collection period. Pulses observed within the one-half hour depolarizing stimulus period were not included in pulse analysis. One-way ANOVA was used to determine significant differences in pulse frequency, amplitude, and cumulative area among untreated, control-transfected, Clock-Δ19-transfected, and mCry1-transfected groups. Mean pulse amplitude variation (MPAV) was calculated by first normalizing mean peak amplitude within each sampling period to 0. Within each treatment group, the mean of the absolute differences of pulse peaks from normalized mean pulse amplitudes represent MPAV.
Results
Circadian clock genes are expressed and oscillate in cultured GT1-7 cells
To confirm the expression of circadian clock genes in the hypothalamic GT1-7 cell line, RT-PCR was performed as described. As shown in Figure 1, the mouse GnRH-secreting cell line, GT1-7, expresses many characterized mammalian circadian clock genes, including Bmal1, clock, mCry1, mCry2, mPer1, mPer2, mPer3, and CKIϵ. Expression of these genes has been observed in both the SCN and peripheral cell types and suggests that the transcriptional feedback loops that form the basis of the circadian clock may function within this cell line.
Figure 1.
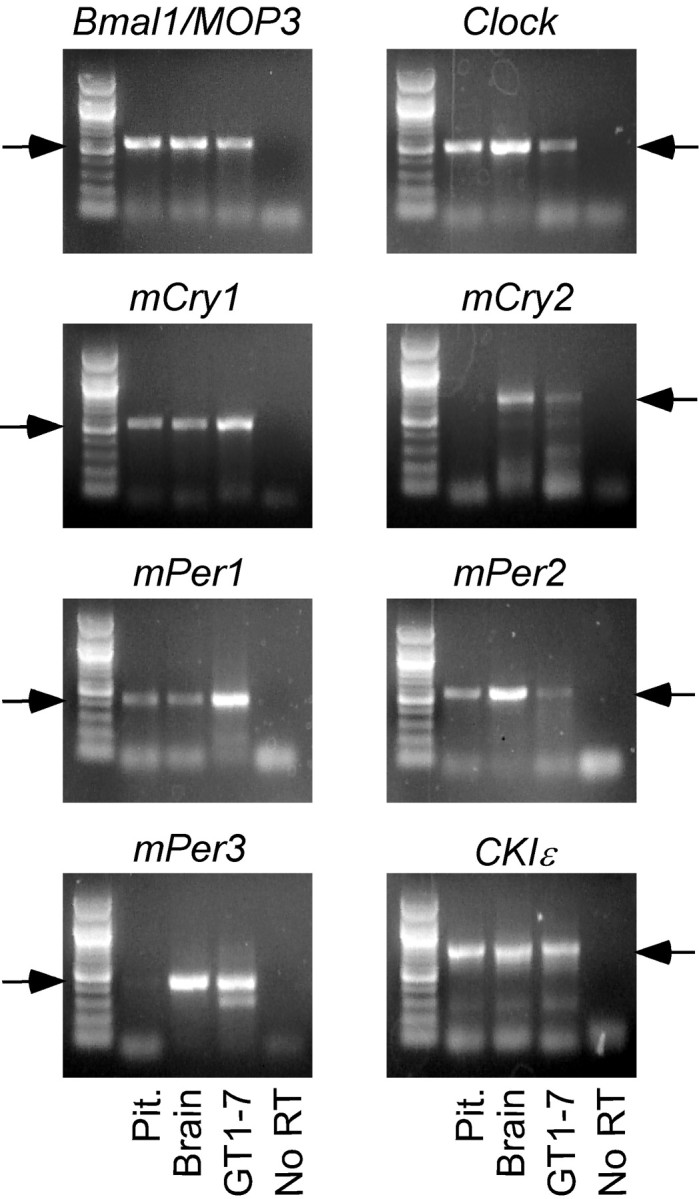
Multiple core circadian clock genes are expressed in cultured GT1-7 cells. RT-PCR bands (arrows) corresponding to murine circadian clock genes Bmal1, Clock, mCry1, mCry2, mPer1, mPer2, mPer3, and CKIϵ are present in tissue preparations of pituitary and total brain, as well as in cultured GT1-7 cells. No RT (far right lane) served as a negative control. A 100 bp ladder (Promega, Madison, WI) is shown in the far left lane.
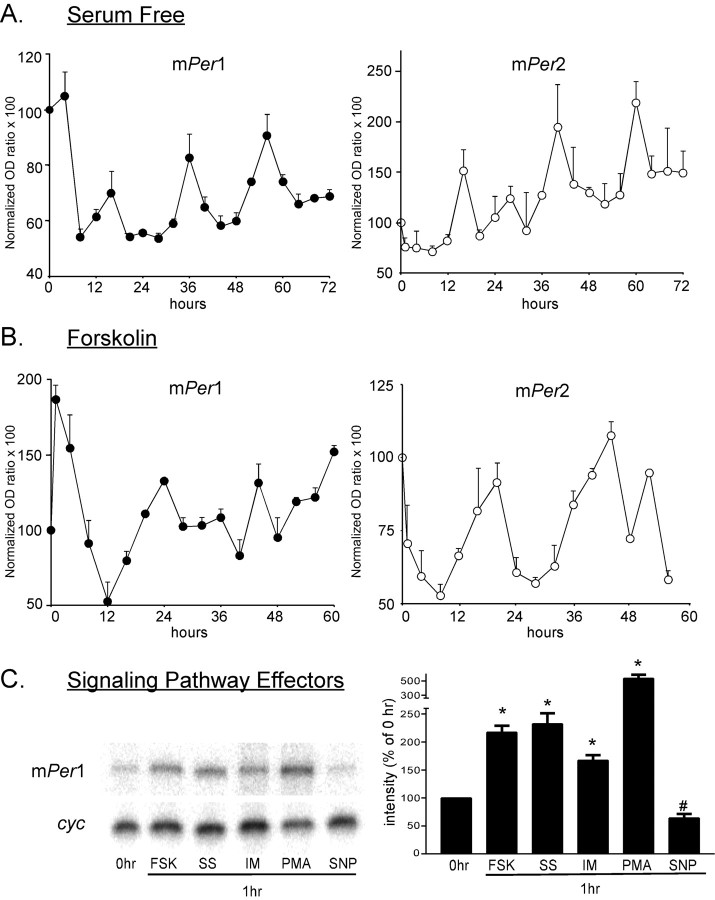
To determine whether these gene products are capable of exhibiting cyclic mRNA accumulation rhythms in GT1-7 cells, as observed previously in rat (rat-1) and mouse (NIH3T3) fibroblasts (Balsalobre et al., 1998; Yagita et al., 2001; Hirota et al., 2002), we performed RPA on total cellular RNA isolated over time. As shown in Figure 2A, mRNA levels of mPer1 and mPer2 oscillate with ∼20-24 hr periodicity in this cell line, as well as in NIH3T3 cells (data not shown), after a shift to SF media conditions, as well as after a 15-30 min exposure to 1.0 μm FSK (Fig. 2B). Under both conditions, the peaks in expression levels of mPer2 lag slightly (∼4 hr) behind mPer1 peak levels, as observed previously in other cell lines (Yagita et al., 2001). Serum-shock and FSK stimulation of clock cycling has been characterized previously in rat-1 and NIH3T3 fibroblasts (Balsalobre et al., 2000b; Yagita and Okamura, 2000), whereas oscillation of clock gene transcripts after a change to SF media has been observed very recently in rat fibroblasts (Hirota et al., 2002), associated with a transient downregulation of many clock genes followed by low-amplitude cycling. This effect has also been observed in the neuronal SCN2.2 cell line, in which minor perturbations such as a depolarizing concentration of KCl or low (2%) levels of serum are sufficient to initiate gene expression oscillation (Allen and Earnest, 2002). In addition, mCry1 and mCry2 mRNA expression levels were observed to oscillate in a rhythm similar to that of mPer1 and mPer2 (data not shown), with mCry2 exhibiting considerably damped amplitudes in comparison with mCry1. Plates of GT1-7 cells cultured and harvested in 10% FBS DMEM for the duration of the experiment (no perturbation) demonstrated no significant oscillations in mRNA accumulation (data not shown).
Figure 2.
Multiple stimuli induce transient increases in expression and oscillation of circadian clock gene expression in cultured GT1-7 cells, as measured by RNase protection assay. A, Circadian gene expression of mPer1 and mPer2 oscillates in GT1-7 cells after a shift to SF media. A, B, Fluctuations in mRNA band intensity are presented as percentage change from 0 hr mPer1/cyc or mPer2/cyc optical density (OD) ratios. Traces indicate the mean of three RNA collections per time point. B, mPer1 and mPer2 expression oscillates after a 15 min stimulation by 10.0 μm FSK, followed by a shift to SF for 60 hr. GT1-7 cells remaining in 10% FBS media (no perturbation) demonstrated no significant mRNA oscillations. C, Representative RNase protection assay of mPer1 and cyclophilin mRNA level after 1 hr of exposure of GT1-7 cells to media containing 10.0 μm FSK (15 min exposure; lane 2), 50% horse serum (lane 3), 1.0 μm Ionomycin (lane 4), 100.0 nm PMA (lane 5), and 100.0 μm SNP (lane 6). The bar graph on the right shows mean changes in mPer1 expression at 1 hr after indicated treatments. Each bar represents the mean of four experiments (*p < 0.01, increase above 0 hr level; #p < 0.05, decrease below 0 hr level).
Activation of the cAMP and mitogen-activated protein kinase pathways, in addition to their characterized effects on circadian gene expression (Balsalobre et al., 2000a), has been shown previously to play a role in the regulation of both GnRH transcription and secretion (Wetsel et al., 1993; Eraly and Mellon, 1995), and changes in intracellular cAMP levels have been implicated in the generation of the pulse rhythm of GnRH secretion (Vitalis et al., 2000). Expression of mPer1 (Fig. 2C), but not mPer2 (data not shown), was transiently stimulated by 15-30 min treatment with 1.0 μm FSK and 1 hr treatment with 50% horse serum, 10 μm ionomycin, and 0.1 μm PMA, phenomena observed previously in rat and mouse fibroblasts (Balsalobre et al., 1998; Yagita et al., 2001), whereas expression of both mPer1 and mPer2 was transiently repressed by 1 hr exposure to the nitric oxide donor SNP (*p < 0.01, increase above 0 hr; #p < 0.05, decrease below 0 hr mRNA band intensity). Transient increases in mPer1 mRNA levels in GT1-7 cells after FSK treatment have been correlated recently with an increase in GnRH secretion in static culture (Olcese et al., 2003).
The transcription factor Bmal1 binds with its heterodimeric partner, Clock, to increase transcription of the Cry and Per genes, forming the “positive limb” of the circadian clock feedback loop. We show in Figure 3A that Bmal1 protein levels oscillate with ∼24 hr rhythm in nuclear extracts of serum-shocked GT1-7 cells, demonstrating that a functional clock appears to exist within this reproductive neuronal cell line, similar to results shown previously in other cell lines (Balsalobre et al., 1998). Exposure to a high concentration of serum (50%) leads to an increase in Clock:Bmal1 heterodimers binding to mPer1 promoter E-boxes, increasing transcription of the mPer1 gene (Jung et al., 2003). As shown in Figure 3B, mPer1 protein levels in GT1-7 nuclear extracts also cycle, exhibiting peaks 8-12 hr out of phase with Bmal1 protein levels.
Figure 3.
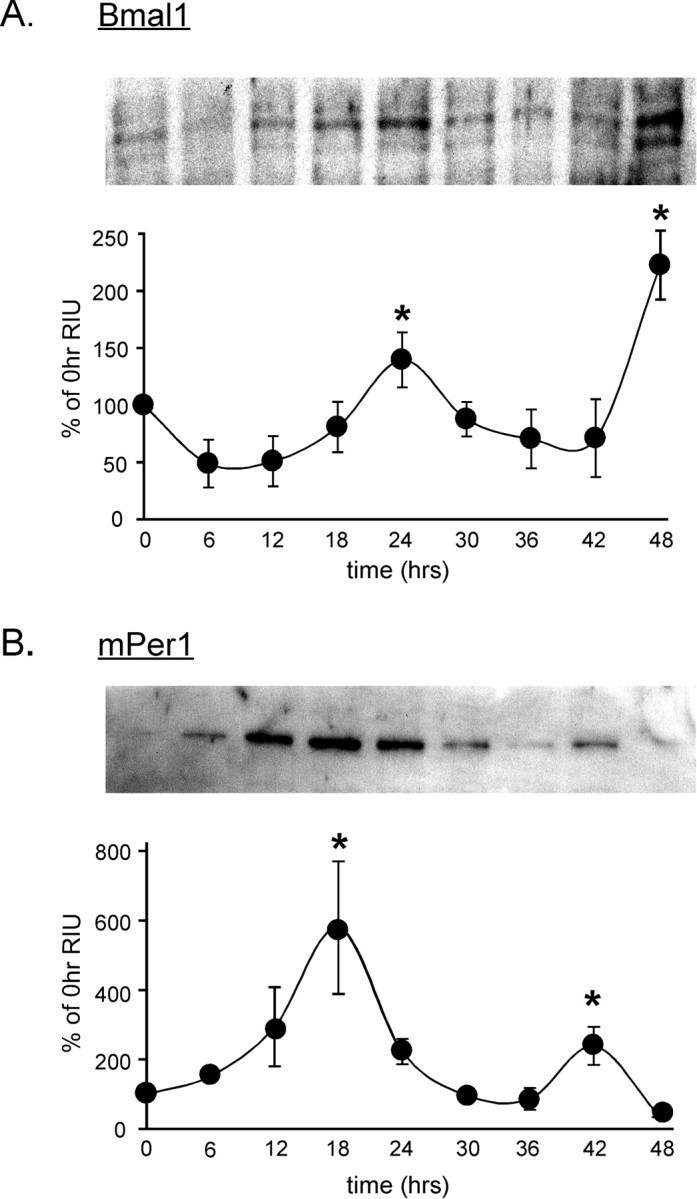
Per1 and Bmal1 proteins exhibit circadian oscillations in cultured GT1-7 cells. A, Protein levels of Bmal1 oscillate in GT1-7 cells after a serum shock. A representative Western blot (top) reveals an ∼60 kDa band present in GT1-7 nuclear extracts exhibiting peak levels at 24 and 48 hr. The corresponding graph shows mean protein levels at each time point (n = 3). B, Oscillation of mPer1 protein over 48 hr exhibits a peak at 12-18 hr after serum shock, 6-12 hr out of phase with the Bmal1 protein rhythm (*p < 0.05; significantly different from nadir as determined by ANOVA).
Circadian clock function is required for normal GnRH secretion patterns
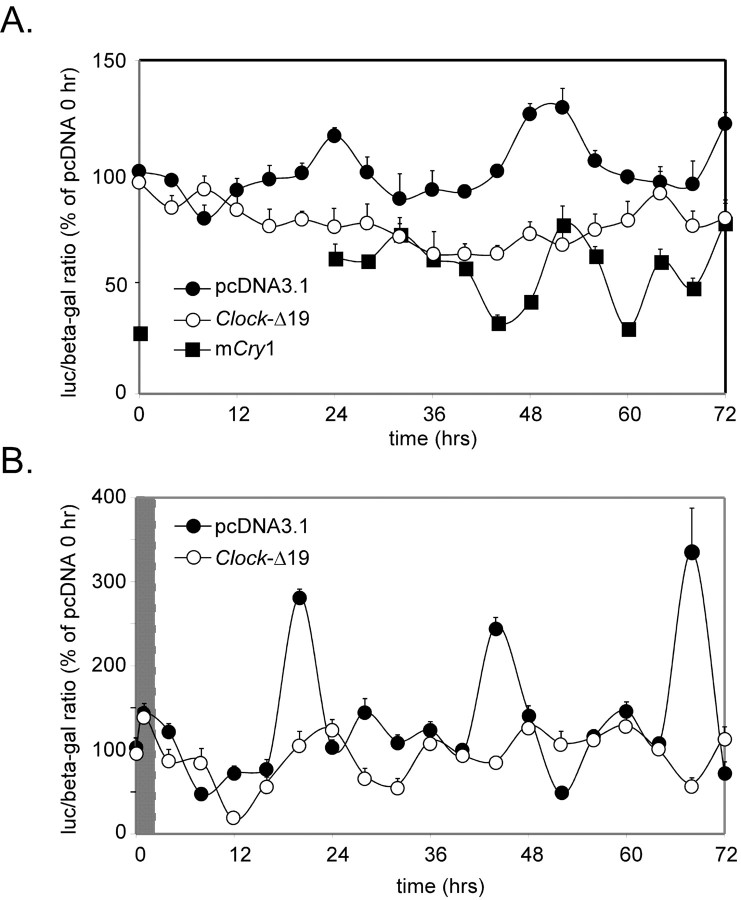
To independently determine both the extent and necessity of clock function in this neuroendocrine cell line, GT1-7 cells were transiently transfected with mPer1-luc (Wilsbacher et al., 2002), with TK-β-gal plasmid used as an internal control. Analysis of normalized luciferase/β-galactosidase ratios indicates that mPer1-luc gene expression oscillates with ∼20-24 hr periods in both serum-free (Fig. 4A) and serum-shocked (Fig. 4B) conditions. Peaks of mPer1-luciferase expression appear to lag 4-8 hr behind mPer1 mRNA peaks observed using RPA (Fig. 2), likely resulting from observed different half-life and degradation rates of luciferase mRNA versus that of mPer1 (Wilsbacher et al., 2002).
Figure 4.
The circadian clock maintains transcriptional regulatory relationships in the GT1-7 cell line. A, B, Oscillation of mPer1-luciferase activity in GT1-7 cells under both SF (A) and serum-shock (B) conditions. Filled circles indicate cells cotransfected with control pcDNA3.1 plasmid. Open circles represent cells cotransfected with a Clock-Δ19 expression plasmid. Filled squares represent cells cotransfected with an mCry1 expression plasmid. Values are presented as percentage changes from 0 hr normalized luciferase/β-galactosidase ratios of pcDNA3.1-transfected cells. Two hr serum-shock in B is indicated by the gray area.
Further suggesting that the molecular clock loop is intact in GT1-7 cells, cotransfection with a plasmid expressing the dominant-negative Clock gene, Clock-Δ19, severely blunted mPer1-luciferase cycling in these cells (Fig. 4). In contrast, cotransfection with the empty control vector, pcDNA3.1, did not affect reporter oscillations, indicating that specific expression of Clock-Δ19 disrupted cyclical regulation of mPer1 transcription in GT1-7 cells, supporting previously published reports of peripheral cell types (Jung et al., 2003). In addition, transient overexpression of mCry1, a potent repressor of mPer1 promoter activity, reduced luciferase expression to ∼20-50% of control levels, although not entirely eliminating oscillations of mPer1-luc reporter activity (Fig. 4A), again demonstrating functionality of circadian clock components in this cell line.
Thus, having established that clock gene products are oscillating in this neuroendocrine cell type in culture, similar to observations in other peripheral cell types, we wished to determine whether this endogenous rhythm is coupled to the ultradian pattern of secretion observed from GnRH neurons. To determine the feasibility of introducing genes into GT1-7 cells while also assaying pulsatile secretion, we asked whether proteins from transiently transfected expression vectors were still expressed after perifusion procedures. GT1-7 cells were transfected with either a rat 5′-GnRH-eGFP or the parent CMV-eGFP vector and perifused for 48 hr (Fig. 5A). Visualization of robust GFP expression in clusters of GT1-7 cells on beads (Fig. 5B) after perifusion indicates that transient gene expression remains high even after 72 hr of combined static incubation and perifusion after transfection. Six columns of GFP-positive cells were counted after perifusion, revealing a transfection efficiency range of ∼18-36% for GnRH-eGFP and ∼25-60% for CMV-eGFP (a stronger promoter). To investigate the effect of Clock-Δ19 expression on pulsatile GnRH secretion, GT1-7 cells cultured on beads in a perifusion system were transiently transfected with Clock-Δ19 16-24 hr before column loading and fraction collection. After loading into columns, cells were perifused in KRB for 6-24 hr, followed by either 1 or 5 min sample collection for an additional 10-12 hr. Significant pulse peaks of GnRH were differentiated from baseline noise using cluster analysis parameters to determine peak and nadir values and significant differences among them (see Materials and Methods).
Figure 5.
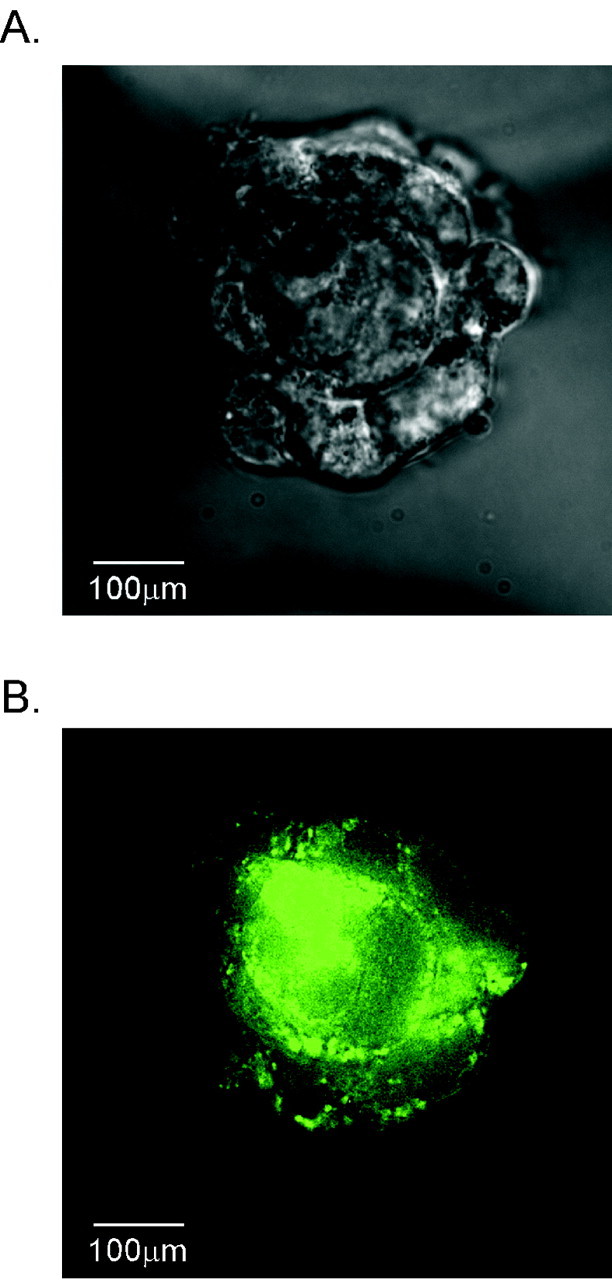
GFP fluorescence reveals robust transgene expression in GT1-7 cells grown on beads after 48 hr of perifusion. A, Bright-field micrograph of a cluster of GT1-7 cells on Cytodex beads after 24 hr static incubation in DMEM/10% FCS followed by 48 hr of perifusion with KRB. Clusters of multiple cells are visible, adhering multiple beads together. B, Flourescence imaging reveals robust GFP expression in the same cell cluster 72 hr after transfection and perifusion. Scale bar, 100 μm. Ranges of transfection efficiencies for rat GnRH-eGFP and CMV-eGFP are ∼18-36 and ∼25-60%, respectively.
Strikingly, whereas GnRH secretion profiles of either untransfected (Fig. 6A) or pcDNA3.1 (control)-transfected (Fig. 6B) GT1-7 cells exhibited secretory inter-pulse intervals of ∼25-30 min, similar to previously published accounts (Krsmanovic et al., 1992; Wetsel et al., 1992), cells transiently expressing Clock-Δ19 demonstrated markedly disrupted patterns of secretion often characterized by large irregular bursts of GnRH release followed by long quiescent periods of minimal secretion (Figs. 6C, 7C). Because of this altered secretory pattern, mean GnRH pulse frequency was significantly decreased by expression of the dominant-negative Clock, to 1.61 ± 0.10 pulses/hr as compared with 2.04 ± 0.03 pulses/hr observed in control-transfected cells (Fig. 7A,B). As shown in Figure 7D, mean GnRH pulse amplitude, although exhibiting an increased trend in perifused Clock-Δ19-expressing cells (71.2 ± 18.5 pg/ml), was not significantly higher than mean pulse amplitude observed in control-transfected perifused cells (63.8 ± 4.1 pg/ml). Although mean pulse amplitude did not significantly differ between Clock-Δ19- and control-transfected perifused cells, pulse amplitudes were highly variable in Clock-Δ19-transfected cells because of irregular patterns of pulse release demonstrated by the representative profiles shown in Figure 7C. This increased variability in GnRH pulse amplitude is depicted by Figure 7E, where the abscissa represents the mean absolute peak variation from mean peak amplitudes within each sample period. Although some perifusion columns of Clock-Δ19-transfected GT1-7 cells exhibited few detectable pulses, others displayed high-amplitude bursts lasting up to 25-30 min, followed by prolonged low-amplitude secretion occasionally near the level of assay detection (Fig. 7C). These variable pulse profiles could be attributable to differential transfection efficiency of Clock-Δ19 in certain columns, resulting in a more or less disrupted pulse rhythm. Interestingly, monolayer cultures of GT1-7 cells transfected with Clock-Δ19, as well as cells on Cytodex 3 beads kept in static incubation, secreted significantly more GnRH into the media over 24 hr (128.3 ± 1.05 pg/ml) than control-transfected cells (36.0 ± 5.99 pg/ml; data not shown). These results suggest that cell configuration and proximity may also play a role in constructing patterns of GnRH pulse release, such that pools of GnRH available for secretion may increase in dispersed cells transfected with Clock-Δ19. These data further demonstrate that a relatively disorganized pulse pattern does not preclude GnRH secretion altogether but rather affects only the mode of release. Interestingly, time of day relative to collection start time had no effect on GnRH pulse profiles of either untreated or transfected GT1-7 cells (data not shown). However, GnRH secretion could not be detected until at least 3-5 hr after column loading, demonstrating that the initiation of pulsatile GnRH release after exposure to perifused KRB is not immediate.
Figure 6.
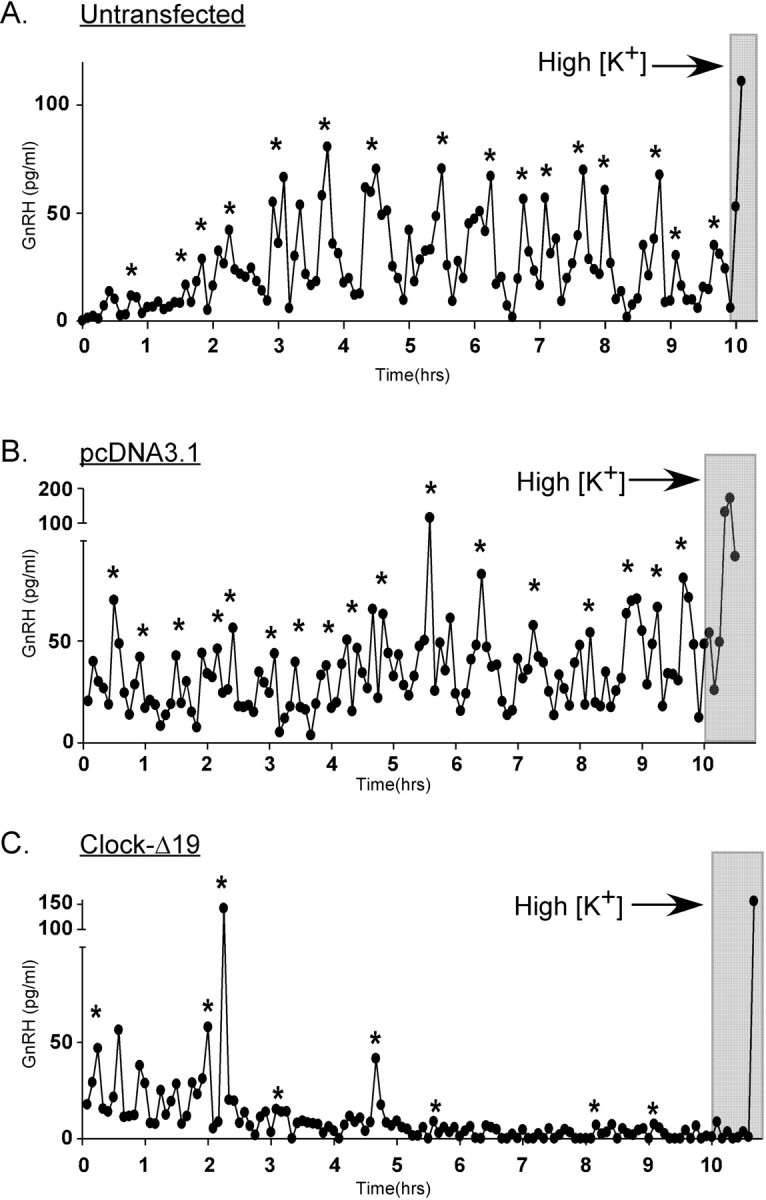
Perturbation of the circadian clock by expression of Clock-Δ19 disrupts patterns of GnRH pulse release from perifused GT1-7 cells. A-C, Representative GnRH pulse release profiles from perifused GT1-7 cells that were untransfected (A), transfected with the control vector pcDNA3.1 (B), or transfected with Clock-Δ19 (C). Significant pulses are determined by CLUSTER and PeakFit pulse analysis programs and are indicated by asterisks. Depolarization is indicated by the gray bars.
Figure 7.
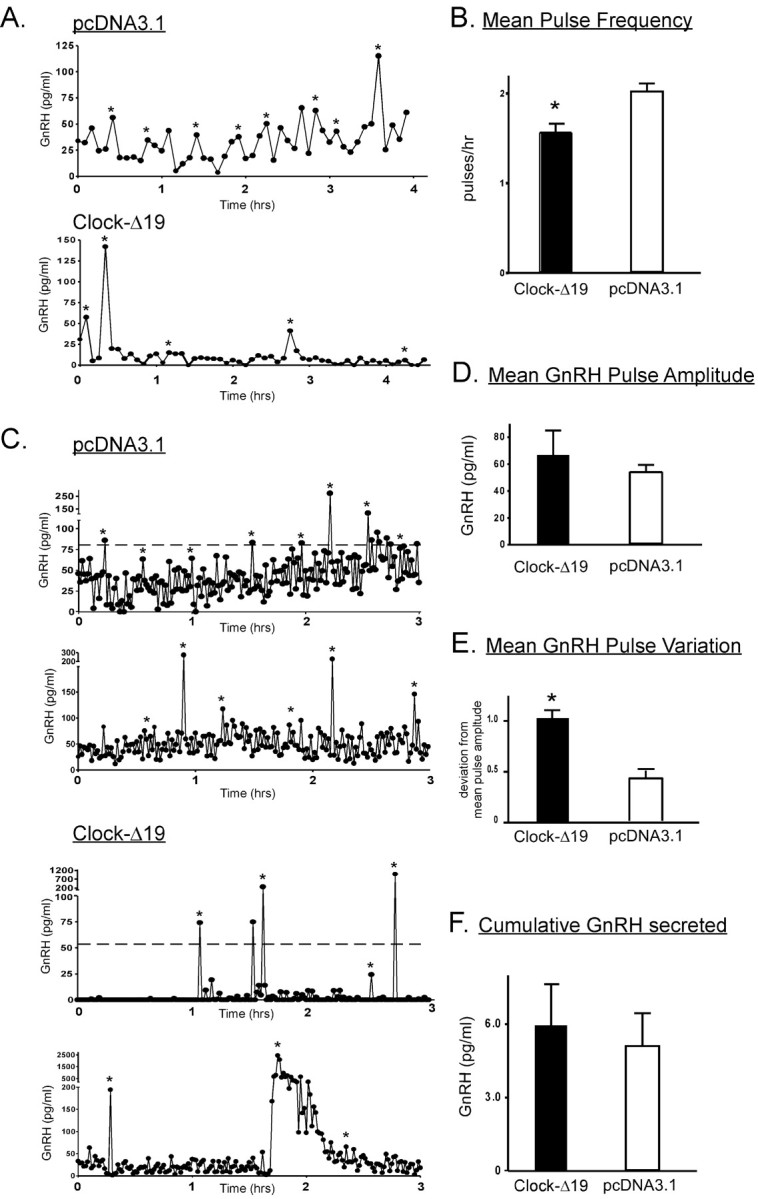
Expression of Clock-Δ19 results in alteration of GnRH pulsatile secretory patterns. A, Representative 5 min GnRH pulse profiles of perifused pcDNA3.1-transfected (top) and Clock-Δ19-transfected (bottom) GT1-7 cells. B, Mean pulse frequency of all sample periods was significantly (*p < 0.01) reduced in perifused Clock-Δ19-transfected cells. C, Representative 1 min GnRH pulse profiles of pcDNA3.1-transfected cells and Clock-Δ19-transfected GT1-7 cells. D, Mean pulse amplitude of Clock-Δ19-transfected cells, although highly variable, was not significantly different from control-transfected cells. Mean pulse amplitude variability, calculated as mean variation from normalized mean pulse amplitude within each 10 hr sampling period, is shown in E and was significantly (*p < 0.01) different between treatment groups. The bar graph in F indicates cumulative GnRH released over the 10 hr sampling period. Although control cells exhibited a stable baseline of release (top), Clock-Δ19-transfected cells displayed high-amplitude bursts of secretion, followed by a return to baseline. Bar graphs in B, D, E, and F represent mean pulse frequency, pulse amplitude, amplitude variability, and cumulative GnRH secretion, respectively, with n = 8 in each experimental and control group, and were derived from both 5 and 1 min sampling data.
To determine whether manipulation of another circadian clock gene could also affect GnRH secretion patterns, GT1-7 cells on beads were transfected with a vector expressing mCry1 under the control of the CMV promoter. As observed in Figure 4, this has the effect of diminishing baseline mPer1 promoter activity, demonstrating that Cry1 maintains a similar functional role in the core molecular clock loop in GT1-7 cells to that observed in the SCN. Strikingly, as demonstrated in Figure 8, GT1-7 cells overexpressing mCry1 exhibit significantly (*p < 0.01) higher mean pulse amplitudes (77.6 ± 3.6 pg/ml) in comparison with cells transfected with the control vector (33.5 ± 3.1 pg/ml), without an appreciable difference in mean pulse frequency. Because mCry1 levels oscillate in a circadian manner in many tissues in vivo, these results suggest that normal circadian increases in mCry1 protein in GnRH neurons could potentially stimulate cellular neurosecretory mechanisms, possibly linking circadian clock cycling to GnRH surge secretion.
Figure 8.
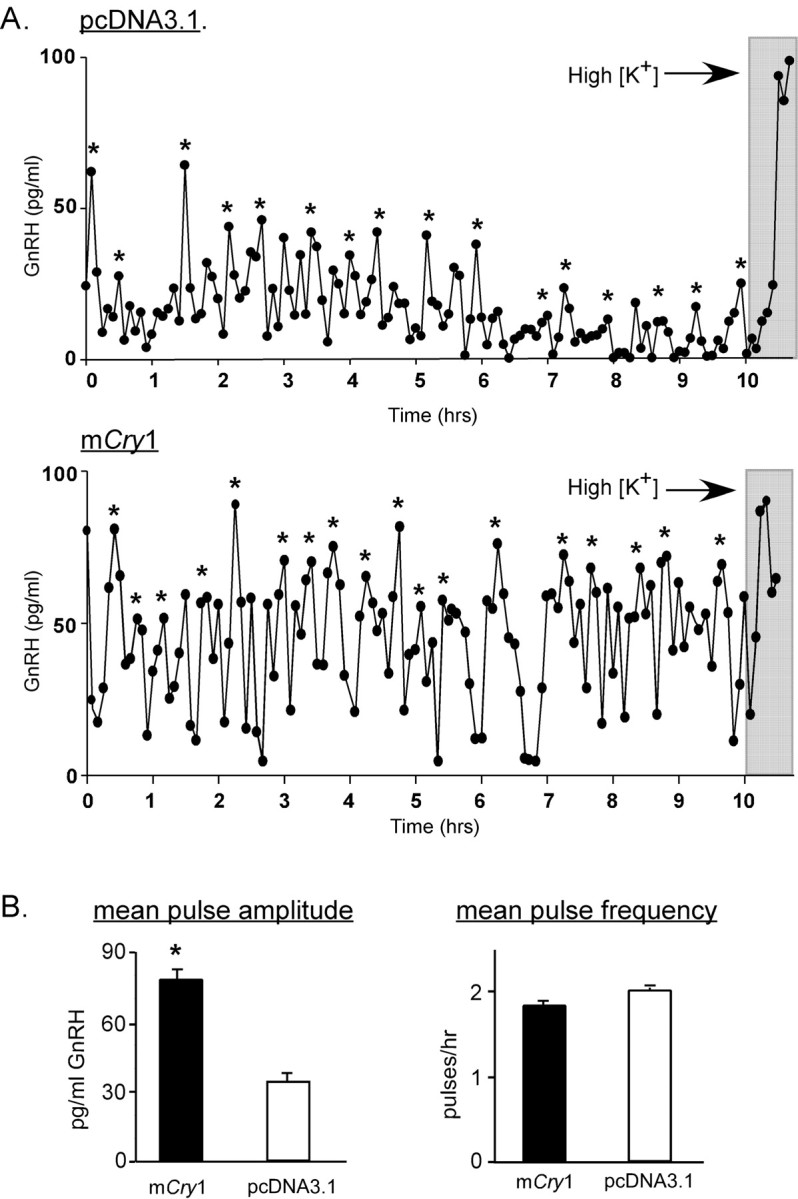
Overexpression of mCry1 in GT1-7 cells results in a significant increase in mean GnRH pulse amplitude. A, Representative GnRH pulse profiles of perifused GT1-7 cells transiently tranfected with a pcDNA3.1 (top) or a Cry1 expression vector (bottom). B, Mean pulse amplitude of Cry1-transfected GT1-7 cells is significantly (*p < 0.01; n = 6) greater than that of control (pcDNA3.1)- transfected cells (left), whereas pulse frequency is not different between Cry1-transfected or pcDNA-transfected cells (right). Gray bars indicate depolarization.
Clock gene regulation of reproductive function in vivo
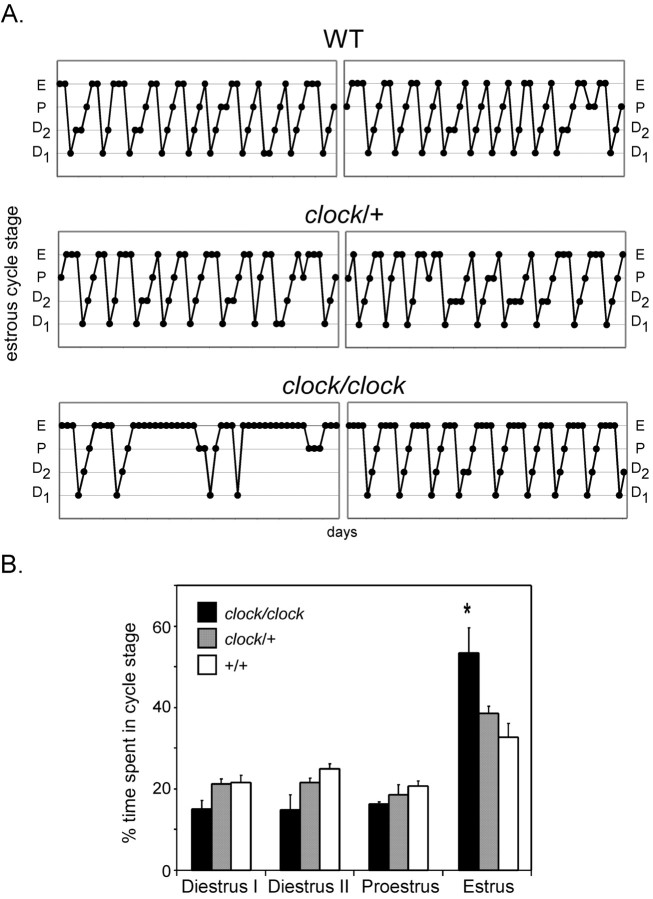
Mice harboring a mutation in the Clock gene (clock/clock mice) exhibit dramatic defects in circadian locomotor rhythms in free-running DD conditions but are able to entrain to a standard LD cycle. These mice also show blunted expression rhythms of many molecular clock components, including Per, Cry, and Bmal1 (Kume et al., 1999). Interestingly, homozygotes also demonstrate smaller litter size and lower frequency of successful matings (Table 1). To determine whether reproductive status is fundamentally altered in these animals, we examined daily vaginal cytology from female clock/clock mice, as well as from their heterozygous (clock/+) and wild-type (+/+) littermates (Fig. 9) ranging in age from 2 to 5 months. Interestingly, whereas wild-type mice demonstrated typical 4 to 5 d estrous cycles, clock/clock mice exhibited prolonged estrous cycles averaging ∼8 d, with a characteristic predominance in the estrus phase. This effect persisted in mice exposed to either LD or DD (data not shown) conditions and occurred irrespective of age. These data indicate that whereas locomotor rhythmicity is uncompromised in clock/clock mice in LD, reproductive function is diminished even in the presence of an entraining light cycle. Although it is likely that a somatic clock mutation may affect multiple sites within the hypothalamic-pituitary-gonadal axis, the above results suggest the possibility that the circadian oscillator may normally be responsible for facilitating transitions between estrous cycle phases, providing an intrinsic signal for the rapid modulation of ultradian GnRH secretion during the infradian oscillations of gonadal hormone secretion in females. In sum, the results of the above studies demonstrate that circadian clock function is linked to the secretory machinery governing timed GnRH pulse release from GT1-7 cells in culture, a link that appears to be preserved in vivo.
Table 1.
Homozygous (clock/clock) and heterozygous (clock/+) male and female mutant mice (n = 4 breeding pairs/combination) exhibit reduced fertility rates compared with breeding pairs of wild-type mice
|
Male |
|
Female |
Breeding success rate |
|---|---|---|---|
| WT | X | WT | 100% |
| clock/+ | X | WT | 60% |
| clock/clock | X | WT | 20% |
| WT | X | clock/+ | 50% |
| clock/+ | X | clock/+ | 50% |
| clock/clock | X | clock/+ | 25% |
| WT | X | clock/clock | 50% |
| clock/+ | X | clock/clock | 0% |
| clock/clock
|
X
|
clock/clock
|
25%
|
Wild-type and mutant littermates, 2-4 months of age, were bred in 12 hr LD cycles. Breeding success rate among genotypic pairings is shown at the far right, indicating the percentage of females bred that gave birth to litters of live pups. Also included in this measure were females carrying pregnancies to term without entering labor, as confirmed postmortem. When possible, females of each genotype were paired with differing representative males from each genotype, such that success rates are indicative of multiple attempts. WT, Wild-type mice.
Figure 9.
Clock/clock mutant mice exhibit prolonged estrous cycles. A, Representative estrous cycles as measured by vaginal cytology from 2- to 5-month-old female wild-type (top), clock/+ (middle), and clock/clock (bottom) mice. D1, Diestrus-I phase; D2, diestrus-II; P, proestrus; E, estrus. B, Time spent in each phase of the estrous cycle by genotype, represented as percentage of total time. Clock/clock mice spend a significantly greater (*p < 0.05; one-way ANOVA) amount of time in estrus than do the other genotypes. Data shown represent mice housed in 12 hr LD conditions (n = 6 per genotype).
Discussion
Recent studies indicate that almost all cells contain molecular clocks that can be synchronized to master clocks residing in the SCN, such that circadian signals can be effectively distributed to multiple tissues (Balsalobre, 2002). Our results demonstrate that GnRH neurons are no exception in this regard, and that circadian clock genes and protein products oscillate in cultured GT1-7 cells after exposure to various stimuli, suggesting a functionality that may impact reproductive rhythms. In addition, our data show that the molecular clock may be coupled to the fundamental mechanism of pulsatile GnRH secretion, because perturbation of normal clock oscillation by transfection of Clock-Δ19 disrupts pulsatile patterns of GnRH secretion.
Because Clock-Δ19 binds in a heterodimeric manner to Bmal1 and will also bind DNA but not activate mPer and mCry transcription (Gekakis et al., 1998; Jung et al., 2003), transient overexpression of this dominant-negative protein decreases transcription of Per and Cry, resulting in a subsequent blunting of clock gene oscillations. Results from the above experiments suggest that cycling levels of Per and Cry in GnRH neurons are required for normally observed patterns of GnRH release. Prevention of these oscillations by expression of Clock-Δ19 leads to an apparent disorganization of GnRH secretion, dramatically slowing pulse frequency and increasing pulse amplitude variability. It is of considerable interest that GnRH pulse patterns were disrupted, although only a percentage of GT1-7 cells expressed Clock-Δ19, suggesting that synchronous secretion of GnRH relies on homogeneity of gene expression within these neurons. The pattern of circadian gene expression in GnRH neurons in vivo has not yet been explored; thus, it remains unknown whether stimuli sufficient to reset the clock in these cells is also responsible for maintenance of secretory pulsatility. In fibroblasts and neuronal cell lines, including the GT1-7 cells, a media change is enough to elicit gene expression cycling, suggesting that shifting cultured GT1-7 cells into KRB perifusion buffer may act to not only reset the clock but also to synchronize pulsatile GnRH secretion.
Interestingly, augmentation of the “negative limb” of the clock (i.e., overexpression of mCry1) significantly increases GnRH pulse amplitude, suggesting that multiple secretory mechanisms may be influenced by intrinsic clock function. The observed increase in GnRH pulse amplitude suggests that the Cry proteins, in addition to their characterized role in the core clock loop, may also interact as promoters of genes required for modulation of neurosecretion. Consistent with this observation, the disruptive effect of Clock-Δ19 may act by inhibiting transcription of Cry. Increases in GnRH pulse amplitude resulting from overexpression of mCry1 suggest that this central clock component is linked to the GnRH secretory machinery in a way that selectively amplifies secreted GnRH. Cry1 and Cry2 are potent inhibitors of Clock/Bmal1-mediated transactivation (Kume et al., 1999), implying that a constituitive inhibition on GnRH release mediated by the positive limb of the clock may exist. These data also raise the interesting possibility that basal GnRH secretion may be relatively dynamic in vivo, rising slightly in the afternoon or evening in conjunction with peak Cry1 protein levels, consistent with the observation of diurnal changes in LH secretion in vivo (Sisk et al., 2001). The presence of an intrinsic circadian clock within GnRH neurons could thus provide insight into mechanisms mediating the observed circadian window of the LH and precedent GnRH surges. However, at what level the circadian clock exerts control over the secretory process remains unclear, and it is indeed possible that the clock may modulate multiple regulatory pathways at the level of GnRH transcription or by mediating neurosecretion at the cell membrane.
Circadian clock proteins have been shown to regulate transcription of other cycling genes often referred to as “clock-controlled genes” in a circadian manner (Jin et al., 1999). Previous studies demonstrate that Clock/Bmal1 heterodimers bind to E-box motifs on proximal promoter sites of target genes to activate transcription (Gekakis et al., 1998; Jung et al., 2003). Increases in transcription are subsequently inhibited by “negative limb” clock components such as the Period and Cryptochrome proteins that complex with Clock/Bmal1 to prevent transcriptional stimulation. The rat and mouse GnRH proximal promoter regions contain a few consensus E-boxes (CANNTG) but no canonical sites (CACGTGA) found in the promoters of many cycling genes (Panda et al., 2002). In vivo studies in both male and female rats have shown that levels of GnRH mRNA, but not primary transcript, exhibit circadian oscillations (Gore, 1998), suggesting a possible post-transcriptional regulation of GnRH content by the circadian clock. Future studies in GT1-7 cells should provide insight into whether this observed in vivo rhythm may be mediated by an endogenous circadian oscillator within GnRH neurons.
Alternately, perturbations of clock cycling could affect multiple regulatory pathways affecting secretion, such that GnRH transcription may not be directly influenced. Microarray studies performed on mRNA from clock/clock mutant mice demonstrate that, whereas most cycling gene expression patterns are disrupted in these animals, many noncycling genes are also either dramatically upregulated or downregulated by this mutation (Panda et al., 2002). Thus, expression of the dominant-negative Clock gene may alter GnRH secretion patterns via changes in membrane protein levels or at the level of GnRH transcription or post-transcriptional processing in addition to alterations of the core clock mechanism.
In support of this, GnRH secretion patterns, although undoubtedly influenced by the availability of processed decapeptide, are not likely mediated by transcriptional changes alone. A recent study of perifused GT1-1 cells treated with transcriptional and translational inhibitors suggests that rapid transcriptional and translational cycles do not underlie GnRH pulsatility (Pitts et al., 2001). However, perifusion after inhibitor treatment by necessity only lasted for 2 hr; thus it remains unknown in this context whether long-term inhibition of the circadian clock transcriptional loop by blockade of protein synthesis would ultimately be reflected in GnRH pulse release. The current study used perifusion periods of up to 24 hr and collection periods of 10 hr, with clock gene overexpression revealing striking differences in GnRH pulse parameters.
Many studies have focused on the unique electrophysiological properties of GnRH neurons that may be required to elicit synchronized release patterns. Recent work investigating activity rhythms of GnRH neurons in slice preparations revealed a complex pattern of rapid conductance oscillations specific to GnRH neurons (Nunemaker et al., 2001, 2003), suggesting that these particular membrane properties may govern episodic release patterns. In a recent study, the same experimenters observed multiple activity rhythms in GnRH neurons spanning milliseconds to minutes and demonstrated that only certain rhythmic parameters may be affected by exogenous signals (Nunemaker et al., 2003). Thus, there may exist convergent neurosecretory mechanisms that are coupled at various cellular context-dependent strengths to circadian, infradian, and ultradian generators within the same neuron or population of neurons, such that disruption of one of these could lead to arrhythmicity in multiple time domains.
Neuronal firing patterns also play important roles in generating synchronized output rhythms in the SCN. Recent studies in Drosophila demonstrated that functioning ion channels are required for oscillation of the molecular circadian clock (Nitabach et al., 2002), suggesting a fundamental interaction between membrane electrical activity and clock oscillation. GnRH neuronal activity and secretion are also intimately linked, because rapid voltage-sensitive Ca2+ oscillations and K+-mediated action potentials are required for normal pulse release patterns (Charles and Hales, 1995; Costantin and Charles, 1999, 2001). Primary neuronal SCN cultures from wild-type mice, grown on micromultielectrode plates, reveal distinct circadian activity patterns, whereas SCN neurons from clock/clock mice exhibit arrhythmic firing patterns, suggesting that the transcriptional clock is linked to control of neuronal membrane potential (Herzog et al., 1998). We recreated this mutation effect specifically in GT1-7 cells in culture via Clock-Δ19 overexpression, raising the possibility that GnRH secretory machinery may similarly be affected by this dominant-negative Clock protein, such that a normal complement of membrane channels, or modification thereof, is absent, thereby perturbing typical release patterns.
The influence of the circadian clock on the reproductive axis can also be observed in mutant mice. Estrous cycle analysis of clock/clock mutant mice provides insight into their subfertile condition. The presence of persistent estrus smears lasting up to four times longer than typically observed suggests that these animals do not immediately resume cycling after ovulation. The somatic Clock gene mutation likely affects multiple points within the reproductive axis, including a possible disruption of GnRH secretion. Previous rodent work demonstrated that LH secretion, clearly pulsatile in metestrus and diestrus and surging on proestrus, becomes quiescent on estrus, only to resume pulsatile release again on diestrus (Fox and Smith, 1985). Observation of clock/clock mice, in conjunction with the above results from GT1-7 cells, suggests that a functional circadian clock may be required for the resumption of synchronous GnRH secretion after the GnRH surge. Whether this could result from a lack of synchronized activity in the SCN (thus perturbing subsequent output from) or in GnRH neurons themselves is unclear and will require additional studies to determine the extent of the reproductive deficiency of these mice. A previous study revealed a significant decrease in both arginine vasopressin and vasoactive intestinal peptide within the SCN of clock/clock mice throughout postnatal development (Herzog et al., 2000). Whether this decrement alters GnRH expression or secretion in vivo is currently unknown. It is of interest that, in contrast to locomotor activity, reproductive abnormalities in clock/clock mutant mice persist in both LD and DD conditions, indicating that endogenous circadian clock disruption in GnRH neurons themselves may produce this observed reproductive deficiency.
Although it remains unclear what effect resetting of the circadian clock may have on fibroblasts, the GT1-7 cell line provides a neuroendocrine model that produces a distinctly timed output, a secretory pattern that can be modulated by both neuronal and hormonal signals. Indeed, activity of GnRH neuronal perikarya, not confined to discrete nuclei, may be modulated in a synchronous manner by humoral signals. A recent study using SCN2.2 cells, an immortalized line derived from rat SCN, demonstrates that circadian rhythms of gene expression and glucose utilization in NIH3T3 fibroblasts can be driven by the SCN 2.2 cells in the absence of synaptic contacts (Allen et al., 2001). These results demonstrate that an uncharacterized humoral factor secreted by the SCN can synchronize other cell types in a paracrine manner. Also, hormonal stimuli shown to modulate the reproductive axis such as retinoic acid (Cho et al., 1998) and glucocorticoids (DeFranco et al., 1994) are also capable of exerting effects on the phase and amplitude of the circadian clock (Balsalobre et al., 2000a; McNamara et al., 2001). In contrast to fibroblast lines, GT1 cell lines provide a valuable model in which a rhythmic neurosecretory output, albeit ultradian, can be measured after molecular alterations of a circadian transcriptional loop.
In sum, the results of the current study demonstrate that an endogenous circadian clock plays a role in regulating parameters of GnRH pulsatility. Although future studies are required to determine the extent and mode of this regulation, these data provide insight into the fundamental mechanisms underlying GnRH pulsatility, as well as presenting a direct influence of the circadian clock on primary neuronal components of the reproductive axis.
Footnotes
This work was supported in part by National Institutes of Health Grants R01 DK44838, T32 HD07203, and F32 DK10100. We thank Jeff Wong, Mona Janfaza, Bridgette Kirkpatrick, Armando Arroyo, and Nichol Goodman for technical assistance, and Mark Lawson and Varykina Thackray for critical analysis of this manuscript.
Correspondence should be addressed to Dr. Pamela L. Mellon, Department of Reproductive Medicine 0674, 2057 Cellular and Molecular Medicine East, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0674. E-mail: pmellon@ucsd.edu.
Copyright © 2003 Society for Neuroscience 0270-6474/03/2311202-12$15.00/0
References
- Allen G, Rappe J, Earnest DJ, Cassone VM ( 2001) Oscillating on borrowed time: diffusible signals from immortalized suprachiasmatic nucleus cells regulate circadian rhythmicity in cultured fibroblasts. J Neurosci 21: 7937-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, Earnest DJ ( 2002) Real-time analysis of rhythmic gene expression in immortalized suprachiasmatic nucleus cells. NeuroReport 13: 2027-2030. [DOI] [PubMed] [Google Scholar]
- Balsalobre A ( 2002) Clock genes in mammalian peripheral tissues. Cell Tissue Res 309: 193-199. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U ( 1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929-937. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U ( 2000a) Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol 10: 1291-1294. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U ( 2000b) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344-2347. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM ( 2002) Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster Proc Natl Acad Sci USA 99: 2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann DW, Mahesh VB ( 1994) Excitatory amino acids: function and significance in reproduction and neuroendocrine regulation. Front Neuroendocrinol 15: 3-49. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA ( 2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles AC, Hales TG ( 1995) Mechanisms of spontaneous calcium oscillations and action potentials in immortalized hypothalamic (GT1-7) neurons. J Neurophysiol 73: 56-64. [DOI] [PubMed] [Google Scholar]
- Cho S, Cho H, Geum D, Kim K ( 1998) Retinoic acid regulates gonadotropin-releasing hormone (GnRH) release and gene expression in the rat hypothalamic fragments and GT1-1 neuronal cells in vitro Brain Res Mol Brain Res 54: 74-84. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N ( 1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156-159. [DOI] [PubMed] [Google Scholar]
- Clarke I, Moore L, Veldhuis J ( 2002) Intensive direct cavernous sinus sampling identifies high-frequency, nearly random patterns of FSH secretion in ovariectomized ewes: combined appraisal by RIA and bioassay. Endocrinology 143: 117-129. [DOI] [PubMed] [Google Scholar]
- Costantin JL, Charles AC ( 1999) Spontaneous action potentials initiate rhythmic intercellular calcium waves in immortalized hypothalamic (GT1-1) neurons. J Neurophysiol 82: 429-435. [DOI] [PubMed] [Google Scholar]
- Costantin JL, Charles AC ( 2001) Modulation of Ca(2+) signaling by K(+) channels in a hypothalamic neuronal cell line (GT1-1). J Neurophysiol 85: 295-304. [DOI] [PubMed] [Google Scholar]
- DeFranco DB, Attardi B, Chandran UR ( 1994) Glucocorticoid receptor-mediated repression of GnRH gene expression in a hypothalamic GnRH-secreting neuronal cell line. Ann NY Acad Sci 746: 473-475. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Mellon PL ( 1995) Regulation of GnRH transcription by protein kinase C is mediated by evolutionarily conserved, promoter-proximal elements. Mol Endocrinol 9: 848-859. [DOI] [PubMed] [Google Scholar]
- Fox SR, Smith MS ( 1985) Changes in the pulsatile pattern of luteinizing hormone secretion during the rat estrous cycle. Endocrinology 116: 1485-1492. [DOI] [PubMed] [Google Scholar]
- Funabashi T, Shinohara K, Mitsushima D, Kimura F ( 2000) Gonadotropin-releasing hormone exhibits circadian rhythm in phase with arginine-vasopressin in co-cultures of the female rat preoptic area and suprachiasmatic nucleus. J Neuroendocrinol 12: 521-528. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ ( 1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564-1569. [DOI] [PubMed] [Google Scholar]
- Gore AC ( 1998) Diurnal rhythmicity of gonadotropin-releasing hormone gene expression in the rat. Neuroendocrinology 68: 257-263. [DOI] [PubMed] [Google Scholar]
- Gray GD, Soderstein P, Tallentire D, Davidson JM ( 1978) Effects of lesions in various structures of the suprachiasmatic-preoptic region on LH regulation and sexual behavior in female rats. Neuroendocrinology 25: 174-191. [DOI] [PubMed] [Google Scholar]
- Herbison AE ( 1997) Noradrenergic regulation of cyclic GnRH secretion. Rev Reprod 2: 1-6. [DOI] [PubMed] [Google Scholar]
- Herbison AE, Dyer RG ( 1991) Effect on luteinizing hormone secretion of GABA receptor modulation in the medial preoptic area at the time of the proestrous luteinizing hormone surge. Neuroendocrinology 53: 317-320. [DOI] [PubMed] [Google Scholar]
- Herbison AE, Heavens RF, Dyer RG ( 1989) Microdialysis of rat preoptic area: evidence that noradrenaline (NA) stimulates release of GABA. J Neurosci Methods 29: 290. [Google Scholar]
- Herzog ED, Takahashi JS, Block GD ( 1998) Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci 1: 708-713. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Grace MS, Harrer C, Williamson J, Shinohara K, Block GD ( 2000) The role of Clock in the developmental expression of neuropeptides in the suprachiasmatic nucleus. J Comp Neurol 424: 86-98. [DOI] [PubMed] [Google Scholar]
- Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y ( 2002) Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured rat-1 fibroblasts. J Biol Chem 277: 44244-44251. [DOI] [PubMed] [Google Scholar]
- Honma S, Ikeda M, Abe H, Tanahashi Y, Namihara M, Honma K, Nomura M ( 1998) Circadian oscillation of BMAL1, a partner of a mammalian clock gene clock, in rat suprachiasmatic nucleus. Biochem Biophys Res Comm 250: 83-87. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, De Vries GJ, Reppert SM ( 1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96: 57-68. [DOI] [PubMed] [Google Scholar]
- Jung H, Choe Y, Kim H, Park N, Son GH, Khang I, Kim K ( 2003) Involvement of CLOCK:BMAL1 heterodimer in serum-responsive mPer1 induction. NeuroReport 14: 15-19. [DOI] [PubMed] [Google Scholar]
- Krsmanovic LZ, Stojilkovic SS, Merelli F, Dufour SM, Virmani MA, Catt KJ ( 1992) Calcium signaling and episodic secretion of gonadotropin-releasing hormone in hypothalamic neurons. Proc Natl Acad Sci USA 89: 8462-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM ( 1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193-205. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM ( 2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855-867. [DOI] [PubMed] [Google Scholar]
- Legan SJ, Karsch FJ ( 1975) A daily signal for the LH surge in the rat. Endocrinology 96: 57-62. [DOI] [PubMed] [Google Scholar]
- Legan SJ, Coon GA, Karsch FJ ( 1975) Role of estrogen as initiator of daily LH surges in the ovariectomized rat. Endocrinology 96: 50-56. [DOI] [PubMed] [Google Scholar]
- Loudon ASI, Wayne NL, Krieg R, Iranmanesh A, Veldhuis JD, Menaker M ( 1994) Ultradian endocrine rhythms are altered by a circadian mutation in the syrian hamster. Endocrinology 135: 712-718. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS ( 2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288: 483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de la Escalera G, Choi ALH, Weiner RI ( 1992) Generation and synchronization of gonadotropin-releasing hormone (GnRH) pulses: intrinsic properties of the GT1-1 GnRH neuronal cell line. Proc Natl Acad Sci USA 89: 1852-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA ( 2001) Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell 105: 877-889. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC ( 2002) Electrical silencing of drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485-495. [DOI] [PubMed] [Google Scholar]
- Nunemaker CS, DeFazio RA, Geusz ME, Herzog ED, Pitts GR, Moenter SM ( 2001) Long-term recordings of networks of immortalized GnRH neurons reveal episodic patterns of electrical activity. J Neurophys 86: 86-93. [DOI] [PubMed] [Google Scholar]
- Nunemaker CS, Straume M, DeFazio RA, Moenter SM ( 2003) Gonadotropin-releasing hormone neurons generate interacting rhythms in multiple time domains. Endocrinology 144: 823-831. [DOI] [PubMed] [Google Scholar]
- Olcese J, Domagalski R, Bednorz A, Weaver DR, Urbanski HF, Reuss S, Middendorff R ( 2003) Expression and regulation of mPer1 in immortalized GnRH neurons. NeuroReport 14: 613-618. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB ( 2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307-320. [DOI] [PubMed] [Google Scholar]
- Pitts GR, Nunemaker CS, Moenter SM ( 2001) Cycles of transcription and translation do not comprise the gonadotropin-releasing hormone pulse generator in GT1 cells. Endocrinology 142: 1858-1864. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U ( 2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251-260. [DOI] [PubMed] [Google Scholar]
- Sisk CL, Richardson HN, Chappell PE, Levine JE ( 2001) In vivo gonadotropin-releasing hormone secretion in female rats during peripubertal development and on proestrus. Endocrinology 142: 2929-2936. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Keen KL, Mogi K, Claude P ( 1999) Pulsatile release of luteinizing hormone-releasing hormone (LHRH) in cultured LHRH neurons derived from the embryonic olfactory placode of the rhesus monkey. Endocrinology 140: 1432-1441. [DOI] [PubMed] [Google Scholar]
- van der Beek EM, Wiegant VM, van der Donk HA, van den Hurk R, Buijs RM ( 1993) Lesions of the suprachiasmatic nucleus indicate the presence of a direct vasoactive intestinal polypeptide-containing projection to gonadotrophin-releasing hormone neurons in the female rat. J Neuroendocrinol 5: 137-144. [DOI] [PubMed] [Google Scholar]
- van der Beek EM, van Oudheusden HJ, Buijs RM, van der Donk HA, van den Hurk R, Wiegant VM ( 1994) Preferential induction of c-fos immunoreactivity in vasoactive intestinal polypeptide-innervated gonadotropin-releasing hormone neurons during a steroid-induced luteinizing hormone surge in the female rat. Endocrinology 134: 2636-2644. [DOI] [PubMed] [Google Scholar]
- van der Beek EM, Horvath TL, Wiegant VM, van den Hurk R, Buijs RM ( 1997a) Evidence for a direct neuronal pathway from the suprachiasmatic nucleus to the gonadotropin-releasing hormone system: combined tracing and light and electron microscopic immunocytochemical studies. J Comp Neurol 384: 569-579. [DOI] [PubMed] [Google Scholar]
- van der Beek EM, Wiegant VM, van Oudheusden HJ, van der Donk HA, van den Hurk R, Buijs RM ( 1997b) Synaptic contacts between gonadotropin-releasing hormone-containing fibers and neurons in the suprachiasmatic nucleus and perichiasmatic area: an anatomical substrate for feedback regulation? Brain Res 755: 101-111. [DOI] [PubMed] [Google Scholar]
- Veldhuis JD, Johnson ML ( 1986) Cluster analysis: a simple, versatile and robust algorithm for endocrine pulse detection. Am J Physiol 250: E486-E490. [DOI] [PubMed] [Google Scholar]
- Vitalis EA, Costantin JL, Tsai PS, Sakakibara H, Paruthiyil S, Iiri T, Martini JF, Taga M, Choi AL, Charles AC, Weiner RI ( 2000) Role of the cAMP signaling pathway in the regulation of gonadotropin-releasing hormone secretion in GT1 cells. Proc Natl Acad Sci USA 97: 1861-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner RI, Martinez de la Escalera G ( 1993) Pulsatile release of gonadotrophin releasing hormone (GnRH) is an intrinsic property of GT1 GnRH neuronal cell lines. Hum Reprod 2: 13-17. [DOI] [PubMed] [Google Scholar]
- Wetsel WC, Valença MM, Merchenthaler I, Liposits Z, López FJ, Weiner RI, Mellon PL, Negro-Vilar A ( 1992) Intrinsic pulsatile secretory activity of immortalized LHRH secreting neurons. Proc Natl Acad Sci USA 89: 4149-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetsel WC, Eraly SA, Whyte DB, Mellon PL ( 1993) Regulation of gonadotropin-releasing hormone by protein kinases A and C in immortalized hypothalamic neurons. Endocrinology 132: 2360-2370. [DOI] [PubMed] [Google Scholar]
- Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliffe LA, Abe M, Block G, Spitznagel E, Menaker M, Takahashi JS ( 2002) Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo Proc Natl Acad Sci USA 99: 489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Okamura H ( 2000) Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett 465: 79-82. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, van Der Horst GT, Okamura H ( 2001) Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292: 278-281. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM ( 1998) Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20: 1103-1110. [DOI] [PubMed] [Google Scholar]