It is not well-understood how the human Mediator complex, TFIIH, and RNA polymerase II (pol II) work together with activators to initiate transcription. Activator binding alters Mediator structure, yet the functional consequences of such structural shifts remain unknown. The p53 C-terminus and its activation domain (p53AD) interact with different Mediator subunits and we find that each interaction differentially affects Mediator structure; strikingly, distinct p53–Mediator structures differentially affect pol II activity. Only p53AD induces formation of a large pocket domain at the Mediator-pol II interaction site, and this correlates with activation of stalled pol II to a productively-elongating state. Moreover, we define a Mediator requirement for TFIIH-dependent pol II CTD phosphorylation and identify substantial differences in pol II CTD processing that correspond to distinct p53–Mediator structural states. Our results define a fundamental mechanism by which p53 activates transcription and suggest Mediator structural shifts trigger activation of stalled pol II complexes.
Although every gene does not rely upon the same set of regulatory factors for its expression, a subset of general transcription factors—including Mediator and TFIIH—appear to be required to transcribe the majority of protein-coding genes1. Comparatively little is known about the Mediator co-activator complex, due in part to its more recent discovery in human cells2 and a lack of identifiable functional domains within its 26 subunits. Yet it is clear that Mediator is the primary regulator of the Pre-Elongation Complex (PEC, which includes TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Mediator and pol II, see Supplementary Information for description of this terminology), which in various forms is responsible for genome-wide transcriptional regulation of protein-coding genes. Indeed, Mediator physically or functionally interacts with most components of the 3.5 MDa PEC, including a tight association with pol II itself3–6. Thus, potential structural changes within the 1.2 MDa Mediator complex might significantly impact gene expression by altering PEC structure and function.
The p53 transcription factor has been identified as a key regulator of aging in mammals7,8 and is one of the most widely-studied proteins in cancer biology because of its strong tumor suppressor function9. Despite this, the molecular mechanisms by which p53 actually activates transcription are not well-defined. It is not clear how the mere presence of p53 at a promoter translates into gene activation, particularly since many p53 target genes display characteristics of genes poised for activation: their promoters contain pre-loaded pol II but remain inactive10. Thus, one mechanism by which p53 likely activates gene expression is via post-recruitment events that trigger pol II promoter clearance and transcription elongation. Precisely how p53 might control such post-recruitment events remains unclear, yet one plausible means involves Mediator, which appears to regulate post-recruitment events in Elk-1-dependent Egr1 gene activation11.
Past work has revealed p53 binds two different Mediator subunits: Med17 and Med1. The N-terminal activation domain of p53 (p53AD) interacts specifically with Med17, whereas a different domain within p53 binds directly to Med112,13. Interestingly, mutation of two residues in the p53AD (L22Q and W23S in human p53; a.k.a. p53QS) prevents expression of most p53 target genes and leads to tumor formation14–16; thus, p53AD is critical for the tumor suppressor function of p5317. As expected, these p53AD mutations (p53QS) prevent its interaction with Med17; however, the second, p53-Med1 interaction is unaffected by the p53QS mutations13, indicating that p53QS can still bind Mediator (via the Med1 subunit) yet is defective in activating transcription. This suggested the p53AD–Mediator interaction was playing an additional role in gene activation that did not involve Mediator recruitment.
Over the past 40 years, the factors required to initiate gene expression have been identified. A major unanswered question that remains is how these factors work together with transcription factors—such as p53—to activate gene expression. About 8 years ago it was discovered that activator binding could trigger significant structural shifts within the human Mediator complex18, suggesting a simple means to regulate its activity. Yet it was unclear whether such structural shifts might serve any regulatory function. Here we describe a structural and functional analysis of p53–Mediator. Our results suggest p53-induced structural shifts within Mediator are essential for regulating post-recruitment steps in gene activation. In particular, a specific p53AD–Mediator structural shift appears to coordinate activation of TFIIH and pol II within the PEC, whereas alternate Mediator structures maintain stalled pol II in an inactive state. The ability of p53 to indirectly control the activity of TFIIH and pol II via structural changes in Mediator offers fundamental insight into how p53 functions as an activator of transcription. Perhaps more significant, the structural shift linked to activation by p53–Mediator is also observed in other activator-Mediator structures, suggesting a unifying theme in transcription activation.
RESULTS
PEC assembly can occur independently of p53 in vitro or in cells
Although past studies have shown that p53AD binds directly to Med1713, the p53 domain responsible for interaction with Med1 was not known12. To better define this second, Med1-interacting domain within p53, we completed a series of experiments (see Supplementary Information and Supplementary Fig. 1–2) that mapped this domain to 31 residues within its C-terminus (residues 363–393: the p53CTD).
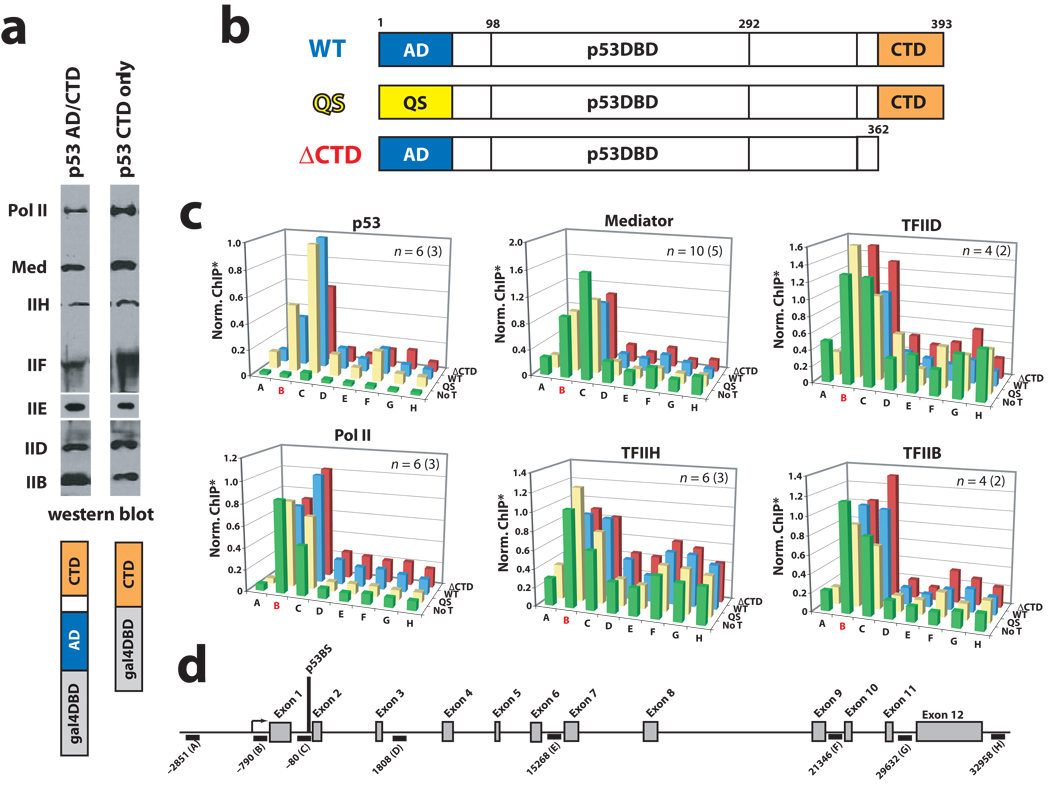
At 1.2 MDa, Mediator represents a major sub-assembly within the PEC. As an independent domain that interacts with Mediator, we examined whether the p53CTD alone might promote PEC assembly on a promoter template. We completed a series of immobilized template assays in which promoter occupancy was examined as a function of p53AD or p53CTD. These experiments were completed essentially as described19. Briefly, we incubated immobilized GAL4 DNA templates with either GAL4–p53CTD or with a GAL4–p53AD/CTD fusion protein, followed by addition of HeLa nuclear extract, which contains the full complement of PEC factors. After a series of washes, promoter-bound factors were detected by immunoblotting. As shown in Figure 1a, the occupancy of PEC components TFIIB, IID, IIE, IIF, IIH, pol II, and Mediator was similar in both experiments. In addition, using purified factors from a reconstituted transcription system20, we examined PEC assembly at the GAL4 promoter in the context of chromatin. Factor binding was assessed with a chromatin sedimentation assay, as described20. As expected, the p53CTD alone enabled assembly of the PEC on chromatin templates, as shown in Supplementary Figure 3. Chromatin sedimentation assays completed at the native HDM2 promoter with wild type and p53QS mutant p53 tetramers revealed similar results. That is, p53QS mutant tetramers, which possess a mutation within p53AD that prevents the p53AD–Med17 interaction but not the p53CTD–Med1 interaction, were also capable of PEC assembly, as was WT p53, on the native HDM2 promoter (data not shown). Thus, the chromatin sedimentation and immobilized template experiments suggested that PEC recruitment did not require p53AD. Importantly, these results were supported by ChIP data in HCT116 cells, as described below.
Figure 1.
The p53AD is not required for PEC assembly in vitro or in cells. (a) Immobilized template assays. Occupancy of PEC components in the presence of promoter-bound GAL4–p53CTD or a GAL4–p53AD/CTD fusion protein. Note that endogenous p53 will not bind the GAL4 promoter. Antibodies—pol II: Rpb1; Mediator: Med1; IIH: ERCC3; IIF: Rap74; IIE: IIEα; IID: TBP. (b) Schematic of WT p53 and mutant p53 proteins used in this study. (c) ChIP assays at HDM2 in p53-null HCT116 cells following transfection with wild-type p53 (WT, blue bars), p53 with a truncated CTD (residues 1–362; ΔCTD, red bars), or p53QS mutant (QS, yellow bars). Data from no transfection controls (No T, green bars) are also shown. The probe location in red (B) represents the promoter/transcription start site. Note that PEC factors appear to be pre-loaded at the promoter, as observed previously21. *ChIP output was normalized to WT p53, typically from C primer. For clarity, error bars are not shown but can be viewed in Supplementary Figure 5. (d) Schematic of the HDM2 locus, showing ChIP probe locations.
To further assess whether PEC occupancy might be dependent upon p53AD, we expressed WT p53, the p53QS mutant, or a p53ΔCTD mutant in p53-null HCT116 cells (Supplementary Fig. 4a). We then performed ChIP assays at the HDM2 gene to determine if differential promoter occupancy would be observed. Each p53 protein tested (WT p53, p53ΔCTD, p53QS, Fig. 1b) occupied the HDM2 promoter after transfection (Fig. 1c). As shown in Figure 1c, Mediator and pol II promoter occupancy was largely similar for each p53 protein examined (p53QS, WTp53, or p53ΔCTD). In fact, each complex appeared to be pre-loaded at the HDM2 gene, consistent with past reports21. Similarly, the general transcription factors TFIIH, TFIID, and TFIIB occupied the HDM2 promoter in the absence of p53 and also in the presence of wild-type or mutant p53 tetramers (Fig. 1c). ChIP assays at the p21 gene yielded similar results (Supplementary Fig. 4b–d). Thus, it appeared that in cells, as observed in vitro, PEC recruitment did not require WT p53.
p53AD–Mediator and post-recruitment activation of transcription
Past in vitro and in vivo studies have demonstrated that p53AD is critical for activating most p53 target genes14–16. Yet functional assays in yeast have suggested Mediator recruitment alone can trigger gene activation in vitro and in vivo22,23.
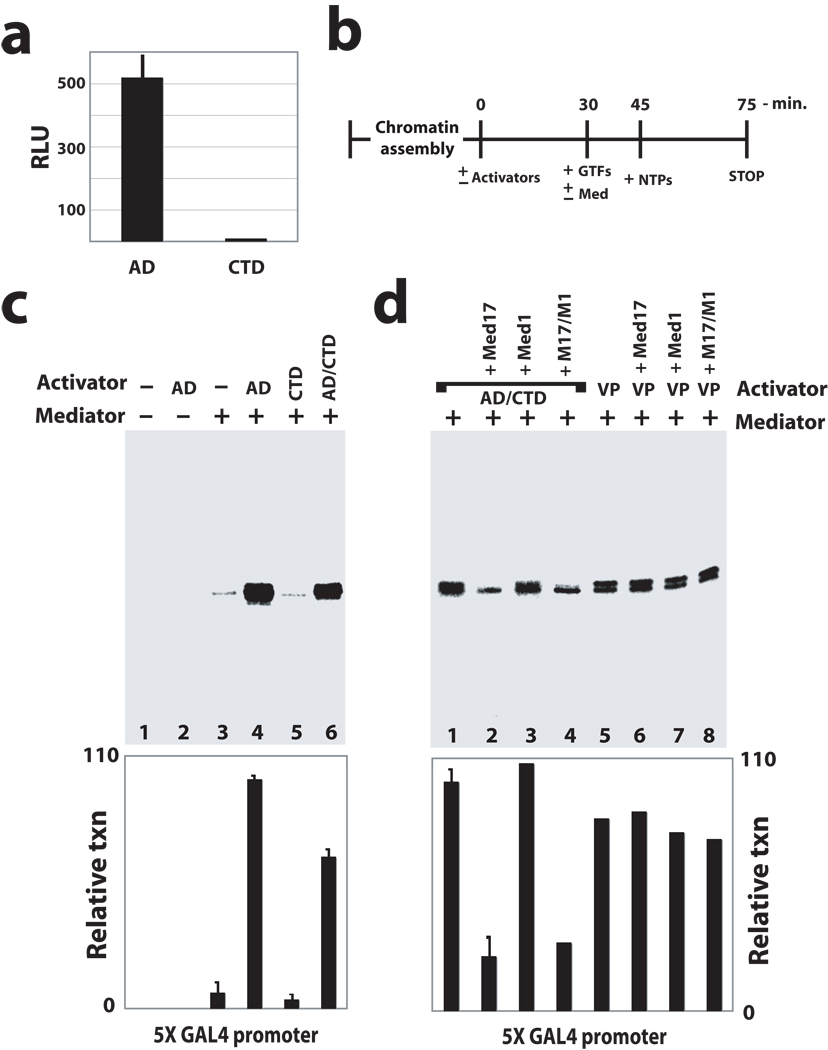
To identify whether p53CTD might functionally resemble p53AD as an activator of transcription, we compared the transcriptional activity of p53AD and p53CTD—two p53 domains capable of recruiting Mediator—in cell-based reporter assays. As shown in Figure 2a, p53AD strongly activated transcription as expected, whereas p53CTD was incapable of activating transcription. To probe this further, we utilized a chromatin-based reconstituted human transcription system (Fig. 2b) consisting of highly purified PEC components, including TFIIA, IIB, IID, IIE, IIF, IIH, pol II, and Mediator (ref. 20; see also Supplementary Fig. 6a–c). Note that Mediator was required for transcription in these assays (Fig. 2c, compare lanes 2 and 4). Strikingly, the p53CTD was incapable of activating transcription in these experiments. In fact, transcript levels with p53CTD matched those without an activator, whereas p53AD displayed potent activity in this assay (Fig. 2c, compare lanes 3–5). The inability of p53CTD to activate transcription was not due to a defect in recruiting Mediator or pol II to the chromatin template, as revealed by chromatin sedimentation assays, described above. Furthermore, primer extension products of just 85 bp are detected from the GAL4 template, indicating that pol II does not clear the promoter without p53AD. A GAL4–p53AD/CTD fusion protein was also capable of activating transcription, indicating the p53CTD does not negatively impact the activation function of p53AD in these assays (Fig. 2c, lane 6). The significance of the p53AD–Mediator interaction was further highlighted by experiments in which recombinant Med17, which will competitively block p53AD–Mediator binding, was able to repress p53AD-dependent transcription (Fig. 2d, compare lanes 1, 2, and 4). However, recombinant Med17 had no effect on transcription activated by VP16 (Fig. 2d, compare lanes 5, 6, and 8), which interacts with Mediator through the Med25 subunit24,25. Thus, Med17 itself is not a general repressor in this assay but can block activation dependent upon p53AD. Incidentally, recombinant Med1, which competes for the p53CTD-Med1 interaction, had little effect on transcript levels (Fig. 2d, compare lanes 1 and 3), suggesting that the p53CTD-Med1 interaction is not critical for transcription activation in this context. By contrast, recombinant Med1 was a potent repressor of VDR-dependent transcription (data not shown), since VDR binds Mediator via the Med1 subunit26.
Figure 2.
p53AD is required to activate stalled pol II in vitro and in cells; PECs assembled in the absence of p53 or with p53CTD remain inactive. (a) The p53CTD cannot activate transcription in cells. Luciferase reporter assays in HEK293 cells transfected with GAL4–p53AD or GAL4–p53CTD; results shown represent mean and standard error from three independent experiments. (b) Outline of reconstituted transcription assay. (c) Reconstituted transcription20 on a promoter containing tandem GAL4 sites; AD: GAL4–p53AD; CTD: GAL4–p53CTD; AD/CTD: GAL4–p53AD/CTD fusion protein. Representative data are shown; plot shows mean and s.e.m. from multiple experiments (n = 4, 10, 6, 4 for lanes 3–6). (d) Recombinant Med17, which blocks the p53AD–Mediator interaction, can block p53-dependent activation in vitro. Transcription is repressed upon addition of recombinant Med17 (lanes 2 and 4); however, Med17 had no detectable impact on VP16-dependent activation (lanes 5–8). VP16 interacts with Mediator through the Med25 subunit24,25. Quantitation of transcripts is shown in the bar graph, and bars represent the s.e.m. for lanes 1 and 2 (n = 10, 2, respectively). Each experiment shown in c and d contained equivalent amounts of chromatin template, general transcription factors (TFIIA, IIB, IID, IIE, IIF, IIH, and pol II), and NTPs. Activators and Mediator added as shown.
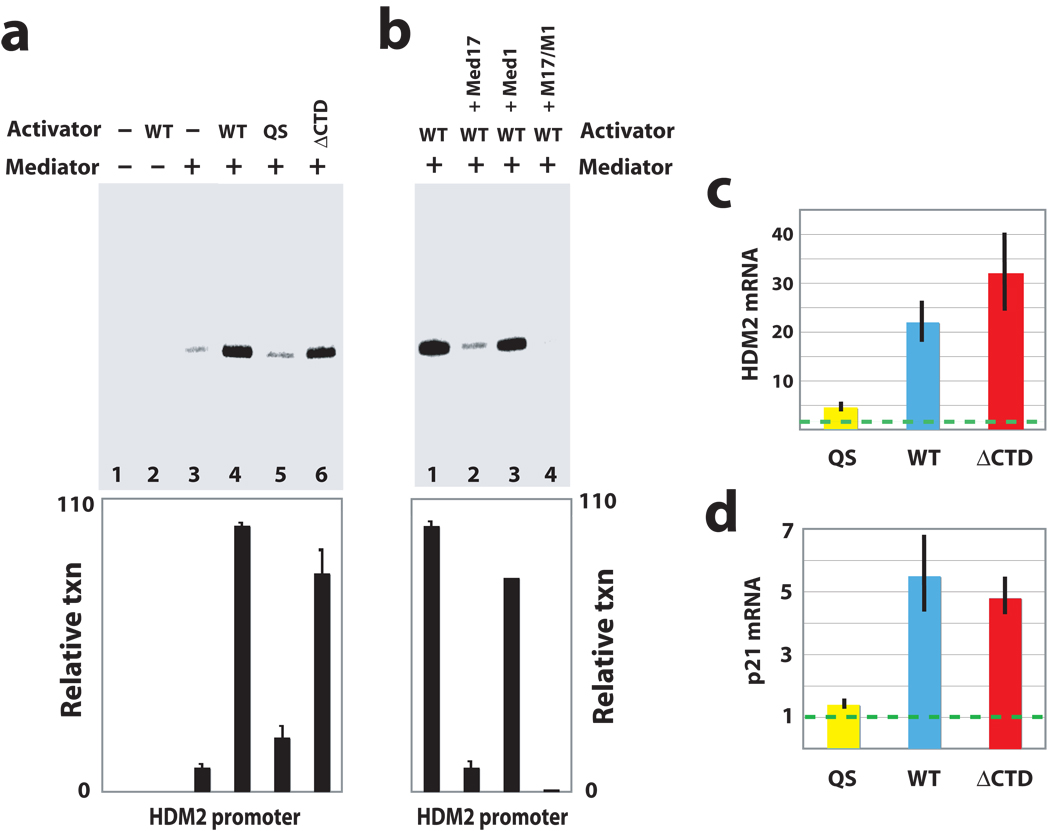
Reconstituted transcription assays with the GAL4-p53 proteins were designed to show what p53AD and p53CTD each contributed to Mediator/PEC function. We next examined each p53 domain within its native context: full-length p53, which binds DNA as a tetramer. Full-length p53 was expressed in insect cells and was isolated as an intact tetramer, based upon gel filtration chromatography and EM analysis (Supplementary Fig. 6d–f). To further define p53AD and p53CTD functions within the tetramer, we also expressed and purified p53 tetramers with a truncated CTD (p53ΔCTD, containing residues 1–362) or with point mutations that inactivate p53AD function (p53QS, with L22Q and W23S mutations). The transcriptional activity of each p53 tetramer was evaluated in the reconstituted transcription system on the native HDM2 promoter, which is strongly induced by p53 in vivo and contains tandem, consensus p53 binding sites. As expected, p53 tetramers containing intact activation domains (wild-type p53 or the p53ΔCTD tetramer) strongly activated transcription from the HDM2 promoter (Fig. 3a, lanes 4 and 6), whereas the p53QS mutant was unable to activate transcription (Fig. 3a, lane 5). In fact, the p53QS mutant was functionally equivalent to “no activator” control experiments (Fig. 3a, lane 3). Furthermore, chromatin sedimentation assays revealed that WT p53 did not differentially recruit Mediator, pol II, or other PEC components to the HDM2 promoter relative to p53QS (data not shown). Thus, similar to experiments with the GAL4 template, it appeared that PEC occupancy at the HDM2 promoter did not correlate with activation and that without an intact p53AD, pol II occupied the promoter but was unable to transition to a productively-elongating state. (In fact, primer extension products of just 155 bp would be detected from the HDM2 template.) As with the GAL4 template experiments, p53-dependent activation at the HDM2 promoter could be blocked with addition of recombinant Med17 (but not Med1, Fig. 3b), again suggesting the p53AD–Mediator interaction was playing a key role in the activation mechanism that did not involve factor recruitment.
Figure 3.
p53AD activates stalled pol II within the context of the native tetramer in vitro and in cells. (a) Reconstituted transcription from the native HDM2 promoter. As with the GAL4 template, transcription was dependent upon Mediator and an intact p53AD. Activators tested were wild-type p53 (WT), p53 with a mutated activation domain (QS), and p53 lacking the CTD (ΔCTD). Each p53 protein was purified as a tetramer. Plot shows mean and s.e.m. from multiple experiments (n = 5, 10, 4, 4 for lanes 3–6). (b) As with the GAL4 template (Fig. 2d), recombinant Med17 could strongly repress p53-dependent transcription at the HDM2 promoter, whereas recombinant Med1 had little effect. The bar graph shows quantitation of transcripts, with bars representing s.e.m. (n = 10, 2 for lanes 1 and 2). Experiments shown in a and b followed the same timeline as shown in Figure 2b and contained the same concentration of HDM2 chromatin template, TFIIA, IIB, IID, IIE, IIF, IIH, pol II, and NTPs. Mediator and activators added as shown. (c) and (d) Summary of results from RT-QPCR experiments measuring HDM2 or p21 mRNA levels in response to the p53 proteins shown. Experiments were completed in p53-null HCT116 cells; dashed line represents mRNA levels measured in non-transfected control cells. Note that similar mRNA levels observed with WT p53 and p53ΔCTD result from their overexpression upon transfection. When expressed at physiological levels, p53ΔCTD is still observed to activate HDM2 and p21, but to a lesser extent relative to WT p53, due to its decreased ability to stably bind DNA55.
We next compared the activity of wild-type p53 with the p53QS or p53ΔCTD mutants in cells. Each protein was expressed in p53-null HCT116 cells (Supplementary Fig. 4a). We compared the ability of each p53 protein to activate transcription at the HDM2 or the p21 gene, each of which has been shown to possess stalled pol II at their promoters21,27–29. As shown in Figure 3c, HDM2 mRNA levels were consistent with our in vitro transcription results from the native HDM2 promoter: p53QS was unable to activate transcription, whereas WT p53 or p53ΔCTD strongly enhanced HDM2 mRNA levels. RT-QPCR assays at the p21 gene yielded similar results (Fig. 3d). Coupled with the data shown in Figure 1 and 2, the data summarized in Figure 3 corroborate numerous in vitro and in vivo studies that link p53AD to activation of p53 target gene expression13–16, yet also demonstrate that p53AD is not required for PEC assembly and functions primarily at a post-recruitment step to activate HDM2 or p21 transcription.
Pol II CTD processing and pol II promoter escape
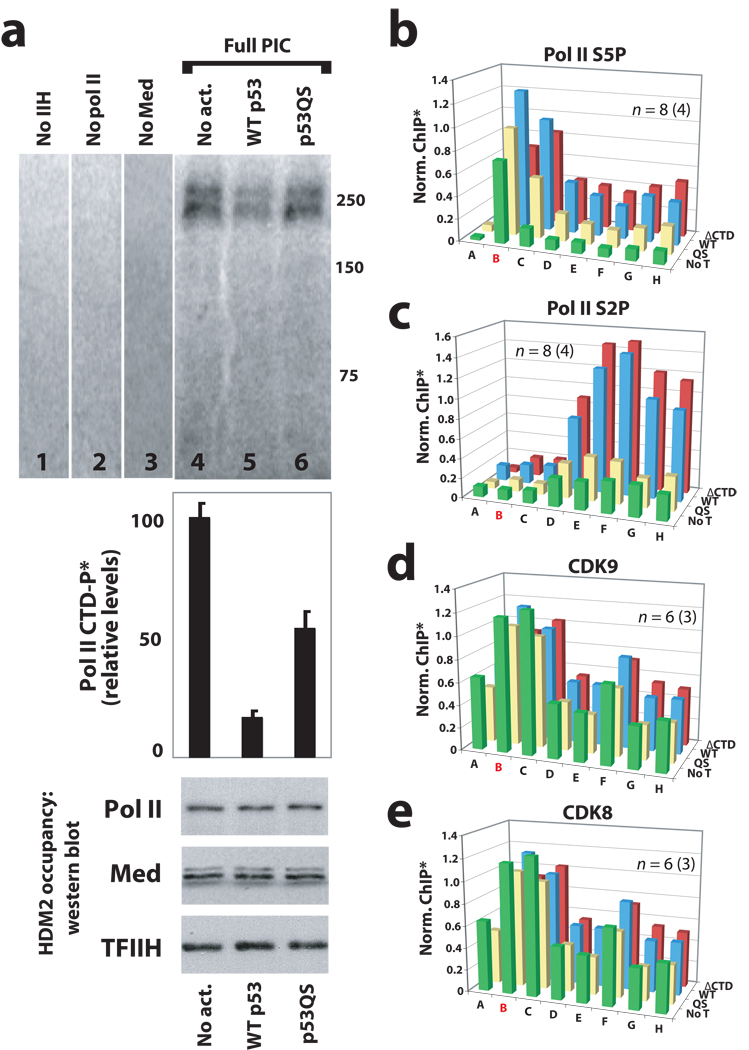
Within the PEC, the pol II CTD is phosphorylated by TFIIH, and this correlates with pol II promoter clearance and transcript elongation. To probe the activity of TFIIH within the PEC, we completed a series of immobilized template kinase assays at the native HDM2 promoter. In these assays, PECs were assembled from purified components—not extracts—so that no contaminating kinase activities would be present. Thus, purified TFIIA, IIB, IID, IIE, IIF, IIH, Mediator and pol II (see ref. 20) were assembled on immobilized HDM2 promoter templates, together with WT p53, the p53QS mutant, or no activator controls. We observed that despite similar PEC–pol II occupancy, levels of phosphorylated pol II CTD were substantially lower in the presence of WT p53 compared with the p53QS mutant or no activator controls (Fig. 4a). These results provided evidence that WT p53 was triggering transcription activation via rapid pol II promoter clearance (and thus less pol II CTD phosphorylation within the PEC), whereas with the p53QS mutant, pol II remained inactive within the PEC. Indeed, the increased levels of pol II CTD phosphorylation observed in lanes 4 and 6 (Fig. 4a) correspond to inactive PECs, as assessed by reconstituted transcription assays (Fig. 3a, lanes 3 & 5). Thus, the increased levels of pol II CTD phosphorylation observed with the p53QS mutant or no activator control experiments (Fig. 4a, lanes 4 & 6) suggest pol II does not efficiently transition to a productively-elongating state in the absence of p53AD. The data in Figure 4a also indicate that pol II CTD phosphorylation did not occur in PECs assembled without pol II itself (Fig. 4a, lane 2) or without the kinase, TFIIH (Fig. 4a, lane 1), as expected. Somewhat unexpected was the complete dependence upon Mediator for TFIIH-dependent phosphorylation of the pol II CTD (Fig. 4a, compare lanes 3 and 5), suggesting that within the PEC, Mediator and TFIIH function synergistically to control pol II CTD phosphorylation. This Mediator requirement for pol II CTD phosphorylation was also observed within PECs assembled on GAL4 promoter templates (data not shown).
Figure 4.
Mediator is required for TFIIH-dependent pol II CTD phosphorylation within the PEC; oncogenic mutations within p53AD (p53QS) prevent activation of stalled pol II. (a) Immobilized template assays with the native HDM2 promoter. Purified PEC factors (TFIIA, IIB, IID, IIE, IIF, IIH, Mediator, and pol II; see ref. 20) were allowed to assemble on the HDM2 promoter; unbound factors were removed by washing, at which time rNTPs were added (including [32P]-ATP) to visualize and quantitate pol II CTD phosphorylation. Data shown follows a 6-minute incubation with rNTPs; radio-labeled bands migrated around 250 kDa, the approximate size of Rpb1. Bar graph represents mean and s.e.m. from multiple independent experiments (n = 6, 10, 4 for lanes 4–6, respectively). Occupancy of PEC components was measured by western blot, several of which are shown below the bar graph. PECs lacking only one specific component (TFIIH, pol II, or Mediator: lanes 1–3) indicate a Mediator requirement for pol II CTD phosphorylation. (b) and (c) ChIP data reveals that phosphorylated forms of pol II are diminished within the HDM2 gene in the absence of an intact p53AD. (d) and (e) Occupancy of pol II CTD kinases CDK9 (a component of P-TEFb) and CDK8 are not negatively impacted by p53AD mutations. Note that occupancy of another major pol II CTD kinase, TFIIH, is also not altered by p53AD mutations, as observed by ChIP in Figure 1c. The probe location in red (B) represents the promoter/transcription start site. *ChIP output was normalized to WT p53, typically from primer C. For clarity, error bars are not shown but can be viewed in Supplementary Figure 8.
Having identified a functional interdependence between human Mediator and TFIIH, we next examined whether the p53QS mutation might affect pol II processing at the HDM2 gene in cells. The human pol II CTD consists of 52 heptapeptide repeats with the consensus sequence YSPTSPS. The major pol II S5 CTD kinase is TFIIH, whereas the major pol II S2 CTD kinase is P-TEFb. The HDM2 gene was probed for S5P and S2P modified forms of the pol II CTD using ChIP. These experiments revealed that cells expressing the p53QS mutant showed reduced pol II occupancy within the HDM2 and p21 genes, despite high pol II occupancy at the promoter. Indeed, major reductions in phosphorylated forms of pol II were observed within the body of the HDM2 and p21 genes (Fig. 4b, c and Supplementary Fig. 7), providing further evidence that pol II was not transitioning to a productively-elongating state in the absence of an intact p53AD. Note these ChIP results were entirely consistent with the reconstituted transcription data (Fig. 2c and Fig. 3a), which clearly demonstrated that p53AD was required for pol II to transition off the promoter to generate a transcript. Differences in pol II CTD phosphorylation did not appear to result from differences in the occupancy of distinct pol II CTD kinases (TFIIH, P-TEFb, or CDK8) at the HDM2 or p21 genes, as evaluated by ChIP (Fig. 1c, Fig. 4d, e, and Supplementary Fig. 4 and 7).
The multi-subunit NELF complex has been shown to regulate post-recruitment events in transcription initiation30 and therefore we also examined the promoter occupancy of NELF at the HDM2 and p21 genes. These results, described further in the Supplementary Information and Supplementary Figure 9, revealed that although NELF may play auxiliary roles in regulating HDM2 and p21 expression, NELF does not appear to be required for controlling p53AD-dependent changes in gene expression.
p53AD triggers a structural shift within Mediator distinct from p53CTD
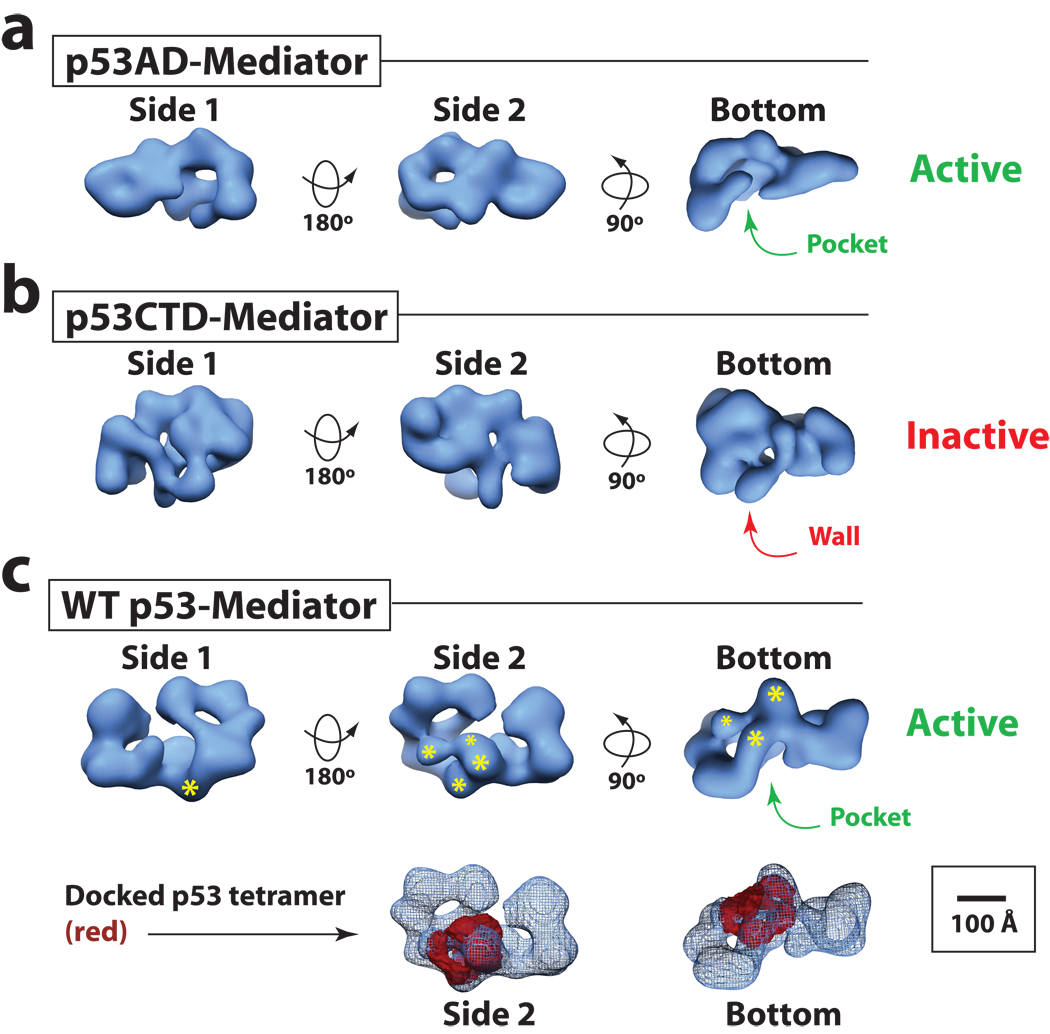
So what is p53AD doing that enables activation of pre-loaded or stalled pol II complexes? Although p53 does not contact pol II directly, p53AD interacts with high affinity to Mediator via the Med17 subunit13. Mediator, in turn, interfaces extensively with the pol II enzyme4,6. Past studies with human Mediator established that its structural state can be altered significantly by transcription factor binding18. In fact, different transcription factors (which can bind different Mediator subunits) are capable of inducing different structural states, establishing a link between the subunit target and the structural state of Mediator. To investigate whether a structural shift might provide a basis for the difference in Mediator activity when bound to p53AD versus p53CTD, we completed EM analysis and single-particle reconstruction of Mediator bound to either p53AD or p53CTD. The 3D structure of p53AD– and p53CTD–Mediator is shown in Figure 5a and 5b (see also Supplementary Information, Supplementary Fig. 10–14 and Supplementary Movie 1–2). In each case, Mediator adopts a dramatically different 3D architecture, indicating that, as expected, different Mediator targets (Med17 for p53AD; Med1 for p53CTD) trigger distinct structural changes within the complex. Strikingly, the p53AD–Mediator structure contains a large “pocket” domain (arrow, Fig. 5a) that is of sufficient size and shape to accommodate the pol II enzyme31. In fact, pol II binds Mediator at this site, as shown in Supplementary Figure 14c; pol II has also been observed to bind at the pocket site within yeast Mediator4. The pocket domain was preserved within the structure of Mediator bound to wild-type p53 tetramers (Fig. 5c and Supplementary Movie 3) and the p53ΔCTD tetramer (Supplementary Fig. 15a and Supplementary Movie 4), each of which show strong activity in vitro and in cells (Fig. 3). By contrast, the p53CTD–Mediator structure lacks this pol II pocket and does not resemble other activator-bound Mediator structures (Supplementary Fig. 14a); instead, p53CTD–Mediator contains a wall of protein density that occupies the pol II site (Fig. 5b) and appears to represent an inactive structural state. Collectively, these structural data establish a link between p53AD-directed structural changes and Mediator-dependent, post-recruitment activation of pol II within the PEC.
Figure 5.
p53AD triggers a structural shift within Mediator distinct from p53CTD. (a) and (b) Different views of Mediator bound to p53AD or p53CTD. Volumes shown at 34 Å and 38 Å resolution, respectively, are rendered to 1.2 MDa, the approximate molecular weight of each complex. A defined pocket domain is evident in p53AD–Mediator (transcriptionally active) whereas in p53CTD–Mediator (transcriptionally inactive), this region is occupied by a wall of protein density. Bar: 100 Å. Note the pocket domain corresponds to the Mediator-pol II interaction site, as shown in Supplementary Figure 14c. (c) The structure of Mediator bound to the wild-type p53 tetramer (34 Å resolution). Like p53AD–Mediator, the pocket domain represents a prominent structural feature. The volume shown is rendered to 1.4 MDa, the approximate molecular weight of the WT p53–Mediator assembly. The general location of the p53 tetramer is highlighted, with asterisks representing the p53 DNA-binding domains. Larger asterisks are closer to the viewer. Lower panels show views of the docked WT p53 tetramer structure47 within the WT p53–Mediator EM map. The p53 tetramer is solid whereas WT p53–Mediator is shown in mesh.
DISCUSSION
Because of its large size and low-abundance, EM is the only structural technique suited to study the 1.2 MDa human Mediator complex. Despite the limited resolution of this technique, EM reconstructions of p53–Mediator clearly define major structural shifts that provide insight into how p53AD coordinately regulates the activity of Mediator, TFIIH, and pol II within the PEC. Interestingly, Mediator, TFIIH, and pol II are each generally required for expression of protein-coding genes1 and the structural and functional data outlined here provide the first demonstration that activator-induced structural shifts within Mediator serve important regulatory functions, including activation of stalled pol II (Fig. 6). Past studies of p53 activation at p21 implied a post-recruitment role for Mediator29; our data confirm this and provide evidence for a conformational switching mechanism whereby p53–Mediator activates stalled pol II complexes at p21 and HDM2. However, not all p53-regulated genes appear to possess promoter-associated pol II prior to activation21. Thus, alternate regulatory mechanisms may predominate at other genes within the p53 network. It will be interesting to compare how Mediator–PEC assembly and structure might vary during activation of these distinct p53 target genes. It will be also be important to define how Mediator might work together with auxiliary factors, such as NELF and P-TEFb30, that likely help enforce the activation of stalled pol II in different promoter contexts.
Figure 6.
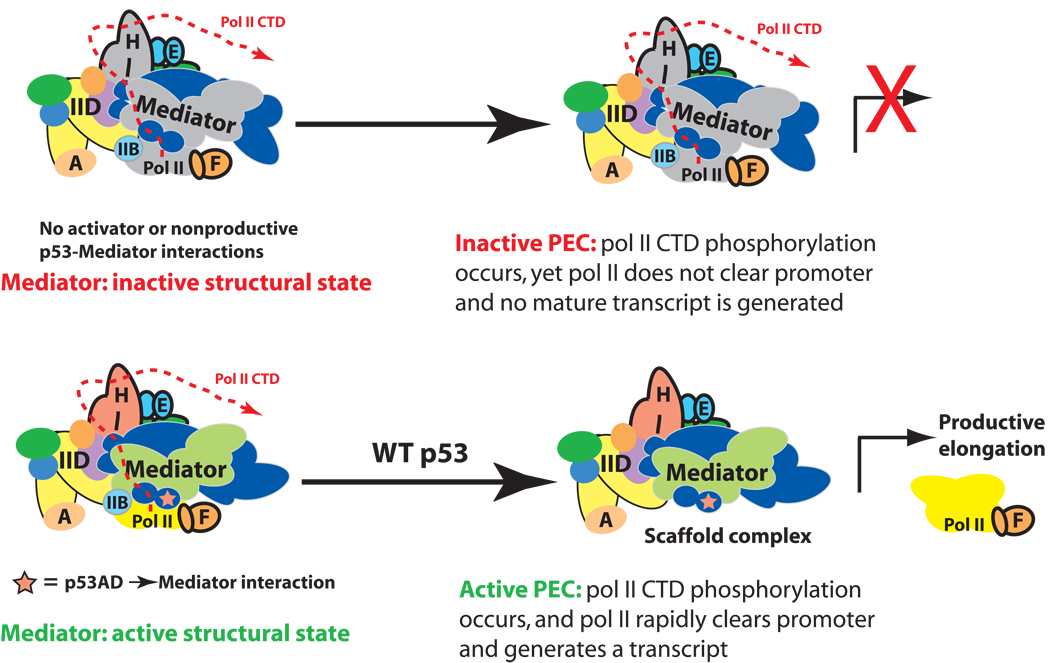
Model outlining how p53AD appears to trigger pol II promoter escape and transcription elongation. Top: Despite full PEC assembly, promoter-proximally stalled pol II remains unable to transition to a productively elongating state in the absence of the p53AD-directed structural shift within Mediator. Bottom: Mediator structural shifts, orchestrated by p53AD, activate stalled pol II to transition to a productively-elongating state. This Mediator-dependent activation appears to be triggered by formation of the pocket domain within Mediator, which interfaces extensively with pol II. Note the location and path of the pol II CTD within the PEC has not been clearly defined.
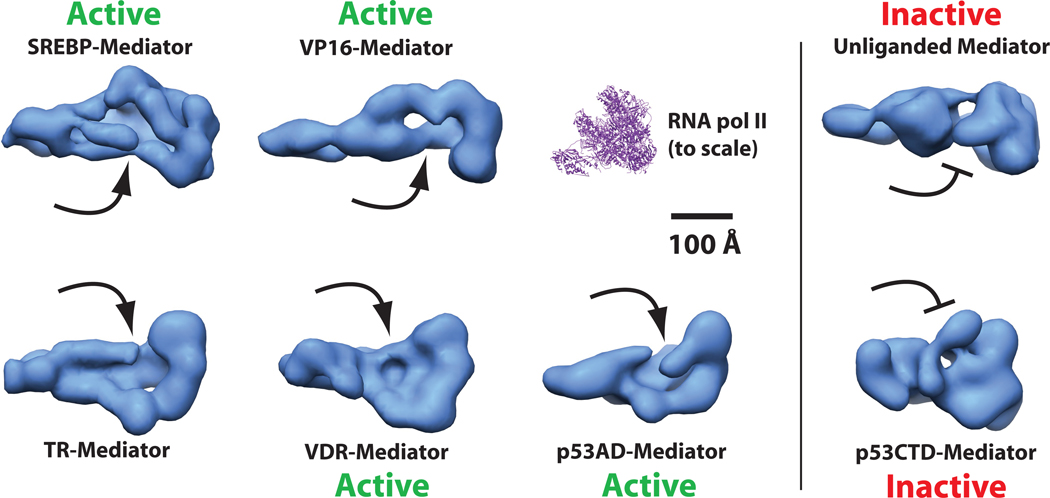
Our results also point to a general mechanism that may govern activator-dependent transcription. Among the activator-bound Mediator structures examined to date, each has been shown to trigger significant structural shifts relative to the unbound, activator-free state18,31. In each case—with the exception of the transcriptionally inactive p53CTD–Mediator structure shown here—a defined, pol II pocket domain is a shared feature, suggesting this structural shift represents a fundamental means by which transcription factors help activate gene expression (Fig. 7). Because Mediator physically or functionally interacts with many PEC components, activator-dependent structural shifts may affect multiple or different steps in transcription initiation in different promoter contexts. The structural plasticity of other general transcription factors (including TFIIB and TFIID) suggests activator-dependent structural changes within the PEC represents a common regulatory strategy32–35. Such a strategy could allow for more diverse transcriptional responses to combinatorial regulatory inputs and provides a straightforward mechanism for regulating transcription after pol II recruitment.
Figure 7.
The pocket domain represents a common structural feature within activator-bound Mediator. Shown are “bottom” views of Mediator complexes bound to different activation domains. The location of the pocket is noted by the arrows. Mediator structures labeled “active” have been shown to activate transcription in a reconstituted system. Note that transcriptionally inactive structures (unliganded Mediator or p53CTD–Mediator structure) lack the pocket domain. The unliganded Mediator structure is not bound to an activator18. Studies with both yeast4 and human Mediator (Supplementary Fig. 14c) reveal that the pocket domain represents the pol II binding site within Mediator. For reference, the structure of the yeast pol II enzyme (pdb 1y1v) is also shown to the same relative scale.
Structural analysis of p53–Mediator has revealed major differences in its conformational state upon binding either p53AD or p53CTD, and these structural shifts were preserved within the context of the p53 tetramer. In particular, formation of a large pocket domain within Mediator was induced upon binding the wild-type p53 tetramer or p53AD alone, whereas the pocket domain was clearly absent in the p53CTD–Mediator structure. Using immobilized template and chromatin sedimentation assays, we observed similar promoter occupancy of PEC factors irrespective of p53AD. Yet reconstituted transcription assays revealed an absolute dependence upon p53AD for activation of transcription. ChIP and RT-QPCR assays in cells suggested a similar phenomenon: PEC factors (including Mediator, pol II, and TFIIH) were pre-loaded at the p53-regulated HDM2 and p21 genes, yet an intact p53AD was required for activation of transcription. We also observed that the presence of p53AD at the promoter had a major impact on pol II promoter clearance and pol II CTD phosphorylation both in vitro and in cells. Coupled with our EM analysis of p53–Mediator, these results suggest that p53AD–Mediator structural shifts enable stalled pol II complexes to transition off the promoter to a productively-elongating state. Notably, p53AD-dependent formation of the pocket domain within Mediator does not appear necessary for pol II recruitment; rather, formation of the pocket domain correlates with changes in pol II processing and promoter escape. How might this occur? What surfaces within Mediator and pol II are involved in this transition, and how might the Mediator-pol II interface change during the p53AD-directed structural shift? Delineation of such questions will require further studies involving cryo-EM analysis of structurally distinct p53–Mediator-pol II assemblies.
Significant differences in TFIIH-dependent pol II CTD phosphorylation were observed within PECs assembled in the presence of WT p53 compared to PECs assembled without an activator or with the oncogenic p53QS mutant. Given the Mediator requirement for pol II CTD phosphorylation within the PEC, these observations, considered together with the functional and structural data, define a mechanism by which p53 precisely coordinates Mediator structural shifts, pol II activation, and TFIIH-dependent pol II CTD phosphorylation. Yeast Mediator is known to physically interact with TFIIH3, hinting that Mediator structural shifts might directly impact TFIIH structure and function within the PEC. Immobilized template assays at the native HDM2 promoter revealed a Mediator requirement for TFIIH-dependent pol II CTD phosphorylation within the fully-assembled PEC. This result indicates that in this context, Mediator must properly orient the pol II CTD for efficient phosphorylation by TFIIH. Immobilized template assays were completed in the presence of WT p53, the oncogenic p53QS mutant, and also in the absence of an activator. Interestingly, pol II CTD phosphorylation occurred in each circumstance, although transcripts were generated only in the presence of WT p53. These results indicate that pol II CTD phosphorylation per se does not correlate with activation of transcription; these data also introduce the intriguing possibility that within these different contexts (Mediator structure will be different in each case), alterations in the pol II CTD phosphorylation pattern might underlie the stark differences in pol II activity.
In addition to Mediator, the p53AD has been shown to interact with other PEC components, including TFIID and TFIIH, and these interactions may play alternate roles in activation36–38. However, a p53AD affinity column purifies only the Mediator complex from the P1M/Q1M fraction, with no detectable co-purifying TFIID or TFIIH (Supplementary Fig. 10a–c). Because the P1M/Q1M fraction is enriched in TFIID and TFIIH as well as Mediator, this suggests that potential p53AD interactions with TFIID or TFIIH are of much lower affinity. It is also notable that there is a Mediator requirement for activated transcription (i.e. not basal transcription) in both yeast and human systems, highlighting Mediator’s essential role in activator-dependent transcription39,40. Yet we cannot exclude the possibility that secondary p53AD interactions with TFIID or TFIIH might play auxiliary roles in activating transcription; moreover, post-translational modifications within p53AD may regulate these interactions in specific promoter contexts41,42. Other co-regulatory factors, including CBP/p300 and HDM2, are targeted by p53AD and clearly serve important roles in regulating p53-dependent gene expression16,43,44. It is currently unclear how the binding of these and other factors might be coordinately regulated; however, because four activation domains could potentially mediate p53-cofactor interactions at the promoter, multiple factors might interact with p53 simultaneously.
Finally, many p53-regulated genes feature pol II and other factors pre-loaded at the promoter21. Since even transient re-activation of p53 can cause tumor regression45,46, a strategy to re-activate certain p53 target genes might involve agents that trigger p53AD-specific structural shifts within Mediator, perhaps by targeting the p53AD-Med17 interface.
METHODS
Purification of p53AD- and p53CTD–Mediator
We isolated Mediator bound to p53AD or p53CTD with a GST-p53AD or GST-p53CTD affinity column from the P1M/Q1M fraction, essentially as described20.
Purification of p53 tetramers
We expressed wild-type p53, p53QS, or p53ΔCTD (residues 1–362) in baculovirus-infected insect cells, as described47.
Isolation of WT p53–Mediator and p53ΔCTD–Mediator
Mediator was purified in an activator-free state using an anti-Med26 antibody resin and eluted with peptide, as described20. Purified p53 tetramers (WT p53 or p53ΔCTD) were added to this activator-free Mediator sample (in approximately 5-fold excess) and mixed for 1 h at 4 °C to allow for p53 tetramer binding to Mediator. These samples were then used for EM experiments.
in vitro transcription
We completed chromatin assembly and in vitro transcription as described20, with a few modifications. For reactions on both the GAL4 and the HDM2 templates, we added PC4 and HMG1 (ProteinOne, Bethesda, MD), to 31 nM and 33 nM, respectively, based upon past studies that showed these factors served as co-activators for p5348,49. Although supplementation with PC4 and HMG1 resulted in small but detectable levels of basal transcription (e.g. see Fig. 2c, lane 3 or Fig. 3a, lane 3), these cofactors increased levels of activated, Mediator-dependent transcription in the reconstituted system (data not shown). For GAL4–p53AD, GAL4–p53AD/CTD, WT p53, and p53ΔCTD activators, we completed control experiments in which the concentration of each was titrated over a 50-fold range to determine the optimum concentration for the assay. Concentrations used in the transcription assays were approximately 5 nM for GAL4–p53 proteins and 20 nM for p53 tetramers. With respect to the transcriptionally inactive proteins (GAL4–p53CTD and p53QS), we titrated these over a 200-fold range to ensure that each did not harbor any significant transactivation function. In all experiments shown with GAL4–p53CTD or p53QS, we added each to a concentration equal to GAL4–p53AD or WT p53, respectively.
RT-QPCR and ChIP
We maintained p53-null HCT116 cells in McCoys 5A media (Gibco-Invitrogen) supplemented with 10% fetal bovine serum and antibiotic/antimycotic mix. We transfected p53 expression vectors (WT p53, p53ΔCTD, or p53QS) into these cells with Lipofectamine 2000 (Invitrogen). Expression levels of each p53 protein were measured by western blot to ensure equal expression among different p53-transfected cell populations (Supplementary Fig. 4a). We performed RT-QPCR and ChIP assays as described50. We obtained antibodies from Santa Cruz (p53, Med1, pol II, TFIIB, TBP, NELF, CDK8, CDK9) and Covance (pol II S2P and pol II S5P). The TFIIH (ERCC3) antibody is a lab stock.
Electron microscopy and image processing
We applied Mediator samples to glow-discharged carbon-coated 400-mesh grids (EM Sciences) and negatively-stained with a 4% uranyl acetate solution following buffer-exchange in a 5% trehalose buffer (0.15M KCl, 20 mM HEPES, 0.1 mM EDTA, pH 7.9). We obtained micrographs on a Tecnai F20 FEG at 29,000× magnification and data was collected on either Kodak SO-163 film or digitized from a Gatan 4k × 4k CCD camera.
We completed image processing with the SPIDER and WEB software package51, essentially as described31. Briefly, we selected and windowed single-particle images into 120 × 120 pixel images (p53AD– or p53CTD–Mediator) or 160 × 160 pixel images (WT p53– or p53ΔCTD–Mediator) prior to multi-reference alignment and classification. A micrograph of the p53AD–Mediator sample is shown in Supplementary Figure 10d. The pixel size was 4.29 Å (film) or 3.71 Å (CCD). We used untilted (0°) micrographs for alignment and classification of single-particle images whereas tilted (25–45°) micrographs were used to generate 3D reference volumes that were cross-correlated against each other to establish a homogenous data set. For p53AD–Mediator, 7166 single-particle images comprised the entire data set, 58% of which were used for the final 3D reconstruction, whereas for p53CTD–Mediator, the data set contained 1738 images, of which 69% were used for angular refinement. EM data for WT p53–Mediator and p53ΔCTD–Mediator contained 3758 and 3090 single-particle images, respectively, of which 76% and 45% were merged, based upon cross-correlation, for subsequent angular refinement. Based upon the 0.5 Fourier shell correlation coefficient52, we low-pass filtered structures to 34 Å resolution (p53AD–Mediator and WT p53–Mediator), 36 Å resolution (p53ΔCTD–Mediator), or 38 Å resolution (p53CTD–Mediator). Angular distribution of single-particle images reveals a good representation of Mediator orientations in 3D space (Supplementary Fig. 11 and 12).
For cryo-EM analysis of p53AD–Mediator, we applied the sample to a glow-discharged grid, followed by blotting and buffer exchange (to remove glycerol) with a 0.1M KCl/HEPES buffer containing 5% trehalose. We then floated the grid on a solution of 16% ammonium molybdate, blotted, and vitrified by rapid plunging into liquid ethane, as described53,54. We collected cryo-EM data on a Tecnai F20 FEG at 29,000× magnification at defocus values ranging from 2.0 to 3.0 µm underfocus. No CTF correction was needed at these defocus values, based upon the resolution of the final 3D reconstruction (see below). After selection of p53AD–Mediator particles from the cryo-EM micrographs, we rotationally and translationally aligned single-particle images (2297 from 84 micrographs) against a series of noise-free 2D projection views obtained from a p53AD–Mediator structure filtered to very low resolution (60 Å). Following a series of angular refinement steps (with a total of 798 reference projections for the final alignment and back projection), we filtered the final 3D reconstruction to 38 Å resolution (based upon 0.5 Fourier Shell Correlation) and rendered to a volume consistent with its predicted molecular weight of 1.2 MDa.
p53 tetramer docking
We completed structure docking of the WT p53 tetramer47 into the WT p53–Mediator or the p53ΔCTD–Mediator structure with both Chimera and SITUS with similar results. The p53 tetramer structure was obtained from the macromolecular structure database (EMD-1141).
Supplementary Material
Acknowledgments
We thank Aaron Donner and Joaquin Espinosa for help with the ChIP assays, RT-QPCR experiments, and other useful advice; Cindi Schwartz and Mary Morphew for assistance with EM data collection. The HDM2 promoter fragment was obtained from Jeremy Blaydes (University of Southampton). Protein expression in insect cells was completed at the Tissue Culture Core Facility at the University of Colorado Cancer Center. We thank Jim Goodrich and Joaquin Espinosa for helpful comments on the manuscript. This work was supported by the NCI (R01 CA127364) and the Ellison Medical Foundation. K.D.M. and C.B. were supported in part by NIH grant T32 GM065103.
Footnotes
Author contributions: K.M. designed, analyzed, and performed most experiments and helped write the paper; S.L. completed immobilized template assays; C.B. assisted with cryo-EM, purified pol II and provided Mediator–pol II EM data; Y.G. mapped Med1 binding domain to p53CTD; D.T. designed and performed experiments and helped write the paper.
References
- 1.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 2.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esnault C, et al. Mediator-dependent recruitment of TFIIH modules in Preinitiation Complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes & Development. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes & Development. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 8.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes & Development. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 10.Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–4023. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Drane P, Barel M, Balbo M, Frade R. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene. 1997;15:3013–3024. doi: 10.1038/sj.onc.1201492. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, et al. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez GS, et al. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat Genet. 2000;26:37–43. doi: 10.1038/79152. [DOI] [PubMed] [Google Scholar]
- 15.Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37:145–152. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes & Development. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 17.Nister M, et al. p53 must be competent for transcriptional regulation to suppress tumor formation. Oncogene. 2005;24:3563–3573. doi: 10.1038/sj.onc.1208354. [DOI] [PubMed] [Google Scholar]
- 18.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Structure, Function, and Activator-Induced Conformations of the CRSP Coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 19.Johnson KM, Wang J, Smallwood A, Carey M. The immobilized template assay for measuring cooperativity in eukaryotic transcription complex assembly. Methods Enzymol. 2004;380:207–219. doi: 10.1016/S0076-6879(04)80010-7. [DOI] [PubMed] [Google Scholar]
- 20.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator co-activator function. Genes & Development. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa JM, Verdun RE, Emerson B. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 22.Gaudreau L, Adam M, Ptashne M. Activation of transcription in vitro by recruitment of the yeast RNA polymerase II holoenzyme. Mol. Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 23.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, DeBeaumont R, Zhou S, NŠŠr AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittler G, et al. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rachez C, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 27.Li P, et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover-Cutter K, Kim S, Espinosa JM, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 31.Taatjes DJ, Schneider-Poetsch T, Tjian R. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat Struct Mol Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 32.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 33.Horikoshi M, Hai T, Lin YS, Green MR, Roeder RG. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 34.Guermah M, Malik S, Roeder RG. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-kappaB and Sp1. Mol. Cell. Biol. 1998;18:3234–3244. doi: 10.1128/mcb.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts SGE, Green MR. Activator-induced conformational change in general transcription factor TFIIB. Nature. 2002;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 36.Xiao H, et al. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 38.Lu H, Levine AJ. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. TIBS. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Di Lello P, et al. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Li AG, et al. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28:408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The MDM-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 44.Van Orden K, Giebler HA, Lemasson I, Gonzales M, Nyborg JK. Binding of p53 to the KIX domain of CREB binding protein. A potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J Biol. Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 45.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 46.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okorokov AL, et al. The structure of p53 tumor suppressor protein reveals the basis for its functional plasticity. EMBO J. 2006;25:5191–5200. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batta K, Kundu TK. Activation of p53 function by human transcriptional coactivator PC4: role of protein-protein interaction, DNA bending, and posttranslational modifications. Mol Cell Biol. 2007;27:7603–7614. doi: 10.1128/MCB.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinney K, Prives C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol Cell Biol. 2002;22:6797–6808. doi: 10.1128/MCB.22.19.6797-6808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomes NP, et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & Development. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 52.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–153. [Google Scholar]
- 53.De Carlo S, El-Bez C, Alvarez-Rua C, Borge J, Dubochet J. Cryo-negative staining reduces electron-beam sensitivity of vitrified biological particles. J Struct Biol. 2002;138:216–226. doi: 10.1016/s1047-8477(02)00035-7. [DOI] [PubMed] [Google Scholar]
- 54.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification--powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.