Abstract
The hypothalamic-pituitary-gonadal endocrine axis regulates reproduction through estrous phase-dependent release of the heterodimeric gonadotropic glycoprotein hormones, LH and FSH, from the gonadotropes of the anterior pituitary. Gonadotropin synthesis and release is dependent upon pulsatile stimulation by the hypothalamic neuropeptide GnRH. Alterations in pulse frequency and amplitude alter the relative levels of gonadotropin synthesis and release. The mechanism of interpretation of GnRH pulse frequency and amplitude by gonadotropes is not understood. We have examined gene expression in LβT2 gonadotropes under various pulse regimes in a cell perifusion system by microarray and identified 1127 genes activated by tonic or pulsatile GnRH. Distinct patterns of expression are associated with each pulse frequency, but the greatest changes occur at a 60-min or less interpulse interval. The immediate early gene mRNAs encoding early growth response (Egr)1 and Egr2, which activate the gonadotropin LH β-subunit gene promoter, are stably induced at high pulse frequency. In contrast, mRNAs for the Egr corepressor genes Ngfi-A binding protein Nab1 and Nab2 are stably induced at low pulse frequency. We show that Ngfi-A binding protein members inhibit Egr-mediated frequency-dependent induction of the LH β-subunit promoter. This pattern of expression suggests a model of pulse frequency detection that acts by suppressing activation by Egr family members at low frequency and allowing activation at sustained high-frequency pulses.
The neuropeptide hormone GnRH is a hypothalamic peptide essential for normal mammalian reproductive function. The primary target of GnRH is the anterior pituitary gonadotrope, which responds to stimulation by increasing the synthesis and secretion of the gonadotropins, LH and FSH, which in turn regulate gonadal development and function (1, 2). The gonadotropins are members of the glycoprotein hormone family that is characterized by a shared α-subunit and a unique, hormone-defining β-subunit (LHβ and FSHβ). GnRH is also essential for gonadotrope proliferation and development, because the hpg mouse, which harbors a deletion in the mouse GnRH gene, has fewer gonadotropes in the anterior pituitary, decreased circulating levels of LH and FSH, and is infertile (3). Cases of hypogonadotropic hypogonadism in humans can be traced to mutations in the GnRH receptor gene on chromosome 4q21 that cause GnRH resistance (4–6).
A unique feature of the hypothalamic-pituitary-gonadal axis is the dependence on episodic release of GnRH into the portal vasculature targeting the gonadotropes of the anterior pituitary (7). The resulting pulsatile GnRH exposure, in turn, causes episodic release of the gonadotropins into the circulation and is essential for the proper function of gonadotropes. Chronic exposure to GnRH or its analogs leads to a reversible suppression of gonadotropin release. During the estrous cycle, the surge of LH secretion essential for promoting ovulation is associated with a rise in GnRH pulse amplitude and frequency. Disruption of this hypothalamic pulsatility disrupts LH secretion (8). At the level of gene expression, studies in primary pituitary cells and pituitary fragments have shown that pulsatile administration of GnRH is necessary for LHβ and FSHβ transcriptional activation. More recent studies in rats have also shown that pulsatile GnRH stimulation is required for activation of gonadotropin β-subunit gene expression and suggest that an interpulse frequency of 30 min is an important component in the activation of LHβ subunit gene expression (9).
Studies with individual promoters in cultured cells have suggested that the effects of GnRH on LHβ and FSHβ are distinct. The induction of the LHβ promoter by GnRH is mediated by steroidogenic factor 1 (SF1) and early growth response 1 (Egr1) (10–12). SF1 is essential for adrenal and gonadal development and SF1 null mice are infertile (13–15); however, a pituitary-specific deletion of SF1 causes hypogonadotropic hypogonadism with decreased LH and FSH in both male and female mice (16). Female Egr1 null mice are sub-fertile due to LH deficiency but have normal FSH levels (17). Male Egr1-deficient mice, however, are fertile due to partial compensation by Egr4 in the pituitary (18). This Egr isoform appears to be more important in males, because female Egr4 null mice are fertile but males are infertile due to testicular abnormalities (19). A different Egr1 null mouse demonstrates a more severe phenotype with both male and female infertility (20). The phenotype of the Egr1 null mice is less severe than LH receptor null mice, which demonstrate underdeveloped gonads and external genitalia (21, 22), because basal expression of LHβ by the gonadotrope appears to be required for normal gonadal development. In contrast, the FSHβ promoter is induced by the activator protein-1 family of factors (23, 24). Multiple complexes of c-fos, FosB, c-jun, and JunB have been detected on the mouse FSHβ promoter (25). However, mice null for various activator protein 1 subunits do not have reproductive phenotypes due to redundant functions within the family (26). Additionally, activin signaling is essential for FSHβ expression (24, 27, 28). The activin receptor signals via both Sma- and Mad-related protein (Smad)2/4 and Smad3/4 heterodimers to regulate the FSHβ promoter through multiple sites (29–31).
It has been established in vivo that LH and FSH synthesis and secretion are differentially sensitive to GnRH pulse frequency (32). Generally, slow GnRH pulse frequencies favor FSH production, whereas rapid frequencies favor LH production. However, the mechanistic basis for the ability of gonadotropes to distinguish pulse frequency is not well described, and the majority of studies of GnRH signaling are performed in static culture using tonic treatment of GnRH. Modeling of the GnRH signal transduction network suggests that cellular systems can interpret signal frequencies by exploitation of the time scales of gene expression activation and activation of signal-transducing proteins (33). Recent examination of the response of LβT2 gonadotropes to GnRH stimulation indicates that gonadotropes exhibit a graduated response to increases in amplitude (34). However, the role of pulse frequency was not examined in depth over longer treatment times.
It is clear from previous work in vivo and in primary culture of pituicytes that gonadotropin gene expression is differentially regulated by GnRH pulse frequency. Genomic surveys of GnRH-stimulated gene expression have been conducted in LβT2 cells, but none have directly addressed the role of pulsatility (35–38). Therefore, we examined the genome-wide impact of pulsatile GnRH to provide a clear assessment of the role of GnRH pulsatility in gonadotrope gene expression. Using a perifusion system to administer pulsatile GnRH without mechanical, thermal, or atmospheric disturbance, we have examined the frequency-dependent changes in LβT2 gonadotrope cell gene expression. This analysis has revealed that stable activation of Egr mRNAs and proteins, immediate early genes responsible for activation of the LHβ promoter, requires fast GnRH pulse frequencies. In contrast, the Ngfi-A-binding protein (Nab) family of Egr corepressors is readily activated by a single pulse or low-frequency stimulation. Additionally, the SF1 corepressor, Dax1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita-critical region on the X-chromosome gene 1), is also stimulated by low-frequency GnRH pulses but is reduced at high frequency. Repression of the frequency-dependent increase in LHβ promoter activity by Nab and Dax suggests that they contribute to the lack of response at low pulse frequencies. The different sensitivity of Egr and Nab to GnRH pulses suggests a model of frequency-dependent transcriptional modulation in which gonadotropes exhibit resistance to low frequency or random signaling input but respond to high-frequency input by increasing LH β-subunit mRNA synthesis.
RESULTS
The Effect of GnRH Pulse Frequency on Gene Expression in LβT2 Gonadotrope Cells
Cells were placed in perfusion culture in serum-free medium for 1 h and then pulsed with 10 or 100 nm GnRH for 5 min. Pulses were repeated every 30, 60, 120, or 240 min for 4 h or cells were treated with tonic GnRH for 4 h. An example pulse profile is shown in Fig. 1A. Total RNA isolated from two independently performed experiments was hybridized to Affymetrix MU74Av2 oligonucleotide chips, which sample 12,000 genes and expressed sequence tags. Analysis of differential expression using the VAMPIRE Bayesian variance modeling approach (39) identified 1127 genes that were altered with GnRH, either by 4 h of pulses or tonic treatment, compared with basal, unstimulated cells in perfusion using Bonferroni multiple testing correction (αBonf < 0.05) (supplemental Table S1 published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Analysis of data from the 10 nm GnRH pulses identified 497 genes that were altered by GnRH (supplemental Table S2 published as supplemental data on The Endocrine Society’s Journals Online web site). Using a less stringent false discovery rate (FDR) correction increases the number of significantly altered genes to 3688 and 2393 for 100 nm and 10 nm GnRH pulses, respectively. We observe a significant 1.5-fold increase in LHβ at a GnRH pulse interval of 60 min using a 5% FDR cutoff, and 1.3-fold increase at longer pulse intervals that is not statistically significant at this cutoff. This increase is consistent with reports from primary cultures and in vivo studies showing an approximately 40% induction in LHβ steady-state mRNA levels in response to pulsatile GnRH treatment. Similar to reports by others, FSHβ expression was below the reliable sensitivity of the microarray assay.
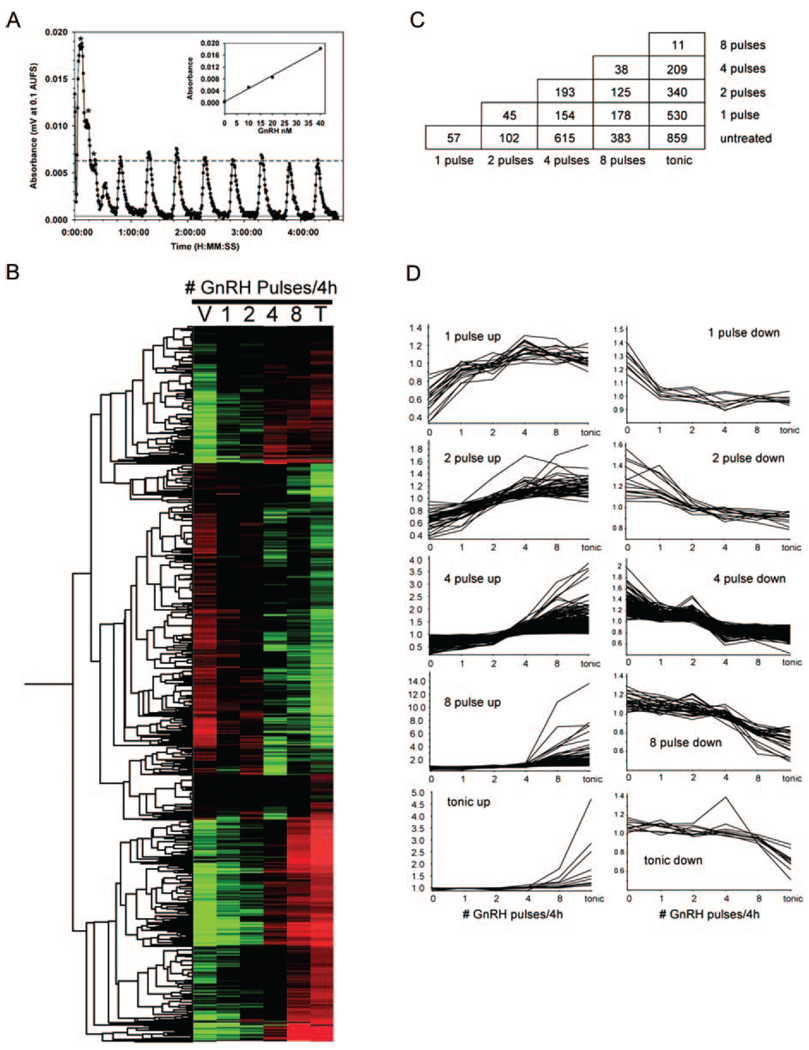
Fig. 1. Frequency-Dependent Gene Expression by GnRH Pulses in LβT2 Cells.
A, Profile of the pulse pattern obtained in cell perifusion system by absorption of phenol red tracer dye at 405 nm. Peaks marked by asterisks represent a calibration with 100%, 50%, and 25% solutions of medium containing 40 nm GnRH. Dotted line represents mean peak height of 13 nm GnRH. The first minor peak after the calibration curve indicates clearance of medium from cells cultured on microcarrier beads. The inset is a regression of calibration peak values against GnRH concentration, showing linearity of peak measurements. B, Duplicate sets of RNA isolated from perifused LβT2 cells were pulsed with 10 or 100 nm GnRH at one, two, four, or eight pulses/4 h, treated with tonic GnRH, or vehicle treated were assayed on Affymetrix MU72Av2 microarrays and compared using VAMPIRE. A heat map showing the results of unsupervised hierarchical clustering of 1127 genes regulated by 100 nm GnRH pulses by Pearson correlation is shown. Green indicates below and red indicates above median level of expression. The list of genes regulated by 10 nm and 100 nm GnRH are provided in supplemental Tables S1 and S2, respectively. C, Pairwise comparison of regulated genes in each treatment group under pulsatile 100 nm GnRH treatment. Genes in each treatment group determined to be significantly regulated in the VAMPIRE analysis were compared in each paired combination. The number of genes found to be different between each pair is reported in the chart and is found by cross-indexing of treatment groups. The comparison of untreated cells with each pulse frequency is shown in the bottom row. The comparison of a single pulse with the other frequencies is shown in the second row from the bottom. Comparisons of each subsequent group proceeds upward in the chart. A list of genes different in each pairwise comparison is provided in supplemental Table S3. D, Multiple patterns of gene expression are present under GnRH pulse regimes. Artificially constructed gene expression profiles were used to identify groups of genes with similar patterns of expression under various GnRH pulse regimes. Genes exhibiting a Pearson coefficient of 0.90 or greater are identified, and their patterns of expression are displayed in the panels under each pattern description. Consistent with the pairwise analysis in panel C, the greatest number of genes are identified under the four pulse per 4 h treatment regime, 208 genes are found to increase in expression at four pulses, whereas 138 genes re-down-regulated at the same frequency. Genes exhibiting each unique pattern of expression are identified in supplemental Table S7. T, Tonic; V, vehicle.
The median normalized expression profiles of the altered genes were grouped using hierarchical clustering (Fig. 1B). A large cluster of genes is evident at the bottom of the figure for which expression increases with increasing number of pulses to a maximum at eight pulses or tonic. A second large cluster in the center of the Fig. 1B contains genes the expression of which decreases with GnRH treatment. The third largest cluster at the top of the figure contains genes the expression of which increases with pulses of GnRH but appears to reach a maximum at four pulses with no further increase at eight pulses or with tonic treatment. This suggested the presence of genes that are induced maximally at a particular pulse frequency. Because these genes might be involved in pulse decoding by the gonadotrope, we were interested in identifying genes with such profiles. Initially, we performed a pairwise comparison of genes altered at each pulse (Fig. 1C and supplemental Table S3 published as supplemental data on The Endocrine Society’s Journals Online web site). These pairwise comparisons are useful for determining the frequency at which a gene is first seen to change. A single pulse of GnRH followed by 4 h of serum-free medium altered 57 genes. Adding more pulses during the 4-h perfusion causes an increase in the number of altered genes. There was a noticeable jump in expression when pulses were given every 60 min compared with every 120 min (193 genes). There were only 11 genes for which expression was increased by tonic GnRH treatment compared with pulses given every 30 min, suggesting that exposure to GnRH at this frequency provides maximal stimulation.
The genes altered by GnRH were mapped to Gene Ontology (GO) classifications to determine whether particular groups were overrepresented (supplemental Table S4 published as supplemental data on The Endocrine Society’s Journals Online web site). One or two pulses of 100 nm GnRH altered the smallest number of genes; therefore, the GO terms contained few genes. The only significant terms were apoptosis and cyclin-dependent kinase inhibition. As the number of pulses increased, additional terms, including kinase regulation, protein dimerization, and cell cycle regulation, became significant. Finally, cytoskeletal actin rearrangements became significant with tonic GnRH. A similar progression of terms was observed with pulses of 10 nm GnRH with the notable exception that steroid metabolic enzymes were significantly altered with 10 nm GnRH pulses every 30 min. We also mapped the dataset using GeneSet Enrichment Analysis. This analysis used all 12,000 probes rather than the list of GnRH-dependent genes. We focused on a set of 1137 curated gene sets and pathways. The gene sets that are enriched in our dataset are listed using a 5% FDR cutoff (supplemental Tables S5 and S6 published as supplemental data on The Endocrine Society’s Journals Online web site). As we had observed for the GO mapping, few gene sets were enriched with one or two pulses of GnRH. Increasing the number of pulses increased the number of significant gene sets. Ribosomal proteins are elevated consistent with the known effects of GnRH on translation, as are components of the TGFβ pathway and immediate early genes induced by adipocyte differentiation. At higher pulse frequencies, a set of genes, identified as phorbol ester target genes, are elevated, consistent with the known activation of protein kinase C by GnRH. Similar gene sets are enriched with 10 and 100 nm GnRH.
We took a different approach to identify genes for which expression showed a stepped change in expression; that is, expression increased at a particular frequency and stayed constant thereafter. To accomplish this, we created 10 synthetic expression profiles that showed the desired stepped increased and decreased expression at one, two, four, and eight pulses or tonic treatment over the 4-h treatment time. We then looked for genes for which expression profiles correlated with each synthetic profile. Genes with correlation coefficients above 0.9 were considered matching the synthetic profiles and examined. Expression profiles for these groups of genes are shown in Fig. 1D, and the list of genes with their correlation coefficients is provided in supplemental material (supplemental Table S7 published as supplemental data on The Endocrine Society’s Journals Online web site). Inspection of these genes revealed a number of transcription factors that regulate the LHβ promoter. The Egr1 and Egr2 genes are increased at fast pulse frequencies (30 and 60 min or four and eight pulses/4 h). SF1 is decreased with tonic treatment. Two Egr corepressors, Ngfi-A binding protein (Nab) 1 and 2, are induced at slow pulse frequencies (one and two pulses/4 h). Indeed, the Nab2 gene shows the highest correlation (r = 0.979) to the synthetic profile that shows a stepped increase in gene expression at pulse frequency of 120 min. Similarly, Nab1 increases with a single pulse of GnRH and shows a good correlation with the synthetic profile (r = 0.923) for a stepped increase with a single pulse. The Nab proteins are corepressors of the Egr family of transcription factors. Because of the obvious importance of these genes to the regulation of LHβ expression, we verified their expression patterns by quantitative PCR (Q-PCR). All genes showed similar changes by Q-PCR and microarray (Fig. 2). It should be noted that Egr1 and Egr2 are significantly activated at the 1 h frequency, but maximal activation occurs at the 30 min frequency for Egr1 and at the hourly frequency for Egr2. Both mRNAs exhibit sensitivity to increased pulse frequency. The Nab mRNAs appear to have peak activation levels at the hourly frequency. The single pulse and tonic treatments are significantly lower than the 1-h pulse frequency for Nab1, and the tonic treatment is lower than the 1-h frequency for Nab2. The differences in activation levels between the 2-h, 1-h, and 30-min pulse frequencies are not statistically significant for Nab 1 and Nab 2, indicating they are not sensitive to increased pulse frequency.
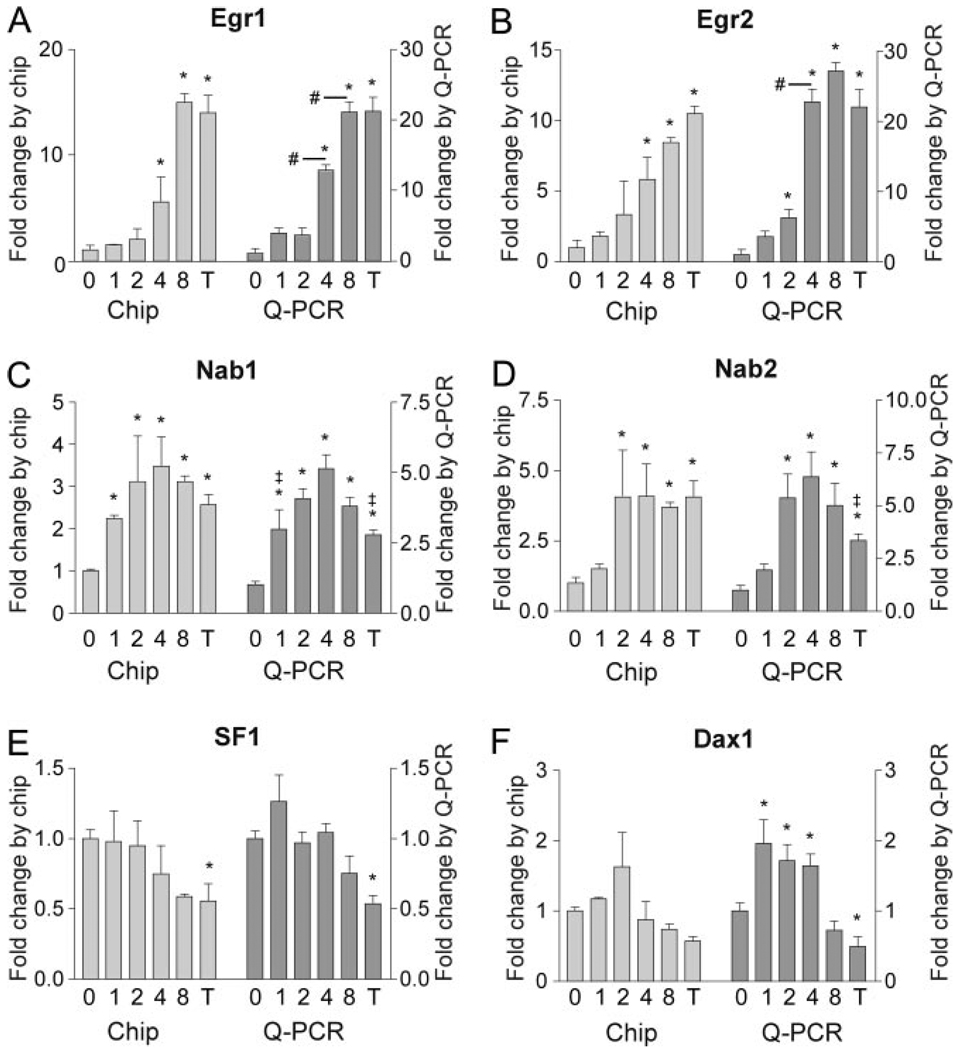
Fig. 2. Confirmation of Gene Expression Patterns Identified Using VAMPIRE Analysis of Affymetrix Microarray by Real-Time Q-PCR.
Patterns of gene expression under zero, one, two, four, and eight pulses/4 h or tonic treatment regimes with 100 nm GnRH are compared from the microarray analysis and from confirmatory Q-PCR of independent samples. The immediate early genes Egr1 (panel A) and Egr2 (panel B) and their corepressors Nab1 (panel C) and Nab2 (panel D) are confirmed to be sensitive to high- and low-frequency pulses, respectively. The regulation of the transcription factor SF1 (panel E) and its corepressor Dax 1 (panel F) are also shown. Dax 1 expression was not identified as significant in the microarray analysis; however, analysis by real time Q-PCR identified a significant increase under low-frequency pulses. *, Significant difference (P ≤ 0.05) from vehicle-treated control; #, significant difference from the preceding treatment group in panels A and B; ‡, significant difference or GnRH-treated groups from four pulse/4 h treatment in panels C and D. chip, Chromatin immunoprecipitation; T, tonic.
Another significant activator of LHβ gene expression, SF1, was found to be down regulated at the highest pulse frequency. This was confirmed by Q-PCR (Fig. 2E). Because of the association of Nab1 and Nab2 gene expression with Egr1 and Egr2 gene induction, we looked for a similar relationship between SF1 and its well-known corepressor, Dax1. In the Q-PCR analysis Dax1 showed a 2-fold increase in gene expression in response to GnRH pulses, but tonic stimulation caused a significant decline in mRNA level, similar to the decline in SF1 mRNA (Fig. 2F).
The Kinetics of Gene Expression after a Single Pulse of GnRH
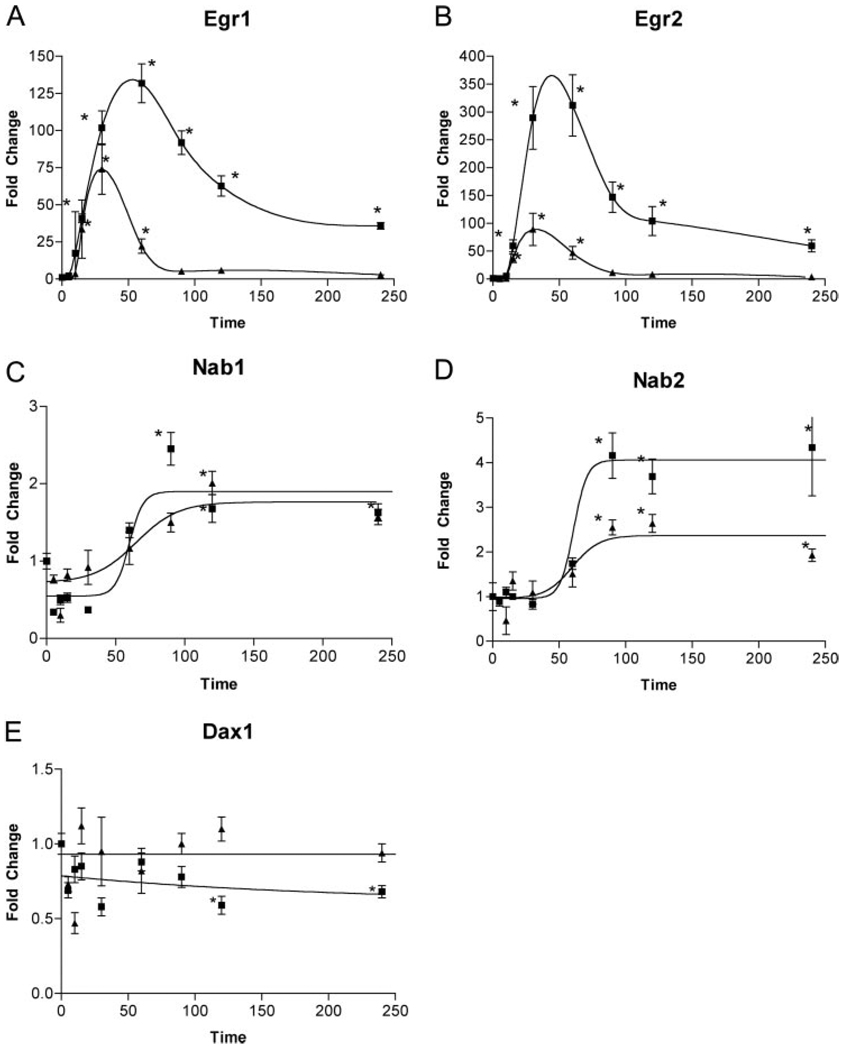
In the pulse regimes examined, mRNAs increased by a single pulse of GnRH may subsequently rapidly decay, such that after 4 h, little evidence of their change in expression is detectable. By this argument, genes that have a short half-life (typically the immediate early genes) are rapidly induced but also rapidly decay, allowing the levels of the mRNA to be regulated acutely. Therefore, we subjected cells in monolayer culture to a single 5-min pulse of either 100 nm or 10 nm GnRH and then harvested RNA at various times thereafter. Expression of Egr1, Egr2, Nab1, Nab2, and Dax1 were measured by Q-PCR. SF1 was not measured because it did not show pulse sensitivity in the initial experiments. As expected, the immediate early genes Egr1 and Egr2 were induced rapidly, within 15 min, showed maximal expression around 60 min, and then decreased rapidly by 120 min (Fig. 3, A and B). At the higher dose of GnRH, both Egr1 and Egr2 maintained elevated expression even at 4 h. At the lower dose of GnRH, expression levels showed the same rapid onset, but reached a lower maximum at 30 min and had returned to basal levels by 90 min. This result explains why Egr1 and Egr2 are significantly elevated at pulse intervals of 30 or 60 min. At the two lower frequencies, 120 or 240 min have elapsed since the previous pulse; therefore, Egr1 and Egr2 levels have returned to near basal levels when RNA is harvested. In contrast, Nab1 and Nab2 show a delayed increase after a single pulse of GnRH (Fig. 3, C and D). No expression is detected until 90 min, and expression is then constant until 4 h. The kinetics of induction are similar at the two doses of GnRH, but the maximal induction is less at the lower dose. Dax1 did not show an induction with a single pulse of GnRH, but did show a small but significant decrease in expression after 120 and 240 min at the 100 nm GnRH dose (Fig. 3E).
Fig. 3. Single-Pulse Kinetics of Egr, Nab, and Dax mRNA Expression in LβT2 Cells Shows the Basis for Differential Pulse Sensitivity.
LβT2 cells were treated for 5 min with a single pulse of 10 nm (▲) or 100 nm (■) GnRH. RNA was harvested at the times indicated, and specific mRNA content was determined by Q-PCR. The results of at least three independent determinations are displayed with error bars indicating sem. Asterisks indicate significant change in mRNA content in comparison with control untreated values as determined by permutation testing. Significance was declared at P ≤ 0.05. Egr mRNAs exhibit a rapid decay after a single pulse, whereas Nab mRNAs are increased and remain stable up to 4 h after treatment.
Induction of Egr, Nab, and Dax1 Proteins by GnRH
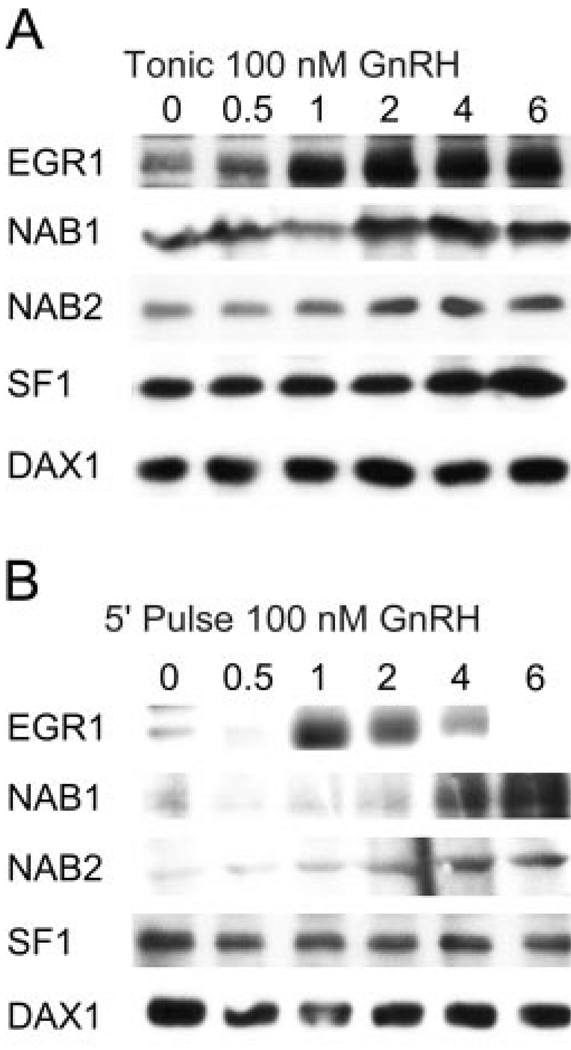
Egr1 is a well-documented regulator of LHβ gene expression, and the GnRH-dependent increase in LHβ promoter activity is partly dependent on the binding and activation of the LHβ promoter by Egr1. Mutation of the Egr binding site in the LHβ promoter or blockade of Egr activation inhibits the GnRH-dependent increase in LHβ promoter activity (11, 40). Additionally, the rapid and transient increase in Egr1 gene expression in response to GnRH treatment suggests a mode of Egr1 regulation of LHβ gene expression consistent with the dependence on pulsatile stimulation found in vivo. Interestingly, the increase in Nab gene expression in response to low GnRH pulse frequencies can be interpreted as either attenuating, in the better understood role as corepressors of Egr activity, or as synergistic, as proposed based on studies performed in CV1 cells (41). However, any model of Egr and Nab protein interaction requires the determination of protein expression levels under similar GnRH stimulatory conditions. Although both microarray and Q-PCR evaluation of Egr and Nab gene expression clearly show differential activation in response to GnRH pulses, this may not be reflected in protein expression levels. GnRH stimulation of LβT2 cells activates cap-dependent translational activity through activation of the cap-binding proteins eIF4E, eIF4G, and Mnk1 (42). Maximal activation of cap-dependent translation initiation proteins occurs within 15–30 min after stimulation with GnRH. Through this activation, GnRH can cause more efficient utilization of mRNA already present in the cell, and therefore increases in protein synthesis may not be closely linked with changes in mRNA production. In this case, although mRNA synthesis may display strong dependence on GnRH pulse regime, such differences may not be reflected in actual protein levels. To address this, we examined the induction of Egr1, Nab1, Nab2, SF1, and Dax protein levels in response to either a single GnRH pulse or to tonic treatment (Fig. 4A). Western blot analysis of LβT2 cells treated with tonic 100 nm GnRH for up to 6 h shows a strong induction of Egr1 beginning at 60 min after treatment, coincident with maximal induced level of mRNA shown in Fig. 3. Induction of Nab1 and 2 occurs later, 2 h after GnRH treatment, in agreement with the mRNA levels in Fig. 3. The appearance of Egr1 precedes the induction of the Nab2 gene at both the RNA and protein level. Egr1 has been shown to positively regulate Nab2 expression as part of a negative feedback loop (43). The increase in both protein levels is delayed approximately 30–60 min after the increase in mRNA (Fig. 3), which accentuates the differential rate of mRNA expression by GnRH. At the protein level, SF1, and Dax1, a corepressor of SF1, do not appear to change significantly in response to GnRH treatment, although the Dax mRNA levels show small changes, as determined by the response to tonic or single pulse GnRH treatment in Fig. 3.
Fig. 4. Egr1 and Nab Proteins Show Patterns of Expression Consistent with Observed Changes in mRNA under Tonic or Single-Pulse Treatment.
LβT2 cells were stimulated with tonic 100 nm GnRH for 6 h or stimulated with a single 5-min pulse of 100 nm GnRH and harvested after 6 h. Whole-cell extracts were analyzed for Egr1, Nab1, Nab2, SF1, and Dax1 content by Western blotting and chemiluminescent detection. Transient induction of Egr1 was observed under GnRH pulse conditions, whereas tonic treatment causes a stable increase in protein. In contrast, both tonic and pulse GnRH stimulation caused similar changes in Nab expression, indicating a lack of pulse sensitivity. Quantification of fold stimulation of Nab proteins is provided in supplemental Fig. S1
Changes in protein levels are striking and persistent in the presence of tonic GnRH, especially for the immediate early gene Egr1. However, transient stimulation by GnRH may lead to a distinct response of protein production. The mRNA response of Egr1 to pulsatile GnRH suggests that sustained increases in Egr1 require high-frequency pulses, whereas the Nab corepressors are induced at a single or low-frequency pulse and remain steady. To follow the kinetics of changing protein levels in response to a single GnRH pulse, plated LβT2 cells were treated with a single 100 nm pulse of GnRH for 5 min and followed for 4 h post treatment. Under this paradigm, the level of Egr1 protein is transiently induced to maximal levels lower than seen for the corresponding tonic GnRH treatment (Fig. 4B), but in a pattern consistent with that seen for induction of mRNA under similar treatment conditions (Fig. 3A). Nab1 and Nab2 are also induced in a manner consistent with induction of mRNA levels, approximately 4-fold (supplemental Fig. S1 published as supplemental data on The Endocrine Society’s Journals Online web site), similar to maximal mRNA levels achieved under the same treatment conditions (Fig. 3D). SF1 and Dax1 protein levels do not change in response to a single GnRH pulse (Fig. 3E).
In general, the levels of the proteins mediating GnRH-induced activation of the LHβ promoter follow patterns of induction consistent with the induction patterns of their corresponding mRNAs. Most significantly, Nab mRNA and protein are induced by a single GnRH pulse (Fig. 3) and are induced at both low and high pulse frequencies (Fig. 2). In contrast, Egr1 mRNA and protein levels are increased transiently by a single GnRH pulse (Fig. 3) and are maintained at high levels by high pulse frequencies (Fig. 2).
The Nab and Dax Proteins Repress LHβ Promoter Activation by GnRH in LβT2 Cells
The role of the immediate early protein Egr1 in activation of the LHβ promoter is well established. Studies of steady-state LHβ mRNA levels in vivo and in microarray studies of endogenous LHβ mRNA expression in LβT2 cells indicate that LHβ mRNA increases approximately 1.5-fold in response to GnRH treatment. Examination of promoter activity by nuclear run-on assay or by measurement of the primary unspliced LHβ transcript suggests that the LHβ induction by GnRH is greater than the 1.5-fold change in mRNA. In studies of the GnRH induction of LHβ promoter activity, it has been clearly shown that Egr1 binding to the LHβ promoter is essential for activation (11, 40), but a role for Nab corepressor proteins remains less clear (41). Despite being known as corepressors, the Nab proteins have been demonstrated to augment the effect of Egr1 and 2 on the LHβ promoter in CV-1 cells (41).This is not the case in gonadotropes, where Nab proteins are repressors of LHβ promoter activity (40). We have confirmed this previously described activity in both LβT2 and in CV-1 cells (data not shown; supplemental Fig. S2 published as supplemental data on The Endocrine Society’s Journals Online web site).
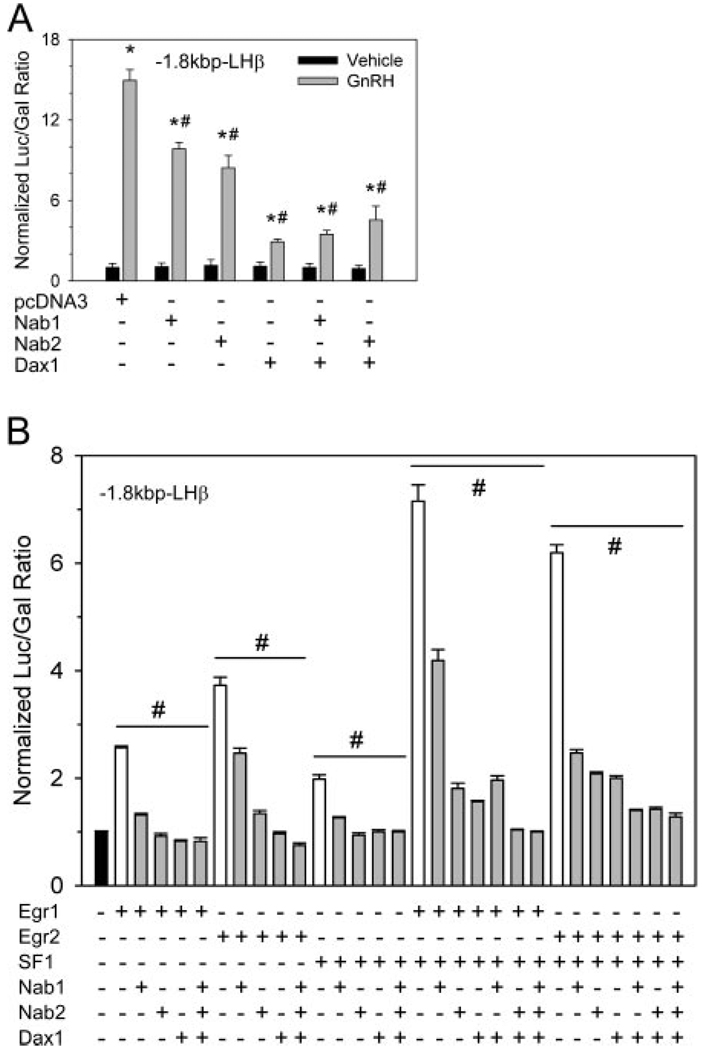
Additionally, the orphan nuclear receptor SF1 is essential for the appropriate expression of LHβ and for GnRH-induced promoter activity, but the role of Dax1 (44) is not clearly understood in the context of gonadotropin gene expression. To firmly establish the roles of both Nab and Dax proteins in activity of the LHβ promoter, we examined the ability of Nab1, Nab2, or Dax1 to repress GnRH-induced LHβ promoter activity. We cotransfected the 1.8-kb rat LHβ promoter driving the luciferase reporter gene into LβT2 gonadotrope cells with either an empty pcDNA3 expression vector or with vectors encoding Nab1, Nab2, and/or Dax1 (Fig. 5A). Treatment of transfected cells with 100 nm GnRH for 6 h resulted in a strong induction of promoter activity. Induction of LHβ promoter activity by GnRH was significantly attenuated by cotransfection of Nab2 or Dax1. Attenuation of the GnRH response was greatest by Dax1. Combination of Dax1 with Nab1 or Nab2 did not cause greater attenuation of GnRH-induced promoter activity than that seen with Dax1 alone.
Fig. 5. Both Dax1 and Nab Family Members Inhibit GnRH-Induced Activation of the −1.8-kb LHβ Promoter.
A, LβT2 cells were cotransfected with the rat −1.8-kb LHβ promoter reporter vector and expression vectors encoding Dax1, Nab1, or Nab 2 and incubated for 48 h. Cells were serum starved overnight and subsequently treated with 100 nm GnRH for 6 h. The ratio of luciferase to cotransfected CMV-β-galactosidase control reporter vector normalized to untreated, pcDNA3 null-expression vector-transfected controls is shown. Mean values are plotted with error bars representing sem. Each bar marked with * indicates significant difference between GnRH treatment and its respective vehicle-treated control (P ≤ 0.05). Bars marked with # indicate significant difference from pcDNA3-transfected, GnRH-treated control (P ≤ 0.05). B, Dax1 and Nab family members attenuate Egr- and SF1-mediated activation of the −1800-bp LHβ promoter. LβT2 cells were cotransfected with the −1.8-kb LHβ promoter reporter plasmid and an expression plasmid expressing Egr1 or Egr2, or SF1 either singly or in combination. Addition of Dax1, Nab 1, or Nab2 singly or in combination attenuated the level of activation. Groups with bars designated with # indicate significant differences (P ≤ 0.05) between activator-repressor cotransfected groups marked in gray from their respective transcriptional activator-only group marked in white.
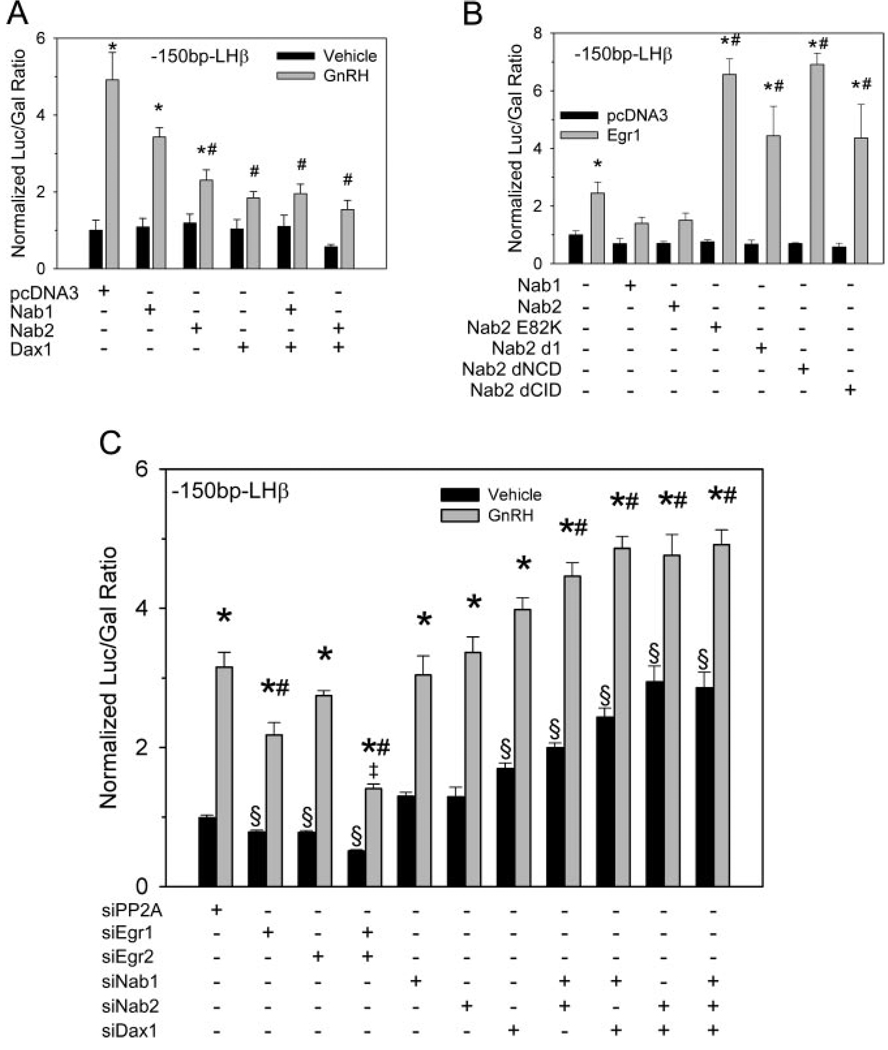
To demonstrate that overexpression of corepressor proteins directly affects the action of the cognate activators, we also examined the ability of the Nab proteins and Dax1 to directly block activation of the LHβ promoter by Egr1 and SF1. The 1.8-kb LHβ promoter was cotransfected with empty expression vector or with expression vectors encoding Egr1 and SF1 (Fig. 5B). Coexpression of Egr1 and SF1 caused a 7-fold induction of LHβ promoter activity. Coexpression of Egr1 and SF1 with Nab1, Nab2, or Dax1 alone reduced LHβ promoter activity more than 50%. Combinations of corepressors did not cause further repression. Taken together these data indicate that, in the context of the LβT2 gonadotrope cell line, Nab proteins inhibit Egr1-mediated induction of the LHβ promoter, as does Dax1. Similar results were obtained using a reporter plasmid bearing the mouse LHβ promoter truncated at bp −150 (Fig. 6A). This promoter bears only the SF1, Egr1, and homeobox binding sites of the promoter (45), therefore allowing direct examination of these regulatory sites independently of other regulatory elements found upstream. As in the rat −1.8 kb promoter study, Nab2 overexpression caused greater suppression of GnRH-induced activity than Nab1, and Dax1 caused the greatest attenuation of GnRH induction. Combination of Dax1with Nab1 and Nab2 did not cause a further suppression of GnRH induction.
Fig. 6. Basal and GnRH-Induced Activity of the −150-bp LHβ Promoter Is Dependent on Egr, Nab Proteins.
A, LβT2 cells were cotransfected with the rat −150-bp LHβ promoter reporter vector and expression vectors encoding Nab1, Nab 2, and Dax1 and incubated for 48 h. Cells were serum starved overnight and subsequently treated with 100 nm GnRH for 6 h. Each bar marked with * indicates significant difference between GnRH treatment and its respective vehicle-treated control (P ≤ 0.05). Bars marked with # indicate significant difference from pcDNA3-transfected, GnRH-treated control (P ≤ 0.05). B, LβT2 cells were transfected with the −150-bp LHβ promoter and either pcDNA3 or an Egr1 expression plasmid. Groups were also cotransfected with expression vectors encoding Nab1, Nab2, or Nab2 mutants. Bars marked with * indicate significant difference from respective pcDNA3 cotransfected control (P ≤ 0.05). Bars marked with # indicate significant difference (P ≤ 0.05) from Egr1 expression vector alone. C, Cotransfection with siRNA directed against Egr, Nab, and Dax1 mRNAs inhibits GnRH induction and increases basal activity of the LHβ promoter. LβT2 cells cotransfected with the −150-bp LHβ promoter and the siRNA noted were treated with vehicle or GnRH for 6 h and assayed for relative luciferase activity. Bars marked with * indicate significant difference (P ≤ 0.05) of GnRH treatment from respective vehicle-treated control. Bars marked with # indicate significant difference from GnRH-treated PP2A control (P ≤ 0.05). Bars marked with § indicate significant difference from vehicle-treated PP2A control (P ≤ 0.05). The bar marked with ‡ indicates significant difference from the GnRH-treated, Egr1-cotransfected group. “si” Prefix indicates small interfering. dNCD, Deleted nuclear colocalization domain; dNCID, deleted CH4 interacting domain.
Transfection of dominant-negative Nab 2 mutant E82K, which does not bind Egr proteins but dimerizes with endogenous Nabs (46), causes an increase in GnRH-stimulated promoter activity by interfering with repression by the endogenous Nab proteins (Fig. 6B). Transfection of the Nab2 deletion mutants d1, NabΔNCD2, or NabΔCID also increases the GnRH-stimulated promoter activity. These mutants lack the YRVLQRANL motif in the NCD-1 Egr-binding domain, the entire NCD2 repression domain, or the entire CID domain that interacts with the CHD4 subunit of the nucleosome-remodeling complex (46, 47).
We examined the direct role of Egr1 and Egr2 in GnRH induction of the LHβ promoter by cotransfection with small interfering RNAs (siRNAs) targeted against their mRNAs. Cotransfection of Egr1 siRNA with the −150-bp promoter reduced GnRH induction to 64% of control. Cotransfection with Egr1 siRNA did not significantly reduce GnRH induction. However, cotransfection with both Egr1 and Egr2 siRNAs in combination further reduced GnRH-induced promoter activity to 41% of control, suggesting that both factors may mediate GnRH effects (Fig. 6C). All Egr combinations caused a decrease in basal promoter activity compared with control, suggesting that both factors are positive regulators of LHβ promoter activity. Cotransfection with Nab1 or Nab2 siRNAs alone does not alter promoter activity but in combination leads to an increase in both basal and GnRH-stimulated activity of the promoter, supporting their inhibitory roles in regulating LHβ promoter activity and suggesting redundancy among the corepressors as well as the activators. Cotransfection with Dax 1 siRNA leads to an increase in promoter activity in comparison to control, and all combinations of Nab1, Nab2, and Dax1 cause increased basal and GnRH-induced promoter activity, supporting their role as negative regulators of promoter activity.
The Nab2 and Dax1 Proteins Block GnRH Pulse Sensitivity of the LH Promoter in LβT2 Cells
At the genomic level, GnRH pulsatility is essential for transcriptional activation of LHβ, and both LHβ and FSHβ gene promoters show differential sensitivity to GnRH pulse regimes. In our perifusion system, we have demonstrated a genome-wide response to GnRH pulse regimes that is unique to each pulse frequency and amplitude. Although steady-state levels of gonadotropin mRNAs increase only moderately in vivo and in cultured cells, the changes are consistent and dependent on the action of Egr1 and SF1. The pulse-dependent increases in Egr and Nab family members, as well as Dax1, suggest a coordinated role in the regulation of gonadotropin gene expression.
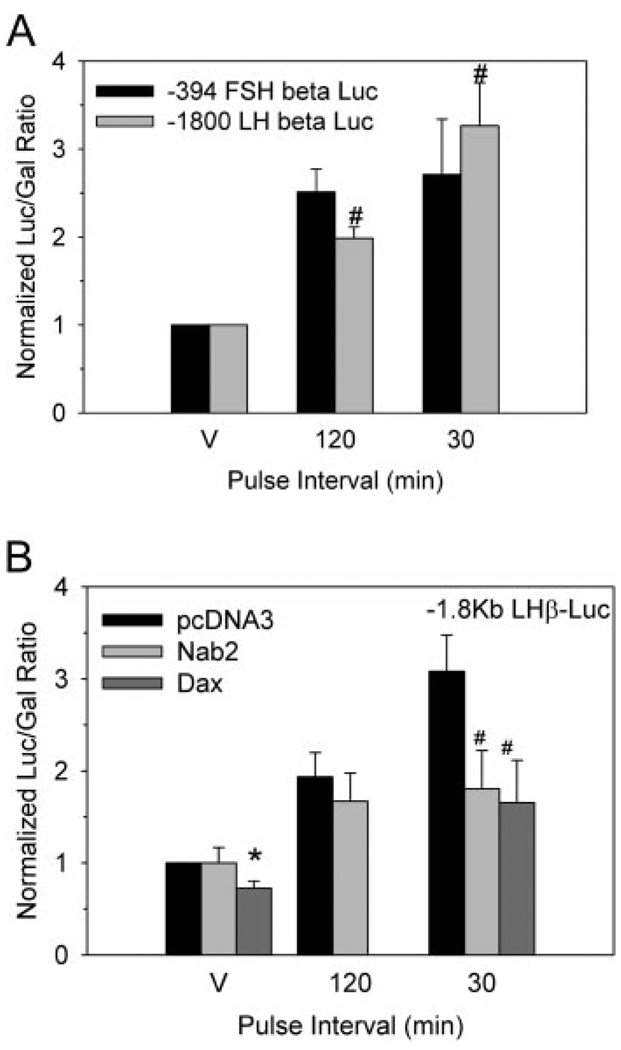
To test this model of coordinated pulse frequency-dependent activation of LHβ promoter activity, we first determined the ability of our perifusion system to recapitulate the pulse-frequency dependence of LHβ promoter activation. We transfected reporter genes under control of the −1.8-kb rat LHβ promoter or the −394-kb mouse FSHβ promoter into LβT2 cells and subjected them to 4 h of GnRH pulses at a frequency of 30 and 120 min. After 6 h, cells were harvested and assayed for reporter gene activity relative to a cotransfected β-galactosidase reporter gene under control of the cytomegalovirus (CMV) promoter. Using high-amplitude 100 nm GnRH pulses, we were unable to detect a frequency-dependent activation of LHβ reporter gene activity (data not shown). However, under lower amplitude, 10 nm GnRH pulses, we were able to show a significant pulse frequency-dependent induction of LH promoter activity, 2-fold occurring at the 120 min frequency and 3-fold occurring at the 30 min frequency, whereas the FSHβ promoter maintained similar activity at either frequency (Fig. 7A). We then examined the ability of Nab2, the family member showing the greatest repression of Egr1 activity in our model system, to inhibit frequency-dependent activation of the LHβ promoter. Consistent with the model that the high-frequency activation of the LHβ promoter is due to the increased stimulation of immediate-early gene expression, cotransfection of Nab2 abrogates the high-frequency activation of the LHβ promoter, but does not affect the level of induction occurring at low pulse frequencies (Fig. 7B). Additionally, the level of LHβ promoter induction seen at high pulse frequency in the presence of overexpressed Nab2 is similar to the level seen at the low pulse frequency. We also examined the ability of Dax1 overexpression to inhibit LHβ-promoter activation in response to GnRH pulses and found, as in the case of tonic administration of GnRH, that overexpressed Dax1 partially inhibits the induction of LHβ promoter activity (Fig. 7B). Therefore, activation of LHβ promoter activity at high pulse frequencies can be attributed to both the increase in Egr expression to levels sufficient to overcome inhibition by Nab and Dax1 inhibition of SF1.
Fig. 7. Nab2 and Dax1 Inhibit GnRH Pulse Sensitivity of the LHβ Promoter.
LβT2 cells cultured on microcarrier beads as in Fig. 1 were transfected with the −1.8 kb LHβ or −398 bp FSHβ promoter reporter vectors and serum starved for 16 h. Cells were then perifused while pulsed with vehicle or 100 nm GnRH at 120- or 30-min pulse intervals. After 6 h, cells were harvested and assayed for reporter vector activity. A, The LHβ promoter exhibits a pulse-dependent increase in activity that is not exhibited by the FSHβ promoter. #, Significant difference (P ≤ 0.05) between 120- and 30-min pulse. B, Cotransfection with Nab2 or Dax1 blocks frequency-dependent activation of the LHβ promoter. Cells transfected as above except with the addition of pcDNA3 null expression vector or vector encoding Nab2 or Dax1 were serum starved 16 h and perifused for 6 h while pulsed with vehicle or 10 nm GnRH at 120-min or 30-min pulse intervals. Luciferase activity normalized to non-pulsed null vector control is plotted. Mean values are plotted with error bars representing sem. *, Significant difference (P ≤ 0.05) of Dax1-transfected, vehicle-pulsed cells from pcDNA3 transfected vehicle-pulsed cells. #, Significant difference (P ≤ 0.05) between Nab2- and Dax1-transfected, 30-min pulsed cells from 30-min null vector-transfected cells. Both Dax1 and Nab2 attenuated the activation at 30-min pulse intervals, and Nab2 had no effect on activation at 120-min pulse intervals. V, Vehicle.
DISCUSSION
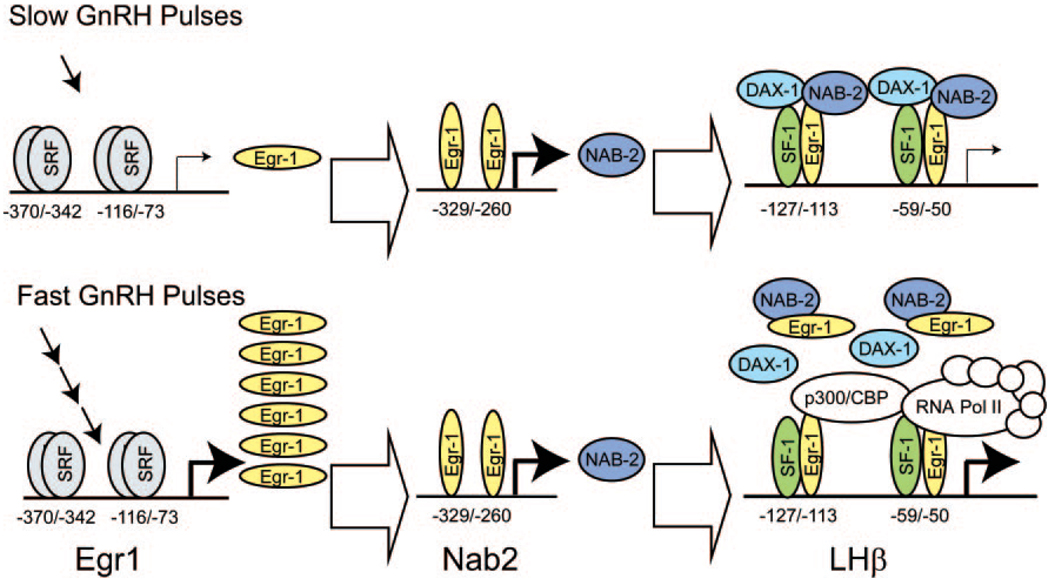
We have presented data showing that GnRH pulse frequency and amplitude differentially activate gene expression in LβT2 gonadotropes. This observation indicates that the gonadotropes are capable of interpreting different pulse regimes and are sensitive to alterations in GnRH pulse frequency and amplitude. The complement of genes increased and decreased in expression under each condition reflects the current status of hypothalamic GnRH input and, ultimately, may influence the overall status of the reproductive endocrine axis by modulating pituitary output of gonadotropins. Although distinct changes occur at each frequency, the most significant changes in gene expression occur at an interpulse interval of 60 min, where 193 genes are significantly altered over the 120 min frequency, and overall 615 genes are significantly altered relative to basal unstimulated cells. An interpulse interval of 30 min or tonic exposure causes only smaller increments of increased gene expression (38 and 11 genes, respectively), indicating that a maximal level of activation occurs at hourly pulses. Thus, the 60-min pulse frequency is a transition point in gonadotrope response to pulsatile GnRH and may represent a threshold level leading to maximal activation of GnRH-regulated gene expression. This observation is consistent with observations in primary culture (32, 48, 49) and in vivo (9) showing that higher frequencies favor increased LHβ mRNA production and LH secretion. We present data suggesting a mechanism responsible for this preferential response of the LHβ gene, and we propose a model describing how this serves as the basis for discrimination of pulse frequency by gonadotropes (Fig. 8).
Fig. 8. Proposed Model for Differential Regulation of the LHβ Promoter by an Egr/Nab Regulatory Feedback Loop.
Under a low pulse frequency, transient stimulation of Egr1 expression leads to a secondary increase in Nab2 protein. The transiently increased Egr1 level is insufficient for sustained activation of the LHβ promoter due to the repression by Nab2 and Dax1. Under high pulse frequency, high Egr1 expression is sustained but does not lead to further increases in Nab expression. The increased Egr1 level quenches Nab2 activity, allowing increased activity of the LHβ promoter through association with transcription-activating factors. CBP, cAMP response element binding protein-binding protein; Pol II, polymerase II; SRF, serum response factor.
The differential sensitivity of both Egr and Nab to GnRH pulse frequency may be central to determining the frequency-dependent response of LHβ mRNA expression. An essential component of this model is that Nab2 is induced directly by Egr1 in a negative feedback loop (43). The Nab1 and Nab2 mRNAs are increased by a single pulse of GnRH and are sustained independently of pulse frequency (Fig. 2, C and D). On the other hand, Egr1 and Egr2 mRNAs are only transiently activated at low pulse frequencies (Fig. 3, A and B). At low amplitude (10 nm), Egr mRNA levels quickly return to basal and at high amplitude (100 nm), mRNA levels are sustained at about 25% of maximal activation at 4 h. This transient increase in Egr mRNA and protein is sufficient to induce a negative feedback loop via induction of Nab2 expression. At the low and transient levels of expression induced by slow 10 nm pulses, Egr action is thus inhibited by the presence of the costimulated Nab proteins. However, at higher pulse frequency, Egr mRNA is strongly induced by GnRH and sustained at a high level that may exceed the capacity of Nab to suppress transcriptional activation (Fig. 2, A and B). The high level of Egr protein induced by GnRH also suggests that such an action can be governed by simple mass action equilibrium between Egr and Nab.
Activation of Nab1 and Egr1 by GnRH has been reported in αT3-1 cells, a cell line of the gonadotrope lineage that does not express LHβ or FSHβ (40). In αT3-1 cells, Egr1 is only transiently stimulated under tonic conditions, but Nab 1 is stimulated and remains stable. Further, Nab1 was shown to suppress Egr1 activity, consistent with their complementary roles in vivo regulating hindbrain development (50). Our observations in LβT2 cells differ from αT3-1 cells in that Egr1 is stably activated under tonic stimulation and is consistent with the observation in αT3-1 cells that Nab1 is stably activated. Others have noted increased Egr1 and Nab1 expression in LβT2 cells, but their treatments were also made under tonic stimulatory conditions (36, 37), and they did not note the sustained activation of Egr1.
In contrast to our results and that of others, Nab2 can induce expression of a truncated LHβ promoter in CV1 monkey kidney fibroblasts, and specificity of action to the LHβ promoter was demonstrated (41). The status of other factors such as SF1, smads, and homeodomain proteins known to interact at the promoter proximal to the Egr binding sites is not known, and that study was conducted outside the context of GnRH stimulation of gonadotropes. Moreover, evidence for such an action of Nab has not been demonstrated in vivo. Our results showing inhibition with Nab1 and 2 is consistent with other studies examining the role of Nab proteins in vivo (50).
The pattern of SF1 gene expression and its corepressor Dax1 also suggests a role in basal promoter regulation. LHβ gene expression is dependent on SF1, which is important in the development and differentiation of the anterior pituitary gonadotropes (51). In contrast to Egr1, SF1 is important for the basal activity of the GnRH promoter. Expression of SF1 is not significantly increased in response to GnRH, but trends toward suppression at high pulse frequencies and is significantly suppressed under tonic stimulation. Similarly, SF1 protein levels do not appear to be markedly changed by GnRH either under tonic or pulsed GnRH administration (Fig. 4, A and B). Although this would imply a decrease in LHβ promoter activity, the decrease in SF1 mRNA coincides with maximal Egr mRNA induction.
The SF1 corepressor Dax1 does show a frequency-dependent regulation of expression that appears at the mRNA level but is not seen at the protein level. The pattern of Dax1 expression predicts that overexpression of Dax1 will decrease basal levels of LHβ promoter activity. We found this to be the case in our perifusion experiments, although the suppression was only moderate, to approximately 70% of control levels (Fig. 7B). Interestingly, Dax1 overexpression also suppressed GnRH-mediated induction of LHβ promoter activity, limiting it to 1.6-fold over unstimulated control levels compared with 3.8-fold for control levels. The constant levels of Nab and Dax1 proteins synthesized in response to low-frequency stimulation may cooperatively provide a threshold level of suppression that must be exceeded by constant, high-frequency stimulation by GnRH inducing stable levels of Egr family members. This causes a transition of promoter activity from a basal mode of transcriptional activation supported by SF1 and Dax1, to a dynamic mode of regulation supported by the interaction of Egr and Nab family members. Additionally, the activation of corepressor proteins in response to single or low-frequency stimulation suggests a role of these proteins analogous to a high-pass filter providing noise suppression. The Nab and Dax corepressors may limit activation of the LHβ promoter by random inputs from uncoordinated GnRH stimulation or from other inputs that also utilize signaling intermediates that may induce Egr gene expression.
In conjunction with pulse frequency, pulse amplitude is also an important component of this regulatory model of GnRH action. It is important to note that at the 10 nm pulse amplitude the activation of Egr and Nab family members is similar in pattern to that observed at the 100 nm GnRH. Although similar levels of Nab mRNA were observed at both 10 nm and 100 nm pulse amplitudes, Egr mRNAs were induced at 10 nm to only 50% or less of the 100 nm level (supplemental material). More importantly, at the lower pulse amplitude, Egr mRNA quickly returns to basal levels whereas at the 100 nm pulse amplitude, a stable level of mRNA is maintained (Fig. 3). Therefore, at lower pulse amplitude, a sustained level of Egr mRNA is highly dependent on pulse frequency.
Others have noted the different activation rates of genes by GnRH using tonic stimulation of gonadotropes in monolayer culture (37, 40). Although generally comparable to tonic stimulation in perifusion culture, it is notable that high amplitude stimulation with GnRH causes sustained activation of Egr1 in perifusion, which was not reported previously. We have also compared tonic stimulation to various pulse frequencies and found that high-frequency pulses are capable of stable Egr induction in perifusion culture, but that the Nab corepressor proteins do not exhibit the same degree of dependence on pulse frequency. It is notable that the overall magnitude of induction is dependent on pulse amplitude, confirming earlier observations that the biosynthetic response to GnRH is coupled to amplitude of stimulation (37). Of interest is the observation that pulse frequency sensitivity is independent of amplitude. A recent study of the GnRH response at the level of individual cells suggested that gonadotropes exhibit a response to increasing amplitude through a hybrid analog-digital mechanism that could serve to suppress noise caused by variable stimulation by the hypothalamus (34). An important difference in our observations is that under pulsatile conditions in perifusion culture, GnRH signaling exhibits hysteresis in the form of stable Egr1 stimulation at high amplitude. We have not determined whether this occurs at the level of Egr1 itself or further upstream in the signaling network activated by GnRH. Current evidenced suggests that ERK signaling pathways do not exhibit hysteresis in response to GnRH stimulation, but it has been demonstrated in another ERK signaling system (52). Some aspects of the Egr1/Nab2 feedback model, e.g. the simultaneous induction of positive and negative regulators, are consistent with the parallel redundant model of response to pulsatile hormone stimulation proposed by others modeling the GnRH signaling network (33). Interestingly, both the hybrid analog-digital model of ERK activation and the model of Egr1/Nab2 feedback loop regulation serve to minimize noise in response to GnRH signaling and are not mutually exclusive explanations of regulation by pulsatile stimulation.
The Egr1/Nab2 feedback model of pulse frequency-dependent regulation of the LHβ promoter is consistent with two major aspects of gonadotropin gene expression. First, in a juvenile and peripubertal state in which GnRH release is sporadic or limited in amplitude, no significant activation of the LHβ promoter will occur due to the preferential activation of Nab and Dax. However, FSH production can occur at low pulse frequencies and can still promote gonadal maturation. With the establishment of coordinated, high-level pulsatile release of GnRH from the hypothalamus, possibly established by gonadal-steroid feedback at the level of the hypothalamus, activation of the LHβ promoter can be achieved through increased Egr expression sufficient to overcome Nab repression, and potentially, loss of Dax-mediated suppression. This presents an overall shift of the mode of LHβ gene expression from basal expression mediated by SF1, to a dynamic mode of expression mediated by Egr1 and ultimately controlled by external GnRH input. Second, such a model of LHβ gene regulation may play a role in the estrous cycle itself. Low pulse frequency GnRH stimulation may favor FSH production by gonadotropes due to active Nab and Dax suppression of LHβ gene expression. The increased GnRH pulse amplitude and frequency preceding the LH surge may then favor increased LH production by the gonadotrope by increasing Egr expression and subsequent activation of the LHβ promoter.
In summary, we have shown that GnRH pulse frequency causes distinct changes in gene expression by activating and suppressing genes in a pattern distinctive for each pulse regime. Examination of the patterns of gene expression has revealed a unique relationship in the regulation of immediate early Egr1 and Egr 2 genes responsible for GnRH-induced transcriptional activation of the LHβ promoter and their cognate corepressors, Nab1 and Nab2. The pattern of expression of these factors predicts that frequency-dependent activation of the LHβ promoter is inhibited by the Nab proteins, and we have shown this in a perifusion system. Therefore we have identified a potential mechanism by which gonadotropes can distinguish GnRH pulse frequency. The differential activation of Egr and Nab genes provides the basis for the frequency-dependent activation of the LHβ gene and, ultimately, regulates the response of the gonadotrope to altered hypothalamic GnRH input.
MATERIALS AND METHODS
Materials
GnRH was purchased from Sigma Chemical Co. (St. Louis, MO). Rabbit polyclonal anti-Egr-1, anti-Nab1, anti-Nab2, anti-SF1, anti-Dax1, and horseradish peroxidase-linked anti-rabbit antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). DMEM and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA). All other reagents were purchased from either Sigma or Fisher Scientific (Pittsburgh, PA).
Cell Culture
LβT2 cells were maintained in monolayer cultures in DMEM supplemented with 10% fetal bovine serum and antibiotics in a humidified 10% CO2 atmosphere at 37 C. For perfusion studies, cells were plated on cytodex 3 microcarrier beads (GE Healthcare, Buckinghamshire, UK) at a density of 1.5 × 107 cells/ml bed volume. After culture for 5 d, cells were pelleted by spinning at 1000 × g for 1 min, resuspended in 10/ml serum-free DMEM with antibiotics, repelleted, and placed in 10 ml serum-free DMEM 16 h. Cells were then loaded into perifusion columns and equilibrated for 1 h in serum-free DMEM supplemented with penicillin and streptomycin at a flow rate of 300 µl/min. Subsequently, cells were pulsed for 2 min once or at 28-, 58-, and 118-min intervals or given tonic 10 or 100 nm GnRH for 4 h for the microarray studies, or 6 h for the transfection studies. Perifusion chambers were constructed from C10 chromatography columns (GE Healthcare) with modified flow adapters and cells were maintained by perifusion with DMEM equilibrated in an atmosphere of 5% CO2 using a minipulse 3 peristaltic pump (Gilson, Middleton, WI). LabView (National Instruments, Austin, TX) virtual instrumentation software and a KPCI-3108 (Keithley Instruments, Cleveland, OH) analog-digital I/O card were used to control perifusion system functions and monitor instrumentation. Pump rate and output from a UV1 chromatography monitor (GE Healthcare) measuring 405 nm absorbance of phenol red tracer dye in GnRH-containing medium was accomplished using an STP-36 (Keithley Instruments) analog interface. Programmed actuation of SV-50 solenoid valves (GE Healthcare) drawing on vehicle or GnRH-containing medium was accomplished using STP-3108D1, ERA-01 relay, and an ADP 5047 interface (Keithley Instruments). Pulse profile analysis, charting, and regressions of calibration curves were performed using PeakFit and Sigma-Plot (Systat, Point Richmond, CA).
Gene Expression Profiling in LβT2 Gonadotrope Cells
Total RNA was harvested from perifused cells using Trizol LS and purified on RNeasy spin columns (QIAGEN, Valencia, CA). The RNA was quantified and its integrity checked by agarose gel electrophoresis. RNA (10 µg) from each condition was analyzed on Affymetrix mouse MU74Av2 gene chips. Duplicates were run for each condition with independently isolated RNA from independent experiments.
The raw expression values derived from Affymetrix Microarray Suite software (MAS 5) were imported into VAMPIRE without prior normalization. We have previously shown that the statistical approach implemented in VAMPIRE finds the majority of genes found by standard ANOVA approaches as well as many genes that are missed by the other methods (38). VAMPIRE is also more robust than ANOVA-based procedures at low sample number. Initially we compared cells perifused with serum-free medium to cells pulsed with GnRH at each frequency. A Bonferroni multiple testing correction (αBonf < 0.05) was applied to identify only those genes with the most robust changes. The genes altered at each frequency were combined to give a complete list of pulse-regulated genes. This approach was then used for pairwise comparisons for each frequency of GnRH treatment against each other frequency. The list of pulse-regulated genes was imported into GeneSpring (Agilent Technologies, Palo Alto, CA). The genes were clustered by hierarchical clustering using Pearson correlation as a measure of similarity. Data were median normalized for visualization with green indicating below-median and red indicating above-median expression. To identify genes that show a stepped increase or decrease in expression at a particular pulse frequency, we designed synthetic genes with the hypothetical expression profiles. We then used these profiles to search for genes the expression profiles of which correlated with the synthetic gene with r > 0.90 using GeneSpring. The expression profiles of these stepped expression clusters were generated, and lists of genes with their correlation coefficients were saved. Lists of genes altered at each pulse frequency generated by VAMPIRE were mapped to gene ontology terms using the program GOby to determine whether any GO classifications were overrepresented (39). Multiple testing correction (αBonf < 0.05) was applied to minimize GO terms arising as false positives. Gene Set Enrichment Analysis was performed using the GeneSet Enrichment Analysis software on the complete dataset as described (53).
Verification of RNA Expression by Quantitative RT-PCR
Total RNA was reverse transcribed and amplified by Q-PCR using 6-carboxy-fluorescine-labeled fluorescent locked nucleic acid probes (Exiqon, Vedbaek, Denmark). Ct values were extracted by manually setting the threshold midway between basal and maximal fluorescence on a log10 scale. Serial dilutions of plasmid or positive control RNA were run in parallel to determine amplification efficiency for each gene, and statistically significant changes were determined using the software REST (54).
Western Blotting
LβT2 cells were grown to confluence in six-well plates, washed once with PBS, and incubated in serum-free medium overnight. Cells were stimulated with agonists for various times at 37 C. Thereafter, cells were washed with ice-cold PBS, and then lysed on ice in sodium dodecyl sulfate sample buffer (50 mm Tris; 5% glycerol; 2% sodium dodecyl sulfate; 0.005% bromophenol blue; 84 mm dithiothreitol; 100 mm sodium fluoride; 10 mm sodium pyrophosphate; and 2 mm sodium orthovanadate, pH 6.8), boiled for 5 min to denature proteins, and sonicated for 5 min to shear the chromosomal DNA. Equal volumes (30–40 µl) of these lysates were separated by SDS-PAGE on 7.5 or 10% gels, electrotransferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore Corp., Bedford, MA). The membranes were blocked with 5% nonfat dried milk in Tris-buffered saline-Tween (50 mm Tris-HCl, pH 7.4; 150 mm NaCl; 0.1% Tween 20). Blots were incubated with primary antibodies in blocking buffer for 60 min at room temperature and then incubated with horseradish peroxidase-linked secondary antibodies followed by chemiluminescent detection.
Transfection and Promoter Activity Assays
In the perifusion experiments, 12–15 × 107 cells were plated on a 1-ml bed volume of cytodex 3 microcarrier beads in a 10-cm Petri dish with DMEM supplemented with 10% fetal bovine serum, penicillin, and streptomycin and grown for 5 d in an atmosphere of 5% CO2. Subsequently, cells were washed once in fresh DMEM and plated in serum-free DMEM supplemented with penicillin-streptomycin and transfected with 4 µg of a 1:1:2 ratio of pGL3-rLHβ-1.8 reporter plasmid (42): either pCMV-Sport 6 or pCMV-Sport 6 encoding the noted cDNAs, and pGL3-CMV-βGal, an internal control reporter bearing the CMV immediate early promoter and a substitution of the pGL3 luciferase coding sequence with the coding sequence of Escherichia coli β-galactosidase (42). Transfections were performed using Fugene 6 according to the manufacturer’s recommendations. After 16 h, cells were loaded into the perifusion chambers as described above and pulsed with GnRH. After the perifusion, Cells were lysed in 100 mm PBS containing 0.1% Triton X-100, vortexed for 30 sec, and clarified by centrifugation at 14,000 × g. Cell lysates were assayed directly for luciferase (Luciferase Assay System, Promega) and β-galactosidase (Galacto-Light Plus, Tropix, Bedford, MA) activity according to manufacturer’s instructions in a 96-well plate using a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA)
LβT2 cells were maintained in 10-cm diameter dishes in DMEM-supplemented cell lysates with 10% FBS at 37 C with 10% CO2. On the day before transfection experiment LβT2 cells (3 × 105 cells per well) were plated in 12-well plates (BD Biosciences, San Jose, CA). For transfection, the media were changed to fresh DMEM with 10% FBS. Transfection was performed using Fugene 6 reagent (Roche Applied Science, Indianapolis, IN) following manufacturer’s protocol. Each well was transfected with 500 ng of −150LHβ or pGL3-rLHβ1.8 reporter plasmid, 50 ng of tk-lacZ, and 50 ng of each expression vector. Empty vectors were used to balance DNA mass as necessary. The following day, the cells were switched to serum-free DMEM supplemented with 0.1% BSA. After incubation for 24 h, the cells were treated with vehicle or 100 nm GnRH for 6 h. Cell lysates were assayed for luciferase and β-galactosidase activity as above.
RNA Interference
siRNAs were purchased from Ambion, Inc. (Austin, TX). LβT2 cells were seeded into 12-well culture plates 1 d before transfection. On d 1, the cells (60–70% confluence) were transfected with 100 nm Egr1, Egr2, NAB1, NAB2, DAX1, or scrambled siRNA using 2 µl FuGene 6 (Roche). The cells were incubated with siRNAs for 48 h. After 24 h the medium was replaced with fresh serum-free medium. Cells were harvested after 48 h and were assayed for luciferase and β-galactosidase activity as above. siRNAs were directed against Egr-1 (siRNA identification no. 157281), 5′-CCUUUCCUACUCCCAACACtt-3′ (sense) and 5′-GUGUUGGGAGUAGGAAAGGtg-3′ (antisense); Egr-2 (siRNA identification no. 240542) 5′-CCUAGAAACCAGACCUUUCAtt-3′ (sense) and 5′-UGAAGGUCUGGUUUCUAGGtg-3′ (antisense); Nab1 (siRNA identification no. 155510), 5′-GCUAGUGUGAUACCAUUUAtt-3′ (sense) and 5′-UAAAUGGUAUCACACUAGCtc-3′ (antisense); Nab2 (siRNA identification no. 155511), 5′-CCUCCUCUCUUACGAGtt-3′ (sense) and 5′-CUCGUAGUAAGAGAGGAGGtt-3′ (antisense); Nr0b1 (siRNA identification no. 162201), 5′-GAUCACCUGCACUUCGAGAtt-3′ (sense) and 5′-UCUCGAAGUGCAGGUGAUCtt-3′ (antisense), or Nr5a1 (siRNA identification no. 188983), 5′-CCUUGUGUCACUACCCACAtt-3′ (sense) and 5′-UGUGGGUAGUGACACAAGGtg-3′ (antisense).
Statistical Analysis
Microarray data were analyzed using VAMPIRE as described in Results; confirmatory Q-PCR data were analyzed by permutation testing for the Q-PCR data. Other data were analyzed using JMP version 5.1 (SAS, Carey, NC). Data were obtained from at least three independent experiments. If necessary, data were corrected for heteroscedasticity and nonnormal residual error by optimal Box-Cox transformation and then subjected to ANOVA using the linear modeling platform. Significance was determined using appropriate post hoc tests at a critical value of α = 0.05. Student’s t test was used for specific pairwise comparisons and Dunnett’s Comparison to Control or Tukey’s honestly significant difference was used for multiple group comparisons. Significance was declared at P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank Drs. L. Jameson (Northwestern University, Evanston, IL), J. Milbrandt (Washington University, St. Louis, MO), and J. Svaren (University of Wisconsin, Madison, WI) for expression plasmids.
This work was supported by R01 HD43758 and K02 HD40803 (to M.A.L.), R01 HD047400 (to N.J.G.W.), R37 HD020377 (to P.L.M.), and by National Institute of Child Health and Human Disease/National Institutes of Health through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction Research (M.A.L., N.J.G.W., P.L.M.). H.Z. was supported by National Institutes of Health Grant T32 HD007203.
Abbreviations
- CMV
Cytomegalovirus
- Dax1
dosage-sensitive sex reversal-adrenal hypoplasia congenita-critical region on the X-chromosome gene 1
- Egr
early growth response
- FDR
false discovery rate
- GO
Gene Ontology
- Nab
Ngfi-A binding protein
- Q-PCR
quantitative PCR
- SF1
steroidogenic factor 1
- siRNA
small interfering RNA
- Smad
Sma- and Mad-related protein
Footnotes
Disclosure Statement: The authors have nothing to disclose.
REFERENCES
- 1.Conn PM, Crowley WFJ. Gonadotropin-releasing hormone and its analogs. Annu Rev Med. 1994;45:391–405. doi: 10.1146/annurev.med.45.1.391. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser UB, Conn PM, Chin WW. Feb Studies of gonadotropin-releasing hormone (GnRH) action using GnRH receptor-expressing pituitary cell lines. Endocr Rev. 1997;18:46–70. doi: 10.1210/edrv.18.1.0289. [DOI] [PubMed] [Google Scholar]
- 3.McDowell IF, Morris JF, Charlton HM. Characterization of the pituitary gonadotroph cells of hypogonadal (hpg) male mice: comparison with normal mice. J Endocrinol. 1982;95:321–330. doi: 10.1677/joe.0.0950321. [DOI] [PubMed] [Google Scholar]
- 4.Achermann JC, Jameson JL. Advances in the molecular genetics of hypogonadotropic hypogonadism. J Pediatr Endocrinol Metab. 2001;14:3–15. doi: 10.1515/jpem.2001.14.1.3. [DOI] [PubMed] [Google Scholar]
- 5.Bedecarrats GY, Linher KD, Janovick JA, Beranova M, Kada F, Seminara SB, Michael Conn P, Kaiser UB. Four naturally occurring mutations in the human GnRH receptor affect ligand binding and receptor function. Mol Cell Endocrinol. 2003;205:51–64. doi: 10.1016/s0303-7207(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 6.Beranova M, Oliveira LM, Bedecarrats GY, Schipani E, Vallejo M, Ammini AC, Quintos JB, Hall JE, Martin KA, Hayes FJ, Pitteloud N, Kaiser UB, Crowley WFJ, Seminara SB. Apr prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2001;86:1580–1588. doi: 10.1210/jcem.86.4.7395. [DOI] [PubMed] [Google Scholar]
- 7.Knobil E. On the control of gonadotropin secretion in the rhesus monkey. Recent Prog Horm Res. 1974;30:1–46. doi: 10.1016/b978-0-12-571130-2.50005-5. [DOI] [PubMed] [Google Scholar]
- 8.McGarvey C, Cates PA, Brooks A, Swanson IA, Milligan SR, Coen CW, O’Byrne KT. Phytoestrogens and gonadotropin-releasing hormone pulse generator activity and pituitary luteinizing hormone release in the rat. Endocrinology. 2001;142:1202–1208. doi: 10.1210/endo.142.3.8015. [DOI] [PubMed] [Google Scholar]
- 9.Burger LL, Dalkin AC, Aylor KW, Haisenleder DJ, Marshall JC. GnRH pulse frequency modulation of gonadotropin subunit gene transcription in normal gonadotropes-assessment by primary transcript assay provides evidence for roles of GnRH and follistatin. Endocrinology. 2002;143:3243–3249. doi: 10.1210/en.2002-220216. [DOI] [PubMed] [Google Scholar]
- 10.Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem. 1999;274:13870–13876. doi: 10.1074/jbc.274.20.13870. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser UB, Halvorson LM, Chen MT. Aug Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-β gene promoter: an integral role for SF-1. Mol Endocrinol. 2000;14:1235–1245. doi: 10.1210/mend.14.8.0507. [DOI] [PubMed] [Google Scholar]
- 12.Mouillet JF, Sonnenberg-Hirche C, Yan X, Sadovsky Y. p300 regulates the synergy of steroidogenic factor-1 and early growth response-1 in activating luteinizing hormone-β subunit gene. J Biol Chem. 2004;279:7832–7839. doi: 10.1074/jbc.M312574200. [DOI] [PubMed] [Google Scholar]
- 13.Morohashi KI, Omura T. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 1996;10:1569–1577. doi: 10.1096/fasebj.10.14.9002548. [DOI] [PubMed] [Google Scholar]
- 14.Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X, Wong M, Bakke M, Zhao L, Frigeri C, Hanley NA, Stallings N, Schimmer BP. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res. 2002;57:19–36. doi: 10.1210/rp.57.1.19. [DOI] [PubMed] [Google Scholar]
- 15.Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 1995;92:10939–10943. doi: 10.1073/pnas.92.24.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, Bakke M, Hanley NA, Majdic G, Stallings NR, Jeyasuria P, Parker KL. Tissue-specific knockouts of steroidogenic factor 1. Mol Cell Endocrinol. 2004;215:89–94. doi: 10.1016/j.mce.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 18.Tourtellotte WG, Nagarajan R, Bartke A, Milbrandt J. Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Mol Cell Biol. 2000;20:5261–5268. doi: 10.1128/mcb.20.14.5261-5268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tourtellotte W, Nagarajan R, Auyeung A, Mueller C, Milbrandt J. Infertility associated with incomplete spermatogenic arrest and oligozoospermia in Egr4-deficient mice. Development. 1999;126:5061–5071. doi: 10.1242/dev.126.22.5061. [DOI] [PubMed] [Google Scholar]
- 20.Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- 21.Lei ZM, Mishra S, Zou W, Xu B, Foltz M, Li X, Rao CV. Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene. Mol Endocrinol. 2001;15:184–200. doi: 10.1210/mend.15.1.0586. [DOI] [PubMed] [Google Scholar]
- 22.Zhang FP, Poutanen M, Wilbertz J, Huhtaniemi I. Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice. Mol Endocrinol. 2001;15:172–183. doi: 10.1210/mend.15.1.0582. [DOI] [PubMed] [Google Scholar]
- 23.Huang HJ, Sebastian J, Strahl BD, Wu JC, Miller WL. Transcriptional regulation of the ovine follicle-stimulating hormone-β gene by activin and gonadotropin-releasing hormone (GnRH): involvement of two proximal activator protein-1 sites for GnRH stimulation. Endocrinology. 2001;142:2267–2274. doi: 10.1210/endo.142.6.8203. [DOI] [PubMed] [Google Scholar]
- 24.Miller WL, Shafiee-Kermani F, Strahl BD, Huang HJ. The nature of FSH induction by GnRH. Trends Endocrinol Metab. 2002;13:257–263. doi: 10.1016/s1043-2760(02)00614-8. [DOI] [PubMed] [Google Scholar]
- 25.Coss D, Jacobs SBR, Bender CE, Mellon PL. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone β gene by gonadotropin-releasing hormone. J Biol Chem. 2004;279:152–162. doi: 10.1074/jbc.M304697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 27.Gregory SJ, Kaiser UB. Regulation of gonadotropins by inhibin and activin. Semin Reprod Med. 2004;22:253–267. doi: 10.1055/s-2004-831901. [DOI] [PubMed] [Google Scholar]
- 28.Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang HJ, Miller WL, Mellon PL. Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- 29.Bernard DJ. Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone β subunit in mouse gonadotrope cells. Mol Endocrinol. 2004;18:606–623. doi: 10.1210/me.2003-0264. [DOI] [PubMed] [Google Scholar]
- 30.Dupont J, McNeilly J, Vaiman A, Canepa S, Combarnous Y, Taragnat C. Activin signaling pathways in ovine pituitary and LβT2 gonadotrope cells. Biol Reprod. 2003;68:1877–1887. doi: 10.1095/biolreprod.102.012005. [DOI] [PubMed] [Google Scholar]
- 31.Suszko MI, Lo DJ, Suh H, Camper SA, Woodruff TK. Mar Regulation of the rat follicle-stimulating hormone β-subunit promoter by activin. Mol Endocrinol. 2003;17:318–332. doi: 10.1210/me.2002-0081. [DOI] [PubMed] [Google Scholar]
- 32.Haisenleder D, Dalkin A, Ortolano G, Marshall J, Shupnik M. A pulsatile gonadotropin-releasing hormone stimulus is required to increase transcription of the gonadotropin subunit genes: evidence for differential regulation of transcription by pulse frequency in vivo. Endocrinology. 1991;128:509–517. doi: 10.1210/endo-128-1-509. [DOI] [PubMed] [Google Scholar]
- 33.Krakauer DC, Page KM, Sealfon S. Module dynamics of the GnRH signal transduction network. J Theor Biol. 2002;218:457–470. [PubMed] [Google Scholar]
- 34.Ruf F, Park MJ, Hayot F, Lin G, Roysam B, Ge Y, Sealfon SC. Mixed analog/digital gonadotrope biosynthetic response to gonadotropin releasing hormone. J Biol Chem. 2006;281:30967–30978. doi: 10.1074/jbc.M606486200. [DOI] [PubMed] [Google Scholar]
- 35.Kakar SS, Winters SJ, Zacharias W, Miller DM, Flynn S. Identification of distinct gene expression profiles associated with treatment of LβT2 cells with gonadotropin-releasing hormone agonist using microarray analysis. Gene. 2003;308:67–77. doi: 10.1016/s0378-1119(03)00446-3. [DOI] [PubMed] [Google Scholar]
- 36.Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem. 2001;276:47195–47201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- 37.Yuen T, Wurmbach E, Ebersole BJ, Ruf F, Pfeffer RL, Sealfon SC. Coupling of GnRH concentration and the GnRH receptor-activated gene program. Mol Endocrinol. 2002;16:1145–1153. doi: 10.1210/mend.16.6.0853. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Bailey JS, Coss D, Lin B, Tsutsumi R, Lawson MA, Mellon PL, Webster NJ. Activin modulates the transcriptional response of LβT2 Cells to gonadotropin-releasing hormone and alters cellular proliferation. Mol Endocrinol. 2006;20:2909–2930. doi: 10.1210/me.2006-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsiao A, Ideker T, Olefsky JM, Subramaniam S. VAMPIRE microarray suite: a web-based platform for the interpretation of gene expression data. Nucleic Acids Res. 2005;33:W627–W632. doi: 10.1093/nar/gki443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfe MW, Call GB. Early growth response protein 1 binds to the luteinizing hormone-β promoter and mediates gonadotropin-releasing hormone-stimulated gene expression. Mol Endocrinol. 1999;13:752–763. doi: 10.1210/mend.13.5.0276. [DOI] [PubMed] [Google Scholar]
- 41.Sevetson BR, Svaren J, Milbrandt J. A novel activation function for NAB proteins in EGR-dependent transcription of the luteinizing hormone β gene. J Biol Chem. 2000;275:9749–9757. doi: 10.1074/jbc.275.13.9749. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen KA, Santos SJ, Kreidel MK, Diaz AL, Rey R, Lawson MA. Acute regulation of translation initiation by gonadotropin-releasing hormone in the gonadotrope cell line LβT2. Mol Endocrinol. 2004;18:1301–1312. doi: 10.1210/me.2003-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumbrink J, Gerlinger M, Johnson JP. Egr-1 induces the expression of its corepressor nab2 by activation of the nab2 promoter thereby establishing a negative feedback loop. J Biol Chem. 2005;280:42785–42793. doi: 10.1074/jbc.M511079200. [DOI] [PubMed] [Google Scholar]
- 44.Iyer AK, McCabe ER. Molecular mechanisms of DAX1 action. Mol Genet Metab. 2004;83:60–73. doi: 10.1016/j.ymgme.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Coss D, Thackray VG, Deng CX, Mellon PL. Activin regulates luteinizing hormone β-subunit gene expression through Smad-binding and homeobox elements. Mol Endocrinol. 2005;19:2610–2623. doi: 10.1210/me.2005-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svaren J, Sevetson BR, Golda T, Stanton JJ, Swirnoff AH, Milbrandt J. Novel mutants of NAB corepressors enhance activation by Egr transactivators. EMBO J. 1998;17:6010–6019. doi: 10.1093/emboj/17.20.6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2006;281:15129–15137. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- 48.Shupnik MA. Effects of gonadotropin-releasing hormone on rat gonadotropin gene transcription in vitro: requirement for pulsatile administration for lutenizing hormone-β gene stimulation. Mol Endocrinol. 1990;4:1444–1450. doi: 10.1210/mend-4-10-1444. [DOI] [PubMed] [Google Scholar]
- 49.Shupnik MA, Fallest PC. Pulsatile GnRH regulation of gonadotropin subunit gene transcription. Neurosci Biobehav Rev. 1994;18:597–599. doi: 10.1016/0149-7634(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 50.Mechta-Grigoriou F, Garel S, Charnay P. Nab proteins mediate a negative feedback loop controlling Krox-20 activity in the developing hindbrain. Development. 2000;127:119–128. doi: 10.1242/dev.127.1.119. [DOI] [PubMed] [Google Scholar]
- 51.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–154. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 52.Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 53.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 54.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.