Abstract
Purpose
To identify the genes and noncoding RNAs (ncRNAs) involved in the neuroprotective actions of a dietary antioxidant (saffron) and of photobiomodulation (PBM).
Methods
We used a previously published assay of photoreceptor damage, in which albino Sprague Dawley rats raised in dim cyclic illumination (12 h 5 lux, 12 h darkness) were challenged by 24 h exposure to bright (1,000 lux) light. Experimental groups were protected against light damage by pretreatment with dietary saffron (1 mg/kg/day for 21 days) or PBM (9 J/cm2 at the eye, daily for 5 days). RNA from one eye of four animals in each of the six experimental groups (control, light damage [LD], saffron, PBM, saffronLD, and PBMLD) was hybridized to Affymetrix rat genome ST arrays. Quantitative real-time PCR analysis of 14 selected genes was used to validate the microarray results.
Results
LD caused the regulation of 175 entities (genes and ncRNAs) beyond criterion levels (p<0.05 in comparison with controls, fold-change >2). PBM pretreatment reduced the expression of 126 of these 175 LD-regulated entities below criterion; saffron pretreatment reduced the expression of 53 entities (50 in common with PBM). In addition, PBM pretreatment regulated the expression of 67 entities not regulated by LD, while saffron pretreatment regulated 122 entities not regulated by LD (48 in common with PBM). PBM and saffron, given without LD, regulated genes and ncRNAs beyond criterion levels, but in lesser numbers than during their protective action. A high proportion of the entities regulated by LD (>90%) were known genes. By contrast, ncRNAs were prominent among the entities regulated by PBM and saffron in their neuroprotective roles (73% and 62%, respectively).
Conclusions
Given alone, saffron and (more prominently) PBM both regulated significant numbers of genes and ncRNAs. Given before retinal exposure to damaging light, thus while exerting their neuroprotective action, they regulated much larger numbers of entities, among which ncRNAs were prominent. Further, the downregulation of known genes and of ncRNAs was prominent in the protective actions of both neuroprotectants. These comparisons provide an overview of gene expression induced by two neuroprotectants and provide a basis for the more focused study of their mechanisms.
Introduction
The photoreceptors (rods and cones) of mammalian retina are the most specialized, metabolically active and fragile of the nerve cells of the retina [1–3]. Photoreceptors are also the most vulnerable of retinal cells to genetic stress, induced by mutations in genes whose expression is specific to photoreceptors, and in ubiquitously expressed genes [4,5]. The breakdown of photoreceptor stability is a major element of age-related retinal disease, and therefore of age-related blindness [6].
The stress-induced death of photoreceptors is accompanied by damage to the survivors [7–9]. Both death and damage appear to be caused by oxidative stress, i.e., by the damaging effects of partially reduced forms of oxygen, often called reactive oxygen species. Absorption of light (the normal function of photoreceptor outer segments) increases oxidation of their lipids, creating morphological and functional damage as light exposure is increased [10–12]. The idea that light-induced damage is caused by oxidative stress is supported by evidence that levels of endogenous antioxidants increase following light damage [13–15], and that exogenous antioxidants are protective [15–21], for cones [22,23] as well as rods.
We have explored the neuroprotective potential of the ancient spice saffron, which shows a strong protective effect against light-induced damage of photoreceptors [24]. The stigmata of Crocus sativus contain powerful antioxidants (crocin, crocetin) in biologically high concentrations [25]; their multiple C=C bonds give the stigmata their color, fragrance, taste, and antioxidant potential. Their concentration in saffron may be an evolutionarily special case, as the plant is a sterile triploid bred by vegetative propagation for its fragrance, taste, color, and medicinal properties. In a recent double blind clinical trial [26], saffron (2 μg/day over 12 weeks) induced a partial but consistent recovery of the electroretinogram elicited from the macula, and of visual acuity. We have also pioneered the use of photobiomodulation (PBM) as a retinal neuroprotectant. Red to infrared (600–1,000 nm) light at low intensities promotes wound healing in skin and oral mucosa [27], and protects photoreceptors from toxin- [28], genetic- [29], and light-induced [30] damage. Furthermore, it reduces laser-induced retinal scarring. PBM delivered transcranially reduces cerebral pathology in animal models of brain damage [31–33] and in human ischemic stroke [34]. PBM acts partly by repairing mitochondrial function and upregulating oxidative phosphorylation [35]. Again, no harmful side effects have been reported at the doses used in this in vivo work (daily doses of 5 J/cm2 or less). To develop the understanding of these neuroprotective effects, we have used microarray techniques to identify the genes regulated by saffron and PBM in their protective actions.
Methods
Experimental organization
The protective potential of dietary saffron, and of PBM, was tested using a light damage assay. Animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, and with protocols approved by the ANU Animal Ethics Committee. Young adult Sprague Dawley rats aged P80–120 were reared in 5 lux cyclic light, and prepared in six groups. Each group comprised two males and two females.
Control
These animals were raised in 5 lux cyclic light, as above. They were routinely fed a vegetable (potato or rice) matrix, developed as a biodegradable packaging material, and we used the same matrix as vehicle for feeding them with saffron.
Saffron-exposed only
Animals were fed saffron at 1 mg/kg/day for 3 weeks. Saffron (stigmata of Crocus sativus, from the Abbruzzo region in Italy) was soaked in water (at 2 mg of spice/ml H2O) and 12 h was allowed for the major antioxidants, which are water-soluble [25], to dissolve fully. The solute was then fed to the rats by injecting a small volume into a piece of the vegetable matrix, which the animal readily ingested. The volume for each daily feed was calculated to provide the solutes from 1 mg of saffron/kg bodyweight. Tissue was collected 24 h after the last feed.
Photobiomodulation-exposed only
Animals were exposed to 670 nm red light from a WARP 75 source (60mW/cm2, Quantum Devices Inc., Barneveld, WI). Animals were handled gently over several days until they were adapted to handling. Each was then gently restrained with a towel and held under a Plexiglas platform with the head ~2.5 cm below the platform. The WARP75 device was placed on top of the platform and turned on for 3 min. This arrangement provided a fluence of 9 J/cm2 at the eye. The animals did not hide from or appear agitated by the red light. Animals were treated in this way once daily for 5 days at 9:00 AM. Tissue was collected 24 h after the last treatment.
Light-damaged only
The animals were kept individually in Plexiglas cages, with food kept on the floor of the cages and water offered from transparent containers, to ensure uniform exposure. After overnight dark adaptation, animals were exposed to bright (1,000 lux) light for 24 h, from a white fluorescent source. Exposure began and ended at 9:00 AM
Saffron light damaged
Animals in this group were fed saffron for 3 weeks, as above. At 9:00 AM on the last day of feeding, they were exposed to damaging light for 24 h, as above. Tissue was collected at the end of this 24 h period.
Photobiomodulation light damaged
Animals in this group were exposed to PBM, as above, for 5 days. Beginning at 9:00 AM on the last day of treatment, they were exposed to damaging light for 24 h, as above. Tissue was collected at the end of this 24 h period.
Tissue collection
At the points in the protocol specified above, animals were euthanized with Lethabarb (60 mg/kg intraperitoneally). The retina from one eye of each animal was dissected free immediately, and placed in an individual tube containing RNAlater (Ambion Biosystems, Austin, TX), and stored at 4 °C overnight. The following day, tubes were transferred to –80 °C. The fellow eye was fixed by immersion in 4% (W/V) paraformaldehyde for examination of morphology and immunohistochemistry.
Fellow eyes were marked on the superior aspect with indelible pen for future orientation, enucleated and immersion-fixed in 4% (W/V) paraformaldehyde for 3 h, washed in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 at pH of 7.4) thrice, then cryoprotected by immersion in 15% (W/V) sucrose overnight. Eyes were sectioned at 12 μm on a cryostat in the superior-inferior axis.
RNA extraction and analysis
RNA was extracted and purified using previously published methods [36]. To determine the quantity and purity of the sample, RNA was analyzed on an ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE) and a 2100-Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA samples were used only if the A260/A280 ratio was above 1.8 and the RNA integrity number was greater than 8.5.
Microarray analysis
To study the changes in gene expression induced in the six experimental groups, we used 18 Affymetrix (Santa Clara, CA) Rat Genome ST arrays. These microarrays contain over 700,000 twenty-five-mer oligonucleotide features representing 27,342 genes. Labeling, hybridization, washing, and scanning of the microarray were performed at the Australian Cancer Research Foundation (ACRF) Biomolecular Resource Facility at the John Curtin School of Medical Research, Australian National University, following the manufacturers’ specifications. The arrays were scanned on the Affymetrix GeneChip 3000 7G high resolution scanner and analyzed using the GeneSpring GX v10 software (Agilent Technologies) and Partek Genomic Suite 6.4 Software (Partek Inc., St. Louis, MO). The hierarchical clustering was performed using GeneSpring on the full entity list (genes plus noncoding RNA [ncRNA]) for each of the six groups. Normalization was performed using the Robust Multichip Average (RMA) algorithm and only gene expression levels with statistical significance (p<0.05) were recorded as being “present” above background levels. Genes with expression levels below this statistical threshold were considered “absent.” For the box and whisker plot, we first ran a multivariate ANOVA (ANOVA) analysis on the six groups to identify genes whose expression was significantly varied (p<0.05, fold-change >2). This yielded a list of 187 entities, from which the box and whisker plot was generated.
The Partek Genomic Suite was used to identify genes and ncRNAs whose expression differed between experimental groups, typically between one experimental group and one control group. Data in the form of a computerized version of the .DAT file (CEL) files were imported and gene expression values were derived using the RMA algorithm on the “core” metaprobe list, which represents RefSeq genes and full-length GenBank mRNAs. For each comparison between treatment and control group, two-sample Student t tests were used to calculate the probability P that the expression of a gene had not changed. Genes and ncRNAs whose expression was significantly changed by treatment were selected using the criteria that p<0.05 and the fold-change in expression >2. The microarray data discussed in this publication have been uploaded to the National Center for Biotechnology Information (NCBI’s) Gene Expression Omnibus [37] and are accessible through gene expression omnibus (GEO) Series accession number GSE22818.
Quantitative polymerase chain reaction
RNA for quantitative polymerase chain reaction (qPCR) was handled in the same way as RNA extracted for the GeneChip® experiments. Three biologic groups were used, with one animal in each treatment group. Superscript III and the accompanying standard protocol (Invitrogen, Carlsbad, CA) were used to convert 1 µg of retinal RNA to cDNA (cDNA). TaqMan® (Applied Biosystems, Foster City, CA) Gene Expression Mastermix (Cat# 4369514) and probes (Table 1) were used to assess the validity of gene expression changes identified in the microarray experiment using a StepOne Plus qPCR machine and StepOne software v2.1 (Applied Biosystems). Assays were performed in duplicate (to account for individual sample variability) and biologic triplicate (to account for biologic variability), with fold changes determined using comparative cycle threshold (Ct; delta-delta ct). Both glyceraldehyde 3-phosphate dehydrogenase (Gapdh) and β-actin (Actb) were used as reference genes in all qPCR experiments.
Table 1. TaqMan Probes used for qPCR.
| Name | Gene symbol | TaqMan assay ID |
|---|---|---|
| angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
Agt |
Rn00593114_m1 |
| Beta actin |
Actb (Control) |
Rn00667869_m1 |
| carnitine O-octanoyltransferase |
Crot |
Rn00583174_m1 |
| chemokine (C-C motif) ligand 2 |
Ccl2 |
Rn01456716_g1 |
| endothelin 2 |
Edn2 |
Rn00561135_m1 |
| fatty acid binding protein 5, epidermal |
Fabp5 |
Rn00821817_g1 |
| fibroblast growth factor 2 |
Fgf2 |
Rn00570809_m1 |
| glyceraldehyde-3-phosphate dehydrogenase |
Gapdh (Control) |
Rn99999916_s1 |
| glial fibrillary acidic protein |
Gfap |
Rn00566603_m1 |
| glutathione peroxidase 3 |
Gpx3 |
Rn00673916_g1 |
| heme oxygenase (decycling) 1 |
Hmox1 |
Rn01536933_m1 |
| optineurin |
Optn |
Rn00595346_m1 |
| signal transducer and activator of transcription 3 |
Stat3 |
Rn00562562_m1 |
| suppressor of cytokine signaling 3 |
Socs3 |
Rn00585674_s1 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Smarcd1 | Rn01533317_m1 |
Listing of all TaqMan probes used in this project including the reference genes Gapdh and Beta Actin.
TdT-mediated dUTP nick end labeling and quantification
Cell death was assessed by the TdT-mediated dUTP nick end labeling (TUNEL) technique to identify the fragmentation of DNA characteristic of apoptotic cells, following a previously published protocol [38] but using a fluorophore, Alexa 594, to visualize the enzymatic reaction. TUNEL-labeled sections were scanned from superior to inferior edge in 1 mm steps and the number of TUNEL-positive profiles in each 1 mm of the outer nuclear layer (ONL) was recorded. The frequency of TUNEL-positive profiles per mm of ONL was averaged from at least two sections per animal, and three or four animals were analyzed for each condition. The Student t test was used to compare the effects of different treatment conditions.
To demonstrate cell survival, the DNA-specific dye bisbenzimide (Calbiochem, La Jolla, CA) was used. Sections were incubated in the dye, diluted 1:10,000 in 1× PBS for 2 min at room temperature.
Results
Saffron and photobiomodulation (PBM) reduced photoreceptor death
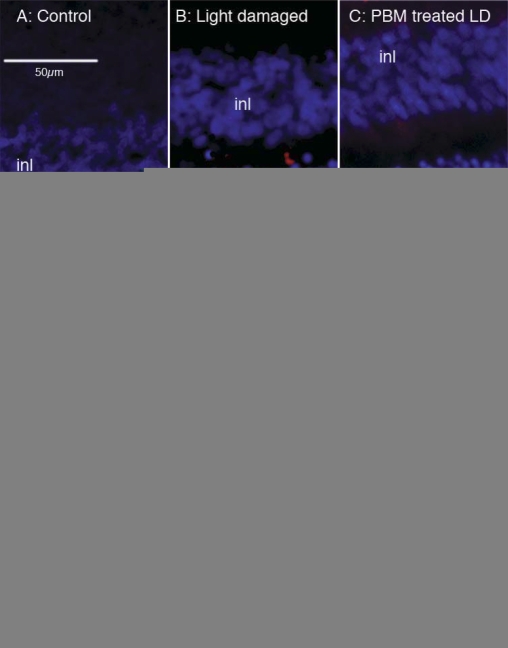
Figure 1 shows the protection of light-stressed photoreceptors in rat retina achieved in the current work, confirming previous reports for saffron [24] and PBM [30]. Light stress caused the death of photoreceptors, shown as TUNEL-labeling of cells in the ONL (Figure 1B). Pretreatment with saffron or PBM reduced the number of TUNEL-positive cells in the ONL (Figure 1C, for PBM), as well as reducing the light-induced thinning of the ONL (data not shown). When quantitative data were pooled (Figure 1D), significant differences were apparent between the LD group on the one hand, and the saffron-treated and PBM-treated groups on the other (control versus LD, p<0.002 on two-tailed t test; LD versus saffron LD, p<0.0025; LD versus PBMLD, p<0.002).
Figure 1.
Photoreceptor rescue by saffron and photobiomodulation. Images show inner and outer nuclear layer of the retina, and the extent of damage caused by light damage to a control animal (A), to a animal subjected to light damage (LD; B) and to an animal pre-treated with photobiomodulation and then subjected to LD (C). The red label, applied with the TdT-mediated dUTP nick end labeling (TUNEL) technique, marks cells whose DNA is undergoing the fragmentation characteristic of apoptotic death. TUNEL-positive cells are confined to the ONL, i.e., they are the somas of photoreceptors. The number of TUNEL-positive cells is reduced by PBM pretreatment. D: Mean numbers of TUNEL-positive cells per mm of outer nuclear layer, for control, LD, SafLD, and PBMLD groups. The reductions in cell death caused by pretreatment with saffron and PBM were statistically significant.
Global analyses of gene expression
Four approaches were used to gain an overview of entity (gene and ncRNA) expression changes in the present data.
Hierarchical clustering analysis
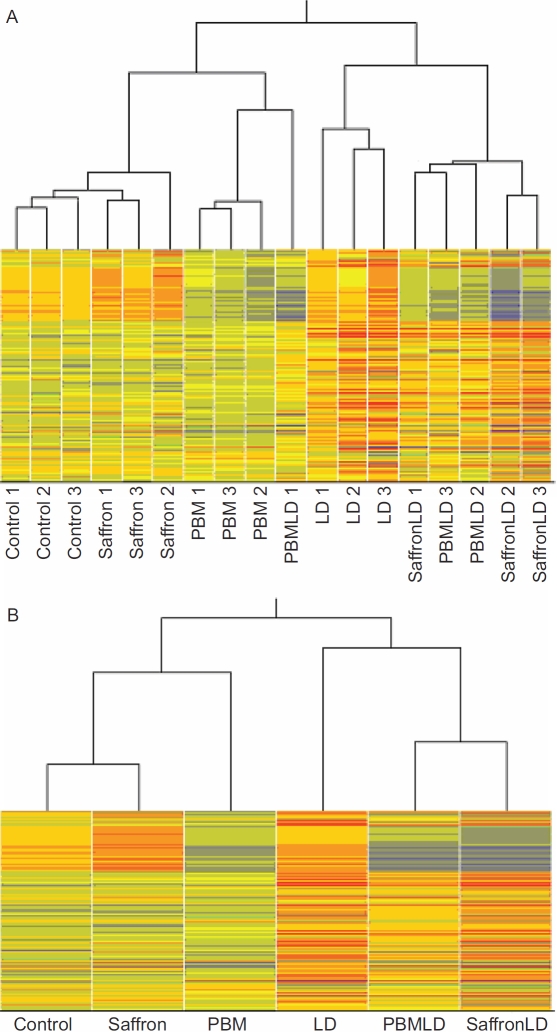
The hierarchical clustering of individual replicates (Figure 2A) indicates that the patterns of gene expression in the three samples of each group were highly reproducible. Of the 18 samples (3 samples in each of 6 groups), 16 clustered most closely with samples from the same group. One exception was PBMLD1, which clustered with the PBM samples; the other was saffronLD1 (SafLD1), which clustered with two of the PBMLD samples. Because the saffron and PBM samples clustered closely within their respective groups, the two exceptions suggest some variability in the impact of LD on gene expression.
Figure 2.
Hierarchical clustering diagram. This diagram shows the degree of similarity/difference between the 18 samples used in this study. Each column represents a sample; there were three control samples, three samples from retinas (each retina from a different animal) treated only with saffron, three from retinas/animals treated only with photobiomodulation (PBM), three from retinas/animals treated only with light damage (LD), three from retinas/animals treated with PBM and LD, and three from retinas/animals treated with saffronLD. The columns are arranged so that the most similar ones are next to each other. The branching lines at the top indicate in more detail the columns/samples that are most similar/different. A: With two exceptions, the three samples from each experimental group resembled each other more than samples in other experimental groups. The exceptions were PBMLD1, which resembled the PBM samples more closely than the other two PBMLD groups; saffronLD1, which resembled the PBMLD samples more closely than the other saffron LD groups. Of the three treatments used (PBM, saffron, LD), LD induced the most variable response by all assessments used. B: When expression values in the three samples of each of the six experimental groups were averaged, a distinct pattern of similarities emerged. The three saffron-only samples were closer to control than the PBM-only, suggesting that saffron by itself regulates fewer genes/entities than PBM. The LD-treated groups clustered together, with the two treated groups (PBMLD and SaffronLD) resembling each other more closely than the LD group. That is, treatment by PBM and Saffron before LD had broadly similar effects on the LD-induced regulation of genes/entities.
The pattern of clustering obtained when the group replicas were averaged is shown in Figure 2B. The three samples exposed to LD cluster together, separate from the three groups not exposed, indicating that LD has a strong impact on retinal gene expression. In the three non-LD groups, the saffron-treated sample clustered closer to control retina, suggesting that PBM alone has a stronger effect on retinal gene expression than saffron alone. Within the three LD-exposed groups, the retinas also exposed to photoreceptor-protective treatment (PBMLD, SafLD1) show gene expression closer to each other than to the LD group, suggesting that PBM and saffron modify the gene expression induced by LD in broadly similar ways.
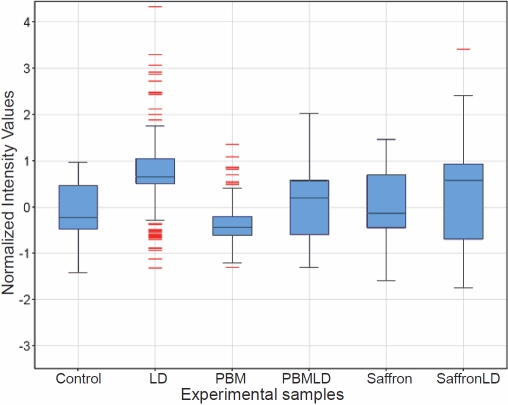
Distributions of gene expression in the six averaged samples—the box and whisker plot
An overview of gene expression in our six experimental groups is gained from the “box and whisker” plot in Figure 3. There were 187 genes included in these analyses; these were selected by a multi-ANOVA analysis of the six experimental groups (p<0.05, fold change [FC]>2).
Figure 3.
“Box and whisker” plots of the distributions of entity expression in the six experimental groups (replicates averaged). There were 187 genes included in these analyses; these were selected by a multi-ANOVA analysis of the six experimental groups (p<0.05, FC > 2). For each sample, the plot shows the median expression value of these genes as the horizontal line across the box. The upper and lower ends of the box mark the first and third quartile values, so that the box “contains” half of the sample value; the extensions show 1.5xIQR, where IQR is the interquartile range for the sample. The red lines indicate “outliers,” genes or ncRNAs whose expression level was greater or less than 1.5xIQR from the median.
For each sample, the plot shows the median expression value of these genes as the horizontal line across the box. The upper and lower ends of the box mark the first and third quartile values, so that the box “contains” half of the sample value; the extensions show 1.5xIQR, where IQR is the interquartile range for the sample. Expression values outside the extensions are considered outlying values, and are shown in red.
LD caused the median expression value to rise from the control value, with the expression of many entities (genes or ncRNAs) lying in outlier regions (12 above, 16 below). Saffron has relatively little effect on the distribution of gene expression levels, but PBM narrows the distribution and creates outliers. These two protective treatments thus seem to have distinctive effects. Finally, the effect of PBM and saffron given before LD was to reduce the LD-induced increase of the median and to reduce the number of outliers (to none in PBMLD, one in saffron LD).
Venn diagram analysis: entities associated with neuroprotection
A third overview of entity regulation associated with the neuroprotective actions of PBM and saffron is given by a Venn diagram analysis (Figure 4); numbers are shown separately for known genes and ncRNAs. The diagram is applied to three sets of regulated entities—those regulated by LD (compared to control); those regulated by LD when preceded by PBM (compared to control): and those regulated by LD when preceded by saffron feeding (compared to control). LD regulated 175 entities. Of these, 50 (44 known genes, 6 ncRNAs) were not regulated beyond criterion when LD was preceded by conditioning with PBM (PBMLD) or with saffron (SafLD). That is, the expression of these 50 entities (listed in Table 2) was suppressed by both PBM and saffron conditioning. Their suppression may be important in the protective actions of PBM and saffron.
Figure 4.
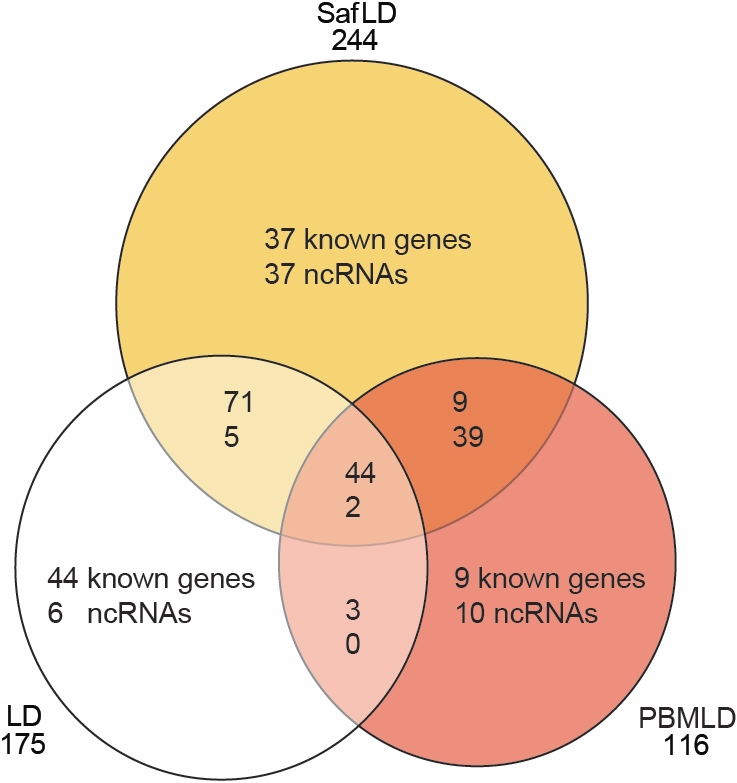
Venn Diagram showing similarity and differences between genes. The diagram is applied to three sets of regulated entities: those regulated by light damage (LD; compared to control); those regulated by LD when preceded by photobiomodulation (PBM; compared to control); and those regulated by LD when preceded by saffron feeding (compared to control). These sets were selected by two-sample Student t test analysis (p<0.05) and fold-change (FC>2).
Table 2. Genes and ncRNA significantly regulated by treatment with photobiomodulation and saffron.
| Probeset ID | Gene assignment | Gene symbol | RefSeq | p-value | FC (LD/C) |
|---|---|---|---|---|---|
| 10901166 |
angiopoietin-like 4 |
Angptl4 |
NM_199115 |
0.046613 |
2.20489 |
| 10738477 |
ADP-ribosylation factor 4-like |
Arf4l |
NM_001107052 |
0.027718 |
−2.12332 |
| 10865442 |
complement component 1, s subcomponent |
C1s |
NM_138900 |
0.027086 |
2.0311 |
| 10847761 |
Cd44 molecule |
Cd44 |
NM_012924 |
0.017188 |
2.40207 |
| 10771649 |
chemokine (C-X-C motif) ligand 11 |
Cxcl11 |
NM_182952 |
0.041843 |
2.71709 |
| 10827231 |
cysteine-rich, angiogenic inducer, 61 |
Cyr61 |
NM_031327 |
0.024324 |
2.07091 |
| 10890654 |
estrogen receptor 2 (ER beta) |
Esr2 |
NM_012754 |
0.009894 |
2.32768 |
| 10714890 |
Fas (TNF receptor superfamily, member 6) |
Fas |
NM_139194 |
0.01695 |
2.9447 |
| 10886031 |
FBJ osteosarcoma oncogene |
Fos |
NM_022197 |
0.008085 |
2.43039 |
| 10797527 |
growth arrest and DNA-damage-inducible, gamma |
Gadd45 g |
NM_001077640 |
0.01519 |
2.2081 |
| 10784120 |
gap junction protein, beta 6 |
Gjb6 |
NM_053388 |
0.033887 |
−2.2151 |
| 10849841 |
interleukin 1 beta |
Il1b |
NM_031512 |
0.046205 |
2.18761 |
| 10862908 |
interleukin 23 receptor |
Il23r |
XM_001067609 |
0.03848 |
2.92018 |
| 10733553 |
interferon regulatory factor 1 |
Irf1 |
NM_012591 |
0.005382 |
2.70651 |
| 10867306 |
hypothetical protein LOC683514 |
LOC683514 |
NM_001127569 |
0.028303 |
2.1278 |
| 10934056 |
moesin |
Msn |
NM_030863 |
0.031765 |
2.04966 |
| 10896814 |
myelocytomatosis oncogene |
Myc |
NM_012603 |
0.007411 |
2.7792 |
| 10920860 |
myeloid differentiation primary response gene 88 |
Myd88 |
NM_198130 |
0.014129 |
2.34728 |
| 10926588 |
nuclear factor of kappa light polypeptide gene enhancer i |
Nfkbie |
NM_199111 |
0.014731 |
2.17613 |
| 10750848 |
nuclear factor of kappa light polypeptide gene enhance |
Nfkbiz |
NM_001107095 |
0.00217 |
2.32035 |
| 10823365 |
purinergic receptor P2Y, G-protein coupled 12 |
P2ry12 |
NM_022800 |
0.017115 |
−2.38721 |
| 10792421 |
plasminogen activator, tissue |
Plat |
NM_013151 |
0.004342 |
2.38492 |
| 10911484 |
protogenin homolog (Gallus gallus) |
Prtg |
NM_001037651 |
0.027196 |
2.04521 |
| 10842475 |
protein tyrosine phosphatase, non-receptor type 1 |
Ptpn1 |
NM_012637 |
0.002949 |
2.18916 |
| 10821581 |
similar to hypothetical protein MGC42105 |
RGD1308116 |
ENSRNOT00000021964 |
0.013255 |
−2.2474 |
| 10710930 |
similar to hypothetical protein DKFZp434I2117 |
RGD1308215 |
NM_001106296 |
0.016796 |
−2.2475 |
| 10803006 |
similar to hypothetical protein B230399E16 |
RGD1559694 |
ENSRNOT00000020858 |
0.034301 |
−2.49615 |
| 10882514 |
RGD1560224 |
RGD1560224 |
ENSRNOT00000009292 |
0.048646 |
−2.31446 |
| 10855681 |
similar to hypothetical protein |
RGD1562590 |
ENSRNOT00000015469 |
0.006687 |
2.09732 |
| 10800434 |
ring finger protein 125 |
Rnf125 |
NM_001108424 |
0.037824 |
2.3249 |
| 10893918 |
strawberry notch homolog 2 (Drosophila) |
Sbno2 |
NM_001108068 |
0.007136 |
2.23202 |
| 10765195 |
selectin, platelet |
Selp |
NM_013114 |
0.03163 |
2.27171 |
| 10704505 |
solute carrier family 1 (neutral amino acid transporter), |
Slc1a5 |
NM_175758 |
0.028834 |
2.70661 |
| 10736795 |
schlafen 2 |
Slfn2 |
NM_001107031 |
0.048056 |
2.03465 |
| 10717935 |
superoxide dismutase 2, mitochondrial |
Sod2 |
NM_017051 |
0.039527 |
2.05701 |
| 10781273 |
stanniocalcin 1 |
Stc1 |
NM_031123 |
0.048406 |
2.26891 |
| 10869149 |
T-cell acute lymphocytic leukemia 2 |
Tal2 |
NM_001109462 |
0.012626 |
2.32825 |
| 10783880 |
transglutaminase 1, K polypeptide |
Tgm1 |
NM_031659 |
0.012214 |
2.7564 |
| 10887306 |
tumor necrosis factor, alpha-induced protein 2 |
Tnfaip2 |
NM_001137633 |
0.038454 |
2.32289 |
| 10858967 |
tumor necrosis factor receptor superfamily, member 1a |
Tnfrsf1a |
NM_013091 |
0.013832 |
2.36185 |
| 10829313 |
transient receptor potential cation channel, subfamily |
Trpm2 |
NM_001011559 |
0.009704 |
−2.04814 |
| 10802422 |
tubulin, beta 6 |
Tubb6 |
NM_001025675 |
0.015119 |
2.4974 |
| 10802995 |
zinc finger protein 516 |
Znf516 |
ENSRNOT00000021768 |
0.000278 |
2.01399 |
| 10813949 |
zinc finger protein 622 |
Znf622 |
ENSRNOT00000014423 |
0.011767 |
2.11 |
| 10821585 |
— |
|
— |
0.001925 |
−2.09732 |
| 10802710 |
— |
|
— |
0.005024 |
2.05529 |
| 10857403 |
— |
|
— |
0.007403 |
2.00653 |
| 10813885 |
— |
|
— |
0.00781 |
2.73034 |
| 10859195 |
— |
|
— |
0.025583 |
2.49356 |
| 10752799 | — | — | 0.026753 | 3.52422 |
Genes and ncRNAs (44 known genes, 6 ncRNAs) whose expression was significantly regulated by light damage (LD), and whose regulation was reduced below criterion when the retina was conditioned by photobiomodulation (PBM) and by saffron. These reductions in regulation may be important for the protective effects of PBM and saffron.
When saffron was given to the animal before light damage (SafLD), the expression of a large number of entities (48 in common with PBM and 74 unique to saffron) were regulated, and were not regulated by LD; i.e., their regulation can be attributed to saffron and may be important in its protective effect. Similarly, when the retina was conditioned by PBM before exposure to LD, the expressions of 67 entities (48 in common with saffron and 19 unique to PBM) was regulated, which were not regulated by LD. Their regulation can be attributed to PBM and may be important in the protective effect of PBM. The entities regulated by saffron and PBM given before LD, and not by LD, are listed in Table 3.
Table 3. Genes and ncRNA regulated by photobiomodulation and saffron during light damage but not regulated by light damage alone.
| Probeset ID | Gene_assignment | Gene symbol | RefSeq | p-value | FC (PBMLD/C) | FC (SafLD/C) |
|---|---|---|---|---|---|---|
| 10786710 |
biotinidase |
Btd |
NM_001012047 |
0.002593 |
−2.58996 |
−2.00118 |
| 10727084 |
cysteinyl-tRNA synthetase |
Cars |
NM_001106319 |
0.000472 |
2.04587 |
2.27983 |
| 10754902 |
discs, large homolog 1 (Drosophila) |
Dlg1 |
NM_012788 |
0.007578 |
2.02011 |
2.20452 |
| 10814281 |
fatty acid binding protein 12 |
Fabp12 |
NM_001134614 |
0.03715 |
−2.09977 |
−2.09843 |
| 10797597 |
isoleucyl-tRNA synthetase |
Iars |
NM_001100572 |
0.01099 |
2.21568 |
2.14011 |
| 10796326 |
optineurin |
Optn |
NM_145081 |
0.001857 |
−2.04049 |
−2.16666 |
| 10810322 |
similar to calmegin |
RGD1310572 |
BC097408 |
0.011866 |
2.57427 |
2.03946 |
| 10753017 |
similar to Putative protein C21orf45 |
RGD1310778 |
BC167102 |
0.000554 |
−2.95657 |
−2.95334 |
| 10758134 |
ubiquitin C |
Ubc |
NM_017314 |
0.006248 |
−2.2729 |
−2.14387 |
| 10765728 |
— |
|
— |
0.000316 |
−2.05098 |
−3.55913 |
| 10722459 |
— |
|
— |
0.000643 |
−4.83475 |
−5.7297 |
| 10722449 |
— |
|
— |
0.000759 |
−4.77591 |
−4.75198 |
| 10722435 |
— |
|
— |
0.000872 |
−4.53877 |
−3.87361 |
| 10838282 |
— |
|
— |
0.001154 |
−3.51509 |
−4.57167 |
| 10703224 |
— |
|
— |
0.001264 |
−3.60718 |
−4.66873 |
| 10722429 |
— |
|
— |
0.00139 |
−3.95495 |
−4.50258 |
| 10722465 |
— |
|
— |
0.00144 |
−4.49582 |
−5.45683 |
| 10722473 |
— |
|
— |
0.00149 |
−2.89842 |
−2.9913 |
| 10862359 |
— |
|
— |
0.00159 |
−4.16679 |
−6.62332 |
| 10894268 |
— |
|
— |
0.001622 |
−3.30485 |
−2.65319 |
| 10932269 |
— |
|
— |
0.001633 |
−2.51937 |
−3.05085 |
| 10722423 |
— |
|
— |
0.001744 |
−3.54235 |
−2.91368 |
| 10722461 |
— |
|
— |
0.002014 |
−3.20546 |
−3.46585 |
| 10722437 |
— |
|
— |
0.002127 |
−3.81721 |
−4.06358 |
| 10722481 |
— |
|
— |
0.002143 |
−4.18134 |
−4.50928 |
| 10855946 |
— |
|
— |
0.002276 |
−4.8482 |
−5.97919 |
| 10722433 |
— |
|
— |
0.002438 |
−2.97454 |
−3.52979 |
| 10722471 |
— |
|
— |
0.002612 |
−4.01264 |
−4.18106 |
| 10722453 |
— |
|
— |
0.002883 |
−2.32871 |
−2.48323 |
| 10721700 |
— |
|
— |
0.002935 |
−2.66889 |
−2.35149 |
| 10839872 |
— |
|
— |
0.003128 |
−3.58942 |
−4.13962 |
| 10722431 |
— |
|
— |
0.003286 |
−2.28992 |
−2.57576 |
| 10722443 |
— |
|
— |
0.003413 |
−3.36447 |
−3.16986 |
| 10722451 |
— |
|
— |
0.00356 |
−3.75565 |
−3.75383 |
| 10932228 |
— |
|
— |
0.003868 |
−2.39044 |
−2.32665 |
| 10722425 |
— |
|
— |
0.004662 |
−2.86663 |
−3.06065 |
| 10722427 |
— |
|
— |
0.004665 |
−4.23993 |
−4.9848 |
| 10722479 |
— |
|
— |
0.004887 |
−2.12953 |
−2.2299 |
| 10870039 |
— |
|
— |
0.005491 |
−2.02844 |
−2.04155 |
| 10722467 |
— |
|
— |
0.006418 |
−2.28354 |
−2.11141 |
| 10867318 |
— |
|
— |
0.007967 |
−3.77635 |
−2.33207 |
| 10722409 |
— |
|
— |
0.012757 |
−2.28142 |
−2.24389 |
| 10820008 |
— |
|
— |
0.014354 |
−2.331 |
−3.03034 |
| 10722383 |
— |
|
— |
0.017588 |
−2.07462 |
−2.12019 |
| 10722379 |
— |
|
— |
0.017621 |
−2.03053 |
−2.08741 |
| 10707643 |
— |
|
— |
0.020362 |
−3.27802 |
−3.23128 |
| 10722417 |
— |
|
— |
0.024253 |
−2.16311 |
−2.33892 |
| 10722387 | — | — | 0.028341 | −2.62369 | −2.64182 |
Genes and ncRNAs regulated by photobiomodulation (PBM) and saffron conditioning, but not by light damage (LD) alone (9 known genes, 39 ncRNAs). Their regulation by PBM and saffron conditioning suggests that they are important in the protective effects of both PBM and saffron
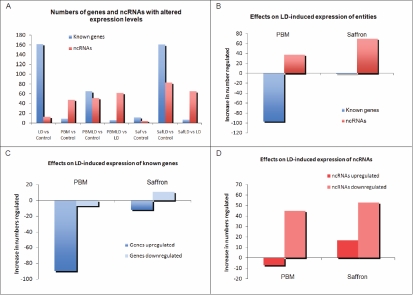
By separating known genes from ncRNAs, the Venn diagram analysis draws attention to the prominence of ncRNAs among the entities regulated by both saffron and PBM when they are exerting their protective actions. For example, LD regulated 175 entities, of which only 13 (7.5%) were ncRNAs. Saffron preceding LD regulated 244 entities, of which 83 (34%) were ncRNAs; while PBM preceding LD regulated 116 entities, of which 51 (44%) were ncRNAs. Among the 48 entities regulated by PBM and saffron, but not by LD, and which are therefore potentially neuroprotective entities, 39 (81%) were ncRNAs.
Expression changes: identified genes and noncoding RNA
Given the prominence of ncRNAs among the entities regulated by saffron and PBM when conditioning LD, we surveyed the relative numbers of genes and ncRNAs in the seven comparisons shown in Figure 5A. As already noted, LD regulated a large number of known genes, but few ncRNAs. Conversely, ncRNAs outnumber known genes in the action of PBM on the control retina (PBM versus control); in the action of PBM when exerting its protective action against LD (PBMLD versus LD); and in the protective action of saffron (saffronLD versus LD). It seems likely that the regulation of ncRNAs accounts for a significant part of the protective effect.
Figure 5.
Analysis of entities regulated (known genes versus ncRNAs) and direction of regulation. A: Numbers of genes and ncRNAs regulated in seven comparisons among the experimental groups. SaffLD is the group given saffron before light damage (LD). B: Effects of saffron and photobiomodulation (PBM) on the numbers of LD-induced expression changes of known genes and ncRNAs. C: Direction of regulation of known genes by PBM and saffron when given as pretreatments to LD. D: Direction of regulation of ncRNAs by PBM and saffron when given as pretreatments to LD.
This suggestion is supported by the difference comparison in Figure 5B. Measuring only changes in the numbers of genes and ncRNAs whose expression was significantly regulated by saffron or PBM before LD, the protective actions of saffron and PBM are both associated with increases in the number of ncRNAs regulated, and decreases in the numbers of identified genes whose expression was regulated.
As a final step, we considered the directions of entity expression changes in these several conditions (Figure 5C, Figure 4D). The most striking outcome of this separation is that the protective effects of PBM and saffron are associated with a decrease in the number of known genes upregulated, and an increase in the number of ncRNAs downregulated.
Validation by real-time PCR
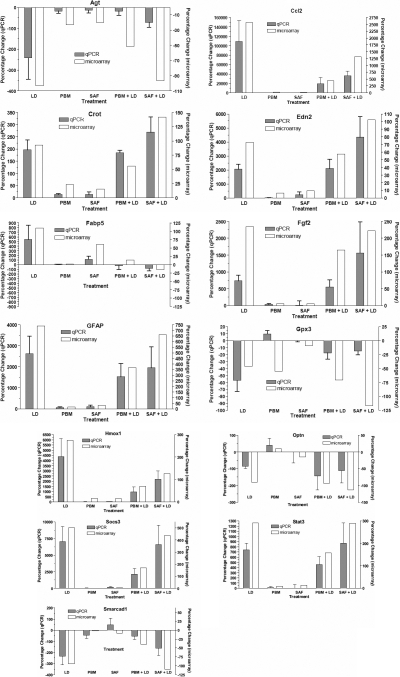
Thirteen genes were chosen for RT–PCR validation of the microarray outcomes; those chosen were strongly regulated and/or retina-relevant. Five genes (Crot, Optn, Edn2, Smarcad1, Gpx3) were significantly regulated by saffron in the LD assay. Crot and smarcad1 are involved in fatty acid metabolism, Edn2 in retinal signaling in response to injury, and Gpx3 in antioxidative activity. Optn acts as an mgluR1 receptor on retinal bipolar cells. Fabp5 is also saffron-regulated, and related to fatty acid metabolism. Fgf and GFAP are proteins upregulated by stress; Stat3 and Socs3 are related to transduction pathways, ccl2 to inflammatory responses, and Agt and heme oxygenase 1 (Hmox1) to cardiovascular control.
Figure 6 shows a comparison for each of the 13 genes between its regulation as assessed by the microarray procedure and its regulation as assessed by RT–PCR. The correlation between the two techniques appears particularly close for ccl2, Socs3, Stat2, Cro, Edn2, Hmox1, Fabp5, and smarcad. Common trends, with quantitative differences at some sample points, are evident for Optn, GFAP, Agt, Fgf2, and Gpx3. Overall, the correlation between the two techniques seems strong.
Figure 6.
Comparisons, for thirteen selected genes, of expression changes in the six experimental groups, assessed by qPCR and microarray analysis.
Entities associated with the protective actions of saffron and photobiolmodulation listed
Light damage–induced regulation inhibited by photobiolmodulation or saffron
The genes and ncRNAs whose regulation by LD was inhibited by PBM or saffron are listed in Table 2; as noted above, this inhibition affected principally (88%) known genes (44 known genes, 6 ncRNAs). All 50 entities were upregulated by LD; they are therefore candidates for genes and regulatory elements whose upregulation is damaging to photoreceptors.
Regulation by photobiolmodulation and saffron, but not LD
Table 3 lists genes and ncRNAs that were not regulated by LD but were regulated by PBM and saffron when conditioning (protecting) photoreceptors challenged by LD. Figure 7 shows that the effects of PBM and saffron on their regulation were highly correlated. The entity regulation shown in Table 3 contrasts in two ways with the pattern of regulation in Table 2: Most of the entities whose regulation was changed by saffron and PBM conditioning were ncRNAs (81%), and all the ncRNAs and half the known genes were downregulated.
Figure 7.
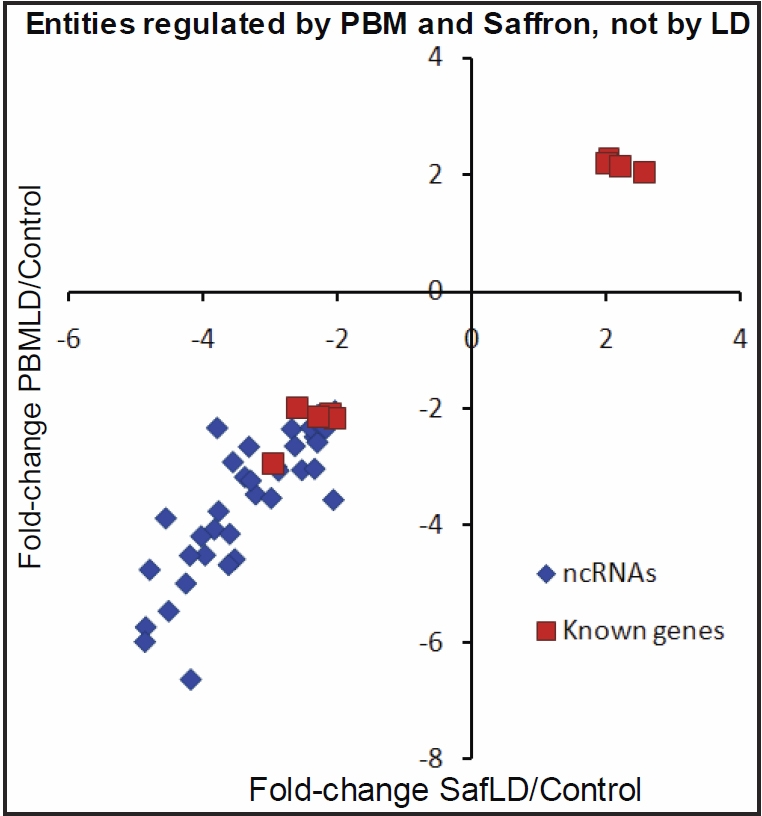
Evidence of similarity in the actions of saffron and photobiomodulation. This graph shows the correlation for 48 entities (9 known genes and 39 ncRNAs) between the change in gene expression associated with photobiomodulation (PBM) and saffron pre-treatments.
Regulation by PBM or saffron, but not light damage
Further candidates for genes and ncRNAs protective to photoreceptors can be found in 74 entities (37 known genes, 37 ncRNAs) regulated by saffron (but not by PBM) when conditioning/protecting photoreceptors (Table 4), and in the 19 entities (9 known genes, 10 ncRNAs) regulated by PBM (but not by saffron) when conditioning/protecting photoreceptors (Table 5).
Table 4. Genes ncRNA regulated by saffron pre-conditioning but not photobiomodulation or light damage.
| Probeset ID | Gene_assignment | Gene symbol | RefSeq | p-value | FC (SafLD/C) |
|---|---|---|---|---|---|
| 10808041 |
alanyl-tRNA synthetase |
Aars |
NM_001100517 |
0.009419 |
2.12845 |
| 10920371 |
coiled-coil domain containing 72 |
Ccdc72 |
NM_001126048 |
0.000362 |
−2.15161 |
| 10753771 |
Cd47 molecule |
Cd47 |
NM_019195 |
0.004538 |
2.18358 |
| 10840895 |
cytochrome c oxidase subunit IV isoform 2 |
Cox4i2 |
NM_053472 |
0.006367 |
−2.04762 |
| 10860548 |
carnitine O-octanoyltransferase |
Crot |
NM_031987 |
0.000412 |
2.42699 |
| 10871623 |
endothelin 2 |
Edn2 |
NM_012549 |
0.003864 |
2.39144 |
| 10791631 |
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
Enpp6 |
NM_001107311 |
0.048871 |
−3.40078 |
| 10740135 |
fascin homolog 2, actin-bundling protein, retinal (Stro |
Fscn2 |
NM_001107072 |
0.002804 |
2.03501 |
| 10938219 |
glycerol kinase |
Gk |
NM_024381 |
0.000123 |
2.16424 |
| 10732439 |
guanine nucleotide binding protein (G protein), gamma 1 |
Gng13 |
NM_001135918 |
0.006565 |
−2.23127 |
| 10733680 |
glutathione peroxidase 3 |
Gpx3 |
NM_022525 |
0.00182 |
−2.17933 |
| 10715200 |
helicase, lymphoid specific |
Hells |
NM_001106371 |
0.000882 |
2.2618 |
| 10863430 |
hexokinase 2 |
Hk2 |
NM_012735 |
0.029119 |
−2.24022 |
| 10733056 |
interferon gamma inducible protein 47 |
Ifi47 |
NM_172019 |
0.001063 |
2.31444 |
| 10714903 |
interferon-induced protein with tetratricopeptide repea |
Ifit3 |
NM_001007694 |
0.004673 |
3.02663 |
| 10753784 |
intraflagellar transport 57 homolog (Chlamydomonas) |
Ift57 |
NM_001107093 |
0.000101 |
2.67228 |
| 10815873 |
interleukin 12a |
Il12a |
NM_053390 |
0.003212 |
−2.04998 |
| 10804187 |
leucyl-tRNA synthetase |
Lars |
NM_001009637 |
0.000471 |
2.05987 |
| 10932310 |
mediator complex subunit 14 |
Med14 |
XM_228713 |
0.02603 |
2.23949 |
| 10923270 |
oligonucleotide/oligosaccharide-binding fold containin |
Obfc2a |
NM_001014216 |
8.85E-05 |
2.0681 |
| 10855114 |
olfactory receptor 820 |
Olr820 |
NM_001000974 |
0.025967 |
2.01366 |
| 10830003 |
pterin-4 alpha-carbinolamine dehydratase/dimerization c |
Pcbd1 |
NM_001007601 |
0.009857 |
−2.05998 |
| 10708281 |
phosphodiesterase 8A |
Pde8a |
NM_198767 |
0.005571 |
−2.00121 |
| 10889475 |
peroxidasin homolog (Drosophila) |
Pxdn |
ENSRNOT00000060139 |
0.00349 |
−2.6691 |
| 10803138 |
RNA binding motif, single stranded interacting protein |
Rbms2 |
NM_001025403 |
0.002005 |
−2.15568 |
| 10716415 |
similar to enolase (46.6 kDa) (2J223) |
RGD1308333 |
NM_001134505 |
0.015495 |
2.04964 |
| 10820002 |
similar to Ac1147 |
RGD1563254 |
XM_577969 |
0.029198 |
−2.23707 |
| 10771190 |
similar to ATP-binding cassette, sub-family G (WHI |
RGD1564709 |
NM_001107205 |
0.044266 |
2.04858 |
| 10797566 |
sphingosine-1-phosphate receptor 3 |
S1pr3 |
ENSRNOT00000019473 |
0.00326 |
2.25911 |
| 10750282 |
solute carrier family 5 (inositol transporters), member 3 |
Slc5a3 |
NM_053715 |
0.002179 |
2.1573 |
| 10842440 |
solute carrier family 9 (sodium/hydrogen exchanger), m |
Slc9a8 |
NM_001025281 |
0.000656 |
2.12805 |
| 10899174 |
SWI/SNF related, matrix associated, actin dependent r |
Smarcd1 |
NM_001108752 |
0.002766 |
−2.04334 |
| 10831606 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
Tap1 |
NM_032055 |
0.011581 |
2.37179 |
| 10902375 |
TBC1 domain family, member 15 |
Tbc1d15 |
ENSRNOT00000005207 |
0.000945 |
2.0265 |
| 10858370 |
ubiquitin specific peptidase 18 |
Usp18 |
NM_001014058 |
0.019532 |
2.19315 |
| 10907681 |
zinc finger protein 385A |
Zfp385a |
NM_001135088 |
0.000479 |
−2.14878 |
| 10846652 |
zinc finger protein 385B |
Zfp385b |
NM_001107736 |
0.001897 |
−2.11791 |
| 10840061 |
— |
|
— |
0.047419 |
2.15179 |
| 10924441 |
— |
|
— |
0.044499 |
2.20978 |
| 10891487 |
— |
|
— |
0.042933 |
2.01488 |
| 10886190 |
— |
|
— |
0.040809 |
−2.64706 |
| 10898158 |
— |
|
— |
0.040237 |
−2.11912 |
| 10930226 |
— |
|
— |
0.039217 |
−2.1554 |
| 10915105 |
— |
|
— |
0.033403 |
−2.30406 |
| 10886854 |
— |
|
— |
0.032145 |
2.23693 |
| 10731193 |
— |
|
— |
0.031476 |
2.4581 |
| 10843907 |
— |
|
— |
0.030096 |
2.01264 |
| 10875117 |
— |
|
— |
0.027972 |
2.25392 |
| 10801781 |
— |
|
— |
0.026332 |
2.81764 |
| 10825167 |
— |
|
— |
0.022483 |
−2.8835 |
| 10722375 |
— |
|
— |
0.019218 |
−2.05422 |
| 10803001 |
— |
|
— |
0.018227 |
2.34996 |
| 10819500 |
— |
|
— |
0.017801 |
2.16908 |
| 10766880 |
— |
|
— |
0.015099 |
2.10145 |
| 10938891 |
— |
|
— |
0.013364 |
2.13311 |
| 10776604 |
— |
|
— |
0.012016 |
−2.84767 |
| 10757702 |
— |
|
— |
0.009955 |
−2.06188 |
| 10867008 |
— |
|
— |
0.009246 |
−2.29587 |
| 10891878 |
— |
|
— |
0.008961 |
−2.10309 |
| 10830454 |
— |
|
— |
0.008641 |
−2.44031 |
| 10827830 |
— |
|
— |
0.006903 |
2.30678 |
| 10897004 |
— |
|
— |
0.006062 |
2.01807 |
| 10742429 |
— |
|
— |
0.005794 |
2.98095 |
| 10834602 |
— |
|
— |
0.005352 |
−2.59778 |
| 10926624 |
— |
|
— |
0.005005 |
2.14084 |
| 10781982 |
— |
|
— |
0.004882 |
2.31978 |
| 10896630 |
— |
|
— |
0.003343 |
−2.13243 |
| 10930622 |
— |
|
— |
0.002504 |
2.29803 |
| 10756086 |
— |
|
— |
0.002238 |
2.23035 |
| 10923938 |
— |
|
— |
0.00188 |
2.22066 |
| 10938893 |
— |
|
— |
0.001545 |
2.49702 |
| 10899788 |
— |
|
— |
0.000365 |
−2.13694 |
| 10766722 |
— |
|
— |
8.76E-05 |
2.60553 |
| 10752219 | — | — | 1.94E-05 | −2.05925 |
Genes and ncRNAs regulated saffron conditioning, but not by photobiomodulation (PBM) and not by light damage (LD) alone (37 known genes, 37 ncRNAs). Their regulation by saffron conditioning suggests that they are important in the protective action of saffron, and not of PBM.
Table 5. Genes and ncRNA regulated by photobiomodulation pre-conditioning but not saffron or light damage alone.
| Probeset ID | Gene_assignment | Gene symbol | RefSeq | p-value | FC (NIRLD/C) |
|---|---|---|---|---|---|
| 10916920 |
ATP synthase, H+ transporting, mitochondrial F0 complex, s |
Atp5l |
NM_212516 |
0.03394 |
−2.14869 |
| 10867731 |
calbindin 1 |
Calb1 |
NM_031984 |
0.03827 |
−2.09147 |
| 10770342 |
epoxide hydrolase 1, microsomal |
Ephx1 |
NM_001034090 |
0.019435 |
−2.16344 |
| 10892381 |
nudix (nucleoside diphosphate linked moiety X)-type mo |
Nudt14 |
NM_001106760 |
0.018954 |
−2.0385 |
| 10847156 |
olfactory receptor 673 |
Olr673 |
NM_001000351 |
0.015388 |
2.0757 |
| 10801260 |
protocadherin gamma subfamily B, 6 |
Pcdhgb6 |
ENSRNOT00000060466 |
6.03E-05 |
2.18939 |
| 10891322 |
polyglutamine-containing protein |
Pqcp |
NM_001012470 |
0.018634 |
2.00327 |
| 10796307 |
similar to calcium/calmodulin-dependent protein ki |
RGD1560691 |
NM_001107365 |
0.019657 |
2.35325 |
| 10817552 |
thioredoxin interacting protein |
Txnip |
NM_001008767 |
0.018274 |
−2.32009 |
| 10886988 |
— |
|
— |
0.000109 |
25.1775 |
| 10718602 |
— |
|
— |
0.000392 |
2.10568 |
| 10721694 |
— |
|
— |
0.011105 |
−2.00257 |
| 10919224 |
— |
|
— |
0.011219 |
−2.57253 |
| 10758033 |
— |
|
— |
0.021858 |
−2.84458 |
| 10840318 |
— |
|
— |
0.028594 |
2.23714 |
| 10878967 |
— |
|
— |
0.031786 |
−2.35082 |
| 10713602 |
— |
|
— |
0.036424 |
−2.41324 |
| 10886894 |
— |
|
— |
0.04513 |
−2.03777 |
| 10797671 | — | — | 0.049361 | −2.07756 |
Genes and ncRNAs regulated by photobiomodulation (PBM) conditioning, but not saffron and not by light damage (LD) alone (9 known genes, 10 ncRNAs). Their regulation by PBM conditioning suggests that they are important in the protective effects of PBM, but not in the protective action of saffron. Entities regulated by PBM, when exerting its protective action (9 known genes, 10 nc RNAs)
Regulation by LD, SaffronLD, and PBMLD
Genes found to be regulated by SaffronLD and LD (Table 6), PBMLD and LD (Table 7), and SaffronLD, PBMLD, and LD (Table 8) are shown in the corresponding tables. These genes are not discussed as the changes in expression levels are likely due to LD and not saffron or PBM.
Table 6. Genes and ncRNA regulated by saffron light damage and light damage.
| Probeset ID | Gene_assignment | Gene symbol | RefSeq | p-value | FC (LD/C) | FC (SafLD/C) |
|---|---|---|---|---|---|---|
| 10752839 |
ADAM metallopeptidase with thrombospondin type 1 motif, |
Adamts1 |
NM_024400 |
0.003178 |
2.63853 |
2.72148 |
| 10765534 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
Adamts4 |
AB042272 |
0.005095 |
2.26079 |
2.2735 |
| 10811900 |
angiotensinogen (serpin peptidase inhibitor, clade A, member |
Agt |
NM_134432 |
0.0037 |
−2.29985 |
−2.04435 |
| 10914799 |
baculoviral IAP repeat-containing 3 |
Birc3 |
NM_023987 |
0.000774 |
3.1982 |
2.21198 |
| 10791652 |
caspase 3, apoptosis related cysteine protease |
Casp3 |
NM_012922 |
0.001718 |
2.62625 |
2.20514 |
| 10736712 |
chemokine (C-C motif) ligand 12 |
Ccl12 |
NM_001105822 |
0.002288 |
4.38236 |
2.10232 |
| 10736697 |
chemokine (C-C motif) ligand 2 |
Ccl2 |
NM_031530 |
0.00058 |
38.8349 |
17.0997 |
| 10745677 |
chemokine (C-C motif) ligand 3 |
Ccl3 |
NM_013025 |
7.62E-05 |
19.8193 |
9.71154 |
| 10736863 |
chemokine (C-C motif) ligand 4 |
Ccl4 |
NM_053858 |
0.001026 |
4.06268 |
3.00096 |
| 10736702 |
chemokine (C-C motif) ligand 7 |
Ccl7 |
NM_001007612 |
0.000248 |
4.68541 |
2.67958 |
| 10729777 |
cholesterol 25-hydroxylase |
Ch25h |
NM_001025415 |
5.47E-05 |
4.58918 |
3.28027 |
| 10764069 |
chitinase 3-like 1 |
Chi3l1 |
NM_053560 |
0.010591 |
2.88704 |
2.22024 |
| 10912908 |
cytokine inducible SH2-containing protein |
Cish |
NM_031804 |
0.003605 |
10.9118 |
7.35678 |
| 10712853 |
cardiotrophin-like cytokine factor 1 |
Clcf1 |
NM_207615 |
0.009706 |
3.56151 |
2.0363 |
| 10814430 |
ceruloplasmin |
Cp |
NM_012532 |
0.003256 |
2.38102 |
2.53086 |
| 10825869 |
colony stimulating factor 1 (macrophage) |
Csf1 |
NM_023981 |
0.000858 |
2.30889 |
2.15895 |
| 10775900 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulat |
Cxcl1 |
NM_030845 |
0.012601 |
4.11514 |
3.36565 |
| 10771655 |
chemokine (C-X-C motif) ligand 10 |
Cxcl10 |
NM_139089 |
0.003066 |
21.4607 |
7.85562 |
| 10784355 |
emopamil binding protein-like |
Ebpl |
NM_001108381 |
0.001569 |
−2.26226 |
−2.09366 |
| 10873706 |
Eph receptor A2 |
Epha2 |
NM_001108977 |
0.002851 |
2.39137 |
2.33589 |
| 10860231 |
fibrinogen-like 2 |
Fgl2 |
NM_053455 |
0.000414 |
4.1065 |
3.1288 |
| 10713045 |
fos-like antigen 1 |
Fosl1 |
NM_012953 |
0.000264 |
5.88589 |
4.16491 |
| 10819523 |
guanylate binding protein 2 |
Gbp2 |
NM_133624 |
0.000294 |
14.8983 |
7.64439 |
| 10819489 |
guanylate binding protein 5 |
Gbp5 |
NM_001108569 |
0.004085 |
7.65404 |
4.22902 |
| 10915843 |
galactosidase, beta 1-like 2 |
Glb1l2 |
ENSRNOT00000037790 |
0.002801 |
−3.04631 |
−2.17671 |
| 10806122 |
heme oxygenase (decycling) 1 |
Hmox1 |
NM_012580 |
0.008369 |
5.70686 |
2.9304 |
| 10908319 |
intercellular adhesion molecule 1 |
Icam1 |
NM_012967 |
0.00315 |
5.74547 |
3.35268 |
| 10831077 |
immediate early response 3 |
Ier3 |
NM_212505 |
0.005316 |
3.02458 |
2.20576 |
| 10845708 |
interferon induced with helicase C domain 1 |
Ifih1 |
NM_001109199 |
0.001642 |
3.36324 |
2.75239 |
| 10936365 |
interleukin 13 receptor, alpha 1 |
Il13ra1 |
NM_145789 |
0.004069 |
2.21356 |
2.08254 |
| 10789857 |
interleukin 17 receptor B |
Il17rb |
NM_001107290 |
0.021732 |
2.29043 |
2.26483 |
| 10813007 |
interleukin 6 signal transducer |
Il6st |
NM_001008725 |
0.018115 |
2.16052 |
2.03731 |
| 10806585 |
jun B proto-oncogene |
Junb |
NM_021836 |
0.000634 |
4.24227 |
3.86837 |
| 10844331 |
lipocalin 2 |
Lcn2 |
NM_130741 |
0.001852 |
11.4978 |
5.73872 |
| 10773853 |
leukemia inhibitory factor (cholinergic differentiation fact |
Lif |
NM_022196 |
0.004038 |
6.05907 |
6.27293 |
| 10751793 |
leucine rich repeat containing 15 |
Lrrc15 |
NM_145083 |
0.021229 |
3.28743 |
2.22908 |
| 10880293 |
mitogen-activated protein kinase kinase kinase 6 |
Map3k6 |
NM_001107909 |
0.018572 |
3.52464 |
2.64741 |
| 10903013 |
methionine-tRNA synthetase |
Mars |
NM_001127659 |
0.00072 |
2.00698 |
2.2033 |
| 10898561 |
myo-inositol oxygenase |
Miox |
NM_145771 |
0.002983 |
−2.24009 |
−2.20493 |
| 10815806 |
myeloid leukemia factor 1 |
Mlf1 |
NM_001107680 |
0.002256 |
2.96741 |
2.28383 |
| 10907881 |
matrix metallopeptidase 3 |
Mmp3 |
NM_133523 |
0.0008 |
12.1207 |
5.47872 |
| 10809392 |
metallothionein 1a |
Mt1a |
NM_138826 |
0.00235 |
3.86934 |
3.13553 |
| 10827989 |
metallothionein 2A |
Mt2A |
NM_001137564 |
0.002073 |
9.97229 |
5.45083 |
| 10715787 |
nuclear factor of kappa light polypeptide gene enhancer |
Nfkb2 |
NM_001008349 |
0.001701 |
3.01772 |
2.0803 |
| 10727717 |
neuronal PAS domain protein 4 |
Npas4 |
NM_153626 |
0.002788 |
4.27359 |
2.5593 |
| 10821698 |
oncostatin M receptor |
Osmr |
NM_001005384 |
0.000642 |
6.0085 |
4.12968 |
| 10881293 |
podoplanin |
Pdpn |
NM_019358 |
0.001418 |
3.99897 |
2.61301 |
| 10930660 |
protein S (alpha) |
Pros1 |
NM_031086 |
0.000526 |
2.09544 |
2.68498 |
| 10939498 |
RNA binding motif protein 41 |
Rbm41 |
NM_001109420 |
0.003826 |
2.27706 |
2.07025 |
| 10874929 |
retinol dehydrogenase 10 (all-trans) |
Rdh10 |
NM_181478 |
0.00379 |
2.9943 |
2.32697 |
| 10900511 |
receptor accessory protein 6 |
Reep6 |
NM_001013218 |
8.44E-05 |
−3.21858 |
−2.57593 |
| 10883071 |
similar to hypothetical protein MGC38716 |
RGD1304963 |
ENSRNOT00000011832 |
0.005265 |
2.07828 |
2.02776 |
| 10816879 |
RGD1564171 |
RGD1564171 |
NM_001109186 |
0.003087 |
2.42088 |
2.40646 |
| 10906926 |
Rho family GTPase 1 |
Rnd1 |
NM_001013222 |
0.020945 |
2.01484 |
2.03486 |
| 10889399 |
radical S-adenosyl methionine domain containing 2 |
Rsad2 |
NM_138881 |
0.016232 |
12.7868 |
4.6819 |
| 10765173 |
selectin, endothelial cell |
Sele |
NM_138879 |
0.003772 |
2.68122 |
2.22527 |
| 10910406 |
sema domain, immunoglobulin domain (Ig), and GPI membr |
Sema7a |
NM_001108153 |
0.000297 |
−2.20198 |
−2.08698 |
| 10744687 |
solute carrier family 13 (sodium-dependent citrate trans |
Slc13a5 |
NM_170668 |
0.00314 |
4.69584 |
3.06774 |
| 10805335 |
solute carrier family 14 (urea transporter), member 1 |
Slc14a1 |
NM_019346 |
0.003031 |
2.6531 |
2.87021 |
| 10804672 |
solute carrier family 26 (sulfate transporter), member 2 |
Slc26a2 |
NM_057127 |
0.015372 |
2.17668 |
2.02817 |
| 10823057 |
solute carrier family 7 (cationic amino acid transpor |
Slc7a11 |
NM_001107673 |
0.000438 |
2.11226 |
2.16576 |
| 10935997 |
SFRS protein kinase 3 |
Srpk3 |
NM_184045 |
0.002203 |
2.15515 |
2.54938 |
| 10927842 |
signal transducer and activator of transcription 1 |
Stat1 |
NM_032612 |
0.000288 |
3.0351 |
2.42253 |
| 10794345 |
sushi domain containing 3 |
Susd3 |
NM_001107341 |
8.31E-06 |
−2.26654 |
−2.62958 |
| 10821959 |
threonyl-tRNA synthetase |
Tars |
NM_001006976 |
0.000161 |
2.33103 |
2.32162 |
| 10936482 |
TIMP metallopeptidase inhibitor 1 |
Timp1 |
NM_053819 |
0.002114 |
6.19945 |
3.69766 |
| 10919694 |
transmembrane protein 108 |
Tmem108 |
ENSRNOT00000014519 |
0.002274 |
−2.05022 |
−2.23734 |
| 10762108 |
transmembrane protein 116 |
Tmem116 |
NM_001159625 |
0.000345 |
−2.39288 |
−2.23365 |
| 10874198 |
tumor necrosis factor receptor superfamily, member 9 |
Tnfrsf9 |
NM_001025773 |
0.001067 |
4.29073 |
3.16758 |
| 10774171 |
uridine phosphorylase 1 |
Upp1 |
NM_001030025 |
0.001233 |
3.84779 |
2.06033 |
| 10720215 |
zinc finger protein 36 |
Zfp36 |
NM_133290 |
0.00113 |
4.38522 |
3.47823 |
| 10935061 |
— |
|
— |
0.000426 |
2.08993 |
2.27601 |
| 10766724 |
— |
|
— |
0.001338 |
3.06184 |
3.89387 |
| 10815496 |
— |
|
— |
0.003018 |
2.12199 |
2.05157 |
| 10802706 |
— |
|
— |
0.004766 |
2.01494 |
2.94154 |
| 10937867 | — | — | 0.006416 | 2.30821 | 2.21384 |
Genes and ncRNA regulated by both Saffron light damage (LD) and LD when compared to control.. The change in expression indicates that these genes (76 genes in total including 71 coding and 5 noncoding RNAs) change in response to light damage and not the treatment paradigm.
Table 7. Genes and ncRNA regulated by photobiomodulation light damage and light damage.
| Probeset ID | Gene_assignment | Gene symbol | RefSeq | p-value | FC (LD/C) | FC (PBMLD/C) |
|---|---|---|---|---|---|---|
| 10855701 |
aquaporin 1 |
Aqp1 |
NM_012778 |
0.000296 |
−2.4813 |
−2.26193 |
| 10761128 |
heat shock protein 1 |
Hspb1 |
NM_031970 |
0.040669 |
5.7848 |
2.67409 |
| 10834613 | RGD1307355 | RGD1307355 | NM_001107822 | 0.008949 | 2.02622 | 2.00283 |
Genes regulated by both photobiomodulation (PBM) light damage (PBMLD) and LD when compared to control. The change in gene expressions indicate that these genes (3 genes in total) change in response to light damage and not the treatment paradigm.
Table 8. Genes and ncRNA regulated by all groups exposed to light damage.
| Probeset ID | gene_assignment | Gene Symbol | RefSeq | p-value | FC (LD/C) | FC (PBMLD/C) | FC (SafLD/C) |
|---|---|---|---|---|---|---|---|
| 10889660 |
aryl hydrocarbon receptor |
Ahr |
NM_013149 |
0.000824 |
3.58244 |
2.55846 |
3.37031 |
| 10860951 |
asparagine synthetase |
Asns |
NM_013079 |
0.000175 |
3.65332 |
3.59864 |
3.94323 |
| 10770710 |
activating transcription factor 3 |
Atf3 |
NM_012912 |
0.000312 |
14.6342 |
7.50088 |
12.4135 |
| 10906024 |
ceramide kinase |
Cerk |
NM_001134861 |
0.002068 |
−2.29855 |
−2.22234 |
−2.5033 |
| 10832934 |
carbohydrate sulfotransferase 3 |
Chst3 |
NM_053408 |
0.001969 |
2.29346 |
4.51694 |
4.59235 |
| 10707832 |
chondroitin sulfate synthase 1 |
Chsy1 |
NM_001106268 |
0.00196 |
2.19289 |
2.26058 |
2.38502 |
| 10847932 |
DEP domain containing 7 |
Depdc7 |
NM_001029916 |
0.000836 |
6.30169 |
3.61235 |
5.17396 |
| 10800919 |
early growth response 1 |
Egr1 |
NM_012551 |
0.000563 |
3.67302 |
2.5765 |
3.72215 |
| 10886121 |
estrogen-related receptor beta |
Esrrb |
NM_001008516 |
0.001754 |
−5.15267 |
−3.39191 |
−5.11574 |
| 10815026 |
fibroblast growth factor 2 |
Fgf2 |
NM_019305 |
0.007852 |
3.69169 |
2.80573 |
3.69923 |
| 10787517 |
growth differentiation factor 15 |
Gdf15 |
NM_019216 |
0.001794 |
3.10005 |
2.16424 |
2.90564 |
| 10747948 |
glial fibrillary acidic protein |
Gfap |
NM_017009 |
0.000262 |
9.85794 |
5.10179 |
8.53628 |
| 10853554 |
guanine nucleotide binding protein (G protein), gamma 11 |
Gng11 |
NM_022396 |
0.005371 |
2.18311 |
2.27589 |
2.84574 |
| 10910562 |
GRAM domain containing 2 |
Gramd2 |
ENSRNOT00000036798 |
0.002852 |
−2.09097 |
−2.0499 |
−2.15488 |
| 10780433 |
interferon regulatory factor 9 |
Irf9 |
NM_001012041 |
0.000286 |
5.11681 |
2.54387 |
3.98553 |
| 10878112 |
Jun oncogene |
Jun |
NM_021835 |
9.06E-05 |
4.10045 |
3.36326 |
4.42475 |
| 10715078 |
kinesin family member 11 |
Kif11 |
ENSRNOT00000022555 |
0.001127 |
6.71624 |
2.93282 |
6.83808 |
| 10778179 |
kringle containing transmembrane protein 1 |
Kremen1 |
NM_053649 |
0.00126 |
3.11252 |
2.52286 |
2.76134 |
| 10869693 |
ladinin 1 |
Lad1 |
NM_001107942 |
0.00464 |
3.41587 |
2.87979 |
2.67287 |
| 10766809 |
laminin, beta 3 |
Lamb3 |
ENSRNOT00000008440 |
0.00104 |
−2.13971 |
−2.06332 |
−2.29965 |
| 10731493 |
lipopolysaccharide-induced TNF factor |
Litaf |
NM_001105735 |
0.000499 |
3.76351 |
3.31786 |
4.20289 |
| 10832646 |
similar to protocadherin 15 |
LOC687745 |
ENSRNOT00000000744 |
3.36E-05 |
2.16788 |
2.11147 |
2.13739 |
| 10797062 |
MAK10 homolog, amino-acid N-acetyltransferase subunit |
Mak10 |
NM_133324 |
0.000456 |
2.14755 |
2.06752 |
2.46397 |
| 10863512 |
methylenetetrahydrofolate dehydrogenase (NADP+ depende |
Mthfd2 |
NM_001109398 |
0.000137 |
3.72708 |
3.3414 |
4.70708 |
| 10855650 |
pleckstrin homology domain containing, family A (phos |
Plekha8 |
NM_001109235 |
0.002149 |
3.46919 |
2.28802 |
3.33038 |
| 10935064 |
proteolipid protein 1 |
Plp1 |
NM_030990 |
0.000277 |
2.15787 |
2.17668 |
2.84846 |
| 10894100 |
phosphatidic acid phosphatase type 2c |
Ppap2c |
NM_139252 |
0.000148 |
−3.99548 |
−3.52564 |
−3.53511 |
| 10933559 |
protein phosphatase, EF-hand calcium binding domain 1 |
Ppef1 |
NM_001034935 |
0.000332 |
−2.01839 |
−2.93703 |
−3.60683 |
| 10719616 |
poliovirus receptor |
PVR |
NM_017076 |
0.003604 |
3.29581 |
2.1509 |
2.47285 |
| 10730098 |
pyrroline-5-carboxylate synthetase (glutamate gamma-semi |
Pycs |
NM_001108524 |
7.37E-07 |
2.22935 |
2.00815 |
2.70701 |
| 10910805 |
similar to c-myc promoter binding protein |
RGD1562639 |
XR_009072 |
0.001046 |
2.29737 |
2.17402 |
2.70157 |
| 10779602 |
similar to hypothetical protein |
RGD1563070 |
NM_001134541 |
0.000934 |
3.13062 |
3.00334 |
3.19235 |
| 10705553 |
similar to F-box only protein 27 |
RGD1563982 |
BC091204 |
0.007204 |
2.97912 |
2.13711 |
3.00821 |
| 10703144 |
ribosomal protein S6 kinase polypeptide 2 |
Rps6ka2 |
ENSRNOT00000017809 |
0.000693 |
2.66827 |
2.6238 |
2.72578 |
| 10886621 |
serine (or cysteine) peptidase inhibitor, clade A, mem |
Serpina3n |
NM_031531 |
0.003527 |
15.3578 |
5.40098 |
9.53535 |
| 10778620 |
solute carrier family 1 (glutamate/neutral amino acid tra |
Slc1a4 |
NM_198763 |
0.000242 |
2.19034 |
2.4281 |
3.04173 |
| 10785326 |
solute carrier family 25, member 30 |
Slc25a30 |
NM_001013187 |
0.00073 |
6.93878 |
4.0298 |
7.26229 |
| 10831976 |
solute carrier family 26, member 8 |
Slc26a8 |
NM_001107614 |
0.016312 |
2.42603 |
2.62488 |
2.11542 |
| 10906608 |
solute carrier family 38, member 2 |
Slc38a2 |
NM_181090 |
0.000719 |
2.11022 |
2.05736 |
2.24878 |
| 10756393 |
solute carrier family 7 (cationic amino acid transporter, |
Slc7a1 |
NM_013111 |
0.001003 |
2.25745 |
2.49319 |
2.63527 |
| 10749372 |
suppressor of cytokine signaling 3 |
Socs3 |
NM_053565 |
0.000246 |
8.40314 |
3.67692 |
7.52167 |
| 10747506 |
signal transducer and activator of transcription 3 |
Stat3 |
NM_012747 |
0.000753 |
4.09848 |
2.61028 |
3.97928 |
| 10773496 |
TNFAIP3 interacting protein 2 |
Tnip2 |
NM_001024771 |
0.000385 |
3.20709 |
2.40442 |
4.38234 |
| 10850775 |
tribbles homolog 3 (Drosophila) |
Trib3 |
NM_144755 |
0.000162 |
2.59849 |
2.31869 |
2.98085 |
| 10859764 |
— |
|
— |
0.000806 |
3.363 |
2.50985 |
3.02341 |
| 10768826 | — | — | 0.005642 | 2.21021 | 2.02298 | 2.28881 |
Genes and ncRNA regulated by all groups light damage (LD), Saffron LD and photobiomodulation (PBM) LD when compared to control. The change in expression indicates that these genes (46 genes in total, including 44 coding and 2 noncoding RNAs) change in response to light damage and not the treatment paradigm.
Discussion
The present results provide an overview of gene and ncRNA regulation associated with the neuroprotective actions of PBM and saffron. The analyses used were chosen partly to provide validation of the method, for example the hierarchical clustering analysis in Figure 2 and the microarray-PCR comparison in Figure 6. In addition, they allow a compare-and-contrast discussion of the possible actions of saffron and PBM.
The box-and-whisker presentation in Figure 3 suggests that PBM and saffron acting on the retina in the absence of a light challenge have distinct effects. Saffron has relatively little effect on the expression of genes by the retina, but when given as pretreatment to LD, saffron reduced the large changes in gene expression induced by LD. PBM by itself had a much more significant effect on retinal gene expression than saffron, narrowing the distribution of entity expression changes and generating many “outliers.” PBM given as pretreatment to LD reduced the gene expression caused by LD toward control levels.
The Venn diagram analysis allowed a logical separation of lists of genes and ncRNAs whose regulation appears to contribute to neuroprotection; it also draws attention to the prominence of ncRNAs (rather than known genes) among the entities regulated during the protective action of PBM and saffron.
Possible mechanisms of protection against light damage
Our study builds upon previous work showing that there are global changes in gene expression due to LD [39–42] and that antioxidants can play a role in ameliorating this stress [15,17,43,61]. A direct example is Hmox1, which has been previously found to be a marker for light-induced stress in the retina and could be controlled by the antioxidant dimethylthiourea [43]. Our results also show a reduction in the expression of Hmox1in both the LD saffron and PBMLD treated samples. In contrast to these findings, a study by Sun and colleagues reported that overexpression of Hmox1 is protective to the retina [44]. This suggests that Hmox1 act as a marker for light-induced stress rather than playing a role in the etiology of the degeneration.
Tissue antioxidant proteins have been reported to be upregulated [13,14] or their activity increased [15] following light exposure; among others, glutathiones (Gpx1), thioredoxin-1, glutathione peroxidase, glutathione-S-transferase, and glutathione reductase have been identified in these findings. In the present study, we found Gpx3 gene expression showed a reduction in the LD animals. Both saffron and PBM mitigated the changes in gene expression following LD, suggesting that both saffron and PBM have a direct regulatory effect on tissue oxidative protection.
Another possible protective mechanism involved in saffron and PBM treatment is through the reduction of inflammation due to the downregulation of chemokine (C-C motif) ligand 2 (ccl2). CCL2 has been found to play an important role in inflammation by inducing leukocyte recruitment and activation [45] [46]. It has been shown to be elevated in many degenerative diseases of the central nervous system, such as multiple sclerosis [47], Alzheimer disease [48], Parkinson disease [49], and amyotrophic lateral sclerosis [50]. In the eye, ccl2 has been shown to play a role in the development of retinal degeneration; ccl2-deficient mice develop age related macular degeneration (AMD) like symptoms [51]. Our results suggest that reducing ccl2 levels to near control levels has a direct correlation with the amount of cell death. Further investigation into the role of ccl-2 in LD in the retina is required.
Different forms of neuroprotection: contrasts in entity expression
LD was used in this study as an assay of the protected/vulnerable status of photoreceptors. It is relevant to recall, however, that exposure to light also involves a neuroprotective action [52,53]. Prior light experience regulates photoreceptor vulnerability to light; both ambient light experienced over long periods and a briefer exposure to very bright light upregulate mechanisms that protect the photoreceptors from a subsequent light challenge.
Recently, we [54] drew a distinction among preconditioning pretreatments that make photoreceptors resistant to LD. The distinction was between pretreatments that damage photoreceptors (examples being light [above] or hypoxia [55]) but nevertheless protect surviving photoreceptors against subsequent stress, and pretreatments that are protective without themselves damaging photoreceptors (examples being saffron [24] and PBM [28,29]). The present results show that the regulation of entity expression associated with light is very different from that associated with a nondamaging pretreatment in at least two ways. First, light regulates principally known genes, upregulating them; by contrast, PBM and saffron regulate large numbers of ncRNAs, mainly downregulating them.
How does saffron act?
The data provide some insight into how saffron acts to protect photoreceptors against LD in the present experiments. A simple, “direct action” hypothesis for the action of an antioxidant is that it does not interact with cells, but rather acts as a direct antioxidant, shortening the lifespan of reactive oxygen species, and reducing the damage they cause. This hypothesis would predict that saffron has little effect on retinal gene expression, and this prediction is not contradicted by the list of entities (data not shown) whose expression was regulated significantly by saffron without LD. The list is short (12 known genes, 5 ncRNAs), and only one entity (an ncRNA) was regulated more than threefold. The “direct action” hypothesis appears to be contradicted, however, by the large number of genes and ncRNAs which were significantly regulated by LD, and whose regulation was reduced significantly by saffron preconditioning (Table 2); and by the large number of genes and (especially) ncRNAs whose expression was significantly regulated by saffron when given as pretreatment to LD (Table 3 and Table 4). As already noted (Figure 5), a large proportion of the entities regulated in these two ways by saffron are ncRNAs, and further understanding of the protective action of saffron will require understanding of the roles of these sequences.
With known genes, the present data allow mechanisms of saffron-induced protection to be postulated for further study. As an example, one of the genes whose expression is upregulated specifically by saffron as part of its protective action against LD (Table 4) is endothelin 2. Expression of this gene is associated with the upregulation of the protective/trophic factor fibroblast growth factor-2 (FGF-2), which is known to be protective against photoreceptors [56–58]. Upstream from endothelin 2, leukemia inhibitory factor is known to upregulate endothelin 2 as part of the Jak/Stat pathway [59]; leukemia inhibitory factor expression has recently been shown to be protective to photoreceptors in the rat LD model [59]. Given the number of genes/entities involved, much detailed work will be required to define the mechanisms of the saffron-induced protection of photoreceptors.
How does photobiolmodulation act?
Previous analyses of the neuroprotective action of PBM [29,35,60] have suggested that the energy of the radiation is absorbed by the mitochondrial enzyme cytochrome oxidase, which serves the key role of sequestering oxygen from the tissue for oxidative phosphorylation pathways, and the production of adenosine-5'-triphosphate (ATP). The result includes restoration of toxin-induced loss of ATP production and increased cell viability. Several studies suggest that the absorption of PBM upregulates intracellular pathways governing the redox state of the cell (reviewed [35]).
The present results confirm that PBM, given without LD, changes retinal gene expression in a significant number of entities, and that, given as a pretreatment to LD, PBM (like saffron) changes the expression of a large numbers of entities, reducing the LD-induced regulation of many (Table 2 and Table 3) and regulating many not affected by LD (Table 5). PBM, like saffron, appears to regulate many intracellular pathways when given as a pretreatment. As with saffron, a large proportion of the entities regulated by PBM are ncRNAs, and further understanding of the protective action of saffron will require understanding to the roles of these sequences.
Neuroprotection: multiple pathways
The present results add to the knowledge of the mechanisms by which photoreceptors, and presumably other neurons, can be protected from degeneration. The present analysis of the action of saffron suggests that its action is more than that of a direct antioxidant; rather, saffron appears to interact very significantly with gene expression. Saffron is a complex of molecules [25] that includes powerful antioxidants, as well as a range of bioactive molecules. Which of these potentially active molecules, or which combination of them, accounts for the neuroprotective action of saffron remains to be determined.
PBM seems to act through at least two pathways, by reducing inflammation and by reducing oxidative damage. Future investigation of the ncRNAs regulated by PBM and saffron could reveal further clues to their mechanism of protection.
Acknowledgments
The authors are grateful to Ms R. Albarracin for her help in the animal experiments. This research was supported by the Australian Research Council through the ARC Centre of Excellence in Vision Science (CE0561903), by a grant-in-aid from the Sir Zelman Cowen Universities Fund and by Foundation Fighting Blindness (FFB) grants (TA-NE-0606–0348-UWI, TA-NP-0709–0465-UWI, and TA-NP-0709–0465-UWI).
References
- 1.Lamb TD. Evolution of vertebrate retinal photoreception. Philos Trans R Soc Lond B Biol Sci. 2009;364:2911–24. doi: 10.1098/rstb.2009.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winkler BS. Glycolytic and oxidative metabolism in relation to retinal function. J Gen Physiol. 1981;77:667–92. doi: 10.1085/jgp.77.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graymore C. Metabolism of the developing retina. Br J Ophthalmol. 1959;43:34–9. doi: 10.1136/bjo.43.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancho-Pelluz J, Arango-Gonzalez B, Kustermann S, Romero FJ, van Veen T, Zrenner E, Ekstrom P, Paquet-Durand F. Photoreceptor cell death mechanisms in inherited retinal degeneration. Mol Neurobiol. 2008;38:253–69. doi: 10.1007/s12035-008-8045-9. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann H. Mechanisms of cell specialization. Invest Ophthalmol. 1969;8:17–25. [PubMed] [Google Scholar]
- 6.Stone J, Maslim J, Valter-Kocsi K, Mervin K, Bowers F, Chu Y, Barnett N, Provis J, Lewis G, Fisher SK, Bisti S, Gargini C, Cervetto L, Merin S, Peer J. Mechanisms of photoreceptor death and survival in mammalian retina. Prog Retin Eye Res. 1999;18:689–735. doi: 10.1016/s1350-9462(98)00032-9. [DOI] [PubMed] [Google Scholar]
- 7.Jozwick C, Valter K, Stone J. Reversal of functional loss in the P23H–3 rat retina by management of ambient light. Exp Eye Res. 2006;83:1074–80. doi: 10.1016/j.exer.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Chrysostomou V, Stone J, Stowe S, Barnett NL, Valter K. The Status of Cones in the Rhodopsin Mutant P23H–3 Retina: Light-Regulated Damage and Repair in Parallel with Rods. Invest Ophthalmol Vis Sci. 2008;49:1116–25. doi: 10.1167/iovs.07-1158. [DOI] [PubMed] [Google Scholar]
- 9.Valter K, Kirk DK, Stone J. Optimising the structure and function of the adult P23H–3 retina by light management in the juvenile and adult. Exp Eye Res. 2009;89:1003–11. doi: 10.1016/j.exer.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Wiegand RD, Giusto NM, Rapp LM, Anderson RE. Evidence for rod outer segment lipid peroxidation following constant illumination of the rat retina. Invest Ophthalmol Vis Sci. 1983;24:1433–5. [PubMed] [Google Scholar]
- 11.Tanito M, Yoshida Y, Kaidzu S, Ohira A, Niki E. Detection of lipid peroxidation in light-exposed mouse retina assessed by oxidative stress markers, total hydroxyoctadecadienoic acid and 8-iso-prostaglandin F2alpha. Neurosci Lett. 2006;398:63–8. doi: 10.1016/j.neulet.2005.12.070. [DOI] [PubMed] [Google Scholar]
- 12.Organisciak DT, Darrow RM, Jiang YI, Marak GE, Blanks JC. Protection by dimethylthiourea against retinal light damage in rats. Invest Ophthalmol Vis Sci. 1992;33:1599–609. [PubMed] [Google Scholar]
- 13.Penn JS, Naash MI, Anderson RE. Effect of light history on retinal antioxidants and light damage susceptibility in the rat. Exp Eye Res. 1987;44:779–88. doi: 10.1016/s0014-4835(87)80041-6. [DOI] [PubMed] [Google Scholar]
- 14.Gosbell AD, Stefanovic N, Scurr LL, Pete J, Kola I, Favilla I, de Haan JB. Retinal light damage: structural and functional effects of the antioxidant glutathione peroxidase-1. Invest Ophthalmol Vis Sci. 2006;47:2613–22. doi: 10.1167/iovs.05-0962. [DOI] [PubMed] [Google Scholar]
- 15.Xie Z, Wu X, Gong Y, Song Y, Qiu Q, Li C. Intraperitoneal injection of Ginkgo biloba extract enhances antioxidation ability of retina and protects photoreceptors after light-induced retinal damage in rats. Curr Eye Res. 2007;32:471–9. doi: 10.1080/02713680701257621. [DOI] [PubMed] [Google Scholar]
- 16.Ranchon I, Gorrand JM, Cluzel J, Droy-Lefaix MT, Doly M. Functional protection of photoreceptors from light-induced damage by dimethylthiourea and Ginkgo biloba extract. Invest Ophthalmol Vis Sci. 1999;40:1191–9. [PubMed] [Google Scholar]
- 17.Tomita H, Kotake Y, Anderson RE. Mechanism of protection from light-induced retinal degeneration by the synthetic antioxidant phenyl-N-tert-butylnitrone. Invest Ophthalmol Vis Sci. 2005;46:427–34. doi: 10.1167/iovs.04-0946. [DOI] [PubMed] [Google Scholar]
- 18.Logvinov SV, Plotnikov MB, Varakuta EY, Zhdankina AA, Potapov AV, Mikhulya EP. Effect of ascovertin on morphological changes in rat retina exposed to high-intensity light. Bull Exp Biol Med. 2005;140:578–81. doi: 10.1007/s10517-006-0029-z. [DOI] [PubMed] [Google Scholar]
- 19.Yilmaz T, Aydemir O, Ozercan IH, Ustundağ B. Effects of vitamin e, pentoxifylline and aprotinin on light-induced retinal injury. Ophthalmologica. 2007;221:159–66. doi: 10.1159/000099295. [DOI] [PubMed] [Google Scholar]
- 20.Stahl W, Sies H. Bioactivity and protective effects of natural carotenoids. Biochim Biophys Acta. 2005;1740:101–7. doi: 10.1016/j.bbadis.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Costa BL, Fawcett R, Li GY, Safa R, Osborne NN. Orally administered epigallocatechin gallate attenuates light-induced photoreceptor damage. Brain Res Bull. 2008;76:412–23. doi: 10.1016/j.brainresbull.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Komeima K, Rogers BS, Lu L, Campochiaro PA. Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc Natl Acad Sci USA. 2006;103:11300–5. doi: 10.1073/pnas.0604056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen J, Yang X, Dong A, Petters RM, Peng YW, Wong F, Campochiaro PA. Oxidative Damage is a Potential Cause of Cone Cell Death in Retinitis Pigmentosa. J Cell Physiol. 2005;203:457–64. doi: 10.1002/jcp.20346. [DOI] [PubMed] [Google Scholar]
- 24.Maccarone R, Di Marco S, Bisti S. Saffron supplement maintains morphology and function after exposure to damaging light in mammalian retina. Invest Ophthalmol Vis Sci. 2008;49:1254–61. doi: 10.1167/iovs.07-0438. [DOI] [PubMed] [Google Scholar]
- 25.Giaccio M. Crocetin from saffron: an active component of an ancient spice. Crit Rev Food Sci Nutr. 2004;44:155–72. doi: 10.1080/10408690490441433. [DOI] [PubMed] [Google Scholar]
- 26.Falsini B, Piccardi M, Minnella A, Savastano C, Capoluongo E, Fadda A, Balestrazzi E, Maccarone R, Bisti S.Saffron Supplementation Improves Retinal Flicker Sensitivity in Early Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci.2010 [DOI] [PubMed] [Google Scholar]
- 27.Eells JT, Wong-Riley MT, VerHoeve J, Henry M, Buchman EV, Kane MP, Gould LJ, Das R, Jett M, Hodgson BD, Margolis D, Whelan HT. Mitochondrial signal transduction in accelerated wound and retinal healing by near-infrared light therapy. Mitochondrion. 2004;4:559–67. doi: 10.1016/j.mito.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 28.Eells JT, Henry MM, Summerfelt P, Wong-Riley MT, Buchmann EV, Kane M, Whelan NT, Whelan HT. Therapeutic photobiomodulation for methanol-induced retinal toxicity. Proc Natl Acad Sci USA. 2003;100:3439–44. doi: 10.1073/pnas.0534746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eells JT, DeSmet KD, Kirk DK, Wong-Riley M, Whelan HT, Ver Hoeve JT, Nork M, Stone J, Valter K. Photobiomodulation for the Treatment of Retinal Injury and Retinal Degenerative Diseases. Proceedings of Light-Activated Tissue Regeneration and Therapy Conference. 2008 [Google Scholar]
- 30.Qu C, Cao W, Fan Y, Lin Y. Near-infrared light protect the photoreceptor from light-induced damage in rats. Adv Exp Med Biol. 2010;664:365–74. doi: 10.1007/978-1-4419-1399-9_42. [DOI] [PubMed] [Google Scholar]
- 31.Oron A, Oron U, Chen J, Eilam A, Zhang C, Sadeh M, Lampl Y, Streeter J, DeTaboada L, Chopp M. Low-level laser therapy applied transcranially to rats after induction of stroke significantly reduces long-term neurological deficits. Stroke. 2006;37:2620–4. doi: 10.1161/01.STR.0000242775.14642.b8. [DOI] [PubMed] [Google Scholar]
- 32.Liang HL, Whelan HT, Eells JT, Wong-Riley MT. Near-infrared light via light-emitting diode treatment is therapeutic against rotenone- and 1-methyl-4-phenylpyridinium ion-induced neurotoxicity. Neuroscience. 2008;153:963–74. doi: 10.1016/j.neuroscience.2008.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw VE, Spana S, Ashkan K, Benabid AL, Stone J, Baker GE, Mitrofanis J. Neuroprotection of midbrain dopaminergic cells in MPTP-treated mice after near-infrared (NIr) light treatment. J Comp Neurol. 2010;518:25–40. doi: 10.1002/cne.22207. [DOI] [PubMed] [Google Scholar]
- 34.Lampl Y, Zivin JA, Fisher M, Lew R, Welin L, Dahlof B, Borenstein P, Andersson B, Perez J, Caparo C, Ilic S, Oron U. Infrared laser therapy for ischemic stroke: a new treatment strategy: results of the NeuroThera Effectiveness and Safety Trial-1 (NEST-1). Stroke. 2007;38:1843–9. doi: 10.1161/STROKEAHA.106.478230. [DOI] [PubMed] [Google Scholar]
- 35.Hamblin M, Demidova N. Mechanisms of low-light therapy. In: Hamblin M, Waynant R, Anders J, Editors. Mechanisms for Low-Light Therapy. Bellingham, WA: The International Society for Optical Engineering, Proc SPIE; 2006. p. 1–12. [Google Scholar]
- 36.Natoli R, Provis J, Valter K, Stone J. Expression and role of the early-response gene Oxr1 in the hyperoxia-challenged mouse retina. Invest Ophthalmol Vis Sci. 2008;49:4561–7. doi: 10.1167/iovs.08-1722. [DOI] [PubMed] [Google Scholar]
- 37.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maslim J, Valter K, Egensperger R, Holländer H, Stone J. Tissue oxygen during a critical developmental period controls the death and survival of photoreceptors. Invest Ophthalmol Vis Sci. 1997;38:1667–77. [PubMed] [Google Scholar]
- 39.Chen L, Wu W, Dentchev T, Zeng Y, Wang J, Tsui I, Tobias JW, Bennett J, Baldwin D, Dunaief JL. Light damage induced changes in mouse retinal gene expression. Exp Eye Res. 2004;79:239–47. doi: 10.1016/j.exer.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Rohrer B, Guo Y, Kunchithapautham K, Gilkeson GS. Eliminating complement factor D reduces photoreceptor susceptibility to light-induced damage. Invest Ophthalmol Vis Sci. 2007;48:5282–9. doi: 10.1167/iovs.07-0282. [DOI] [PubMed] [Google Scholar]
- 41.Huang H, Frank MB, Dozmorov I, Cao W, Cadwell C, Knowlton N, Centola M, Anderson RE. Identification of mouse retinal genes differentially regulated by dim and bright cyclic light rearing. Exp Eye Res. 2005;80:727–39. doi: 10.1016/j.exer.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 42.Kassen SC, Ramanan V, Montgomery JE. T Burket C, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–31. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- 43.Kutty RK, Kutty G, Wiggert B, Chader GJ, Darrow RM, Organisciak DT. Induction of heme oxygenase 1 in the retina by intense visible light:Suppression by the antioxidant dimethylthiourea. Proc Natl Acad Sci USA. 1995;92:1177–81. doi: 10.1073/pnas.92.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun MH, Pang JH, Chen SL, Kuo PC, Chen KJ, Kao LY, Wu JY, Lin KK, Tsao YP. Photoreceptor protection against light damage by AAV-mediated overexpression of heme oxygenase-1. Invest Ophthalmol Vis Sci. 2007;48:5699–707. doi: 10.1167/iovs.07-0340. [DOI] [PubMed] [Google Scholar]
- 45.Manzo A, Caporali R, Montecucco C, Pitzalis C. Role of chemokines and chemokine receptors in regulating specific leukocyte trafficking in the immune/inflammatory response. Clin Exp Rheumatol. 2003;21:501–8. [PubMed] [Google Scholar]
- 46.Yoshie O. Role of chemokines and chemokine receptors in leukocyte trafficking. Nippon Rinsho. 2005;63(Suppl 4):437–43. [PubMed] [Google Scholar]
- 47.Banisor I, Leist TP, Kalman B. Involvement of beta-chemokines in the development of inflammatory demyelination. J Neuroinflammation. 2005;2:7. doi: 10.1186/1742-2094-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishizuka K, Igata-Yi R, Kimura T, Hieshima K, Kukita T, Kin Y, Misumi Y, Yamamoto M, Nomiyama H, Miura R, Takamatsu J, Katsuragi S, Miyakawa T. Expression and distribution of CC chemokine macrophage inflammatory protein-1 alpha/LD78 in the human brain. Neuroreport. 1997;8:1215–8. doi: 10.1097/00001756-199703240-00031. [DOI] [PubMed] [Google Scholar]
- 49.Sawada M, Imamura K, Nagatsu T. Role of cytokines in inflammatory process in Parkinson's disease. J Neural Transm Suppl. 2006;70:373–81. doi: 10.1007/978-3-211-45295-0_57. [DOI] [PubMed] [Google Scholar]
- 50.Wilms H, Sievers J, Dengler R, Bufler J, Deuschl G, Lucius R. Intrathecal synthesis of monocyte chemoattractant protein-1 (MCP-1) in amyotrophic lateral sclerosis: further evidence for microglial activation in neurodegeneration. J Neuroimmunol. 2003;144:139–42. doi: 10.1016/j.jneuroim.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 51.Chan CC, Ross RJ, Shen D, Ding X, Majumdar Z, Bojanowski CM, Zhou M, Salem N, Jr, Bonner R, Tuo J. Ccl2/Cx3cr1-deficient mice: an animal model for age-related macular degeneration. Ophthalmic Res. 2008;40:124–8. doi: 10.1159/000119862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penn JS, Tolman BL, Thum LA, Koutz CA. Effect of light history on the rat retina: timecourse of morphological adaptation and readaptation. Neurochem Res. 1992;17:91–9. doi: 10.1007/BF00966869. [DOI] [PubMed] [Google Scholar]
- 53.Liu C, Peng M, Laties AM, Wen R. Preconditioning with bright light evokes a protective response against light damage in the rat retina. J Neurosci. 1998;18:1337–44. doi: 10.1523/JNEUROSCI.18-04-01337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Y, Valter K, Stone J. Environmental Damage to the Retina and Preconditioning: Contrasting Effects of Light and Hyperoxic Stress. Invest Ophthalmol Vis Sci. 2010 doi: 10.1167/iovs.09-5050. [DOI] [PubMed] [Google Scholar]
- 55.Grimm C, Wenzel A, Groszer M, Mayser H, Seeliger M, Samardzija M, Bauer C, Gassmann M, Remé CE. HIF-1-induced erythropoietin in the hypoxic retina protects against light-induced retinal degeneration. Nat Med. 2002;8:718–24. doi: 10.1038/nm723. [DOI] [PubMed] [Google Scholar]
- 56.Faktorovich EG, Steinberg RH, Yasumura D, Matthes MT, LaVail MM. Photoreceptor degeneration in inherited retinal dystrophy delayed by basic fibroblast growth factor. Nature. 1990;347:83–6. doi: 10.1038/347083a0. [DOI] [PubMed] [Google Scholar]
- 57.Valter K, Bisti S, Gargini C, Di Loreto S, Maccarone R, Cervetto L, Stone J. Time course of neurotrophic factor upregulation and retinal protection against light-induced damage after optic nerve section. Invest Ophthalmol Vis Sci. 2005;46:1748–54. doi: 10.1167/iovs.04-0657. [DOI] [PubMed] [Google Scholar]
- 58.O'Driscoll C, O'Connor J, O'Brien CJ, Cotter TG. Basic fibroblast growth factor-induced protection from light damage in the mouse retina in vivo. J Neurochem. 2008;105:524–36. doi: 10.1111/j.1471-4159.2007.05189.x. [DOI] [PubMed] [Google Scholar]
- 59.Bürgi S, Samardzija M, Grimm C. Endogenous leukemia inhibitory factor protects photoreceptor cells against light-induced degeneration. Mol Vis. 2009;15:1631–7. [PMC free article] [PubMed] [Google Scholar]
- 60.Wong-Riley MT, Liang HL, Eells JT, Chance B, Henry MM, Buchmann E, Kane M, Whelan HT. Photobiomodulation directly benefits primary neurons functionally inactivated by toxins: role of cytochrome c oxidase. J Biol Chem. 2005;280:4761–71. doi: 10.1074/jbc.M409650200. [DOI] [PubMed] [Google Scholar]
- 61.Tanito M, Kaidzu S, Anderson RE. Delayed loss of cone and remaining rod photoreceptor cells due to impairment of choroidal circulation after acute light exposure in rats. Invest Ophthalmol Vis Sci. 2007;48:1864–72. doi: 10.1167/iovs.06-1065. [DOI] [PubMed] [Google Scholar]