Abstract
Protein evolution is not simply a race toward improved function. Because organisms compete for limited resources, fitness is also affected by the relative economy of an organism’s proteome. Indeed, many abundant proteins contain relatively high percentages of amino acids that are metabolically less taxing for the cell to make, thus reducing cellular cost. However, not all abundant proteins are economical, and many economical proteins are not particularly abundant. Here we examined protein composition and found that the relative synthetic cost of amino acids constrains the composition of microbial extracellular proteins. In Escherichia coli, extracellular proteins contain, on average, fewer energetically expensive amino acids independent of their abundance, length, function, or structure. Economic pressures have strategically shaped the amino acid composition of multicomponent surface appendages, such as flagella, curli, and type I pili, and extracellular enzymes, including type III effector proteins and secreted serine proteases. Furthermore, in silico analysis of Pseudomonas syringae, Mycobacterium tuberculosis, Saccharomyces cerevisiae, and over 25 other microbes spanning a wide range of GC content revealed a broad bias toward more economical amino acids in extracellular proteins. The synthesis of any protein, especially those rich in expensive aromatic amino acids, represents a significant investment. Because extracellular proteins are lost to the environment and not recycled like other cellular proteins, they present a greater burden on the cell, as their amino acids cannot be reutilized during translation. We hypothesize that evolution has optimized extracellular proteins to reduce their synthetic burden on the cell.
IMPORTANCE
Microbes secrete proteins to perform essential interactions with their environment, such as motility, pathogenesis, biofilm formation, and resource acquisition. However, because microbes generally lack protein import systems, secretion is often a one-way street. Consequently, secreted proteins are less likely to be recycled by the cell due to environmental loss. We demonstrate that evolution has in turn selected these extracellular proteins for increased economy at the level of their amino acid composition. Compared to their cellular counterparts, extracellular proteins have fewer synthetically expensive amino acids and more inexpensive amino acids. The resulting bias lessens the loss of cellular resources due to secretion. Furthermore, this economical bias was observed regardless of the abundance, length, structure, or function of extracellular proteins. Thus, it appears that economy may address the compositional bias seen in many extracellular proteins and deliver further insight into the forces driving their evolution.
Introduction
By enveloping cellular life, membranes separate proteomes into the following two distinct groups: cellular and extracellular. While free-living bacteria secrete extracellular proteins through several dedicated pathways, there are no known systems by which extracellular proteins are imported (1, 2). Consequently, extracellular proteins are less likely to be recycled by the cell or passed down during cell division. Many extracellular proteins are involved in pathogenesis and have been noted for their unique compositional biases (3, 4), which are significant enough to be predictive (3, 5–12). However, identifying and exploiting these biases has received more attention than determining which pressures led to them (13, 14).
Evolution selects for phenotypic changes that increase organismal fitness. At the molecular level, amino acid substitutions that enhance, diversify, or maintain beneficial functions or phenotypes are favored. However, not all substitutions are predicted to affect protein folding or function (15, 16). Nonetheless, such neutral substitutions, particularly in abundant proteins, can affect the metabolic load of an organism and thus be subject to natural selection (17, 18). Accordingly, microbes that thrive in nutrient-restrictive environments have proteins with fewer sulfurs, carbons, or nitrogens (19–23). Similarly, transient nutrient starvation results in expression of proteins with less of a limiting element (24, 25). Furthermore, many amino acid biosynthetic enzymes contain less of the amino acids they produce (26, 27).
Protein composition is also shaped by the energy required to synthesize individual amino acids (21, 28–32). The total synthetic cost of an amino acid includes both the ATPs/GTPs used in biosynthesis and the energy lost to central metabolism from the consumption of precursors (28, 29, 33, 34). The synthetic costs of amino acids vary by over 6-fold in Escherichia coli: Gly costs 11.7 high-energy phosphate bonds (~P) or ATPs, whereas Trp costs 74.3 (see Text S1, p. 5 and 6 in the supplemental material) (28). Numerous studies have found that abundant proteins are often composed of amino acids that require fewer ATPs to produce (21, 28–31). Here, we demonstrate that protein composition and economy are more tightly coupled to location. Compared to cytoplasmic, periplasmic, or membrane proteins, extracellular proteins contain a significantly higher composition of economic amino acids.
RESULTS AND DISCUSSION
Protein location and cost in Escherichia coli.
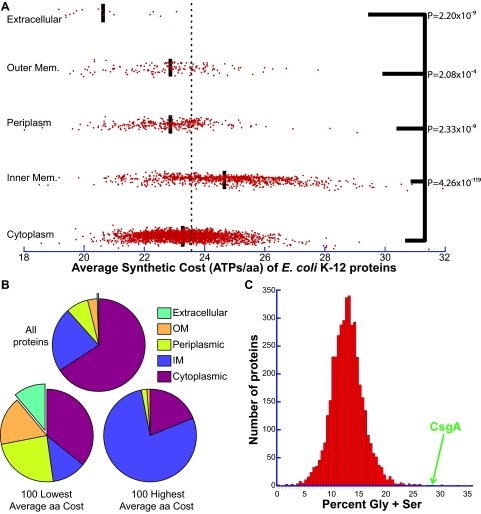
We calculated the average synthetic cost (ASC) of each protein in E. coli K-12 (Fig. 1A; see also Data Set S1, tab A, in the supplemental material) using the amino acid synthetic cost of chemoheterotrophic bacteria (28, 29). Strikingly, 11 of the 100 most economical proteins (those with the lowest ASCs) were extracellular, even though extracellular proteins comprise only 0.37% of total proteins—a 30-fold enrichment (Fig. 1B; see also Text S1, p. 7, in the supplemental material). Extracellular proteins required 2.9 fewer ATPs per residue than an average protein (U test, P = 1.96 × 10−9) (see Text S1, p. 8, in the supplemental material). Thus, for a typical protein in E. coli, these biases would save ~900 ATPs. The ASC was nearly predictive for the location in E. coli, as not a single extracellular protein had an ASC above the global average.
FIG 1 .
Protein location in E. coli is indicative of synthetic cost. (A) Each protein in E. coli is plotted based upon its average synthetic cost (ASC) and cellular location. Dotted line, mean ASC of all proteins in E. coli; black bars, mean ASC of proteins in that location; aa, amino acid. U tests were used to compare the protein ASCs of each location against those of cytoplasmic proteins. (B) Locations of all proteins, the 100 most economical proteins, and the 100 least economical proteins in E. coli, as ranked by ASC. OM, outer membrane; IM, inner membrane. (C) Histogram of the percent Gly plus Ser for all proteins in E. coli.
Periplasmic and outer membrane proteins were enriched by 3- and 5-fold, respectively, among economical proteins; inner membrane proteins were more likely to contain expensive residues. Surprisingly, outer membrane proteins were significantly more economical than inner membrane proteins due to an increased number of expensive amino acids in integral membrane proteins (1.8 ATPs per amino acid; U test, P = 4.21 × 10−31) (see Fig. S1 and Text S1, p. 12–14, in the supplemental material). The ASCs of outer membrane β-barrel and membrane-anchored proteins were similar to those of cytoplasmic proteins; however, outer membrane lipoproteins, many of which have soluble periplasmic domains, had significantly lower ASCs than cytoplasmic proteins (see Fig. S1 and Text S1, p. 12–13).
Protein economics of extracellular appendages.
One of the most abundant extracellular proteins in E. coli is the major subunit of the curli fiber CsgA (35) (see Fig. S2B in the supplemental material). Like some curli-specific gene products, CsgA is rich in Gly and Ser. CsgA is composed of 19.2% Gly (global mean, 7.2%) and 28.5% Gly plus Ser (global mean, 12.8%) (Fig. 1C; see also Text S1, p. 15, in the supplemental material), the sixth largest amount in any E. coli protein. Intriguingly, the curli regulator CsgD increases expression of a gene for the biosynthetic enzyme GlyA, which interconverts Gly and Ser (36). CsgD may increase GlyA to balance Gly and Ser pools, resulting in efficient curli production. More appropriately, as ancient (37), relatively simple amino acids, Gly and Ser are two of the least expensive ones to produce (28, 33, 34) (see Text S1, p. 5, in the supplemental material). Consequently, CsgA has the ninth lowest ASC in E. coli, utilizing 4.17 fewer ATPs per residue than average (see Text S1, p. 16 and 17, in the supplemental material). The major subunits of flagella and type 1 pili have the 6th and 11th lowest ASCs (see Fig. S2 and Text S1, p. 16 and 17, in the supplemental material).
The relative economy of extracellular proteins is not due to enrichment of the same amino acids. Collectively, extracellular proteins in E. coli contain more of the inexpensive amino acids Ala, Asn, Gln, Ser, and Thr and fewer of the expensive residues Arg, His, Met, Phe, and Trp (see Text S1, p. 14, in the supplemental material). When examining amino acid usage in CsgA, FimA, and FliC, we found that all three contain fewer aromatic residues. However, each major fiber subunit had a unique combination of inexpensive amino acids (see Text S1, p. 18, in the supplemental material). Enrichment of Gly and Asn is responsible for 65.8% of CsgA’s energy savings, whereas 54.9% of FimA’s savings are due to enrichment of Ala and Thr. In contrast, FliC is not particularly rich in any one amino acid; 31.7% of its savings are due to enrichment of Asn and Thr. Instead, FliC contains reduced amounts of aromatic amino acids relative to CsgA or FimA. Thus, extracellular proteins are not simply rich in a specific subset of economical amino acids; rather, they contain many combinations of inexpensive amino acids and typically lack expensive ring-structured amino acids.
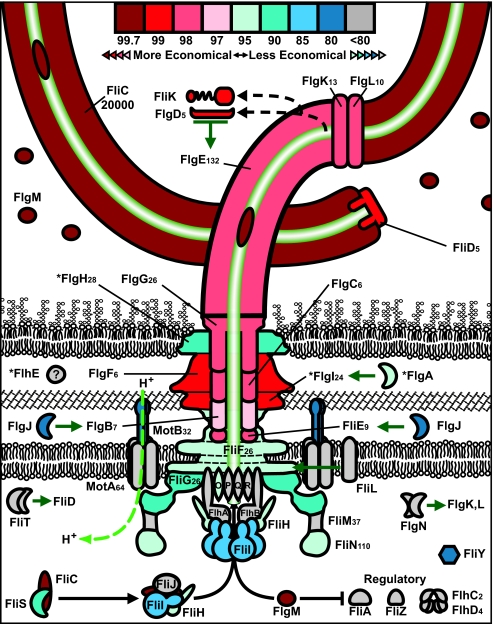
The bacterial flagellum is one of the most complex and well-studied cellular structures in bacteria (38). With multiple proteins in every cellular compartment, the flagellum is an excellent system to analyze the connection between protein location and ASC within a single organelle. Indeed, the cost of flagellar proteins decreases the farther they are from the cytoplasm (Fig. 2, darker reds). Extracellular flagellar proteins have significantly lower ASCs than cytoplasmic flagellar proteins (2.6 ATPs per amino acid; t test, P = 2.62 × 10−6) (see Fig. S3A and Text S1, p. 19, in the supplemental material). Furthermore, curli and type I pilus proteins show economic trends similar to those shown by flagellar proteins (see Fig. S4 and Text S1, p. 16 and 17, in the supplemental material). Because the ASC might be influenced by protein length, abundance, or function (28, 31), we tried to correlate the ASCs of flagellar proteins with these criteria. However, we did not find a significant trend when comparing the ASCs of flagellar proteins with their lengths or abundances (38) (Spearman’s rs = −0.189 and −0.255, P = 0.232 and 0.209, respectively) (see Fig. S3 and Text S1, p. 20, in the supplemental material). Additionally, the function of extracellular flagellar proteins includes structural, assembly, and regulatory roles; thus, function does not appear to affect their relative economy. Intriguingly, cytoplasmic regulatory proteins of flagella are relatively expensive (see Text S1, p. 16, in the supplemental material); however, FlgM, a secreted anti-sigma factor (39), is quite economical. Among regulatory proteins in K-12 (UniProt gene ontology [GO], 65,007), FlgM is by far the most economical, requiring 3.91 fewer ATPs per residue than an average regulatory protein.
FIG 2 .
Protein abundances and costs of flagellar proteins. Flagellum diagram showing colored economic percentiles of each protein. Proteins with lower ASCs (more economical) have higher percentiles and are dark red. ASC increases in the following order: dark red to pink, light to dark blue, gray. *, proteins with Sec secretion sequences. The number of proteins per flagellum is indicated if known (38).
In a more encompassing analysis, we reexamined the correlation between ASC and length, abundance, or function in the E. coli proteome. Although cytoplasmic and periplasmic proteins had significant negative correlations between abundance and cost, outer membrane β-barrel and integral membrane proteins did not (see Text S1, p. 21 and 22, in the supplemental material). Additionally, while protein length and cost were weakly correlated overall (rs = −0.05; P = 0.0009), there were no significant correlations in outer membrane, periplasmic, or extracellular proteins (see Text S1, p. 23 and 24, in the supplemental material). Finally, many extracellular proteins are fibrous in E. coli; therefore, we examined the ASCs of several different fibrous protein polymers. As expected, extracellular protein polymers contained fewer expensive amino acids than their intracellular counterparts (1.7 ATPs per amino acid; t test, P = 4.3 × 10−4) (see Text S1, p. 25, in the supplemental material). Collectively, these results suggest that location has a more significant role on the amino acid composition of proteins than previously appreciated.
How significant are the energy savings garnered by FliC and FimA? Previous studies have suggested that a single neutral substitution can affect cell growth and thus be subject to negative selection (17, 21). Indeed, one Gly-to-Trp substitution in FliC would increase the total cellular ATP requirement of E. coli by 0.031%. We compared the ASCs of FliC and FimA to that of an average cellular protein to calculate how much energy E. coli saves by making these proteins with less expensive amino acids (see Text S1, p. 26, in the supplemental material). The biases in FliC save the cell 4.4 × 107 ATPs per flagellum. If converted to H+, these savings correspond to the energy required to run the flagellum at 100 Hz for 24 min. In a typical E. coli cell, FliC or FimA savings (2.2 × 108 ATPs for five flagella or 300 fimbriae) represent a 1.10% reduction in the overall cellular cost. Accordingly, flagellar mutants rapidly overtake wild-type (WT) strains due to lower metabolic loads (40), constitutive flagellar mutants (flgM and fliD) grow slower due to excess FliC production (41), and flgG mutants but not motAB mutants outcompete WT bacteria on plates (42).
Alternative costs.
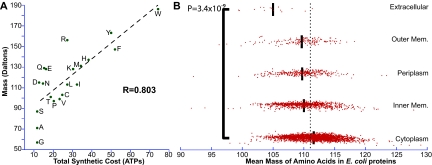
The association between amino acid costs and protein abundance has been explored using different parameters, including amino acid mass and atomic composition (17, 19, 21, 24, 29, 31, 33, 34). A composite of atomic content, mass, has been proposed as a complementary approach to calculate relative costs (31). The synthetic costs and masses of amino acids are highly correlated (R = 0.803, P = 2.02 × 10−5) (Fig. 3A; see also Text S1, p. 6, in the supplemental material). Predictably, we found extracellular proteins have smaller amino acids than those of cytoplasmic proteins (6.7 Da per amino acid for E. coli; U test, P = 3.4 × 10−9) (Fig. 3B; see also Text S1, p. 28–32, in the supplemental material). Subsequently, we looked at their carbon and nitrogen content. Diversion of carbon precursors from central metabolism is the largest component of the synthetic cost of amino acids (28, 33, 34). While the carbon content of amino acids significantly correlates with their synthetic costs, nitrogen content does not (see Fig. S5 and Text S1, p. 6, in the supplemental material). However, extracellular proteins have significantly lower contents of both carbon and nitrogen compared to those of cytoplasmic proteins (8.5%, 6.4% less per amino acid; U test, P = 3.0 × 10−9 and 5.0 × 10−8, respectively) (see Fig. S5 and Text S1, p. 28, in the supplemental material). Amino acids in extracellular proteins also have on average lower sulfur content and Gibbs free energy (43) (46.4%, 8.3% less per amino acid; U test, P = 1.5 × 10−6 and 4.1 × 10−9, respectively) (see Fig. S5 and Text S1, p. 28, in the supplemental material).
FIG 3 .
Molecular masses of amino acids correlate with their synthetic costs; extracellular proteins have simpler amino acids. (A) The synthetic costs of amino acids positively correlates with their masses. Letter codes are used to indicate individual amino acids. (B) Mass has been used as an alternative cost for amino acids (31). Using mass, extracellular proteins of E. coli are significantly more economical; their amino acids are simpler and have less mass than average. Dotted line, mean average mass of amino acids of all proteins in E. coli, black bars, mean average mass of amino acids of proteins in that location. U tests were used to compare mass-based costs.
Protein function and structure.
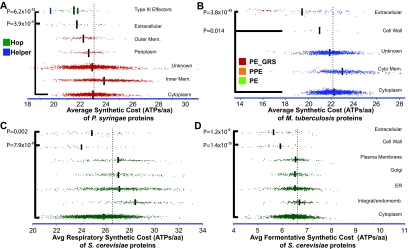
The extracellular environment represents a unique folding environment that may affect amino acid preferences. To explore this possibility, we looked at ASCs in type III secretion effectors, which many pathogenic bacteria secrete directly into eukaryotic cells (44). Although type III effectors are extracellular proteins, they function within the host in an environment that is similar to the bacterial cytoplasm. Despite their potential functional constraints, the type III effectors (HOPs) of Pseudomonas syringae had significantly lower ASCs than cytoplasmic proteins (1.48 ATPs per amino acid; U test, P = 6.22 × 10−15) (Fig. 4A; see also Data Set S1, tab D, and Text S1, p. 33 and 34, in the supplemental material). Type III effectors of other animal and plant pathogens also had significantly lower ASCs (1.27 ATPs per amino acid; U test, P = 4.4 × 10−19) (see Fig. S6, Data Set S1, tab E, and Text S1, p. 33, in the supplemental material). To further investigate if localized protein economy is independent of function, we inspected bacterial serine proteases. On average, extracellular serine proteases cost 0.72 ATPs less per residue than cellular serine proteases (U test, P = 2.7 × 10−9) (see Text S1, p. 35, in the supplemental material). In Escherichia and Bacillus species, the savings were 1.15 and 1.18 ATPs per residue (t test, P = 1.6 × 10−8 and 4.6 × 10−5, respectively) (see Text S1, p. 35, in the supplemental material).
FIG 4 .
The relationship between protein location and cost extends to diverse organisms. (A) Costs and locations of extracellular proteins and type III effectors: Hops (Hrp outer proteins; green) and Hop helpers (blue) from P. syringae. Smaller blue and green bars, mean ASC of Hops and Hop helpers, respectively. (B) Costs and locations of M. tuberculosis proteins. PE-GRS (red), PPE (orange), and PE (green) family proteins are indicated. (C, D) Costs and locations of S. cerevisiae proteins under respiratory and fermentative growth conditions. Dotted lines, each organism’s mean ASC; black bars, the mean ASC of proteins in that location. U tests were used to compare the protein ASCs of each location against those of an organism’s cytoplasmic proteins.
Oxidation and proteolysis are more likely in the harsh environment, where extracellular proteins function. Protein oxidation levels are influenced by amino acid composition, protein structure, and the particular oxidant to which proteins are exposed (H2O2, HOCl, or NO) (45–48). Commonly oxidized residues include Met, Cys, and the aromatic amino acids. With the exception of Tyr, there are fewer of these amino acids in extracellular proteins; however, the majority of the cost savings in extracellular proteins is not due to these biases (see Text S1, p. 14, 18, and 34, in the supplemental material). Besides oxidation, extracellular proteins are also exposed to extracellular peptidases. Within the gastrointestinal (GI) tract, the extracellular proteins of E. coli encounter trypsin, chymotrypsin, and elastase, which cleave after basic, aromatic, and simple amino acids (49). However, the effects that these peptidases would have on ASC are contradictory and are unlikely to be the primary cause of compositional economy in extracellular proteins.
The unique folding environment of the extracellular space may also constrain protein structure. Consequently, we examined the predicted secondary structure and intrinsic disorder of nonmembrane proteins in E. coli and P. syringae. Extracellular proteins in E. coli had significantly less helical structure and significantly more strand content than cytoplasmic proteins (see Text S1, p. 36 and 37, in the supplemental material). However, there was no correlation between the ASCs of E. coli proteins and their helix, strand, or coil content (see Text S1, p. 36, in the supplemental material). Furthermore, strands are by far the most expensive secondary structure (see Text S1, p. 37). Such analysis led us to examine which structures had the most savings relative to cytoplasmic proteins. While all three secondary structures were less expensive in extracellular proteins, coiled regions had the most economical substitutions (see Text S1, p. 37). Finally, extracellular proteins had greater amounts of disordered regions compared to those of cytoplasmic proteins; however, these differences are not significant in E. coli (t test, P = 0.324) (see Text S1, p. 40 and 41). Disordered regions are also a small percentage of the overall structure of extracellular proteins and thus do not significantly alter the relative economy of these proteins (see Text S1, p. 40 and 41, in the supplemental material). We found similar results in P. syringae, except for type III effectors which had higher contents of disordered regions and lower strand content than other extracellular proteins (see Text S1, p. 38–41, in the supplemental material). Disordered regions in type III effectors may assist their function within eukaryotes where such structural disorder is more common and is often associated with protein-protein interactions (50, 51). Collectively, these results suggest that structural differences are not responsible for the economy of many extracellular proteins and that more economical substitutions occur more frequently in less structured regions.
Ubiquity of extracellular protein economy.
A wide variety of other extracellular proteins also contain, on average, fewer expensive amino acids. For example, the elastases and exotoxins of Pseudomonas aeruginosa, the S-layer and holdfast proteins of Caulobacter crescentus, the secreted α-domain of many autotransporters, and the major capsule protein Caf1p of Yersinia pestis are among the most economical proteins in their respective organisms (see Data Set S1, tabs F and G, in the supplemental material). More distantly related organisms, such as the Gram-positive pathogen Mycobacterium tuberculosis and the budding yeast Saccharomyces cerevisiae, showed similar patterns. Both cell wall and extracellular proteins of M. tuberculosis have significantly lower ASCs, partially due to the many cell surface antigens in the PE-GRS protein family (Fig. 4B; see also Data Set S1, tab C, and Text S1, p. 11 and 32, in the supplemental material). Likewise, the cell wall and extracellular proteins of yeast have significantly lower ASCs under both respiratory and fermentative growth conditions (Fig. 4C and D; see also Data Set S1, tab B, and Text S1, p. 9, 10, 30, and 31, in the supplemental material).
To see just how broad this affect was, we initially examined the ASCs of the extracellular proteins in all 717 Gram-negative organisms in PSORTdb. To our knowledge, this is the most extensive examination of protein synthetic cost in bacteria. Overall, the amino acids in extracellular proteins cost 1.3 ATPs less than those in cytoplasmic proteins (U test, Z = 64.1, P << 1 × 10−323). However, savings for individual species such as E. coli are typically greater. This analysis of the proteomes of PSORTdb may underestimate the average cost savings of extracellular proteins by overrepresenting certain species such as E. coli, excluding known extracellular proteins (see Text S1, p. 1, in the supplemental material), overlooking the effects of GC content on amino acid composition and carbon content (52–54), and including species that are obligate anaerobes or amino acid auxotrophs for which synthetic cost are difficult to assess.
Currently, the relative cost of proteins in organisms lacking one or more amino acid synthesis pathways is difficult to assess. Abundant proteins in two different Chlamydia species have been shown to contain either more or less of the amino acids for which they are auxotrophic, perhaps due to metabolic or nutritional differences (29). When comparing the costs of extracellular proteins in Gammaproteobacteria, we found that several insect endosymbionts had relatively expensive extracellular flagellar proteins (see Text S1, p. 42 and 43, in the supplemental material). This increased cost comes from amino acid biases that only partially reflect their metabolism or nutrition (55–57). For example, Buchnera aphidicola has more His, Ile, and Lys and less Gly, Thr, and Val in its extracellular flagellar proteins, despite its capacity to synthesize these amino acids. Other factors, including transport efficiency, host metabolic interdependency, or GC skew may be more relevant (53, 58). More knowledge of how these factors affect amino acid composition is needed to properly study protein economy in auxotrophic organisms.
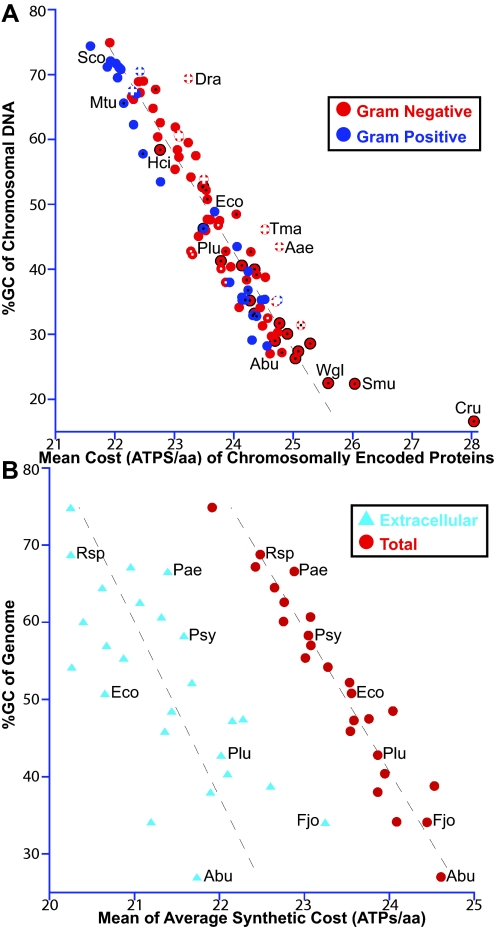
Protein composition is also affected by an organism’s GC content (54). GC-rich codons tend to code for less expensive amino acids (31) (see Text S1, p. 6, in the supplemental material). Accordingly, proteins produced by GC-rich organisms are, on average, less expensive to synthesize than proteins produced by organisms with lower GC content levels (Fig. 5A). For example, the mean cost of M. tuberculosis proteins is less than that of E. coli proteins due to high GC content affecting amino acid preferences (Fig. 5A, compare Mtu and Eco) (59). To overcome these limitations, we looked, individually, at a diverse collection of 25 Gram-negative aerobes representing a wide range of genomic GC contents. Each has retained the ability to synthesize the standard 20 amino acids (aa) (see Data Set S1, tab H, in the supplemental material). In all of these organisms, extracellular proteins had significantly lower ASCs and mean amino acid masses compared to those of other cellular proteins (Fig. 5B; see also Fig. S7 and Text S1, p. 44, in the supplemental material). Given the trends in Fig. 5B, a typical Gram-negative organism with 50% GC content would save 2.05 ATPs per amino acid in its extracellular proteins, an 8.7% reduction in synthetic cost (see Text S1, p. 27, in the supplemental material). Assuming 5 × 105 copies per cell, amino acid biases in extracellular proteins would reduce the total cellular cost by 1.54%. Theoretically, in a direct competition, strains without these savings would be outnumbered by nearly 15-fold within 250 generations.
FIG 5 .
Per residue, proteins in GC-rich organisms cost less to synthesize; however, extracellular proteins are still economical. (A) Chromosomal GC content and mean ASC of chromosomally encoded proteins in 70 Gram-negative (red) and 30 Gram-positive (blue) bacteria. White plus sign, thermophile; white center, psychrophile; black center, host-associated organism; black outline, chromosomal DNA is <1.3 MB, less than that of “Candidatus Pelagibacter ubique” which currently has the smallest genome among free-living organisms (67). Slope = −15.1, R = 0.930. (B) Comparison of the mean ASC of extracellular proteins to that of total proteins in 25 Gram-negative bacteria. Each is capable of aerobic growth and synthesis of all 20 aa (see Data Set S1, tab H, in the supplemental material). Slope = −11.0 and −17.0; R = 0.696 and 0.960, respectively. Eco, Escherichia coli; Cru, “Candidatus Carsonella ruddii”; Smu, “Candidatus Sulcia muelleri”; Wgl, Wigglesworthia glossinidia; Bap, Buchnera aphidicola; Abu, Arcobacter butzleri; Ply, Photorhabdus luminescens; Aae, Aquifex aeolicus; Tma, Thermotoga maritime/petrophila; Hci, “Candidatus Hodgkinia cicadicola”; Mtu, M. tuberculosis; Dra, Deinococcus radiodurans; Sco, Streptomyces coelicolor; Rsp, Rhodobacter sphaeroides; Pae, Pseudomonas aeruginosa; Psy, Pseudomonas syringae pv. tomato; Fjo, Flavobacterium johnsoniae.
Conclusion.
Previous studies have explored the connection between amino acid cost and a variety of attributes (21, 24, 28–34, 60, 61). However, as evidenced in the flagella system of E. coli, cellular location can have a stronger influence on the average cost of amino acids. We found that the synthetic costs of extracellular proteins are significantly reduced in E. coli, P. syringae, M. tuberculosis, S. cerevisiae, and many other organisms. Furthermore, this economic bias seems present despite the abundance, length, function, or structure of extracellular proteins. Understanding these compositional biases in extracellular proteins may improve current prediction methods. In Fig. 5B, 92.3% of extracellular proteins have an ASC below the organism’s mean ASC. Additionally, comprehending the economic selection of amino acids in extracellular proteins may elucidate new pressures upon and constraints of their evolution, particularly in horizontally acquired genomic islands where disparate codon usage and GC content gradually adapt to the host (62–64)
Microbes interact with their environment directly through external structures, leading to possible loss of surface proteins. Besides secretion, extracellular proteins are lost during fiber shedding, outer membrane blebbing, and cell wall damage. This egress of extracellular proteins is likely irreparable (1, 2); consequently, they are less likely to be recycled by the cell’s chaperone and protease systems. Such loss increases the relative cost of extracellular proteins to the cell. Accordingly, excessive production of extracellular proteins results in a decreased growth rate and competitive fitness (40–42, 65). Therefore, we propose that there is a strong selection for less expensive amino acids in extracellular proteins to counteract this loss of cellular resources.
MATERIALS AND METHODS
Calculating ASC and other cost values.
To calculate protein cost, including the ASC (ATPs/amino acid), mean amino acid mass, mean Gibbs free energy (∆G), and average number of atoms (carbon, nitrogen, and sulfur) per amino acid, we used the following equation, protein cost = (∑Ci × Fi)/L, where Ci is the appropriate cost of the amino acid i, Fi is the frequency of the ith amino acid, and L is the total protein length.
The different cost values for amino acids used were either the synthetic cost in ATPs, the amino acid mass in daltons, the ∆G for an amino acid, or the number of carbons, nitrogens, or sulfurs in a given amino acid. For ASC, different synthetic costs were used depending on the organisms. For E. coli K-12, P. syringae pv. tomato strain DC3000, M. tuberculosis H37Rv, and other bacteria, the synthetic costs for amino acids in chemoheterotrophic bacteria were used (28); for S. cerevisiae, the respiratory and fermentative synthetic costs of amino acids in yeast were used (34).
For more information about cost values, see Text S1, p. 5 and 6 and associated notes, in the supplemental material. Similar economic trends were seen using cost values other than synthetic cost, including the atomic composition, Gibbs free energy (43), and mean mass of amino acids (31) (Fig. 3 and 4; see also Fig. S5 and Text S1, p. 28–32, in the supplemental material). Statistical comparisons of cost values between locations were performed primarily using the Mann-Whitney U test; data from many data sets failed the normality test (see detailed statistics in Data Set S1, tabs A to C, in the supplemental material). P values were determined for large Z and t values using R (http://www.r-project.org).
Protein location.
The protein sequences and locations for E. coli K-12 were obtained from EchoBASE EchoLOCATION (http://www.york.ac.uk/res/thomas/index.cfm); YdbA (EB1284) lacked sequence data. The locations of FlgM (39) and FliK (66) were reassigned to extracellular proteins, and FlgJ was reassigned to periplasmic protein (38). The S. cerevisiae protein locations and sequences were downloaded from the Comprehensive Yeast Genome Database (http://mips.helmholtz-muenchen.de/genre/proj/yeast/). Protein sequence and locations for M. tuberculosis, P. syringae, and other bacteria were obtained from PSORTdb (http://db.psort.org/). For M. tuberculosis, Fmt, TrmD, Hns, HupB, and ribosomal proteins (59) were changed to cytoplasmic ones. All bacterial proteins with the GO term “secreted” (keyword 0964 in UniProt) were reassigned to extracellular ones, except the cell wall proteins of M. tuberculosis. Other changes are listed in Text S1, p. 1, in the supplemental material; modified locations in E. coli and M. tuberculosis are in boldface in Data Set S1, tabs A and C, in the supplemental material.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We thank B. Bender, M. Swanson, L. Simmons, J. Zhang, J. Fuentes, R. Frisch, and Chapman laboratory members for helpful discussions and critically reading of the manuscript.
This work was funded by NIH R01 grant AI073847 to M.R.C.
Footnotes
Citation Smith, D. R., and M. R. Chapman. 2010. Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio 1(3):e00131-10. doi:10.1128/mBio.00131-10.
REFERENCES
- 1. Reumann S., Inoue K., Keegstra K. 2005. Evolution of the general protein import pathway of plastids (review). Mol. Membr. Biol. 22:73–86 [DOI] [PubMed] [Google Scholar]
- 2. Saier M. H., Jr. 1994. Protein uptake into E. coli during Bdellovibrio infection. A process of reverse secretion? FEBS Lett. 337:14–17 [DOI] [PubMed] [Google Scholar]
- 3. Nakashima H., Nishikawa K. 1994. Discrimination of intracellular and extracellular proteins using amino acid composition and residue-pair frequencies. J. Mol. Biol. 238:54–61 [DOI] [PubMed] [Google Scholar]
- 4. Turlin E., Pascal G., Rousselle J. C., Lenormand P., Ngo S., Danchin A., Derzelle S. 2006. Proteome analysis of the phenotypic variation process in Photorhabdus luminescens. Proteomics 6:2705–2725 [DOI] [PubMed] [Google Scholar]
- 5. Cedano J., Aloy P., Perez-Pons J. A., Querol E. 1997. Relation between amino acid composition and cellular location of proteins. J. Mol. Biol. 266:594–600 [DOI] [PubMed] [Google Scholar]
- 6. Diaz-Mejia J. J., Babu M., Emili A. 2009. Computational and experimental approaches to chart the Escherichia coli cell-envelope-associated proteome and interactome. FEMS Microbiol. Rev. 33:66–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao Q. B., Wang Z. Z., Yan C., Du Y. H. 2005. Prediction of protein subcellular location using a combined feature of sequence. FEBS Lett. 579:3444–3448 [DOI] [PubMed] [Google Scholar]
- 8. Gardy J. L., Brinkman F. S. 2006. Methods for predicting bacterial protein subcellular localization. Nat. Rev. Microbiol. 4:741–751 [DOI] [PubMed] [Google Scholar]
- 9. Matsuda S., Vert J. P., Saigo H., Ueda N., Toh H., Akutsu T. 2005. A novel representation of protein sequences for prediction of subcellular location using support vector machines. Protein Sci. 14:2804–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinhardt A., Hubbard T. 1998. Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res. 26:2230–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schneider G. 1999. How many potentially secreted proteins are contained in a bacterial genome? Gene 237:113–121 [DOI] [PubMed] [Google Scholar]
- 12. Yu C. S., Lin C. J., Hwang J. K. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13:1402–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrade M. A., O’Donoghue S. I., Rost B. 1998. Adaptation of protein surfaces to subcellular location. J. Mol. Biol. 276:517–525 [DOI] [PubMed] [Google Scholar]
- 14. Nishikawa K., Kubota Y., Ooi T. 1983. Classification of proteins into groups based on amino acid composition and other characters. II. Grouping into four types. J. Biochem. 94:997–1007 [DOI] [PubMed] [Google Scholar]
- 15. King J. L., Jukes T. H. 1969. Non-Darwinian evolution. Science 164:788–798 [DOI] [PubMed] [Google Scholar]
- 16. Suckow J., Markiewicz P., Kleina L. G., Miller J., Kisters-Woike B., Muller-Hill B. 1996. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J. Mol. Biol. 261:509–523 [DOI] [PubMed] [Google Scholar]
- 17. Bragg J. G., Wagner A. 2009. Protein material costs: single atoms can make an evolutionary difference. Trends Genet. 25:5–8 [DOI] [PubMed] [Google Scholar]
- 18. Richmond R. C. 1970. Non-Darwinian evolution: a critique. Nature 225:1025–1028 [DOI] [PubMed] [Google Scholar]
- 19. Baudouin-Cornu P., Surdin-Kerjan Y., Marliere P., Thomas D. 2001. Molecular evolution of protein atomic composition. Science 293:297–300 [DOI] [PubMed] [Google Scholar]
- 20. Elser J. J., Fagan W. F., Subramanian S., Kumar S. 2006. Signatures of ecological resource availability in the animal and plant proteomes. Mol. Biol. Evol. 23:1946–1951 [DOI] [PubMed] [Google Scholar]
- 21. Li N., Lv J, Niu D. K. 2009. Low contents of carbon and nitrogen in highly abundant proteins: evidence of selection for the economy of atomic composition. J. Mol. Evol. 68:248–255 [DOI] [PubMed] [Google Scholar]
- 22. Lv J., Li N., Niu D. K. . 2008. Association between the availability of environmental resources and the atomic composition of organismal proteomes: evidence from Prochlorococcus strains living at different depths. Biochem. Biophys. Res. Commun. 375:241–246 [DOI] [PubMed] [Google Scholar]
- 23. Mazel D., Marliere P. 1989. Adaptive eradication of methionine and cysteine from cyanobacterial light-harvesting proteins. Nature 341:245–248 [DOI] [PubMed] [Google Scholar]
- 24. Bragg J. G., Wagner A. 2007. Protein carbon content evolves in response to carbon availability and may influence the fate of duplicated genes. Proc. Biol. Sci. 274:1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fauchon M., Lagniel G., Aude J. C., Lombardia L., Soularue P., Petat C., Marguerie G., Sentenac A., Werner M., Labarre J. 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9:713–723 [DOI] [PubMed] [Google Scholar]
- 26. Alves R., Savageau M. A. 2005. Evidence of selection for low cognate amino acid bias in amino acid biosynthetic enzymes. Mol. Microbiol. 56:1017–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perlstein E. O., de Bivort B. L., Kunes S., Schreiber S. L. 2007. Evolutionarily conserved optimization of amino acid biosynthesis. J. Mol. Evol. 65:186–196 [DOI] [PubMed] [Google Scholar]
- 28. Akashi H., Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99:3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heizer E. M., Jr., Raiford D. W., Raymer M. L., Doom T. E., Miller R. V., Krane D. E. 2006. Amino acid cost and codon-usage biases in 6 prokaryotic genomes: a whole-genome analysis. Mol. Biol. Evol. 23:1670–1680 [DOI] [PubMed] [Google Scholar]
- 30. Raiford D. W., Heizer E. M., Jr., Miller R. V., Akashi H., Raymer M. L., Krane D. E. 2008. Do amino acid biosynthetic costs constrain protein evolution in Saccharomyces cerevisiae? J. Mol. Evol. 67:621–630 [DOI] [PubMed] [Google Scholar]
- 31. Seligmann H. 2003. Cost-minimization of amino acid usage. J. Mol. Evol. 56:151–161 [DOI] [PubMed] [Google Scholar]
- 32. Swire J. 2007. Selection on synthesis cost affects interprotein amino acid usage in all three domains of life. J. Mol. Evol. 64:558–571 [DOI] [PubMed] [Google Scholar]
- 33. Craig C. L., Weber R. S. 1998. Selection costs of amino acid substitutions in ColE1 and ColIa gene clusters harbored by Escherichia coli. Mol. Biol. Evol. 15:774–776 [DOI] [PubMed] [Google Scholar]
- 34. Wagner A. 2005. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22:1365–1374 [DOI] [PubMed] [Google Scholar]
- 35. Barnhart M. M., Chapman M. R. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chirwa N. T., Herrington M. B. 2003. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology 149:525–535 [DOI] [PubMed] [Google Scholar]
- 37. Higgs P. G., Pudritz R. E. 2009. A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiology 9:483–490 [DOI] [PubMed] [Google Scholar]
- 38. Macnab R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 39. Hughes K. T., Gillen K. L., Semon M. J., Karlinsey J. E. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280 [DOI] [PubMed] [Google Scholar]
- 40. Neidhardt F. C., Curtiss R., III, Ingraham J. L., Lin E. C. C., Low K. B., Jr., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 41. Kutsukake K., Iino T. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 176:3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Easom C. A., Clarke D. J. 2008. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol. 8:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amend J. P., Shock E. L. 1998. Energetics of amino acid synthesis in hydrothermal ecosystems. Science 281:1659–1662 [DOI] [PubMed] [Google Scholar]
- 44. Cornelis G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825 [DOI] [PubMed] [Google Scholar]
- 45. Brandes N., Rinck A., Leichert L. I., Jakob U. 2007. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol. Microbiol. 66:901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hawkins C. L., Pattison D. I., Davies M. J. 2003. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25:259–274 [DOI] [PubMed] [Google Scholar]
- 47. Stadtman E. R., Berlett B. S. 1991. Fenton chemistry. Amino acid oxidation. J. Biol. Chem. 266:17201–17211 [PubMed] [Google Scholar]
- 48. Xu G., Takamoto K., Chance M. R. 2003. Radiolytic modification of basic amino acid residues in peptides: probes for examining protein-protein interactions. Anal. Chem. 75:6995–7007 [DOI] [PubMed] [Google Scholar]
- 49. Chung C. H., Ives H. E., Almeda S., Goldberg A. L. 1983. Purification from Escherichia coli of a periplasmic protein that is a potent inhibitor of pancreatic proteases. J. Biol. Chem. 258:11032–11038 [PubMed] [Google Scholar]
- 50. Dunker A. K., Oldfield C. J., Meng J., Romero P., Yang J. Y., Chen J. W., Vacic V., Obradovic Z., Uversky V. N. 2008. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9(Suppl. 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shimizu K., Toh H. 2009. Interaction between intrinsically disordered proteins frequently occurs in a human protein-protein interaction network. J. Mol. Biol. 392:1253–1265 [DOI] [PubMed] [Google Scholar]
- 52. Baudouin-Cornu P., Schuerer K., Marliere P., Thomas D. 2004. Intimate evolution of proteins. Proteome atomic content correlates with genome base composition. J. Biol. Chem. 279:5421–5428 [DOI] [PubMed] [Google Scholar]
- 53. Clark M. A., Moran N. A., Baumann P. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 16:1586–1598 [DOI] [PubMed] [Google Scholar]
- 54. Singer G. A., Hickey D. A. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17:1581–1588 [DOI] [PubMed] [Google Scholar]
- 55. Sandstrom J., Telang A., Moran N. A. 2000. Nutritional enhancement of host plants by aphids—a comparison of three aphid species on grasses. J. Insect Physiol. 46:33–40 [DOI] [PubMed] [Google Scholar]
- 56. Wilkinson T. L., Adams D., Minto L. B., Douglas A. E. 2001. The impact of host plant on the abundance and function of symbiotic bacteria in an aphid. J. Exp. Biol. 204:3027–3038 [DOI] [PubMed] [Google Scholar]
- 57. Zientz E., Dandekar T., Gross R. 2004. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68:745–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., III, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 60. de Bivort B. L., Perlstein E. O., Kunes S., Schreiber S. L. 2009. Amino acid metabolic origin as an evolutionary influence on protein sequence in yeast. J. Mol. Evol. 68:490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wagner A. 2007. Energy costs constrain the evolution of gene expression. J. Exp. Zool. B Mol. Dev. Evol. 308:322–324 [DOI] [PubMed] [Google Scholar]
- 62. Hacker J., Kaper J. B. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679 [DOI] [PubMed] [Google Scholar]
- 63. Langille M. G., Hsiao W. W., Brinkman F. S. 2010. Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 8:373–382 [DOI] [PubMed] [Google Scholar]
- 64. Lawrence J. G., Ochman H. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383–397 [DOI] [PubMed] [Google Scholar]
- 65. Pintar J., Starmer W. T. 2003. The costs and benefits of killer toxin production by the yeast Pichia kluyveri. Antonie Van Leeuwenhoek 83:89–97 [DOI] [PubMed] [Google Scholar]
- 66. Minamino T., Gonzalez-Pedrajo B., Yamaguchi K., Aizawa S. I., Macnab R. M. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295–304 [DOI] [PubMed] [Google Scholar]
- 67. Giovannoni S. J., Tripp H. J., Givan S., Podar M., Vergin K. L., Baptista D., Bibbs L., Eads J., Richardson T. H., Noordewier M., Rappe M. S., Short J. M., Carrington J. C., Mathur E. J. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.