Abstract
Biosynthesis of B-band lipopolysaccharide (LPS) in Pseudomonas aeruginosa follows the Wzy-dependent pathway, requiring the integral inner membrane proteins Wzx (O-antigen [O-Ag] flippase), Wzy (O-Ag polymerase), and WaaL (O-Ag ligase). For an important first step in deciphering the mechanisms of LPS assembly, we set out to map the membrane topology of these proteins. Random and targeted 3′ wzx, wzy, and waaL truncations were fused to a phoA-lacZα dual reporter capable of displaying both alkaline phosphatase and β-galactosidase activity. The results from truncation fusion expression and the corresponding differential enzyme activity ratios allowed for the assignment of specific regions of the proteins to cytoplasmic, transmembrane (TM), or periplasmic loci. Protein orientation in the inner membrane was confirmed via C-terminal fusion to green fluorescent protein. Our data revealed unique TM domain properties in these proteins, particularly for Wzx, indicating the potential for a charged pore. Novel periplasmic and cytoplasmic loop domains were also uncovered, with the latter in Wzy and WaaL revealing tracts consistent with potential Walker A/B motifs.
IMPORTANCE
The opportunistic pathogen Pseudomonas aeruginosa synthesizes its virulence factor lipopolysaccharide via the Wzy-dependent pathway, requiring translocation, polymerization, and ligation of lipid-linked polysaccharide repeat units by the integral inner membrane proteins Wzx, Wzy, and WaaL, respectively. However, structural evidence to help explain the function of these proteins is lacking. Since membrane proteins are difficult to crystallize, topological mapping is an important first step in identifying exposed and membrane-embedded domains. We mapped the topologies of Wzx, Wzy, and WaaL from P. aeruginosa PAO1 by use of truncation libraries of a randomly fused C-terminal reporter capable of different enzyme activities in the periplasm and cytoplasm. Topology maps were created based directly on residue localization data, eliminating the bias associated with reliance on multiple topology prediction algorithms for initial generation of consensus transmembrane domain localizations. Consequently, we have identified novel periplasmic, cytoplasmic, and transmembrane domain properties that would help to explain the proposed functions of Wzx, Wzy, and WaaL.
Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic bacterial pathogen that often infects compromised individuals, such as those suffering from burn wounds, cancer, AIDS, and cystic fibrosis (1). Essential to its virulence is the presence of lipopolysaccharide (LPS), which is the predominant lipid species in the outer leaflet of the outer membrane (OM) and as such is an integral component of the bacterial cell surface. The importance of LPS has been demonstrated with respect to many virulence traits, including inactivation of surface-deposited C3b complement (2), formation of a layer of protection against serum-mediated lysis (3), and aminoglycoside antibiotic binding (4). More recently, LPS in P. aeruginosa has been shown to affect the interaction of the bacterium with the cystic fibrosis transmembrane (TM) regulator (5), the secretion of type III effectors (6), and the formation of biofilm architecture (7).
P. aeruginosa LPS is composed of three distinct regions, namely, the proximal lipid A moiety (which anchors the molecule in the OM), the core oligosaccharide (which possesses important phosphate substituents), and the distal O-antigen (O-Ag) polysaccharide (8). Two glycoforms of O-Ag, known as A and B band, are synthesized by P. aeruginosa, with the latter being the immunodominant cell surface antigen. Differences in B-band O-Ag composition and structure are responsible for the classification of P. aeruginosa into 20 serotypes (9).
Previous work by our group has provided genetic evidence demonstrating that B-band LPS is synthesized via the Wzy-dependent pathway, in which several integral inner membrane (IM) proteins are believed to function in stepwise assembly of the mature glycoform in P. aeruginosa PAO1 (10, 11). According to the Wzy-dependent model, LPS biosynthesis begins in the cytoplasm, where trisaccharide repeat units are built on undecaprenyl pyrophosphate (Und-PP) in the inner leaflet of the IM (12). Individual Und-PP-linked trisaccharide units are subsequently translocated by the O-Ag flippase Wzx to the outer leaflet of the IM (10). Periplasmic polymerization of trisaccharide repeat units at the reducing terminus is then mediated by the O-Ag polymerase Wzy to preferred modal lengths governed by the chain length regulator proteins Wzz1 and Wzz2 (13). Completed O-Ag chains are ligated to lipid A-core by the O-Ag ligase WaaL to form B-band LPS (14).
The essential nature of the above-mentioned proteins with respect to B-band LPS biosynthesis has been demonstrated for P. aeruginosa, with the loss of function of Wzx (10), Wzy (11), or WaaL (14) correlating with an abrogation of B-band LPS biosynthesis. However, due to the intrinsic difficulties of working with integral IM proteins, structural characterization of these proteins and their homologues in various organisms has primarily been based on in silico analysis of the amino acid sequence. In this study, in order to gain a better understanding of the various domains of each protein with possible functional importance, we mapped the complete IM topology of Wzx, Wzy, and WaaL from P. aeruginosa PAO1 through phoA-lacZα dual-reporter fusion to random-length and targeted 3′ gene truncation libraries.
The dual-reporter approach was employed in order to yield different alkaline phosphatase (AP) and β-galactosidase (BG) enzyme activity ratios contingent on its subcellular localization (15). Detection of only high AP activity would indicate a periplasmically localized truncation, whereas detection of only high BG activity would signify cytoplasmic localization of a given truncation. However, if the terminal residue of the truncation were localized within α-helical transmembrane segments (TMS), a combination of both AP and BG activities would be observed. On the basis of this premise, various truncation clones were initially isolated via growth on dual-indicator agar plates supplemented with both AP- and BG-specific chromogenic substrates, resulting in pigmented colonies indicating potential periplasmic (blue), cytoplasmic (red), or TM (purple) truncations. To determine the length of gene truncation, constructs from pigmented colonies were sequenced, thereby allowing for the construction of a topology map for each protein. Subsequent quantification of AP and BG enzyme activities was carried out for representative residues to substantiate the subcellular localization of various domains. Together, this allowed for the construction of unbiased topology maps for Wzx, Wzy, and WaaL. To independently verify the orientation of each protein within the IM, full-length gene fusions to green fluorescent protein (GFP) were created. This investigation represents the first simultaneous structural characterization of all three integral IM proteins essential to LPS assembly and based directly on experimental evidence. Importantly, our data have revealed previously unknown TM and loop domains and pointed to their potential significance in the stepwise biogenesis of B-band LPS in P. aeruginosa PAO1.
RESULTS
Truncation library screening.
Random 3′ gene truncation libraries of wzx, wzy, and waaL from P. aeruginosa PAO1, fused to phoA-lacZα, were generated as described in Materials and Methods. Following the sequencing of clones from colored colonies, various 3′ truncation fusions were obtained for wzx, wzy, and waaL, which were used to generate topology maps via the HMMTOP 2.0 program (16). Targeted clones were constructed for regions of the proteins lacking sufficient random truncation coverage, yielding combined totals of 76, 105, and 66 unique truncations for wzx, wzy, and waaL, respectively. The subcellular localizations of the various helices and loops within these maps were further verified and validated via quantification of both AP and BG enzyme activity for representative residues by use of established methods (17, 18).
Topology of the O-Ag flippase Wzx.
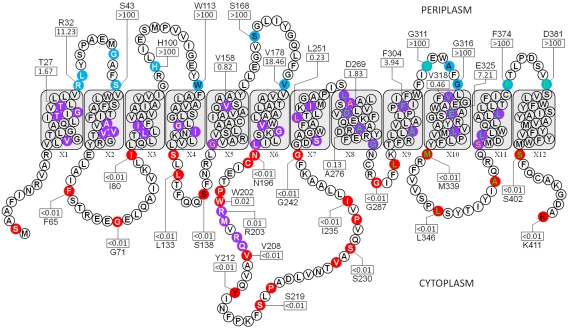
Random truncation libraries were generated for wzx, with the aim of obtaining optimal coverage of the entire protein. However, it was observed that sufficient random coverage was not possible at positions in the protein downstream of I255. As such, to ensure initial unbiased coverage of the protein, an “interval-scanning” approach was taken in which truncations were generated every 7 amino acids downstream of I255, after which targeted truncations were made to elucidate remaining regions of Wzx. The N and C termini of the O-Ag flippase were revealed to be localized in the cytoplasm, with 12 TMS in between (Fig. 1). Three relatively large periplasmic loops flanked by the first six TMS were identified. The cytoplasmic face of the protein was found to contain several large loop regions, constrained by TMS X2 and X3 (20 amino acids), X6 and X7 (47 amino acids), and X10 and X11 (18 amino acids). This is in contrast to results from in silico prediction analysis, which did not reveal the extents of the various cytoplasmic loops, in particular the third, and underestimated the number of charged amino acids within the TM region by over 55% (data not shown).
FIG 1 .
Topological map of Wzx from P. aeruginosa PAO1 based on phoA-lacZα fusion analysis (GenBank accession no. 15598349). Colored residues represent the amino acid positions of each truncation used. Residue colors denote the subcellular localization of a given truncation: blue, periplasm; purple, TM; and red, cytoplasm. Truncation letter colors denote the method of truncation generation: white, random; green, interval scanning; black, targeted (periplasm and cytoplasm); orange, targeted (TM). All TMS are labeled (X1 to X12). The AP/BG enzyme normalized activity ratios (NARs) for representative residues (see Table S1 in the supplemental material) are displayed in rectangles. Amino acid identity is displayed above/below each NAR for quantified residues.
Topology of the O-Ag polymerase Wzy.
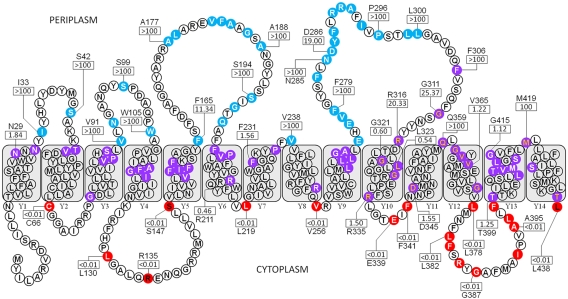
Fourteen TMS were identified in Wzy, with four intervening periplasmic loops and two intervening cytoplasmic loops of possible functional significance (Fig. 2). Of the periplasmic loops characterized, that between TMS Y9 and Y10 is the largest at 42 amino acids, followed by a comparable 36-residue loop localized between TMS Y5 and Y6. Two sizeable cytoplasmic loops were also uncovered, with the first having a length of 24 residues, flanked by TMS Y4 and Y5. The second spans 21 amino acids and is flanked by TMS Y12 and Y13. The former amino acid stretch was partially shifted into a TM helix while the latter was completely absent from the cytoplasm upon analysis of in silico prediction data (data not shown).
FIG 2 .
Topological map of Wzy from P. aeruginosa PAO1 based on analysis of 105 phoA-lacZα fusions (GenBank accession no. 15598350). Colored residues represent the amino acid positions of each truncation used. Residue colors denote the subcellular localization of a given truncation: blue, periplasm; purple, TM; red, cytoplasm. Truncation letter colors denote the method of truncation generation: white, random; black, targeted (periplasm and cytoplasm); orange, targeted (TM). All TMS are labeled (Y1 to Y14). The AP/BG enzyme NARs for representative residues (see Table S2 in the supplemental material) are displayed in rectangles. Amino acid identity is displayed above/below each NAR for quantified residues.
Topology of the O-Ag ligase WaaL.
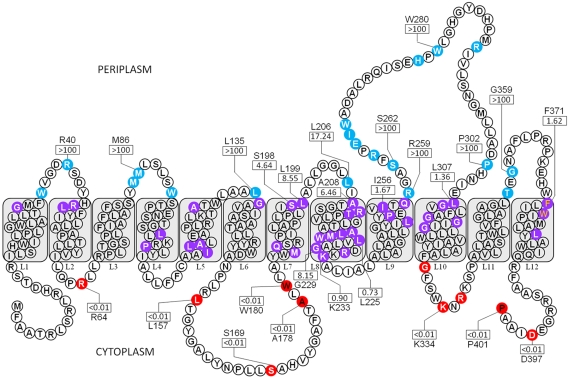
Topology mapping of WaaL from P. aeruginosa PAO1 revealed an integral IM protein with 12 TMS (Fig. 3). One large periplasmic loop domain was identified, localized between TMS L9 and L10, composed of 48 amino acids, and containing the functionally critical H303 residue (19). Adjacent to this main periplasmic loop is a second, smaller loop, containing 14 amino acids, flanked by TMS L11 and L12. The length of TMS L8 was extended, following the software program output (see Materials and Methods), to 26 residues to account for experimental data obtained based on both color scoring and enzyme activity data for the random fusion clone K233. As such, this TMS is only 1 residue longer than the default upper limit of 25 residues per TMS constraining the HMMTOP 2.0 algorithm (16). PhoA-LacZα fusion screening of WaaL truncations also revealed that the cytoplasmic face of the protein contains a sizeable loop of 30 amino acids between TMS L6 and L7. This tract of amino acids was predicted to begin in the periplasm and end within a TM helix when purely de novo prediction algorithm analysis was performed in the absence of experimental input (data not shown).
FIG 3 .
Topological map of WaaL from P. aeruginosa PAO1 based on analysis of 66 phoA-lacZα fusions (GenBank accession no. 15600192). Colored residues represent the amino acid positions of each truncation used. Residue colors denote the subcellular localization of a given truncation: blue, periplasm; purple, TM; red, cytoplasm. Truncation letter colors denote the method of truncation generation: white, random; black, targeted (cytoplasm); orange, targeted (TM). All TMS are labeled (L1 to L12). The AP/BG enzyme NARs for representative residues (see Table S3 in the supplemental material) are displayed in rectangles. Amino acid identity is displayed above/below each NAR for quantified residues.
Protein orientation analysis.
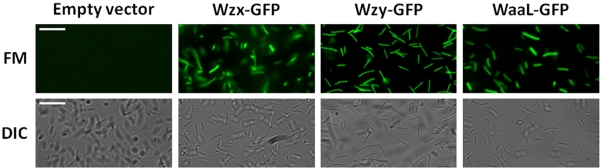
The use of C-terminal GFP tagging has been established as an additional experimental approach for determining the subcellular localization of a given domain in integral IM proteins (20), as GFP will not fluoresce in the periplasm (21). To confirm the IM orientation of Wzx, Wzy, and WaaL, translational fusions to GFP were created and expressed in P. aeruginosa PAO1. All constructs displayed pronounced fluorescence across the entire cell population, indicating cytoplasmic localization of the C terminus in Wzx, Wzy, and WaaL (Fig. 4). These results served to independently verify the orientation determined through PhoA-LacZα topology mapping (Fig. 1, 2, and 3). When expressed in its respective chromosomal knockout in P. aeruginosa PAO1, each GFP fusion was able to restore the synthesis of B-band LPS (see Fig. S1 in the supplemental material). Synthesis of A-band LPS, deficient in the absence of a functional ligase, was also restored upon expression of WaaL-GFP in the waaL mutant background (data not shown).
FIG 4 .
Fluorescence micrographs of P. aeruginosa PAO1 expressing C-terminal GFP fusions of Wzx, Wzy, and WaaL from respective pHERD26T clones. Images were captured at ×400 as described in Materials and Methods. FM, fluorescence micrograph; DIC, differential interference contrast. White bar = 15 µm.
DISCUSSION
Membrane proteins represent over a quarter of all known proteins and play integral roles in a vast array of cellular process, including import/export of substrates, energy production, signal transduction events, and assembly of cellular components. However, due to the inherent difficulties associated with expressing, purifying, and crystallizing membrane proteins, of the more than 61,000 entries in the Protein Data Bank to date, just over 250 represent unique membrane protein structures (White Laboratory [http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html]). In the absence of crystallographic data, topological mapping of membrane proteins can provide valuable information on the sizes and subcellular localizations of various domains potentially contributing to their overall function. This rationale has led us to examine the membrane topologies of Wzx, Wzy, and WaaL from P. aeruginosa PAO1.
A leading approach for examining membrane protein topology involves the fusion of C-terminal reporter tags at various lengths of the amino acid sequence, with independent PhoA (periplasmically active) and LacZ (cytoplasmically active) fusions often created; fusion of LacZ to periplasmic residues may yield low BG activity (22) or toxicity to cells (23). Due to the requirement of separate fusions, the expression levels of reciprocal PhoA and LacZ fusions must be normalized for comparison of enzyme values at the same truncation residue. The activity of either AP (PhoA) or BG (LacZ) can be monitored by the breakdown of enzyme-specific chromogenic substrates, thus allowing for rapid analysis of fusion activity (17). To date, all published data on the full membrane topologies of Wzx (24), Wzy (25), and WaaL (26) proteins from various Gram-negative species, as well as their homologues (27, 28), have employed the use of separate PhoA and LacZ truncation fusions. Furthermore, these studies were invariably aimed at validating the results of consensus TMS localizations based on multiple in silico topology predictions. The generation of random fusions was carried out in only three of these studies, which still relied primarily on purpose-built fusions (24–26). In the remaining studies, targeted fusions were created based exclusively on in silico analyses (27, 28). To overcome the limitations of separate PhoA and LacZ fusions, we employed a chimeric PhoA-LacZα dual-reporter system capable of displaying both AP and BG activity (15) to map the topologies of Wzx, Wzy, and WaaL from P. aeruginosa PAO1. Functional LacZ was reconstituted only by the host-encoded ω fragment of LacZ in the cytoplasm, with no periplasmic toxicity associated with the LacZα moiety. This reporter system was originally developed by Alexeyev and Winkler (15) to map the membrane topology of the ATP/ADP translocase Tlc from Rickettsia prowazekii.
In this investigation, we took the approach opposite to that used in all previously published topology characterizations of homologous O-Ag assembly proteins, i.e., we generated unbiased random and interval-scanning libraries of wzx, wzy, and waaL fused to the phoA-lacZα dual-reporter construct. These were then screened based on both pigmentation phenotype and enzyme activity quantitation. On the basis of the results obtained from these initial fusion libraries, preliminary topology maps were derived using segment localization data via the HMMTOP 2.0 server, which generates outputs based directly on experimental data (16). To remedy ambiguous regions where coverage based on either the random truncation or the “interval-scanning” approach was lacking, we constructed targeted truncations to help clarify their localizations, allowing for the generation of finalized topology maps for Wzx, Wzy, and WaaL.
Targeted fusions generally yielded higher absolute enzyme activity values than their random truncation library-generated counterparts (see Tables S1 to S3 in the supplemental material). The full-length wzx, wzy, and waaL constructs fused to phoA-lacZα, which were used to create their respective random truncations, were cloned into the same pPLE01 source vector as that used for the individual targeted truncations for each gene. However, creation of the targeted truncations introduced 2 amino acids between the terminal residue of the truncation and the reporter motif. These residues were unavoidable, as they were translated from the 3′ PstI restriction endonuclease site used for cloning of the targeted fusions immediately upstream of the reporter. While the same 3′ restriction site was used to linearize the full-length gene fusions in advance of exonuclease III treatment for random library generation, the ensuing enzyme treatment regimen resulted in removal of the PstI site, thus eliminating the two residues described above. As such, compared to the truncations that were actively made, the reporter motif in the randomly generated truncations may have been more sterically constrained; this would have resulted in lower absolute enzyme values upon quantitation due to lowered substrate accessibility by either reporter moiety. Regardless of the method of generation, when the various activities across the three different proteins were normalized as percentages of the maximum activity of a particular set, general trends could be inferred from the AP/BG normalized activity ratio (NAR) for a specific residue. NARs of <0.1, between 0.1 and 10, and >10 were found to coincide with cytoplasmic, TM, and periplasmic residues, respectively, allowing for localization data comparison between Wzx, Wzy, and WaaL.
Wzx.
Wzx and its homologues are members of the polysaccharide transporter (PST) family of proteins, present in both Gram-negative and Gram-positive organisms as well as in Archaea (29). PST family members have 10 to 14 predicted TMS (29), consistent with our experimentally derived model of 12 TMS. The first topological characterization of a Wzx homologue, PssL from Rhizobium leguminosarum bv. trifolii strain TA1, also yielded a model consisting of 12 TMS (28), as did a recent study of Wzx from Salmonella enterica serovar Typhimurium group B (WzxSe) (24). However, these models were based on purpose-built PhoA and LacZ fusions made to support in silico predictions and, as described below, overlooked certain key features.
The cytoplasmic loop residue compositions for Wzx from P. aeruginosa (WzxPa) are such that no motifs resembling conventional ATPase consensus sequences are present. Furthermore, the extent of loop lengths on this face of the protein would suggest an elaborate tertiary-structure architecture in the cytoplasm, which is reinforced by secondary-structure-propensity analysis of the various loop domains (see Fig. S2 in the supplemental material). This would be consistent with the predicted function of Wzx in mediating translocation of lipid-linked oligosaccharides, a process which would likely be initiated in the cytoplasm. Due to its size, cytoplasmic loop 3 (CL3X) of WzxPa in particular may play an important role during the translocation step, perhaps functioning as part of a gating mechanism governing entry on the cytoplasmic face of the protein. Four of the random truncations near the membrane interface were color scored as purple, but quantitation of their enzyme activities as well as those of flanking residues revealed AP/BG NARs reflective of cytoplasmic localization consistent with all three proteins studied. Background breakdown of the AP-specific substrate was likely due to kinking of the reporter moiety towards the membrane through the secondary structure, as the aforementioned purple clones are immediately downstream of a Pro residue. Conversely, the periplasmic face of the protein contains only 3 loops of possible functional importance, the largest of which is only 16 amino acids in length, alluding to a less intricate tertiary structure for this face of the protein (see Fig. S2 in the supplemental material).
Contrary to the mainly in silico-based topology models of Wzx proteins put forth by others, we observed that 9 of the 12 TMS present in WzxPa contain at least one charged residue, with some possessing as many as seven charged amino acids (see Fig. S5 in the supplemental material). Interestingly, the only other investigation involving the topology of a Wzx O-Ag flippase (WzxSe) proposed a membrane topology model in which only 5 charged amino acids were predicted among the total of 12 proposed TMS (24). Rather, the locations of the various TMS appear to have been constrained by the localization of the various Arg, Asp, Glu, His, and Lys residues. As with the above-mentioned characterization of PssL from R. leguminosarum bv. trifolii, the proposed topology of WzxSe was based on the initial generation of an in silico-derived map, with only 12 reciprocal PhoA and LacZ fusions created to verify the predictions.
The distribution of so many charged residues within TMS of WzxPa provides a possible explanation to further elucidate the mechanism of its function. The negatively charged sugar units of O-Ag repeats in P. aeruginosa, for instance, that of strain PAO1 (serotype O5) containing two mannuronic acid residues and one fucosamine residue (9), would need to be translocated through the hydrophobic environment of the IM bilayer before polymerization by Wzy. This process would be more energetically favorable if the TMS of Wzx were arranged in a pore-like structure with charged residues lining the interior, particularly since the majority of the charged amino acids within the TMS are cationic. With the use of the RHYTHM server (http://proteinformatics.charite.de/rhythm/) to identify potential TMS-TMS and TMS-membrane contact points in membrane proteins, our analysis of TMS X1 to X12 revealed numerous candidates for each interaction environment (see Fig. S5 in the supplemental material). By extension, it is possible to generate a preliminary model of the potential WzxPa channel within the IM and identify the charged residues that may line the interior. While the observations described above are consistent with the predicted role of WzxPa, further investigation is required to fully understand the actual translocation mechanism, whether it be active or passive.
The substrate specificity of Wzx has been proposed to be dependent on the identity of the first sugar of the O-Ag subunit, as evidenced by cross-complementation experiments in which nonnative Wzx proteins were able to restore wild-type LPS synthesis if the organism from which they were derived contained the same sugar as the initial unit of its O-Ag repeat (30). However, the basis for substrate specificity remains unknown. Prior work in an Escherichia coli K-12 system has shown that a full-length O-Ag repeat unit is not required for translocation by Wzx (31). These data suggest that recognition of the initiating sugar is important for flippase function. As described above, the charge property of the potential Wzx channel, as well as the overall amino acid composition of the various loops, may confer substrate specificity. Interestingly, periplasmic loop 2 in WzxPa (PL2X) contains an RX10G tract of amino acids (see Table S4 in the supplemental material). This amino acid motif is discussed in detail below for Wzy and WaaL, with respect to its potential role in interaction with the initiating sugar of the O-Ag repeat unit.
Wzy.
The data from this study indicate that WzyPa (438 amino acids) contains 14 TMS, with two large periplasmic loops flanked either by TMS Y5 and Y6 or by TMS Y9 and Y10 (Fig. 2). Truncations in the tail end of the latter loop were found to yield purple colony coloration (F306, G311, and R316); however, determination of the AP/BG NAR indicated that these residues each displayed a NAR consistent with all periplasmic residues studied and as such were assigned to the periplasm. Initially, this would appear at odds with the proposed 12-TMS model for Wzy from Shigella flexneri (WzySf) (382 amino acids). However, ClustalW alignment results for WzyPa and WzySf indicate that specific subcellular regions of these proteins correspond with each other until the termination point of periplasmic loop 5 (PL5). Although the amino acid sequence of WzyPa is longer than that of WzySf, the positions of analogous TM and loop portions of the proteins were found to align well when compared to one another, with two additional TM domains in WzyPa accounting for the extra amino acids (data not shown). Differences in the numbers of TMS and the sizes of various periplasmic loops between the two homologues may reflect differences in substrate specificities (32), as O-Ag units in all serotypes of S. flexneri (except type 6) are tetrasaccharide repeats beginning with N-acetyl-d-glucosamine (33), whereas those in P. aeruginosa PAO1 are trisaccharide repeats beginning with N-acetyl-d-fucosamine (9).
To date, Wzy protein structure remains poorly characterized, and as such, information relating to catalytic mechanisms of O-Ag polymerization is lacking. It has been proposed that Wzy and WaaL may function in similar ways, as each protein requires O-Ag polysaccharides bound to Und-PP and is subsequently able to transfer O-Ag to a sugar acceptor (26). For Wzy, the sugar acceptor would be another O-Ag repeat unit bound to Und-PP. Conceptually, Wzy would have to interact with the same molecular structure in two different ways, similar to the model proposed by Bastin et al. (34). The first method of interaction would require prolonged docking and retention of the extending O-Ag chain, either still bound to Und-PP or independently associated with the protein. The former scenario would be more plausible, as remaining bound to the lipid carrier would undoubtedly facilitate the retention of the growing chain in the IM before the completion of chain polymerization. The second mode of interaction would entail association with the incoming O-Ag repeat still bound to Und-PP. The latter interaction would need to be more short-term to allow for either (i) the recycling of the new Und-PP and the transfer of the newly arrived O-Ag subunit to the reducing terminus of the growing chain bound to the initial Und-PP or (ii) the shuttling of the incoming Und-PP-linked repeat to the prolonged docking site to extend the O-Ag chain and displace the previous Und-PP carrier. In both of the above-mentioned general interaction scenarios, it is conceivable that similar recognition motifs would be required. Upon comparison against each other via ClustalW alignment, PL3Y and PL5Y are of comparable size and contain many conserved or structurally equivalent residues (see Fig. S6 in the supplemental material). Polymerase proteins such as Wzy have also been found to contain a conserved HX10G so-called “polymerase motif,” though an exact motif such as this is not present in WzyPa. This motif was also identified in WaaL proteins (26). In WaaL from P. aeruginosa PAO1 (WaaLPa), the periplasmic His residue of the HX10G motif (H303) was found to be critically important for function (19). On the basis of complementation experiments, His could be replaced by Arg, creating an RX10G tract and demonstrating the functional equivalency of His and Arg residues in this setting. Our investigation of WzyPa topology has revealed the presence of RX10G motifs in both PL3Y and PL5Y (see Table S4 in the supplemental material). PSIPRED analysis (35) of the PL3Y and PL5Y secondary structures indicates analogous positioning of both RX10G tracts on their respective loops, with each beginning in the longer α-helical domain and running to the flexible linker separating the two α helices of each loop (see Fig. S3 in the supplemental material). Taken together, this conserved tract of amino acids with similar secondary-structure patterning between PL3Y and PL5Y may represent the same motif present in two different tertiary-structure contexts within the same protein.
In addition to the two main periplasmic loops, we have also identified a large, 24-residue cytoplasmic loop (CL2Y) flanked by TMS Y4 and Y5 (Fig. 2). This loop contains the sequence RENQGRRMLVLLS (see Table S4 in the supplemental material), which is remarkably similar to the consensus Walker B ATPase sequence of R/KXXXGXXXLhhhhD (X, any amino acid; h, hydrophobic residue; underlined, consensus match), which forms a β-sheet structure (36). Incidentally, PSIPRED secondary-structure analysis of CL2Y has revealed a high propensity for β-strand formation in this loop domain (see Fig. S3 in the supplemental material). This cytoplasmic domain is connected directly to PL3Y through TMS Y5, a helix with charge properties. The other sizeable cytoplasmic loop in WzyPa (CL6Y) also has a high propensity for the secondary structure, with α helices sandwiched by β-stranded regions (see Fig. S3 in the supplemental material). Functional characterization of both loops is currently under way.
WaaL.
Consistent with WaaL homologues from different bacterial species (26), in silico secondary-structure analysis predicted the presence of a large characteristic periplasmic loop in WaaLPa, in which several amino acids appeared to be conserved; these were subjected to alanine-scanning mutagenesis. Among these mutants, only the H303A mutation was unable to restore the production of B-band LPS in a waaL knockout background. Upon screening of a panel of H303G, H303N, and H303R mutants, only the H303R mutation was able to preserve the function of WaaLPa, likely due to the ability of Arg to carry a positive charge, similar to what was found for His (19). The H303 residue is the leading residue in the HX10G motif described above for Wzy, which could also function in the form of an RX10G motif. Through WaaL topology mapping, we have confirmed the presence of a large, 48-residue periplasmic loop between TMS L9 and L10 (PL5L) containing the catalytically essential H303 residue. Subsequent to the investigation by our group, an analogous catalytically essential His residue (H337) was identified in WaaL from E. coli (WaaLEc) (37). This demonstrates the importance of the conserved positively charged residue in the ligation of O-Ag to the lipid A-core, though the length of the periplasmic loop in WaaLEc on which this residue is located has not been conclusively determined (37). Using computer modeling of amino acids constituting the proposed loop domain as well as 22% of the downstream TMS, the same authors proposed that the conserved acidic residue would form part of a putative “catalytic center” created by the tertiary structure of the extended principal periplasmic loop (as described above), resulting in two pairs of perpendicular α helices. In contrast, PL5L from WaaLPa possesses a high level of propensity for the formation of a single α helix, as well as a partial β-strand character, with a high degree of flexibility (see Fig. S4 in the supplemental material). The validation of either model awaits the detailed biophysical determination of its three-dimensional (3-D) structure.
Abeyrathne and Lam also examined both truncated and chimeric versions of WaaLPa (19). C-terminally truncated versions of WaaLPa with truncations comprising amino acids 1 to 101, 1 to 156, 1 to 232, 1 to 301, and 1 to 353 were generated, but none were able to complement a waaL mutant. On the basis of the topology characterized in our investigation, the version with a truncation comprising amino acids 1 to 353 would have contained the required catalytic H303 residue and intact PL5L, yet this version was nonfunctional. This suggests an important functional role for the region of WaaLPa comprising amino acids 354 to 401, which likely contributes to stabilization of the protein and/or participation in the catalytic mechanism. Chimeric fusions to WaaLEc were also constructed, and the only one able to restore a wild-type level of B-band O-antigen production contained WaaLPa residues 1 to 360 and a C-terminal fragment of WaaLEc residues 389 to 419. As determined from the topology map, the former stretch of amino acids would have encompassed a portion of PL6L that contained a Glu and an Asn residue, with this portion in its entirety containing several charged residues. The latter tract of amino acids from WaaLEc is predicted by an in silico analysis to form an 18-residue TM helix, which would maintain an Arg residue in an analogous loop position and contribute an Asp residue in place of the Lys, Glu, and His residues present in the native WaaLPa PL6L sequence. Since this final periplasmic loop of WaaLPa is charged, sizeable, flexible, and predicted to possess secondary structure (see Fig. S4 in the supplemental material), this loop may be analogous to EL4, a predicted periplasmic loop adjacent to the principal loop from WaaLEc found to contain a charged residue (R216) important for O-Ag ligation (37).
WaaL from Vibrio cholerae O1 (WaaLVc) has a sequence length (398 amino acids) comparable to those of WaaLPa (401 amino acids) and WaaLEc (419 amino acids), with the largest periplasmic loop located from S234 to L327 in a primary-structure position similar to those for both WaaLPa (R259 to E306) and WaaLEc (N259 to E342). However, WaaLVc was proposed to contain only 10 TMS, each at a maximum length of only 8 amino acids, with all intervening loops being relatively large (26). In comparison, the average thickness of the IM in E. coli K-12 and P. aeruginosa PAO1 has been measured at 59 Å (38). Moreover, a higher level of membrane insertion instability was observed for artificial TM helices, as the number of residues decreased incrementally from 19 to 15 within a model lipid bilayer (39). Combined with inconsistent PhoA fusion quantitation values (e.g., periplasmic D79), these data cast doubt on the number and boundaries of TMS described for WaaLVc and suggest that further model refinement is required.
Investigation of the mechanism for O-Ag ligation to lipid A-core also revealed that WaaLPa was able to hydrolyze ATP in vitro at a rate in accordance with Michaelis-Menten kinetics (19). As ATP is not believed to exist in the periplasm (40), this would entail a cytoplasmic domain capable of ATP hydrolysis. However, in silico topology analysis for WaaLPa did not reveal the presence of any cytoplasmic loops having either sufficient size or recognizable ATPase motif sequence conservation (19). The recent in silico topology prediction for WaaLEc yielded analogous results, revealing the lack of a sizeable cytoplasmic motif that would be capable of hydrolyzing ATP. Interestingly, in the present study of WaaLPa, we have discovered the presence of a large, 30-residue cytoplasmic loop flanked by TMS L6 and L7 (Fig. 3) and thus revealed a candidate cytoplasmic motif for the observed ATPase activity of the protein in vitro. As purification of WaaLPa yielded a dimer (19), interaction of the two cytoplasmic domains from monomers of the protein may be required to form a contiguous quaternary structure capable of hydrolyzing ATP. In silico analysis predicted that this cytoplasmic stretch of residues in WaaLPa began in the periplasm and ended within a TM helix, thus demonstrating the limitations of relying solely on in silico topology predictions and reinforcing the benefits of obtaining experimentally based entire-protein topology maps for downstream functional characterization of membrane proteins.
The above-mentioned loop does not possess a high degree of amino acid sequence similarity to conventional ATPase motifs, although a TGYG tract is present at amino acids 158 to 161 (see Table S4 in the supplemental material), corresponding to a conserved portion of the consensus GXS/TGXGKS/TS/T Walker A/phosphate-binding loop (P-loop) sequence (41). Deviant Walker A motifs in which the downstream Lys residue, carrying a positive charge, can be located 2 residues upstream of the TGXG motif have also been found (42). The residue located 2 positions upstream of the TGYG motif is a positively charged Arg residue, possibly analogous in position and function to Lys in deviant Walker A domains. Walker A domains generally contain an uncapped α helix preceded by a loop rich in Gly residues (41). Secondary-structure propensity indicates that CL3L forms a flexible coiled-coil structure in the region of TGYG, followed immediately by a helix motif (see Fig. S4 in the supplemental material), consistent with general secondary-structure characteristics of Walker A motifs. CL3L in WaaLPa is currently being characterized in our laboratory.
Conclusion.
Among all three proteins studied, numerous motifs known to promote oligomerization of TM helices have been identified. The most well known is the GX3G motif, in which two Gly residues, spaced 4 residues apart, promote packing of TMS. Ala and Ser residues have been shown to substitute for Gly in the above-mentioned motif, now referred to as the GASRight motif for right-handed α helices (43). This motif possesses high propensity for the formation of a flat surface capable of docking against grooves created by bulkier amino acids also spaced 4 residues apart in a nearby helix (43). From our investigation, numerous GASRight motifs can be found in Wzx, Wzy, and WaaL (see Table S4 in the supplemental material). Since antiparallel pairs of α helices tend to form between sequential TMS (43), the GASRight motif content of Wzx, Wzy, and WaaL TMS may provide a clue as to the nature of the local packing and domain organization for the various proteins. As such, tracts of amino acids separated by large distances of primary structure may interact through packing and tertiary-structure events to form a functional domain.
While the Wzy-dependent model for LPS biosynthesis generally fits with observed phenotypes for mutants at various stages of the process, the exact functional mechanisms of each stage have yet to be elucidated. In this investigation, we have discovered the presence of novel domains in both TM and soluble subcellular localizations for three essential proteins in this pathway. While Wzx, Wzy, and WaaL catalyze different events during the assembly process, these proteins are conceptually required to interact with an equivalent molecular structure, in the form of O-Ag, albeit in differing capacities. Remarkably, we have uncovered the presence of a common periplasmic motif in all of the proteins that are required for the Wzy-dependent assembly pathway. The first manifestation of the RX10G motif in the Wzy-dependent assembly pathway can be found in PL2X of WzxPa. It is also found in both PL3Y and PL5Y of WzyPa. An RX10G motif is also located on PL5L, the functionally essential loop of WaaLPa (see Table S4 in the supplemental material). In each of these cases, the initial Arg residue is followed immediately by another charged (for Wzx and Wzy) or highly polar (for WaaL) amino acid. Intriguingly, the equivalent motif is present twice in Wzz1 and seven times in Wzz2 from P. aeruginosa PAO1, both proteins that would function to regulate the modal chain length of O-Ag polymerization carried out by WzyPa. While the oligomeric state of full-length Wzz proteins has been determined (44) and X-ray crystallographic data exist for the soluble periplasmic domain (45), the mechanism by which Wzz proteins interact with O-Ag and regulate the chain length is presently unknown. The concept of a common interaction motif for a specific O-Ag subunit would serve as a unifying thread between Wzx, Wzy, Wzz, and WaaL in a given species and merits further investigation.
In conclusion, results from this study have revealed the membrane topology of Wzx, Wzy, and WaaL from P. aeruginosa PAO1 at a resolution unmatched by previous attempts for homologous proteins. Rather than relying on in silico topology predictions to frame ensuing fusion creation, we first generated localization data based on experimental evidence and then used these data to create a final topology map for Wzx, Wzy, and WaaL. This has allowed us to eliminate the inherent bias of predetermined TMS localization and, in doing so, revealed the positions of novel TMS as well as the locations and extents of previously unidentified periplasmic and cytoplasmic loop domains in line with the proposed functions for each protein. This investigation will serve as a springboard for detailed functional characterization of these proteins, which will undoubtedly further our understanding of such a widely conserved yet poorly understood biosynthesis pathway for virulence-associated polysaccharides in bacteria.
MATERIALS AND METHODS
DNA manipulations.
Plasmid DNA was isolated with a GenElute miniprep kit (Sigma). Products of PCR amplifications and restriction digestions were cleaned with a QIAquick PCR cleanup kit (Qiagen). The resultant output from sequenced clones was analyzed via the open reading frame (ORF) finder program created by the National Center for Biotechnology Information (Bethesda, MD). The oligonucleotide primer sequences used in this study are available upon request.
Construction of PhoA-LacZα and GFP translational fusions.
The strains and plasmids used in this study are listed in Table 1. The phoA-lacZα reporter sequence from pMA632 (15) was cloned between the EcoRV and HindIII sites of pBluescript II SK(+) under the control of the lac promoter to create pPLE01. Full-length genes were PCR amplified from P. aeruginosa PAO1 genomic DNA and cloned upstream of the reporter construct in pPLE01 by use of either the XbaI and BamHI sites (for wzx) or the SacI and XbaI sites (for wzy and waaL). Random exonuclease III-generated truncation fusion libraries were created as previously described by Alexeyev and Winkler (15). E. coli DH10B was used as an α-complementing host strain. Fusion junction sequencing for random truncations was carried out at the Laboratory Services division, University of Guelph. Targeted truncation fusions for a respective gene were created in the same manner as for the full-length constructs used for library generation, by cloning of PCR products of various 3′ truncation positions upstream of phoA-lacZα. To construct the GFP fusions, PCR products (amplified from P. aeruginosa PAO1 genomic DNA) were cloned upstream of the gfp-his8 reporter in the pWaldo-GFPd vector (46) for wzx, wzy, and waaL to yield pWaldo-wzx-GFP, pWaldo-wzy-GFP, and pWaldo-waaL-GFP. For fluorescence analysis of P. aeruginosa strains, the pWaldo clones were used as templates for PCR amplification of the GFP fusion constructs prior to pHERD26T cloning (47). All PCR amplifications were performed using KOD Hot Start DNA polymerase (Novagen). All digestions and ligations were performed with enzymes from Invitrogen or NEB.
TABLE 1 .
Bacterial strains and plasmids
| Strain or plasmid | Genotype, phenotype, or relevant characteristic(s)c | Reference or source |
|---|---|---|
| Strain | ||
| P. aeruginosa | ||
| PAO1 | Serotype O5; A+ B+ | Laboratory stock |
| PAO1waaL | waaL::Gmr derived from strain PAO1; A− B− | 14 |
| PAO1wzx | wzx::Gmr derived from strain PAO1; A+ B− | 10 |
| PAO1wzy | wzy::Gmr derived from strain PAO1; A+ B− | 11 |
| E. coli DH10B | F− araD139 Δ(ara leu)7697 ΔlacX74 galU galK rpsL deoR ϕ80dlacZΔM15 endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | Laboratory stock |
| Plasmid | ||
| pBluescript II SK(+) | Escherichia expression vector (Plac); Apr | Fermentas |
| pHERD26T | Pseudomonas expression vector (PBAD araC); Tetr | 47 |
| pMA632 | Plasmid encoding phoA-lacZα dual-reporter construct; Apr | 15 |
| pPLE01a | pBluescript II SK+ with phoA-lacZα from pMA632; Apr | This study |
| pWaldo-GFPd wzx | Plasmid carrying gfp-his8; Kmr | 46 |
| pHERD26T-wzx-GFP | pHERD26T carrying wzx-gfp-his8; Tetr | This study |
| pPLE01-wzxb | Full-length wzx (encoding aa 1-411) fused to phoA-lacZα; Apr | This study |
| pWaldo-wzx-GFP wzy | pWaldo carrying wzx-gfp-his8; Kmr | This study |
| pHERD26T-wzy-GFP | pHERD26T carrying wzy-gfp-his8; Tetr | This study |
| pPLE01-wzyb | Full-length wzy (encoding aa 1-438) fused to phoA-lacZα; Apr | This study |
| pWaldo-wzy-GFP waaL | pWaldo carrying wzy-gfp-his8; Kmr | This study |
| pHERD26T-waaL-GFP | pHERD26T carrying waaL-gfp-his8; TetR | This study |
| pPLE01-waaLb | Full-length waaL (encoding aa 1-401) fused to phoA-lacZα; Apr | This study |
| pWaldo-waaL-GFP | pWaldo carrying waaL-gfp-his8; Kmr | This study |
All targeted truncations for wzx, wzy, and waaL were cloned upstream of phoA-lacZα in pPLE01 and are represented in Fig. 1, 2, and 3.
All random truncation libraries for wzx, wzy, and waaL were generated using the respective full-length fusion vectors and are represented in Fig. 1, 2, and 3.
Superscript “+” or “−” after A or B denotes the presence or absence of the particular O polysaccharide, respectively. Resistance (r) is shown for gentamicin (Gm), ampicillin (Ap), tetracycline (Tet), and kanamycin (Km). aa, amino acids.
Color scoring and enzyme quantitation.
Ligation recovery cultures for truncation libraries were plated on defined-medium agar plates supplemented with ampicillin (100 µg/ml; Sigma), isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM; Roche), 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (80 µg/ml; Sigma), and 6-chloro-3-indolyl-β-d-galactoside (Red-Gal) (100 µg/ml; Research Organics) as previously described (15). Subcultures from overnight inoculations were grown to mid-exponential phase in Miller’s LB broth (Invitrogen) supplemented with ampicillin and IPTG. Quantitation of AP and BG activity was carried out as previously described, including compensation for spectrophotometric absorbance caused by cell debris (17), with cells for Wzy BG assays treated with 50 µL of B-PER II permeabilization reagent (Thermo), instead of chloroform-SDS, to increase permeabilization in order to compensate for low protein expression levels. All NAR values presented are a result of quadruplicate independent enzyme assays. Residue localization data based on color scoring and enzyme activity analysis were entered into the HMMTOP version 2.0 prediction algorithm in order to determine the positions and lengths of TMS on the basis of experimental results (16).
Fluorescence microscopy.
P. aeruginosa PAO1 overnight cultures expressing GFP fusions were subcultured and grown to exponential phase in LB broth at 37°C containing 90 µg/ml tetracycline (Sigma) with 0.1% l-arabinose. Culture aliquots were sedimented, resuspended in 1× phosphate-buffered saline (PBS), and spotted on a glass slide. No fixation to the slide was carried out, as GFP fluorescence may become labile following various fixative treatments. Cells were imaged at ×400 magnification using a Zeiss Axiovert 200 M fluorescence microscope, with excitation at 470 nm and emission collection at 525 nm, using the Improvision Openlab 5 software package (PerkinElmer).
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We are thankful for the kind gifts of plasmid pMA632 from Herbert H. Winkler (University of South Alabama), pWaldo-GFPd from Jan-Willem de Gier (Stockholm University), and pHERD26T from Hongwei D. Yu (Marshall University).
This work was supported by operating grants from the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research (CIHR) (no. MOP-14687). S.T.I. is the recipient of a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship doctoral award. J.S.L. is the holder of a Tier 1 Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
Footnotes
Citation Islam, S. T., V. L. Taylor, M. Qi, and J. S. Lam. 2010. Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. mBio 1(3):e00189-10. doi:10.1128/mBio.00189-10.
REFERENCES
- 1.Lyczak J. B., Cannon C. L., Pier G. B. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 2.Schiller N. L., Hatch R. A., Joiner K. A. 1989. Complement activation and C3 binding by serum-sensitive and serum-resistant strains of Pseudomonas aeruginosa. Infect. Immun. 57:1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dasgupta T., de Kievit T. R., Masoud H., Altman E., Richards J. C., Sadovskaya I., Speert D. P., Lam J. S. 1994. Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect. Immun. 62:809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadurugamuwa J. L., Lam J. S., Beveridge T. J. 1993. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob. Agents Chemother. 37:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder T. H., Reiniger N., Meluleni G., Grout M., Coleman F. T., Pier G. B. 2001. Transgenic cystic fibrosis mice exhibit reduced early clearance of Pseudomonas aeruginosa from the respiratory tract. J. Immunol. 166:7410–7418 [DOI] [PubMed] [Google Scholar]
- 6.Augustin D. K., Song Y., Baek M. S., Sawa Y., Singh G., Taylor B., Rubio-Mills A., Flanagan J. L., Wiener-Kronish J. P., Lynch S. V. 2007. Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa. J. Bacteriol. 189:2203–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau P. C. Y., Lindhout T., Beveridge T. J., Dutcher J. R., Lam J. S. 2009. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J. Bacteriol. 191:6618–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King J. D., Kocincova D., Westman E. L., Lam J. S. 2009. Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15:261–312 [DOI] [PubMed] [Google Scholar]
- 9.Knirel Y. A., Bystrova O. V., Kocharova N. A., Zahringer U., Pier G. B. 2006. Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J. Endotoxin Res. 12:324–336 [DOI] [PubMed] [Google Scholar]
- 10.Burrows L. L., Lam J. S. 1999. Effect of wzx (rfbX) mutations on A-band and B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa O5. J. Bacteriol. 181:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Kievit T. R., Dasgupta T., Schweizer H., Lam J. S. 1995. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5). Mol. Microbiol. 16:565–574 [DOI] [PubMed] [Google Scholar]
- 12.Whitfield C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3:178–185 [DOI] [PubMed] [Google Scholar]
- 13.Daniels C., Griffiths C., Cowles B., Lam J. S. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4:883–897 [DOI] [PubMed] [Google Scholar]
- 14.Abeyrathne P. D., Daniels C., Poon K. K. H., Matewish M. J., Lam J. S. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187:3002–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexeyev M. F., Winkler H. H. 1999. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J. Mol. Biol. 285:1503–1513 [DOI] [PubMed] [Google Scholar]
- 16.Tusnády G. E., Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- 17.Manoil C., Alan M. T. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions, p. 61–75 In Methods in cell biology, vol. 34 Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 18.Manoil C., Boyd D., Beckwith J. 1988. Molecular genetic analysis of membrane protein topology. Trends Genet. 4:223–226 [DOI] [PubMed] [Google Scholar]
- 19.Abeyrathne P. D., Lam J. S. 2007. WaaL of Pseudomonas aeruginosa utilizes ATP in in-vitro ligation of O antigen onto lipid A-core. Mol. Microbiol. 65:1345–1359 [DOI] [PubMed] [Google Scholar]
- 20.Drew D., Sjöstrand D., Nilsson J., Urbig T., Chin C.-N., de Gier J.-W., von Heijne G. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. U. S. A. 99:2690–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feilmeier B. J., Iseminger G., Schroeder D., Webber H., Phillips G. J. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froshauer S., Green G. N., McGovern D. B. K., Beckwith J. 1988. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J. Mol. Biol. 200:501–511 [DOI] [PubMed] [Google Scholar]
- 23.Lee C., Li P., Inouye H., Brickman E. R., Beckwith J. 1989. Genetic studies on the inability of beta-galactosidase to be translocated across the Escherichia coli cytoplasmic membrane. J. Bacteriol. 171:4609–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunneen M. M., Reeves P. R. 2008. Membrane topology of the Salmonella enterica serovar Typhimurium group B O-antigen translocase Wzx. FEMS Microbiol. Lett. 287:76–84 [DOI] [PubMed] [Google Scholar]
- 25.Daniels C., Vindurampulle C., Morona R. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28:1211–1222 [DOI] [PubMed] [Google Scholar]
- 26.Schild S., Lamprecht A.-K., Reidl J. 2005. Molecular and functional characterization of O antigen transfer in Vibrio cholerae. J. Biol. Chem. 280:25936–25947 [DOI] [PubMed] [Google Scholar]
- 27.Mazur A., Krol J. E., Marczak M., Skorupska A. 2003. Membrane topology of PssT, the transmembrane protein component of the type I exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii strain TA1. J. Bacteriol. 185:2503–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazur A., Marczak M., Król J. E., Skorupska A. 2005. Topological and transcriptional analysis of pssL gene product: a putative Wzx-like exopolysaccharide translocase in Rhizobium leguminosarum bv. trifolii TA1. Arch. Microbiol. 184:1–10 [DOI] [PubMed] [Google Scholar]
- 29.Hvorup R. N., Winnen B., Chang A. B., Jiang Y., Zhou X.-F., Saier M. H., Jr 2003. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270:799–813 [DOI] [PubMed] [Google Scholar]
- 30.Marolda C. L., Vicarioli J., Valvano M. A. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095–4105 [DOI] [PubMed] [Google Scholar]
- 31.Feldman M. F., Marolda C. L., Monteiro M. A., Perry M. B., Parodi A. J., Valvano M. A. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129–35138 [DOI] [PubMed] [Google Scholar]
- 32.Samuel G., Reeves P. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519 [DOI] [PubMed] [Google Scholar]
- 33.Lindberg A. A., Kärnell A., Weintraub A. 1991. The lipopolysaccharide of Shigella bacteria as a virulence factor. Rev. Infect. Dis. 13:S279–S284 [DOI] [PubMed] [Google Scholar]
- 34.Bastin D. A., Stevenson G., Brown P. K., Haase A., Reeves P. R. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 7:725–734 [DOI] [PubMed] [Google Scholar]
- 35.Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker J. E., Saraste M., Runswick M. J., Gay N. J. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez J. M., McGarry M. A., Marolda C. L., Valvano M. A. 2008. Functional analysis of the large periplasmic loop of the Escherichia coli K-12 WaaL O-antigen ligase. Mol. Microbiol. 70:1424–1440 [DOI] [PubMed] [Google Scholar]
- 38.Matias V. R. F., Al-Amoudi A., Dubochet J., Beveridge T. J. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185:6112–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnakumar S. S., London E. 2007. Effect of sequence hydrophobicity and bilayer width upon the minimum length required for the formation of transmembrane helices in membranes. J. Mol. Biol. 374:671–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugsley A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 57:50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones P. M., George A. M. 1999. Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol. Lett. 179:187–202 [DOI] [PubMed] [Google Scholar]
- 42.Mitchell M. S., Rao V. B. 2004. Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology 321:217–221 [DOI] [PubMed] [Google Scholar]
- 43.Walters R. F. S., DeGrado W. F. 2006. Helix-packing motifs in membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 103:13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larue K., Kimber M. S., Ford R., Whitfield C. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem. 284:7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tocilj A., Munger C., Proteau A., Morona R., Purins L., Ajamian E., Wagner J., Papadopoulos M., Van Den Bosch L., Rubinstein J. L., Fethiere J., Matte A., Cygler M. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15:130–138 [DOI] [PubMed] [Google Scholar]
- 46.Drew D., Lerch M., Kunji E., Slotboom D.-J., de Gier J.-W. 2006. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3:303–313 [DOI] [PubMed] [Google Scholar]
- 47.Qiu D., Damron F. H., Mima T., Schweizer H. P., Yu H. D. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.