Abstract
Amadori-modified proteins (AMPs) are the products of nonenzymatic glycation formed by reaction of reducing sugars with primary amine-containing amino acids and can develop into advanced glycated end products (AGEs), highly stable toxic compounds. AGEs are known to participate in many age-related human diseases, including cardiovascular, neurological, and liver diseases. The metabolism of these glycated proteins is not yet understood, and the mechanisms that reduce their accumulation are not known so far. Here, we show for Escherichia coli that a conserved glycopeptidase (Gcp, also called Kae1), which is encoded by nearly every sequenced genome in the three domains of life, prevents the accumulation of Amadori products and AGEs. Using mutants, we show that Gcp depletion results in accumulation of AMPs and eventually leads to the accumulation of AGEs. We demonstrate that Gcp binds to glycated proteins, including pyruvate dehydrogenase, previously shown to be a glycation-prone enzyme. Our experiments also show that the severe phenotype of Gcp depletion can be relieved under conditions of low intracellular glycation. As glycated proteins are ubiquitous, the involvement of Gcp in the metabolism of AMPs and AGEs is likely to have been conserved in evolution, suggesting a universal involvement of Gcp in cellular aging and explaining the essentiality of Gcp in many organisms.
IMPORTANCE
Glycated proteins (Amadori-modified proteins [AMPs] and advanced glycated end products [AGEs]) are known to participate in many age-related diseases. Their existence in fast-growing organisms was considered unlikely, as their formation was assumed to be slow. Yet, recent evidence demonstrated their existence in bacteria, and our data suggest a bacterial mechanism that reduced their accumulation. We identify in Escherichia coli a protein, Gcp, which carries out this function. Gcp is conserved in all domains of life and is essential in many organisms. Although it was annotated as a chaperon protease, there were no experimental data to support this function. Our findings are compatible with the annotation and will open up studies of the bacterial metabolism of glycated proteins. Furthermore, the data from the bacterial systems may also be instrumental in understanding the metabolism of glycated proteins, including their toxicity in human health and disease.
Introduction
O-sialoglycoprotein endopeptidase (OSGEP), also called Gcp (glycopeptidase), tops the list of “conserved hypothetical” proteins, which were considered top priority targets for experimental studies by Galperin and Koonin (1). This ubiquitous enzyme is one of few highly conserved proteins which are encoded by every sequenced genome in the three domains of life, with the exception of the highly degraded, organelle-like bacterial genomes of Carsonella ruddii and Sulcia muelleri (2). Gcp was first described as a secreted protein of Mannheimia haemolytica (formerly known as Pasteurella haemolytica), isolated from bovine pneumonia, where it exhibits glycopeptidase activity specificity to O-sialic acid-containing glycoproteins (3). However, the ubiquity of Gcp homologs cannot be explained by this function, as sialic acids are highly uncommon in organisms other than vertebrates (4).
There have been several other publications dealing with the potential role of Gcp homologs. These homologs were found to have an HSP70 actin-like folding and were therefore predicted to be ATP-dependent proteases with chaperon activity (5), but no such function was demonstrated for any Gcp homolog. In Saccharomyces cerevisiae, the Gcp homolog Kae1 was found to be part of a complex involved in telomere uncapping and elongation (6, 7), and the Gcp homolog of the hyperthermophilic archaeon Pyrococcus abyssi exhibits apurinic endonuclease activity in vitro (8). These data suggest that Gcp is involved in DNA metabolism, also supported by the finding that Gcp-depleted Escherichia coli cells had modified nucleoids and that Gcp-deficient yeast mitochondria lose their mitochondrial DNA (9–11). These studies do not provide direct evidence for the molecular function of GCP but indicate that its function affects a broad range of physiological activities.
Gcp therefore represents an evolutionary puzzle: could highly similar and conserved proteins (2) have diverged so much in function across the domains of life, without much divergence in sequence? Alternatively, it is possible that the most conserved roles of Gcp have yet to be identified and that at least some Gcp orthologs have multiple physiological roles.
We assumed that Gcp functions as a glycoprotein-related chaperon protease, as suggested by bioinformatics, structural analysis (5), and enzymatic glycopeptidase activity (3). Therefore, we looked for a sugar-related pathway which is highly conserved in evolution and important for the physiology of all cells. One such conserved role is the handling of toxic glycation products that accumulate in all organisms and are harmful to living cells. On the basis of this assumption, we examined the possible role of Gcp in the metabolism of glycated proteins (Amadori products and advanced glycated end products [AGEs]).
Amadori-modified proteins (AMPs) are the products of nonenzymatic glycation formed by the reaction of reducing sugars with primary amine-containing amino acids in proteins. These glycated proteins are formed via a multistep chemical reaction known as an Amadori rearrangement and can further develop into irreversible, highly stable compounds known as AGEs (12–15).
AGEs participate in the pathophysiology of several human diseases, including cardiovascular, neurological, and liver diseases (16–19). As the formation rate of AGEs depends on the carbohydrate and oxygen concentrations, conditions of chronic hyperglycemia and oxidative stress, such as diabetes, result in increased accumulation of AGEs and consequent toxic effects (20).
The formation of AGEs was shown to be a slow process, taking weeks to months. On the basis of this view, it was assumed that only long-lived proteins are at potential risk for developing these modifications and that nonenzymatic glycation of bacterial proteins is unlikely, due to the relatively short generation time of most bacteria. However, in 2001, Mironova et al. provided the first evidence for the nonenzymatic glycation and presence of AGEs in E. coli, a rapidly growing organism (21).
Here, we provide genetic and biochemical evidence that a function of Gcp is the metabolizing of glycated proteins (Amadori products and AGEs). We show that upon Gcp depletion in an E. coli mutant, growth rate is dramatically reduced and cells accumulate highly active AMPs and subsequently AGEs. We further show by immunoprecipitation that Gcp primarily binds glycated proteins and characterize several candidate substrates, including the elongation factor Tu and the pyruvate dehydrogenase (PDH) complex, which was previously shown to be susceptible to carbonylation in E. coli (22). Moreover, we show a correlation between the internal level of glycation and the inhibitory effect of Gcp depletion.
These findings indicate a direct involvement of Gcp in the metabolism of glycated proteins and support the predicted function of Gcp as a glycoprotein-related chaperon protease. On the basis of these findings, we present a model which is compatible with the complex Gcp-related phenotypes and suggest a conserved role for Gcp activity.
RESULTS
Construction and physiology of Gcp-depleted E. coli.
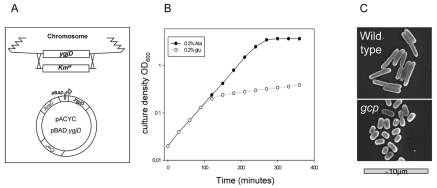
In order to investigate the possible function of Gcp in the metabolism of glycated proteins in E. coli, we constructed a strain in which Gcp levels can be reduced. In this strain, the chromosomal ygjD gene, coding for Gcp, was deleted and complemented by the presence of a low-copy-number plasmid carrying the ygjD coding sequence under the regulation of the arabinose-dependent pBAD promoter (Fig. 1A) (see Materials and Methods). The bacteria grew normally in the presence of 0.2% arabinose but not in the presence of glucose, which repressed the expression of Gcp. When the bacterial cultures were diluted into glucose-containing medium, the growth rate gradually declined, and growth was dramatically reduced after a time period equivalent to about six generations (Fig. 1B). The viability of the bacteria during Gcp depletion was monitored by plating on arabinose-containing LB plates. No reduction of viability was observed, demonstrating that the growth inhibition of Gcp-depleted cells can be restored by activation of ygjD expression. Moreover, although there was no increase in turbidity, there was an increase in cell number, suggesting that Gcp depletion results in smaller bacteria. Indeed, using electron microscopy, we could show that Gcp-depleted cells are much smaller and spherical, resembling the phenotype of stationary-phase bacteria (Fig. 1C). It should be noted that previous reports also demonstrated that Gcp depletion results in altered cell morphology (11). Handford et al. observed under Gcp depletion an unusual morphology, with a varied appearance mostly consisting of enlarged cells. The fact that we saw only small cells may be explained by the use of different E. coli strains (MC4100 versus MG1655), but it is clear that a specific mechanism involved in cell morphology determination is affected under Gcp-restrictive conditions.
FIG 1 .
Construction and growth phenotype of the Gcp-depleted strain. (A) Schematic representation of the Gcp-depleted strain. The chromosomal ygjD gene, coding for Gcp, was deleted and complemented by a low-copy-number plasmid with an arabinose (Ara)-inducible ygjD allele. (B) Growth of the Gcp-depleted strain in the presence of 0.2% arabinose or 0.2% glucose for activation or repression of ygjD expression (respectively). Growth was monitored by measuring absorbance at 600 nm. (C) Scanning electron micrographs of wild-type and Gcp-depleted cells.
Accumulation of AMPs and AGEs in Gcp-depleted cells.
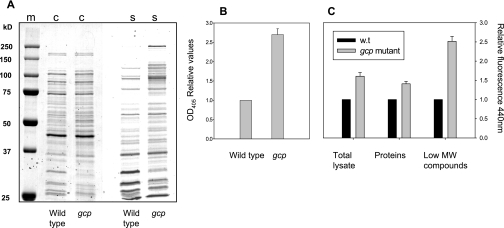
In order to determine the levels of glycated proteins in wild-type and Gcp-depleted cells, lysates were extracted and further separated into soluble proteins and low-molecular-weight compounds by use of size exclusion chromatography. Proteins were separated by SDS-PAGE, and the gels were stained either by Coomassie brilliant blue (for total proteins) or by sugar-specific silver staining of diols (for glycated proteins) (23). The results presented in Fig. 2 indicate that some E. coli proteins showed a specific staining pattern, demonstrating the steady state of glycated proteins under these conditions. Reduction of Gcp levels did not significantly change the total protein pattern but resulted in an accumulation of a subset of glycated proteins (Fig. 2A). Interestingly, the accumulation of glycated proteins was observed mainly at the high-molecular-mass region (~75 to 150 kDa). As the spectrum of proteins in this size range is fairly limited in E. coli, it is possible that these bands represent a mobility shift of lower-molecular-weight proteins that was caused by glycation. In order to quantify the levels of glycated proteins, we used a periodate-based colorimetric assay specific for the quantification of Amadori protein products (23). The results demonstrate that Gcp depletion resulted in about 2.5-fold accumulation of Amadori products in the purified protein fraction (Fig. 2B).
FIG 2 .
Accumulation of glycated proteins and AGEs following Gcp depletion. Lysates were extracted from wild-type and Gcp-depleted cells and further separated into proteins and low-molecular-mass compound fractions, as described in Materials and Methods. (A) Proteins were subjected to SDS-PAGE, followed by staining with Coomassie brilliant blue for total proteins (c) or diol-specific silver stain for glycated proteins (s). (B) Quantification of glycated proteins by use of a periodate-based colorimetric assay. (C) AGE-specific fluorescence (relative; excitation at 370 nm and emission at 440 nm) following Gcp depletion in total lysates and in the fractions of purified proteins and low-molecular-weight (MW) compounds. The results represent three independent experiments.
Amadori products are relatively unstable intermediates in the process of protein glycation. However, these molecules can further develop into the highly stable AGEs. The presence of AGEs was determined by AGE-specific fluorescence, with measurement of emission at 440 nm upon excitation at 370 nm. We observed that in lysates extracted from Gcp-depleted cells, there was a 60% increase in the accumulation of AGEs relative to the levels in lysates of nondepleted cells (Fig. 2C). Lysates were further separated as before, and proteins and low-molecular-weight fractions were examined individually. Our data presented in Fig. 2C demonstrate that the protein fraction derived from Gcp-depleted cells showed about 40% higher levels of AGEs than that derived from wild-type cells, while in the low-molecular-weight fraction, the accumulation of AGEs was 2.5-fold higher. These results demonstrate that Gcp depletion results in accumulation of AGEs and that most of the AGEs accumulate as low-molecular-weight compounds.
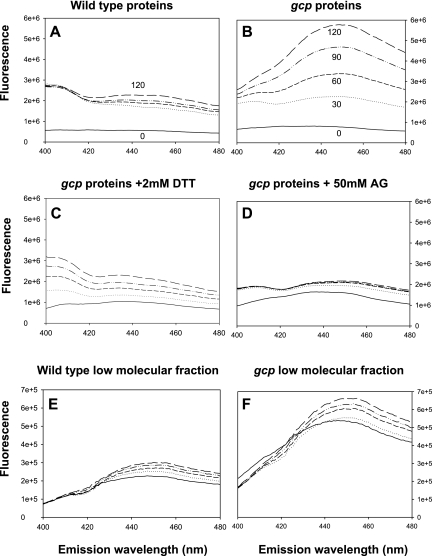
In order to establish the origin of AGEs accumulated upon Gcp depletion, we examined the potential of the separated fractions from wild-type and Gcp-depleted cells to form AGEs in vitro. The kinetics of AGE formation was measured using AGE-specific fluorescence at 30-min intervals during incubation at the E. coli physiological temperature of 37°C. Under these conditions, purified proteins extracted from the Gcp-depleted cells rapidly developed into AGEs and accumulation of AGEs continued for at least 2 h (Fig. 3B). On the other hand, proteins extracted from the wild-type strain showed a reduced potential for development of AGEs, and accumulation reached a low plateau after 30 min of incubation (Fig. 3A). A kinetic analysis of AGE formation in the low-molecular-weight fractions indicated that fractions derived from both the wild-type strains and the Gcp-depleted strains demonstrated low potential for forming AGEs (Fig. 3E and F). The formation of AGEs from AMPs is known to be oxidation dependent. Indeed, in a control experiment with the most potent fraction (a protein fraction from Gcp-depleted cells), addition of 2 mM dithiothreitol (DTT) resulted in a considerable decrease in the rate of AGE formation (Fig. 3C). Moreover, addition of 50 mM aminoguanidine completely abolished AGE accumulation, as this agent in known to prevent the cross-linking which is essential for AGE formation (24) (Fig. 3D).
FIG 3 .
Kinetic studies of AGE formation in proteins and in low-molecular-weight fractions. Samples were kept at 37°C, and AGEs were monitored every 30 min for 2 h by measurement of AGE-specific fluorescence. The emission spectrum from 400 nm to 480 nm upon excitation at 370 nm is presented. Kinetics of AGE formation in the protein fractions (A and B) and low-molecular-weight fractions (E and F) of wild-type and Gcp-depleted (gcp) strains, respectively. Effect of 2 mM DTT (C) or 100 mM aminoguanidine (AG) (D) on the kinetics of AGE formation in the protein fractions extracted from Gcp-depleted bacteria.
Search for potential Gcp substrates.
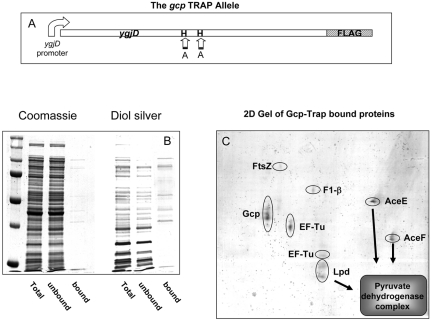
The findings that Gcp depletion results in accumulation of AMPs and subsequently AGEs suggest a direct involvement of Gcp in their metabolism. In order to capture potential Gcp substrates, we used a coimmunoprecipitation assay. Since potential Gcp substrates accumulate primarily in its absence (Fig. 2A) and since enzyme-substrate interaction is generally too rapid to be captured by coimmunoprecipitation, we constructed a “Gcp-Trap” strain, an established method for substrate capturing (25, 26). The method, previously used for identifying substrates of known chaperon proteases, is based on the idea that mutations in the active site can produce proteins that bind the substrates but lack catalytic activity. The Gcp-Trap allele was constructed by changing the essential metal-binding histidine residues at positions 111 and 115 to alanines in the predicted metallopeptidase domain (Fig. 4A). This modified allele was cloned into a pBR322 vector with a sequence coding for a FLAG tag (27) at the 5′ end of the gene. The plasmid carrying the Gcp-Trap allele was introduced into the strain deleted for the chromosomal Gcp-coding gene, and the culture was depleted of Gcp. The allele coding for the Gcp-Trap strain did not compensate for the depletion of the chromosomal ygjD gene, demonstrating the importance of the metal-binding domain for Gcp function. When growth ceased, the cells were disrupted and the inactive Gcp protein was precipitated using an anti-FLAG agarose resin, as described in Materials and Methods. Total proteins, unbound and bound fractions, were separated using SDS-PAGE and stained with either Coomassie brilliant blue or diol-specific silver stain for detection of glycated proteins. The results shown in Fig. 4B demonstrate that the Gcp-Trap strain accumulated glycated proteins, similar to the accumulation observed for the nontagged Gcp-depleted strain, reconfirming the importance of the metal-binding site for enzymatic function. Moreover, the proteins found to be bound to Gcp-Trap were predominantly glycated polypeptides, and their pattern was fairly similar to the pattern of the subset of glycated proteins found to accumulate upon Gcp depletion (compare Fig. 4B and Fig. 2A).
FIG 4 .
Characterization of Gcp substrates by use of immunoprecipitation. Immunoprecipitation was performed by the use of a strain containing an inactive, metal-binding-deficient Gcp (Gcp-Trap). Bacteria were grown as described for Fig. 1, and Gcp was depleted by the addition of glucose. When growth stopped, the cells were lysed, and proteins were purified as described for Fig. 2 and subjected to immunoprecipitation using an anti-FLAG resin as described in Materials and Methods. (A) Schematic representation of the Gcp-Trap allele. The predicted zinc-binding histidines were replaced by alanines, and a FLAG tag was added to the 5′ end. (B) SDS-PAGE of total proteins and of fractions bound and unbound to the anti-FLAG resin, stained with Coomassie brilliant blue for total proteins or a diol-specific silver stain for glycated proteins. (C) Separation of Gcp-suspected substrates on two-dimensional gel electrophoresis gels. Proteins were identified by mass spectrometry, and protein names are indicated next to each spot.
The proteins bound to the Gcp-Trap allele were separated by two-dimensional (2D) PAGE chromatography, and 10 of the most-prominent spots that were visualized on the gel were identified using mass spectrometry. These proteins included the cell division protein FtsZ, the F1-β subunit of the ATP synthase AtpD, and the elongation factor Tu (EF-Tu), all of them essential proteins in E. coli. Moreover, all three components of the pyruvate dehydrogenase (PDH) complex, AceE, AceF, and Lpd, were also among the Gcp-trapped proteins (Fig. 4C). These results are compatible with those obtained in the high-throughput experiments performed by Butland et al. (28), which found interaction between Gcp and both EF-Tu and AceE. It should be noted that the locations of some of these proteins differed from the theoretical location predicted by molecular weight. FtsZ and AtpD appear to have higher molecular weights, while Lpd and EF-Tu were present in multiple spots. These abnormalities suggest that these proteins underwent a posttranslational modification, probably involving glycation at some step.
Some of the potential substrates captured by Gcp-Trap were previously shown to be subjected to carbonylation. PDH, EF-Tu, and the ATP synthase subunits were modified in mammalian mitochondria (29, 30), while PDH and EF-Tu were also found to be carbonylated in E. coli (22).
The finding that FtsZ is glycated in Gcp-depleted cells is interesting and can explain the morphology changes observed to occur under Gcp depletion, as FtsZ is known to participate in septation and to mediate cell wall biosynthesis by interaction with peptidoglycan-modifying enzymes (31).
Lower levels of active PDH complex in Gcp-depleted cells.
The levels of active PDH complex were further investigated and compared between wild-type and Gcp-depleted cells. Active complexes were partially purified using ultracentrifugation. Under these conditions, only stable complexes precipitate, due to their massive molecular weight. The PDH complex was visualized by SDS-PAGE, and the results indicate that there is a 60% decrease in the amount of stable PDH complex in lysates from Gcp-depleted cells, compared with the amount for the wild type (Fig. 5A and B). The precipitated complexes have similar specific activities, as measured by monitoring the accumulation of NADH in the presence of pyruvate and coenzyme A for 60 s (Fig. 5C). These results indicate that a smaller amount of stable PDH complex is found in Gcp-depleted cells and suggest that Gcp is involved in preserving the glycation-prone PDH complex.
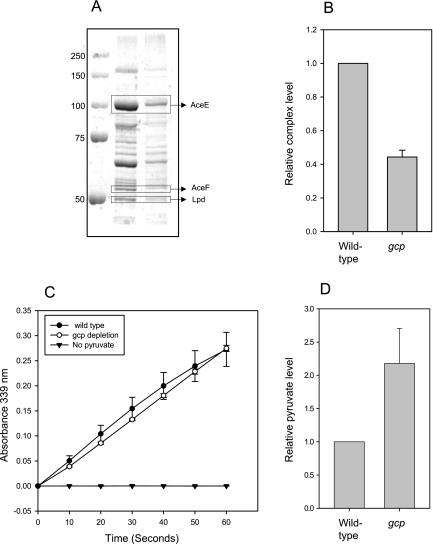
FIG 5 .
Partial purification and activity measurements of the pyruvate dehydrogenase (PDH) complex from wild-type and Gcp-depleted cells. Equal amounts of proteins from wild-type and Gcp-depleted cells were centrifuged for the precipitation of the PDH complex. (A) SDS-PAGE of PDH purification. The complex components are indicated. (B) Relative quantification of the amount of PDH complex from three independent experiments. (C) In vitro activity measurement of the PDH complex from wild-type and Gcp-depleted cells. The graph represents the accumulation of the reaction product NADH by measuring its specific absorption at 339 nm. Specificity was validated by comparison to a negative control lacking the substrate, pyruvate.
In order to determine whether the reduction in active PDH is physiologically significant, we measured the level of its substrate, pyruvate. It was previously shown that pyruvate accumulates in mutants with inactive PDH (32), and therefore, an increase in pyruvate levels represents the inability of PDH to function. Indeed, depletion of Gcp resulted it an approximately 2-fold accumulation of pyruvate, compared to the level for wild-type cells (Fig. 5D), demonstrating the severe physiological effects of reduction in active PDH complexes.
The severity of the Gcp depletion phenotype correlates with the level of protein glycation.
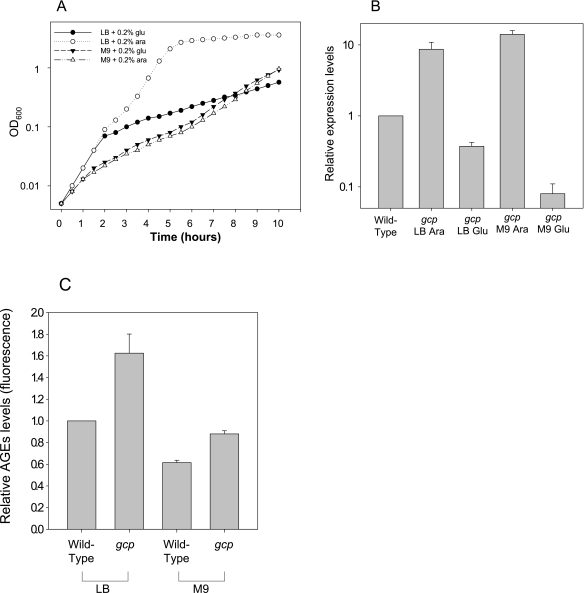
Our data suggest that Gcp prevents accumulation of glycated proteins. Therefore, we assumed that the level of glycation will influence the severity of the effect of Gcp depletion, which results in dramatic growth rate reduction. This assumption could be tested by reducing the intracellular levels of glycated proteins, which is predicted to lessen the effect of Gcp depletion. Unfortunately, no known extracellular agent that can decrease glycation in E. coli is currently available. However, it was reported by Dimitrova et al. (33) that intracellular glycation levels are dramatically reduced when bacteria are grown in minimal M9 medium. We therefore compared the growth rates of our Gcp depletion strain in rich LB medium and minimal M9 medium in the presence of arabinose (for activation of gcp expression) or glucose (for its repression). Cultures were diluted to optical densities at 600 nm (OD600s) of 0.005 (106 cells/ml), and turbidity was monitored for 10 h. The data presented in Fig. 6A demonstrate that when bacteria were grown in rich LB medium, repression of gcp expression resulted in a dramatic reduction of growth rate after about six generations. However, when bacteria were grown in M9 minimal medium to comparable cell concentrations, repression of Gcp expression did not affect growth rate during that time course. Moreover, under Gcp repression conditions, the depleted strains grew at an increased rate in the nutrient-inferior minimal medium, leading to increased turbidity after 8 h, in contrast to the growth phenotype observed under nonrepressing conditions.
FIG 6 .
Gcp depletion growth phenotype in rich LB medium and minimal M9 medium. (A) The Gcp-depleted strain was grown in the presence of 0.2% arabinose or 0.2% glucose for activation or repression of Gcp expression, respectively, in rich LB medium and minimal M9 medium. The cells were diluted from arabinose-containing medium into fresh medium supplemented with arabinose or glucose as indicated to a concentration of 106/ml, and growth was monitored for 10 h by measuring absorption at 600 nm. (B) Relative expression levels of the Gcp-depleted strain grown under different conditions as indicated. Relative mRNA levels measured using real-time PCR are presented. The wild-type expression level represents chromosomal gcp expression levels of wild-type cells grown in LB medium. Wild-type expression levels were similar in both media and were not significantly altered by supplementation with either glucose or arabinose.
These results indicate that depletion of Gcp has a reduced effect on growth rate under conditions that reduce glycation. However, it is also possible that Gcp synthesis is not shut down in minimal medium as efficiently as in rich medium. This possibility was ruled out by the experiment represented in Fig. 6B, showing that the levels of gcp transcripts, measured by real-time PCR, are actually even lower in minimal medium than in LB under repression conditions. Thus, under repression conditions (in the presence of glucose), the gcp transcript level of cells grown in LB medium was reduced to about 35% of the normal expression level found in wild-type cells, while in M9 medium, the repression reduced gcp transcript levels to less than 10% of the normal expression level. In view of this result, showing reduced gcp transcript levels yet no physiological effects of Gcp depletion, we determined the levels of AGEs in the wild type and in the mutant in rich and minimal media. The results shown in Fig. 6C indicate that, indeed, there are lower levels of glycation products in minimal medium, as reported before (33). Gcp depletion resulted in an increase in the level of AGEs even in minimal medium. However, this level was still lower than the level of AGEs in the wild type grown in rich medium, in agreement with the lack of phenotypic manifestation of Gcp depletion in minimal medium.
Taken together, these results demonstrate that although in minimal medium there is less Gcp in depleted bacteria, the effect of the depletion is less severe than in rich medium. These results are compatible with the hypothesis that the severe phenotype of Gcp depletion is due to accumulation of glycated proteins.
DISCUSSION
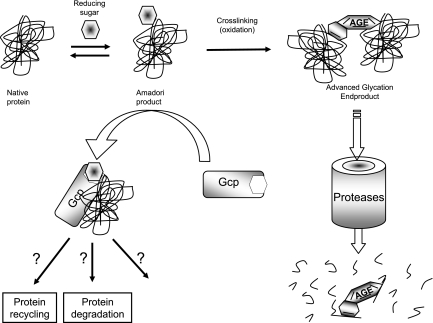
On the basis of the data presented here, our model (Fig. 7) suggests that Gcp is directly involved in AMP metabolism and that its depletion results in accumulation of AMPs, which serve as the potential for development of AGEs. These AGEs are probably further degraded in the cell, as they accumulate in the fraction of low-molecular-weight compounds.
FIG 7 .
Schematic model of Amadori-modified proteins and AGE metabolism in E. coli. The model suggests that Gcp participates in the metabolism of glycated proteins by reducing the level of Amadori products, thereby decreasing the formation of AGEs. Once AGEs are formed, they are broken down to smaller molecules.
The molecular mechanism of Gcp function is not yet fully understood. One possibility is that Gcp is involved in Amadori product neutralization by protein degradation. This hypothesis is compatible with the current annotation of Gcp as a glycopeptidase, as was shown for M. haemolytica (3). Another possibility is that Gcp plays a role in recycling of proteins by assisting deglycation using its putative chaperon activity (5).
Several enzymes, known collectively as “amadoriases,” were found to have deglycating activity, having the ability to reverse nonenzymatic glycation by cleaving the bond between amino acids and corresponding sugars (34, 35). One such enzyme, capable of deglycating fructoselysine, was found in E. coli (36). These enzymes were shown to act only on low-molecular-weight peptides and are unable to work on proteins. Gcp may act by introducing glycated proteins to amadoriases either by allowing access to the sugar moiety on the full-length proteins (using its predicted chaperon activity) or by cleaving the Amadori product in a way that will enable better access of amadoriases to the glycated peptides.
The mechanism which leads to cessation of cell growth after Gcp depletion is likely to be the reason for the essentiality of Gcp in several organisms. Our data suggest that Gcp depletion results in accumulation of glycated protein products which cause growth arrest. Support for this assumption is obtained by the finding that Gcp depletion has a greatly reduced effect on growth under conditions which reduce protein glycation (Fig. 6). Accumulation of AMPs and AGEs can be potentially toxic, as AGEs were found to increase cellular oxidative stress (37). However, we found that addition of antioxidants such as lipoic acid, ascorbic acid, or glutathione to the growth medium did not relieve the Gcp depletion phenotypes. Another possibility is that glycated proteins left untreated can cause metabolic dysfunction. Damaged enzymes can be potentially toxic if they have a dominant negative effect, such as competing for substrates with the normal enzyme or leading to inactivation of complexes. This explanation appears promising, as all of the potential substrates of Gcp identified in this study were parts of multimeric protein complexes.
In mammalian cells, the toxicity of AGEs is facilitated by their interaction with specific receptors, soluble as well as membrane bound. Our bioinformatics search suggests that the E. coli genome does not contain genes coding for homologs of these receptors. It is still possible that a direct interaction of AGEs with specific target proteins is involved in the toxicity. Localization of such putative complexes may shed light on their function but is complicated by the facts that there are many types of AGEs and that the availability of antibodies is limited.
The downstream metabolism of AGEs should also be further investigated. We show that a high fraction of the AGEs is present in the low-molecular-weight fraction. These low-molecular-weight AGEs probably result from a proteolytic breakdown of modified proteins. The enzymatic mechanism responsible for AGE degradation is currently unknown. Yet, we eliminated the possible participation of the three major cytosolic ATP-dependent proteases, as a triple mutant at Lon, ClpX, and HslV still accumulates low-molecular-weight AGEs when Gcp is depleted. However, because in our genetic system Gcp is not completely abolished, we cannot yet rule out the possibility that Gcp is involved in AGE degradation.
The specific molecules that lead to protein glycation in bacteria are currently unknown. Since extracellular concentration of reducing sugars did not correlate with intracellular glycation levels, it was postulated that intracellular metabolic pathways are the source of these agents (33). One potentially active agent shown to be produced in E. coli is methylglyoxal, synthesized by the methylglyoxal synthase MgsA (38). However, deletion of msgA did not relieve the Gcp depletion phenotypes, demonstrating that this agent is not a dominant factor in Gcp metabolic function (C. Katz and E. Z. Ron, unpublished data).
The results presented here demonstrate a role for Gcp in preventing accumulation of glycated proteins, which are ubiquitous substrates in living cells. Thus, this metabolic function is likely to extend to all domains of life and should also be tested for in Archaea and Eukarya. These findings open up the avenue for investigating glycated proteins in bacteria, including their metabolism, functions, and possible toxicity. The ability to study glycated proteins in an easy model organism, such as E. coli, which is amenable to genetic manipulation, will advance investigations of glycation processes in ageing and in human diseases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this work are described in Table 1.
TABLE 1 .
Strains and plasmids
| Plasmid or strain | Feature(s) | Description, genotype, or reference(s) |
|---|---|---|
| Plasmids | ||
| pBAD24 | Plasmid containing an arabinose-inducible promoter | 44 |
| pBAD:ygjD | Ampr; arabinose-dependent ygjD expression | pBAD24 with the ygjD ORF cloned after EcoRI and XbaI digestion |
| pACYC:pBAD:ygjD | Cmr; low-copy-number plasmid for enhanced control of ygjD expression | pBAD:ygjD digested with BspHI and cloned into pACYC184 digested with the same enzyme |
| pBR322 | 45, 46 | |
| pBR322 Gcp-Trap | Ampr; constitutive expression of FLAG-tagged inactive ygjD for immunoprecipitation | 5′ FLAG-tagged ygjD with its endogenous promoter cloned into an EcoRI-digested PBR322 plasmid; the metal-binding histidine residues at positions 111 and 115 were modified to alanines |
| Bacterial strains | ||
| K-12 MG1655 | Wild type | |
| K-12 MG1655 pACYC:pBAD:ygjD | Wild type carrying a plasmid with cloned inducible gcp | K-12 MG1655 pACYC:pBAD:ygjD |
| Gcp depletion strain | Used for Gcp depletion; the deletion is complemented by an inducible ygjD gene on a plasmid | K-12 MG1655 ΔygjD pACYC:pBAD:ygjD |
| Gcp-Trap strain | Used for Gcp immunoprecipitation; this is the Gcp depletion strain with an additional plasmid coding for a peptidase-deficient Gcp protein | K-12 MG1655 ΔygjD pACYC:pBAD:ygjD pBR322 Gcp-Trap |
Growth conditions.
Bacteria were grown in standard LB medium (Difco) or minimal M9 medium at 37°C. The medium was supplemented with arabinose or glucose at 0.2%. Control strains were grown under the same conditions as Gcp-depleted strains to reduce the possible background of the sugar concentrations present in the growth media. Antibiotics were added when required at the following concentrations: for ampicillin (Amp), 100 µg/ml; for kanamycin (Km), 50 µg/ml; and for chloramphenicol (Cm), 25 µg/ml.
Genetic manipulations.
The chromosomal ygjD gene was deleted, as previously described (39), in the presence of the complementation plasmid pACYC:pBAD:ygjD. The Gcp-Trap allele was constructed in two steps. First, the ygjD open reading frame (ORF) and promoter were amplified using the FLAG epitope tag-containing primers and cloned into EcoRI-digested pBR322. Second, the conserved metal-binding domain was manipulated by replacing the two conserved histidines (positions 111 and 115) with alanines by use of PCR-based site-directed mutagenesis.
Preparation of samples for determination of Amadori products and AGEs.
Lysates were extracted using a Qproteome bacterial protein preparatory kit (Qiagen) according to the manufacturer’s recommendation. The lysates were further separated into purified proteins and low-molecular-weight compounds by use of GE Healthcare HITRAP desalting columns, equilibrated with either phosphate-buffered saline (PBS) or H2O for AGE or Amadori product analysis, respectively, according to the manufacturer’s recommendation. These columns possess an exclusion limit of 5,000 g/mol of globular compound; thus, compounds with lower molar masses are here referred to as low-molecular-weight compounds. Nucleic acids were digested as part of the Qproteome extraction procedure and therefore were present mostly in the low-molecular-weight fraction. Separations were performed and evaluated using the Amersham AKTA Prime Plus fast-performance liquid chromatography (FPLC) system.
Analysis of glycated proteins on SDS-PAGE gels.
Proteins were extracted and purified as described above and separated using 10% SDS-PAGE. The gels were stained using Coomassie brilliant blue staining for total protein analysis and using diol-specific silver staining as described in reference 23 for glycated-protein analysis. The Amadori product colorimetric assay was preformed as described in reference 40.
Determination of AGEs.
AGEs were quantified using AGE-specific fluorescence by scanning emissions ranging from 400 nm to 480 nm upon excitation at 370 nm at 37°C with a HORIBA scientific FluoroLog-3 spectrofluorometer. The data presented represent either the full-range spectrum or the 440-nm emission peak.
Gcp-Trap coimmunoprecipitation assay.
The Gcp-Trap strain was grown overnight in the presence of arabinose and diluted 1/200 into fresh LB supplemented with 0.2% glucose. The culture was kept at logarithmic ODs (0.2 to 0.8) by further dilutions until growth ceased. Twenty milliliters of culture at an OD600 of 0.4 was harvested and subjected to total soluble protein purification as described for the sample preparation. A desalting stage was used for buffer replacement, and purified proteins were eluted in Tris-buffered saline (TBS) buffer, pH 7.4, followed by concentration to 0.5 ml using 10K Amicon Ultra (Millipore). The sample was incubated overnight at 4°C with 50 µl of anti-FLAG M2-agarose (Sigma) prewashed with TBS. Samples were washed five times with 1 ml TBS buffer and eluted using 200 µl TBS supplemented with 100 µM 3× FLAG peptide (Sigma). Total proteins, unbound and bound fractions, were subjected to SDS-PAGE and stained either with Coomassie brilliant blue or with a diol-specific silver stain.
2D analysis and protein identification.
Fifty micrograms of proteins obtained by Gcp-Trap coimmunoprecipitation assay was separated using two-dimensional gel electrophoresis with 13-cm IPG strips, pH 4 to 7 (GE Healthcare), and identified using mass spectrometry, as described in reference 41.
Pyruvate dehydrogenase purification and activity measurement.
Ten milliliters of wild-type and Gcp-depleted cultures at an OD600 of 0.4 was harvested, and the pellets were resuspended in 1 ml buffer (50 mM K-phosphate, pH 7.5, 5 mM EDTA, and 2 mM DTT) and disrupted using sonication. Lysates were centrifuged for 30 min at 40,000 × g for membrane precipitation, and equal protein amounts of the membrane-free supernatants were centrifuged again for 1.5 h at 150,000 × g for PDH precipitation. The PDH complexes were analyzed by SDS-PAGE as described above. PDH activity was measured as described in reference 42. Control reactions without pyruvate were used as a negative control for background assessment.
Pyruvate measurements.
Pyruvate extraction was based on the hot-water method described in reference 43. Ten-milliliter samples at an OD600 of 0.4 were collected and centrifuged, and the pellets were washed with distilled water. The pellets were resuspended with 200 µl boiled H2O and incubated at 95°C in a Thermomixer (Eppendorf) with strong agitation for 5 min, the samples were centrifuged for 30 min at −10°C, and the supernatants were immediately used for pyruvate measurement using a BioVision pyruvate assay kit according to the manufacturer’s fluorescence protocol. Fluorescence was measured using a Synergy HT multidetection reader (excitation, 530 nm; emission, 590 nm).
RNA extraction and real-time PCR experiment.
RNA was stabilized using RNAprotect reagent (Qiagen), followed by extraction using an RNeasy minikit (Qiagen) according to the manufacturer’s protocols. Residual DNA was digested using RQ1 RNase-free DNase (Promega), and the samples were purified using the RNeasy cleanup procedure. Five hundred nanograms of total RNA was reverse transcribed using random hexamers (Amersham) and ImPromII reverse transcriptase (Promega). Real-time PCRs for determination of gcp transcript levels were performed using 500 nM primers GCAAATACCATTCGTGACAA and TGCACTTAATCATCAGCGTAT in a 10-µl volume with SYBR green PCR master mix (Applied Biosystems). Reactions were run on a Rotorgene 6000 (Corbett) using the standard cycling parameters. Relative gene expression data analysis was carried out by the ΔCT method as described in the manufacturer’s protocol (Corbett).
ACKNOWLEDGMENTS
We thank Dvora Biran and Aviram Rasouly for productive discussions and suggestions. We are grateful to Sagi Huja and Doerte Becher (Greifswald University) for helping us with the proteomic experiments.
C.K. was supported by a fellowship from the Alfried Krupp von Bohlen and Halbach Foundation graduate school, “Functional Genomics of Pathogens.”
Footnotes
Citation Katz, C., I. Cohen-Or, U. Gophna, and E. Z. Ron. 2010. The ubiquitous conserved glycopeptidase Gcp prevents accumulation of toxic glycated proteins. mBio 1(3):e00195-10. doi:10.1128/mBio.00195-10.
REFERENCES
- 1. Galperin M. Y., Koonin E. V. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galperin M. Y. 2008. Social bacteria and asocial eukaryotes. Environ. Microbiol. 10:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdullah K. M., Lo R. Y., Mellors A. 1991. Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene. J. Bacteriol. 173:5597–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angata T., Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439–469 [DOI] [PubMed] [Google Scholar]
- 5. Aravind L., Koonin E. V. 1999. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287:1023–1040 [DOI] [PubMed] [Google Scholar]
- 6. Mao D. Y., Neculai D., Downey M., Orlicky S., Haffani Y. Z., Ceccarelli D. F., Ho J. S., Szilard R. K., Zhang W., Ho C. S., Wan L., Fares C., Rumpel S., Kurinov I., Arrowsmith C. H., Durocher D., Sicheri F. 2008. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell 32:259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Downey M., Houlsworth R., Maringele L., Rollie A., Brehme M., Galicia S., Guillard S., Partington M., Zubko M. K., Krogan N. J., Emili A., Greenblatt J. F., Harrington L., Lydall D., Durocher D. 2006. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124:1155–1168 [DOI] [PubMed] [Google Scholar]
- 8. Hecker A., Leulliot N., Gadelle D., Graille M., Justome A., Dorlet P., Brochier C., Quevillon-Cheruel S., Le Cam E., van Tilbeurgh H., Forterre P. 2007. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 35:6042–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oberto J., Breuil N., Hecker A., Farina F., Brochier-Armanet C., Culetto E., Forterre P. 2009. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res. 37:5343–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hecker A., Graille M., Madec E., Gadelle D., Le Cam E., van Tilbergh H., Forterre P. 2009. The universal Kae1 protein and the associated Bud32 kinase (PRPK), a mysterious protein couple probably essential for genome maintenance in Archaea and Eukarya. Biochem. Soc. Trans. 37:29–35 [DOI] [PubMed] [Google Scholar]
- 11. Handford J. I., Ize B., Buchanan G., Butland G. P., Greenblatt J., Emili A., Palmer T. 2009. Conserved network of proteins essential for bacterial viability. J. Bacteriol. 191:4732–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baynes J. W., Watkins N. G., Fisher C. I., Hull C. J., Patrick J. S., Ahmed M. U., Dunn J. A., Thorpe S. R. 1989. The Amadori product on protein: structure and reactions. Prog. Clin. Biol. Res. 304:43–67 [PubMed] [Google Scholar]
- 13. Horvat S., Jakas A. 2004. Peptide and amino acid glycation: new insights into the Maillard reaction. J. Pept. Sci. 10:119–137 [DOI] [PubMed] [Google Scholar]
- 14. Njoroge F. G., Monnier V. M. 1989. The chemistry of the Maillard reaction under physiological conditions: a review. Prog. Clin. Biol. Res. 304:85–107 [PubMed] [Google Scholar]
- 15. Singh R., Barden A., Mori T., Beilin L. 2001. Advanced glycation end-products: a review. Diabetologia 44:129–146 [DOI] [PubMed] [Google Scholar]
- 16. Hyogo H., Yamagishi S. 2008. Advanced glycation end products (AGEs) and their involvement in liver disease. Curr. Pharm. Des. 14:969–972 [DOI] [PubMed] [Google Scholar]
- 17. Jandeleit-Dahm K., Cooper M. E. 2008. The role of AGEs in cardiovascular disease. Curr. Pharm. Des. 14:979–986 [DOI] [PubMed] [Google Scholar]
- 18. Takeuchi M., Yamagishi S. 2008. Possible involvement of advanced glycation end-products (AGEs) in the pathogenesis of Alzheimer’s disease. Curr. Pharm. Des. 14:973–978 [DOI] [PubMed] [Google Scholar]
- 19. Takeuchi M., Yamagishi S. 2009. Involvement of toxic AGEs (TAGE) in the pathogenesis of diabetic vascular complications and Alzheimer’s disease. J. Alzheimers Dis. 16:845–858 [DOI] [PubMed] [Google Scholar]
- 20. Goh S. Y., Cooper M. E. 2008. Clinical review: the role of advanced glycation end products in progression and complications of diabetes. J. Clin. Endocrinol. Metab. 93:1143–1152 [DOI] [PubMed] [Google Scholar]
- 21. Mironova R., Niwa T., Hayashi H., Dimitrova R., Ivanov I. 2001. Evidence for non-enzymatic glycosylation in Escherichia coli. Mol. Microbiol. 39:1061–1068 [DOI] [PubMed] [Google Scholar]
- 22. Fredriksson A., Ballesteros M., Dukan S., Nystrom T. 2005. Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J. Bacteriol. 187:4207–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dubray G., Bezard G. 1982. A highly sensitive periodic acid-silver stain for 1,2-diol groups of glycoproteins and polysaccharides in polyacrylamide gels. Anal. Biochem. 119:325–329 [DOI] [PubMed] [Google Scholar]
- 24. Lehman T. D., Ortwerth B. J. 2001. Inhibitors of advanced glycation end product-associated protein cross-linking. Biochim. Biophys. Acta 1535:110–119 [DOI] [PubMed] [Google Scholar]
- 25. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122:209–220 [DOI] [PubMed] [Google Scholar]
- 26. Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683 [DOI] [PubMed] [Google Scholar]
- 27. Thomas P. H., Kathryn S. P., Virginia L. P., Randell T. L., Carl J. M., Douglas P. C., David L. U., Conlon P. J. 1988. A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat. Biotechnol. 6:1204–1210 [Google Scholar]
- 28. Butland G., Peregrin-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537 [DOI] [PubMed] [Google Scholar]
- 29. Rabbani N., Thornalley P. J. 2008. Dicarbonyls linked to damage in the powerhouse: glycation of mitochondrial proteins and oxidative stress. Biochem. Soc. Trans. 36:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schutt F., Bergmann M., Holz F. G., Kopitz J. 2003. Proteins modified by malondialdehyde, 4-hydroxynonenal, or advanced glycation end products in lipofuscin of human retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 44:3663–3668 [DOI] [PubMed] [Google Scholar]
- 31. Varma A., Young K. D. 2004. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 186:6768–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M., Ho P. Y., Yao S., Shimizu K. 2006. Effect of lpdA gene knockout on the metabolism in Escherichia coli based on enzyme activities, intracellular metabolite concentrations and metabolic flux analysis by 13C-labeling experiments. J. Biotechnol. 122:254–266 [DOI] [PubMed] [Google Scholar]
- 33. Dimitrova R., Mironova R., Ivanov I. 2004. Glycation of proteins in Escherichia coli: effect of nutrient broth ingredients on glycation. Biotechnol. Biotechnol. Equip. 18:99–103 [Google Scholar]
- 34. Sakai Y., Yoshida N., Isogai A., Tani Y., Kato N. 1995. Purification and properties of fructosyl lysine oxidase from Fusarium oxysporum S-1F4. Biosci. Biotechnol. Biochem. 59:487–491 [DOI] [PubMed] [Google Scholar]
- 35. Takahashi M., Pischetsrieder M., Monnier V. M. 1997. Isolation, purification, and characterization of amadoriase isoenzymes (fructosyl amine-oxygen oxidoreductase EC 1.5.3) from Aspergillus sp. J. Biol. Chem. 272:3437–3443 [DOI] [PubMed] [Google Scholar]
- 36. Wiame E., Delpierre G., Collard F., Van Schaftingen E. 2002. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J. Biol. Chem. 277:42523–42529 [DOI] [PubMed] [Google Scholar]
- 37. Loske C., Neumann A., Cunningham A. M., Nichol K., Schinzel R., Riederer P., Munch G. 1998. Cytotoxicity of advanced glycation endproducts is mediated by oxidative stress. J. Neural Transm. 105:1005–1015 [DOI] [PubMed] [Google Scholar]
- 38. Booth I. R., Ferguson G. P., Miller S., Li C., Gunasekera B., Kinghorn S. 2003. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem. Soc. Trans. 31:1406–1408 [DOI] [PubMed] [Google Scholar]
- 39. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahmed N., Furth A. J. 1991. A microassay for protein glycation based on the periodate method. Anal. Biochem. 192:109–111 [DOI] [PubMed] [Google Scholar]
- 41. Rosen R., Buttner K., Schmid R., Hecker M., Ron E. Z. 2001. Stress-induced proteins of Agrobacterium tumefaciens. FEMS Microbiol. Ecol. 35:277–285 [DOI] [PubMed] [Google Scholar]
- 42. Bisswanger H. 1981. Substrate specificity of the pyruvate dehydrogenase complex from Escherichia coli. J. Biol. Chem. 256:815–822 [PubMed] [Google Scholar]
- 43. Hiller J., Franco-Lara E., Weuster-Botz D. 2007. Metabolic profiling of Escherichia coli cultivations: evaluation of extraction and metabolite analysis procedures. Biotechnol. Lett. 29:1169–1178 [DOI] [PubMed] [Google Scholar]
- 44. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. 1977. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene 2:75–93 [DOI] [PubMed] [Google Scholar]
- 46. Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]