Abstract
The diversity of HIV-1 is a confounding problem for vaccine design, as the human immune response appears to favor poor or strain-specific responses to any given HIV-1 virus strain. A significant portion of this diversity is manifested as sequence variability in the loops of HIV-1's surface envelope glycoprotein. Here we show that the most variable sequence positions in the third variable (V3) loop crown cluster to a small zone on the surface of one face of the V3 loop ß-hairpin conformation. These results provide a novel visualization of the gp120 V3 loop, specifically demonstrating a surprising preponderance of conserved three-dimensional structure in a highly sequence-variable region. From a structural point of view, there appears to be less diversity in this region of the HIV-1 “principle neutralizing domain” than previously appreciated.
Introduction
The envelope glycoproteins, gp120 and gp41, found on the surface of infectious HIV-1 are the predominant viral constituents exposed to the human immune system. The gp120 glycoprotein, which mediates the critical binding of virus to cells, consists of a sequence-conserved core, sequence-variable loops, and large numbers of glycosylation sites. The three-dimensional structure of the core of gp120 folds into a compact shape. Of the sequence-variable loops, only the V3 loop has been visualized, pointing outward into the solvent away from the core.1 Overall, the structure suggests an arrangement in which the sequence-variable surface loops form an outer glycosylated shell around the sequence-conserved core of gp120. This outer glycosylated shell shields much of gp120 from antibodies directed against the core whereas the sequence-variable loops display mutations over time that protect them from strain-specific antiloop antibodies.
Sequence variability does not, however, necessarily indicate three-dimensional (3D) structural variability. There are many examples of conserved 3D structures that share no sequence similarity whatsoever.2 We postulated that this could be the case for V3 as, despite its sequence variability, the V3 loop must display structural conservation since it participates in the binding of gp120 to the virus coreceptor, the chemokine receptors CXCR4 and CCR5.3,4 Thus, the functional data suggest that within the sequence variation that characterizes V3, there could be hidden a conserved 3D structure.
Data from the Los Alamos National Database5 show that along the linear sequence of the V3 region, the positions that vary maximally in terms of sequence do not cluster to one linear fragment, but are distributed throughout the V3 loop (Supplemental Fig. 1; see www.liebertonline.com/aid). Furthermore, several crystallographic structures of the V3 loop crown in complex with neutralizing anti-V3 human monoclonal antibodies (mAbs) have been unveiled,6–10 which reveal an epitope consisting of ∼16 amino acids (the V3 crown) with a conserved stem:turn:stem β-hairpin structure. Here, we sought to visualize the primary sequence variability in the V3 loop crown in a new way, i.e., by mapping it into tertiary (3D) structural space.
Materials and Methods
Bioinformatics
V3 loop sequence information was gathered from the Los Alamos National Labs HIV Database5 and organized as follows:
Unfiltered HIV-1 V3 loop sequences were downloaded from the Los Alamos HIV Sequence Database.
Empty sequences (containing no alphanumeric characters), sequences annotated to indicate that they were actually not a V3 loop sequence, and sequences containing a non-amino acid letter, character, or characters were removed from the (1) set.
Only group M sequences were selected from (2) by removing sequences from HIV-1 groups “N,” “O,” and “U.”
Sequences having exactly 35 amino acids were selected from (3). This was necessary to reliably align all the sequences and calculate the positional variability. Sequences with more or less than 35 amino acids are naturally rare and are mostly laboratory strains.
All sequences without a patient ID were excluded and only one sequence per patient was selected. We included this criterion to enforce some measure of clinical relevance, preventing bias toward any one patient.
After filtering, 6010 sequences remained for analysis.
Structures used
The V3 structures used in this chapter are those conformations seen in complex with mAbs 447 (PDB code 1q1j), 2219 (PDB code 2b0s), and F425-B4e8 (PDB Code 2qsc).7–9
Positional sequence variability score
We scored the sequence variability at each position using the equation 1 – (p/n) where p is the number of occurrences of the most common amino acid at that position and n is the total number of sequences analyzed (n = 6010). This score was then converted into a percentage. The scores were normalized by setting the most variable position of the 35 V3 loop positions equal to 100%, and multiplying the remaining values by the same coefficient required to bring that most variable position to 100%. Hydrophobic residues I, L, and V were treated as a single residue since they have very similar volumes, shapes, and physical properties from the point of view of receptor or antibody engagement. Thus, the greater the score, the more amino acid variability there is at that specific position in the V3 loop. The number of occurrences at each position and calculations are shown in Supplemental Table 1 (see www.liebertonline.com/aid).
Visualizing positions of the variable sequences in the 3D structure of V3
The conformation of the 16 amino acid crown of the V3 loop seen in the crystallographic complex of the V3 crown with the 2219 mAb, 8KRKRIHIGPGRAFYTT23, was divided into four structural zones. These four zones are the four distinct protein surfaces of a classical amphipathic ß-hairpin. The left-most Zone 1 is composed of the residues at the base of the ß-hairpin, where the N-terminus of the segment and the C-terminus of the segment start to come together to form the antiparallel ß-strand midsection of the ß-hairpin, in this case, composed of V3 positions 8KRK10 and 23T. Zones 2 and 3 are formed by the opposite faces of the planar, antiparallel ß-strand midsection of the ß-hairpin (as this ß-hairpin is amphipathic). Thus, Zone 2 is hydrophilic and is composed of V3 positions 11R, 13H, 19A, and 22T, and Zone 3 is highly hydrophobic and is composed of V3 positions 12I, 14I, 20F, and 21Y. Zone 4 is the turn of the peptide chain that connects the two antiparallel ß-strands of the ß-hairpin, in this case V3 positions 15GPGR18. The average sequence variability score was derived by calculating the average of the positional sequence variability scores for the residues occurring in each of the four zones.
Cells, plasmids, reagents, and mAbs
The following reagents were obtained through the AIDS Research and Reference Reagent Program: HXB2-env,11 pNL4-3.Luc.R-E-,12,13 pSV-rev,14,15 bicyclam JM-2987 (hydrobromide salt of AMD-3100),16–18 TAK-779,19,20 and U87.CD4.CXCR4 cells.21 The human mAbs used were previously described: anti-CD4bs mAb 1570,22 anti-V3 mAbs 694/98D,23 838-12D,24 and antiparvovirus B19 mAb 1418.25 Recombinant SDF-1α and MIP-1β were purchased as controls (PEPROTECH, Rocky Hiss, NJ). Recombinant HIVIIIB gp120 (Repligen) and HIVSF162 gp120 (courtesy of Dr. Abraham Pinter, The Public Health Research Institute Center, Newark, NJ) were purchased and used as controls.
Site-directed mutagenesis
The 581-bp BglII fragment of the HXB2 env gene was cloned into pCR-Script (Stratagene, La Jolla, CA). The mutagenesis assay was performed on this plasmid with a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The BglII fragment with the mutations was digested from the plasmid and ligated back into the HXB2 env. The mutations were verified by DNA sequencing.
Production of pseudotyped viruses
pNL4-3.Luc.R-E-, pSV-rev, and HXB2-env WT or the vector containing the α-helical V3 mutant were cotransfected into 293T cells using calcium phosphate. Cell-free culture supernatants were collected at 48 h posttransfection and filtered through 0.22-μm filters. The supernatants were centrifuged at 43,000 rpm (Beckman, TL-110 rotor) for 1 h at 4°C. The pelleted virions were resuspended in 1 ml of DMEM and stored at −80°C. The level of p24 in each virus stock was determined using a noncommercial ELISA as described previously.26
Virus infectivity assay
U87.CD4.CXCR4 cells were seeded in 96-well tissue culture plates at a density of 1 × 104 cells/well 1 day prior to the infection. The cells were pretreated with 100 μl of culture medium for 2 h at 37°C prior to infection. The cells were infected with 500 ng p24/well and cultured for 2 days, after which luciferase activity was measured using the Bright-Glo Luciferase assay system (Promega) and a Lumimark Plus luminometer (Bio-Rad, Hercules, CA). A pseudotyped virus, which does not contain the Env protein, was used as a negative control (mock, data not shown). All infectivity assays were done in triplicate.
Results
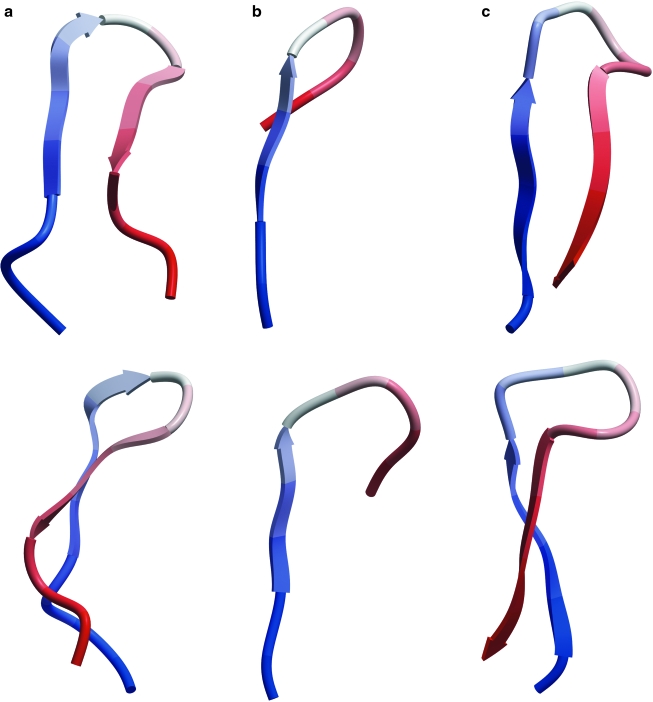
We compared three published 3D conformations of the V3 loop crown that have been visualized by x-ray crystallography. All the structures are ß-hairpins (Fig. 1), as are the structures of the V3 loop crown in situ in gp1201 and at least five newly crystallized x-ray structures of the V3 loop in complex with new anti-V3 mAbs (X-P. Kong, personal communication). Specifically, the conformation of the V3 crown seen in complex with the 2219 mAb exhibits a pure amphipathic ß-hairpin with hydrophobic side chains on one face of the hairpin and hydrophilic residues on the other face.8 The other ß-hairpins are twisted strand versions of this ß-hairpin, as all have a ß-turn at the GPG motif in the V3 loop, including one structure that retains the ß-turn despite substitution of GPG with RPR.8
FIG. 1.
Ribbon diagrams of three HIV gp120 V3 loops complexed with different anti-V3 mAbs, as viewed from the front (top) and side (bottom). Thus, V3 loops are shown in the context of (a) mAb 2219, (b) mAb 447-52D, and (c) mAb F425-B4e8. Loops a and b are from HIV-1 strain MN, whereas c is from strain RP142. Each 3D structure is colored from N-terminal to C-terminal in a blue to red gradient.
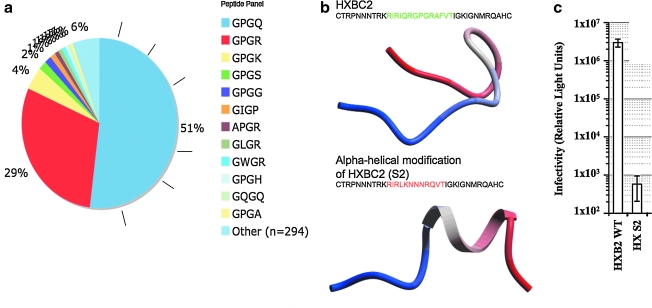
We performed a bioinformatics and protein design analysis to determine that nearly all infective V3 loop crown sequences recorded from HIV-1-infecting patients are ß-hairpin-like and not α-helical (Fig. 2). First, the prediction that all V3 crowns share a ß-hairpin conformation predicts that ß-turn sequence motifs occur almost universally at the location in the V3 crown commonly populated by a GPG motif. In actuality, this is the case (Fig. 2a), so we hypothesized that such a strong selection for a ß-hairpin conformation in the V3 crown must mean that this conformation is somehow required for viral fitness/infectivity. To independently test this hypothesis, we designed a V3 loop with a strong preference for an α-helical fold in its crown region. Indeed, this design was not infective. The near universal signature for a V3 crown ß-hairpin in all recorded HIV-1 sequences, along with the incompatibility of α-helical conformations for viral fitness, suggests that a ß-hairpin conformation is a reasonable 3D visualization of the V3 crown for most, if not all, V3 crown sequences.
FIG. 2.
(a) Plot of the occurrence of the tetrameric motif in the V3 loop crown sequence (at the ß-turn location) in a representative sample of all known V3 loops. Of the recorded motifs occurring in the turn sequence 94% have already been identified in the literature as sequences that strongly predict a ß-turn or ß-hairpin conformation.34 Of the remaining 6%, 5% are sequences that moderately predict a ß-turn and none is a strongly α-helical sequence. (b) Design and testing of a V3 loop with an α-helical crown (designated S2, bottom panel). The ribbon structure shown below each sequence is the ab initio folding of the red or green colored portion of the sequence, a technique previously shown to predict the structural preferences of the V3 loop crown accurately enough to recapitulate crystallographic V3 loop conformations.27 The upper sequence shown in green is the wild-type Hxbc2 V3 crown sequence (HXB2WT). The lower sequence differs only in the amino acids shown in red, which are designed to fold ab initio an α-helical V3 loop sequence (HX S2). (c) Infectivity of HXB2 wild-type (HXB2 WT) and V3 loop engineered HXB2 chimeric pseudotyped (HX S2) viruses. The result shows mean luciferase activity (y-axis) resulting from productive infection of cells with virus ± SD from triplicate wells. The α-helical V3 crown construct HX S2 shows no activity and thus is not infective, whereas HXB2WT shows the full luciferase activity typical of productive infection of cells by virus.
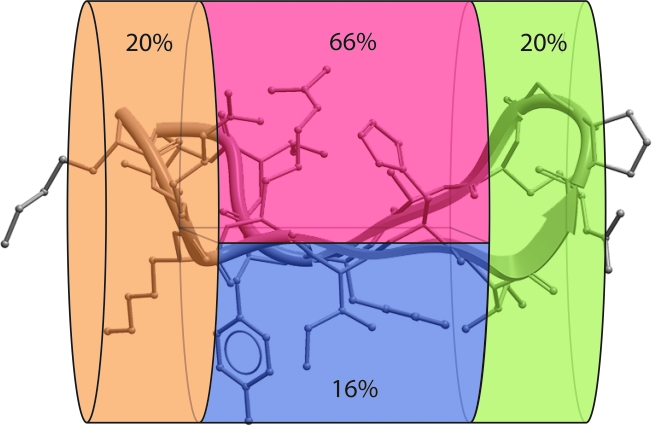
The ß-hairpin conformation seen in complex with Ab 2219 may be the most representative of the various observed V3 loop crown conformations based on two recent observations suggesting that the 2219 ß-hairpin conformation is more energetically stable than the other V3 loop variations observed.27 First, the identical V3 loop sequence of the MN strain of HIV-1 was crystallographically resolved into two different conformations in complex with mAbs 447-52D7 and 2219.8 Ab initio visualization of the dynamic ensemble of conformations of this single sequence recapitulated the observed two crystallographic conformations and demonstrated that the 2219-bound conformation was the lower energy conformation.27 Second, a large set of V3 loop sequences uniformly had greater energetic stability when placed in the 2219-bound conformation as opposed to the conformation seen bound to the mAb 447-52D.27 These observations suggest that the 2219-bound conformation may be the most biologically informative conformation of the several conformations observed in complex with different mAbs or in situ on gp120. Consequently, we mapped the sequence variability of the V3 loop onto the 2219-bound V3 loop crown conformation. Interestingly, when the variable positions found between positions 8 and 23 of the V3 loop crown are mapped onto the 3D structure of the 2219-bound conformation of the V3 loop crown, they cluster into one small continuous protein surface zone on the hydrophilic face of the amphipathic ß-hairpin. The average sequence variability score of the amino acids contributing to this surface is 66% compared to 16–20% for the other surfaces of V3 (Fig. 3).
FIG. 3.
Average sequence variability of V3 loop positions mapped onto the 3D structure of the 2219 mAb-bound conformation of the crown of the V3 loop of clade B strain RP322 consisting of residues 8–23 of the V3 loop 8KRKRIHIGPGRAFYTT23. The V3 loop crown region is divided into four different zones: the left-most division (orange) contains the base of the V3 crown (8KRK10 and 23T). The upper middle division (red) contains the residues pointing their side chains into the hydrophilic face of the amphipathic ß-hairpin (11R, 13H, 19A and 22T). The lower middle division (blue) contains residues that point their side chains into the hydrophobic face of the amphipathic ß-hairpin (12I, 14I, 20F and 21Y). The division on the right (green) contains the conserved ß-turn (15GPGR18). The average sequence variability score, calculated as noted in Materials and Methods, is noted in each region.
Discussion
We show that a backbone ß-hairpin conformation may be nearly universally conserved in the sequence-variable V3 loop crown, and that when mapped onto a canonical ß-hairpin, the side chains of the most sequence-variable positions in the V3 loop crown are restricted to a minority of the overall 3D protein surface of the crown. Indeed, the sequence-variable positions cluster to a single surface zone. Thus, from a 3D structural point of view, the V3 loop is more conserved than variable.
The V3 loop crown is considered to be a flexible structure, flickering between the various ß-hairpin conformations. Alternative conformations may place different positions into the structural zones we have defined here, but our preliminary analysis suggests that the four-dimensional view of the dynamic structure adheres to the understanding we have presented here even better, with at least one of the more variable positions in Zone 4 occupying a location in Zone 2 15% of the time (data not shown). Thus, incorporation of dynamics is expected to further strengthen the view that sequence variability maps strongly to Zone 2.
This new view of the V3 loop as a structurally conserved region of the HIV-1 envelope is not inconsistent with previous studies localizing the ability of virus to bind chemokine receptors to the V3 loop. As the host chemokine receptors do not vary in sequence or structure, the shape within the V3 loop that must interact with these receptors also must not vary. These results show that the amino acids of the V3 loop contributing to this invariant coreceptor-engaging shape are not few, but constitute most of the V3 loop. Indeed, our previous work suggesting that the key residues for coreceptor engagement (11, 24, and 25) cluster to form a single continuous coreceptor binding site reveals how these few positions might depend on a conserved structure throughout the V3 loop crown28: (1) the coreceptor binding cluster of positions 11, 24, and 25 of the V3 loop is located in the stem of the V3 loop crown, which we have found here to be structurally conserved, and (2) these positions are only brought together to form a cluster by a backbone ß-hairpin conformation (including a ß-turn at the tip) that organizes and ramifies through the entire tertiary structure of the V3 loop crown, a ß-hairpin preference that we have here shown is conserved throughout HIV-1 strains. Thus, in this model, the formation of the coreceptor binding site in the V3 loop is dependent on all the atoms of the V3 loop crown except the side chain atoms in Zones 2 and 3 (see Materials and Methods). These are also the atoms comprising most of the conserved structural portion of the V3 loop we have shown in this work, so we can speculate that the reason for the structural conservation is in fact largely coreceptor binding constraints.
Many anti-V3 mAbs are immunologically cross-reactive and display cross-clade neutralizing activity.23,24,29–31 In addition, V3 loops can induce cross-strain neutralizing antibody responses in animals.32,33 These findings demonstrate antigenic conservation in the V3 loop, which is consistent with the new visualization we report here, but at odds with previous reports of the V3 loop giving rise to narrow or type-specific antibodies. Crystallographic structures of broadly neutralizing mAbs 447-52D and 2219 show that these broadly neutralizing mAbs bind to atoms outside the variable zone we have identified,7,8 whereas crystal structures of narrowly or strain-specific antibodies such as 268 bind to at least one side chain within the zone (XP. Kong, personal communication). The obvious implications are that the structurally conserved areas of the V3 loop account for the cross-reactivity of many anti-V3 antibodies: these antibodies bind to an epitope that varies little between strains. Conversely, antibodies binding to the small sequence variable zone are easily escaped by sequence variation and therefore exhibit less cross-reactivity. Thus, structural conservation may coincide with broad neutralization in the V3 loop. If so, many diverse broadly neutralizing antibodies targeting the V3 loop may yet be discovered, as our work shows that the majority of the V3 surface is structurally conserved.
Why, then, are broadly neutralizing anti-V3 loop antibodies not more frequently observed? It is notable that the sequence-variable zone of V3 is populated by polar and charged side chains (the hydrophilic face of the amphipathic ß-hairpin) that, in general, are more solvent exposed in proteins.34 The proclivity of the immune system to first form strain-specific anti-V3 antibodies early after infection or immunization suggests that the small V3 zone of variability may be more exposed to the immune system, perhaps via greater solvent exposure. (The increased occurrence of electrostatic charge in this region may also promote antibody specificity to this region via unknown mechanisms favoring electrostatic charge as a binding glue.) Whatever the mechanism, the implication is that the conserved surfaces of the V3 loop are more antigenically silent, masked, shielded, or buried either by glycans or by the other variable loops, as compared to this sequence variable zone, but that broadly neutralizing mAbs are unavoidable by the virus as the structurally conserved portions must be exposed during the process of infection to engage the chemokine receptors. Thus, this small sequence variable zone is probably selected by viral fitness to be antigenically dominant, resulting in readily observed type-specific mAbs and rare broadly neutralizing antibodies. This view suggests a novel hypothesis that broadly neutralizing antibodies may easily be raised by synthetic V3 loop mimicking immunogens that enhance the antigenicity of the structurally conserved zones, while eliminating or otherwise silencing the antigenicity of the sequence variable zone.
The findings in this work therefore suggest a novel approach for designing immunogens that can induce broadly reactive anti-V3 loop neutralizing antibodies: the new visualization we have described could serve as a blueprint for identifying novel V3 epitopes and/or designing novel V3 mimotopes to the 75% of the V3 loop that structurally appears to be shared across HIV-1 strains. Similarly, the approach we demonstrate here of mapping out the exact locations of sequence variability and structural conservation in “variable regions” may be useful for mining conserved neutralization epitopes in other variable loops of gp120, as well as on surfaces of other antigenically variable pathogens.
Supplementary Material
Acknowledgments
We thank Catarina Hioe for critically reading this manuscript and Jennifer Fuller for manuscript review and contributions to figures. This work was supported by a grant from the Bill and Melinda Gates Foundation (38631 to S.Z-P.) and the NIH (DPOD004631 and R01A1084119 to TC). The NSF provided training funds for the support of D.A. (DGE-0333389).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Huang C. Tang M. Zhang M, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo Conte L. Ailey B. Hubbard T. Brenner S. Murzin A. Chothia C. SCOP: A structural classification of proteins database. Nucleic Acids Res. 2000;28(1):257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trkola A. Dragic T. Arthos J, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 4.Hill C. Deng H. Unutmaz D, et al. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71(9):6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leitner T, editor; Foley B, editor; Hahn B, et al., editors. HIV Sequence Compendium. Los Alamos National Laboratory, NM: Theoretical Biology and Biophysics Group; 2005. No. LA-UR 06-0680. [Google Scholar]
- 6.Burke V. Williams C. Sukumaran M, et al. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 monoclonal antibodies: Potential implications for design of V3 based immunogens. Structure. 2009;17(11):1538–1546. doi: 10.1016/j.str.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanfield R. Gorny M. Williams C. Zolla-Pazner S. Wilson I. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Stanfield R. Gorny M. Zolla-Pazner S. Wilson I. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell C. Pantophlet R. Schiefner A, et al. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J Mol Biol. 2008;375(4):969–978. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhillon A. Stanfield R. Gorny M. Williams C. Zolla-Pazner S. Wilson I. Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set. Acta Crystallogr D Biol Crystallogr. 2008;D64(Pt 7):792–802. doi: 10.1107/S0907444908013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page KA. Landau NR. Littman DR. Construction and use of a human-immunodeficiency-virus vector for analysis of virus infectivity. J Virol. 1990;64(11):5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor RI. Chen BK. Choe S. Landau NR. VPR is required for efficient replication of human-immunodeficiency-virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 13.He J. Choe S. Walker R. Di Marzio P. Morgan D. Landau N. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69(11):6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith AJ. Cho MI. Hammarskjold ML. Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64(6):2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rekosh D. Nygren A. Flodby P. Hammarskjold ML. Wigzell H. Coexpression of human immunodeficiency virus envelope proteins and tat from a single simian-virus 40 late replacement vector. Proc Natl Acad Sci USA. 1988;85(2):334–338. doi: 10.1073/pnas.85.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Declercq E. Yamamoto N. Pauwels R, et al. Highly potent and selective-inhibition of human-immunodeficiency-virus by the bicyclam derivative JM3100. AntimicrobAgents Chemother. 1994;38(4):668–674. doi: 10.1128/aac.38.4.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridger GJ. Skerlj RT. Thornton D, et al. Synthesis and structure-activity-relationships of phenylenebis(methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication––effects of macrocyclic ring size and substituents on the aromatic linker. J Med Chem. 1995;38(2):366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- 18.Hendrix CW. Flexner C. MacFarland RT, et al. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob Agents Chemother. 2000;44(6):1667–1673. doi: 10.1128/aac.44.6.1667-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba M. Nishimura O. Kanzaki N, et al. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96(10):5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dragic T. Trkola A. Thompson DAD, et al. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci USA. 2000;97(10):5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjorndal A. Deng HK. Jansson M, et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71(10):7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeffs SA. Gorny MK. Williams C, et al. Characterization of human monoclonal antibodies selected with a hypervariable loop-deleted recombinant HIV-1(IIIB) gp120. Immunol Lett. 2001;79(3):209–213. doi: 10.1016/s0165-2478(01)00289-9. [DOI] [PubMed] [Google Scholar]
- 23.Gorny MK. Conley AJ. Karwowska S, et al. Neutralization of diverse human-immunodeficiency-virus type-1 variants by an anti-V3 human monoclonal-antibody, 447-52D. J Virol. 1992;66(12):7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorny MK. VanCott TC. Hioe C, et al. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J Immunol. 1997;159(10):5114–5122. [PubMed] [Google Scholar]
- 25.Gigler A. Dorsch S. Hemauer A, et al. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J Virol. 1999;73(3):1974–1979. doi: 10.1128/jvi.73.3.1974-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyambi PN. Nkengasong J. Peeters M, et al. Reduced capacity of antibodies from patients infected with human-immunodeficiency-virus type-1 (HIV-1) group-O to neutralize primary isolates of HIV-1 group-M viruses. J Infect Dis. 1995;172(5):1228–1237. doi: 10.1093/infdis/172.5.1228. [DOI] [PubMed] [Google Scholar]
- 27.Almond D. Tetsuya K. Kong X. Zolla-Pazner S. Cardozo T. Dynamic characterization of the V3 loop crown. Antiviral Ther. 2007;12(Suppl 4):P13–31. [Google Scholar]
- 28.Cardozo T. Kimura T. Philpott S. Weiser B. Burger H. Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23(3):415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- 29.Gorny M. Williams C. Volsky B, et al. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76(18):9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorny M. Xu J. Karwowska S. Buchbinder A. Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150(2):635–643. [PubMed] [Google Scholar]
- 31.Pantophlet R. Aguilar-Sino R. Wrin T. Cavacini L. Burton D. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology. 2007;364(2):441–453. doi: 10.1016/j.virol.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolla-Pazner S. Cohen S. Pinter A, et al. Cross-clade neutralizing antibodies against HIV-1 induced in rabbits by focusing the immune response on a neutralizing epitope. Virology. 2009;392(1):82–93. doi: 10.1016/j.virol.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zolla-Pazner S. Cohen SS. Krachmarov C. Wang SX. Pinter A. Lu S. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology. 2008;372(2):233–246. doi: 10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 34.Kaur H. Raghava GPS. An evaluation of beta-turn prediction methods. Bioinformatics. 2002;18(11):1508–1514. doi: 10.1093/bioinformatics/18.11.1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.