Abstract
SIVsmmFGb is a lentivirus swarm that induces neuropathology in over 90% of infected pigtailed macaques and reliably models central nervous system HIV infection in people. We have previously studied SIVsmmFGb genetic diversity and compartmentalization during acute infection, but little is understood about diversity and intertissue compartmentalization during chronic infection. Tissue-specific pressure appeared to affect the diversity of Nef sequences between tissues, but changes to the Env V1 region and Int diversity were similar across all tissues. At 2 months postinfection, compartmentalization of the SIVsmmFGb env V1 region, nef, and int was noted between different brain regions and between brain regions and lymph nodes. Convergent evolution of the nef and env V1 region, and divergent evolution of int, was noted between compartments and all genes demonstrated intratissue temporal segregation. For the env V1 region and nef, temporal segregation was stronger in the brain regions than the periphery, but little difference between tissues was noted for int. Positive selection of the env V1 region appeared in most tissues at 2 months postinfection, whereas nef and int faced negative selection in all tissues. Positive selection of the env V1 region sequences increased in some brain regions over time. SIVsmmFGb nef and int sequences each saw increased negative selection in brain regions, and one lymph node, over the course of infection. Functional differences between tissue compartments decreased over time for int and env V1 region sequences, but increased for nef sequences.
Introduction
As of December 2007, approximately 33.2 million people worldwide were estimated to be infected with HIV.1–4 Although the immunological and lymphocytic aspects of infection have been extensively studied, the central nervous system (CNS) represents another site of viral pathogenesis. HIV-associated neurocognitive disorders (HAND) are a group of neurological complications caused by direct or indirect viral mechanisms3,5–7 that affect one-third to two-thirds of AIDS patients,3,5,8 with a 6-month mortality rate of 67%.9,10 A common animal model for HIV studies is simian immunodeficiency virus (SIV), a primate lentivirus originally derived from sooty mangabeys, which induces a disease similar to human AIDS in infected pigtailed macaques.4,6,11,12 This model is applicable to neuropathogenesis studies13,14 as infected animals develop SIV encephalitis upon immunosuppression and the development of simian AIDS.11,15,16 Replicable studies with this model are difficult, however, as most SIV strains induce neuropathology in only 25–40% of infected macaques.5,6 Our studies use a reliably neuropathogenic virus strain, SIVsmmFGb, which is neurovirulent in 90–100% of pigtailed macaques.6 Characterization of SIVsmmFGb revealed that this virus is a largely CCR5-tropic swarm rather than a single genetic strain.7,17 As HIV-1 is also a quasispecies in vivo,18,19 SIVsmmFGb could model infection of the primate brain by this genetically diverse, neuropathogenic lentivirus, without the complexities associated with using multiple virus strains.
In our previous study with SIVsmmFGb, we analyzed the genetic diversity and compartmentalization of the env V1 region and the nef and int genes in the CNS and lymph nodes of pigtailed macaques after initial tissue seeding at 1 week postinfection (w.p.i.).17 In that study, we elucidated the genetic composition of our SIVsmmFGb stock virus quasispecies and found that initial seeding of the CNS and peripheral lymphoid tissues reduces the diversity of Env V1 region and integrase amino acid sequences in all tissues, but that changes in the diversity of Nef varied between tissues. In addition, we discovered that all three SIVsmmFGb genes formed separate genetic compartments between different regions within the brain and between the brain regions and the peripheral lymph nodes. For the env V1 region, positive selection was found to play some role in the development of intertissue compartmentalization, whereas negative selection was found to be a factor in the compartmentalization of nef and int.
In that study, we chose to analyze the env V1 region, nef, and int sequences harvested from the CNS at 1 w.p.i., as actively replicating virus is present in many regions of the brain at this early timepoint.20,21 Whereas sequences harvested at 1 w.p.i. represent the virus that initially seeds the tissues, viral load setpoints have yet to be established and the virus is still undergoing active replication.16,20,22–24 With continued replication, the virus may potentially acquire additional mutations and, perhaps, evolve differently in each tissue due to variations in selective pressure between compartments. Other studies have shown that SIV replication in the brain is suppressed by 3 w.p.i. and actively replicating virus cannot be detected again in the CNS until over 9 w.p.i.16,20,25 During this time of heavily suppressed replication, we would not expect the virus to accumulate any more mutations or undergo further evolution.
For this follow-up study, we harvested sequences from the peripheral lymph node tissues and different regions of the brain at 2 months postinfection (m.p.i.) during viral latency in the CNS. Sampling at this timepoint allows for analysis of the genetic diversity and compartmentalization of the SIVsmmFGb envelope (env, Env) V1 region, nef (nef, Nef ), and integrase (int, Int) genes following the establishment of viral setpoints and suppression of replication in the CNS. We analyzed the diversity of the SIVsmmFGb envelope (env, Env V1), nef (nef, Nef ), and integrase (int, Int) sequences harvested from the CNS and lymphoid tissues of infected pigtailed macaques at 2 m.p.i. and compared this diversity to those genes in our SIVsmmFGb stock virus. We also investigated whether intertissue genetic compartmentalization of the SIVsmmFGb env V1 region, nef, and int sequences was present at 2 m.p.i. In addition, we analyzed changes in intertissue compartmentalization of the SIVsmmFGb genes over the course of infection by comparing our results to those obtained from our previous analyses of viral sequences harvested at 1 w.p.i.17
Materials and Methods
Animal inoculation and tissue harvesting
Six juvenile pigtailed macaques from the Yerkes National Primate Research Center were inoculated intravenously with 100 TCID50 of SIVsmmFGb stock virus, produced as described previously.6 Each animal was inoculated and processed individually, with a triaging protocol used during necropsy to reduce variability and ensure tissue collection from matching anatomic sites. All macaques were euthanized at 2 m.p.i. by a barbiturate overdose and were extensively perfused with saline to prevent contamination of the CNS tissues by the blood. Samples of blood and cerebrospinal fluid (CSF) were harvested prior to sacrifice for the analysis of viral loads and for other studies. Tissue samples were collected from the axillary lymph node, mesenteric lymph node, basal ganglia, midfrontal cortex, hippocampus, and cerebellum of each animal, and the samples were quick-frozen on dry ice and stored at −80°C.
DNA extraction and preparation
Small segments were removed from each tissue sample with sterile scalpels, working on ice to minimize tissue decomposition. The excised fragments were placed in cell lysis buffer containing 0.1 M NaCl, 10 mM Tris, pH 8.0, 25 mM EDTA, 0.5% SDS, and ddH2O and then homogenized with a 1.0-ml syringe plunger. The samples were treated with 50 μg proteinase K, incubated overnight at 55°C, and periodically mixed by vortexing. The samples were then brought to room temperature and the DNA harvested by a sequential extraction with Tris-saturated phenol, phenol:chloroform:isoamyl alcohol (25:24:1), chloroform, and 100% ethanol. The resulting DNA was pelleted by a 15,000-rpm centrifugation for 30 min at 4°C, washed three times with 70% ethanol, air dried, and resuspended in ddH2O. The concentration of all DNA samples was quantified using a UV spectrophotometer and the samples were stored at −20°C.
PCR amplification of SIVsmmFGb genes from tissue-derived proviral DNA and cloning strategy
The env V1 region, nef, and int genes were amplified from tissue proviral DNA by nested polymerase chain reaction (PCR) with primers described previously17 using an Expand High Fidelity PCR System kit (Roche), according to the manufacturer's protocol. These PCR products were purified on 0.9% (env) or 1.2% (nef, int) agarose gels, extracted with a QIAquick Gel Extraction Kit (Qiagen), and then ligated into the pGEM-T Vector System I (Promega). After 24 h of incubation at 4°C, ligations were transformed into Invitrogen ElectroMAX DH10B Escherichia coli cells (recA1, endA1) according to the manufacturer's protocol. Single bacterial colonies were used to prepare plasmid DNA containing the viral gene inserts, according to standard techniques. Approximately 2 μg of each p-GEM-T vector/gene insert-positive sample was sent to MWG Biotech for sequencing (MWG Sequencing, Huntsville, AL). The sequencing reactions were performed using pGEM-T vector-specific sequencing primers described previously.17
DNA sequence analysis
The SIVsmmFGb gene sequences were analyzed with EditSeq in the DNAStar v7.1.0 software package (Lasergene) and the following sequences were discarded: poor reads, junk sequences, incomplete nef and int sequences, nef and int clones with deletions or insertions, and env sequences that did not span the complete V1 region. Valid clones were copied into MegAlign in DNAStar v7.10 (Lasergene), translated into amino acid sequences, and those clones with premature stop codons were discarded. At least 20 valid clones for each gene were recovered from each tissue, from each experimental animal.
Tissue-isolated viral gene sequence alignment and grouping
Sequences for the env V1 region, nef, and int were harvested from the SIVsmmFGb stock virus and each gene was broken into groups as described.17 Valid amino acid sequences, determined for each gene from each tissue in each experimental animal, were aligned with the stock virus subgroup consensuses for that gene via the Clustal W method in MegAlign. These alignments were exported to PAUP* 4.0b10 and neighbor-joining phylogenetic trees were generated, using both distance- and parsimony-based analyses, with no differences noted between the two methods (data not shown). The sequences isolated from the tissues were sorted into groups defined by the stock virus analysis and the mean prevalence of each sequence group in each tissue across all animals was determined as described.17 The mean prevalence of each sequence group in the tissues of the animals sacrificed at 2 m.p.i. was compared statistically to the prevalence of that group in the SIVsmmFGb stock virus17 using the Mann–Whitney rank-sum test in SigmaStat Demo 2.03 software (Systat). The prevalence of each sequence group was pooled for all regions of the brain and all lymph node tissues and then averaged to determine the mean prevalence for each sequence group in the CNS and in the lymphoid tissues. The mean prevalences of each group in the lymph node and brain pools were then compared statistically using the Mann–Whitney rank sum test. These values were also compared to the prevalence of each sequence group in the SIVsmmFGb stock virus. The Mann–Whitney test was used due to the nonnormal distributions of group prevalence between animals as well as the unequal variance between groups from the stock virus and those from the tissues. Those Mann–Whitney comparisons with a p-value less than or equal to 0.05 were considered statistically significant.
Phylogenetic and phenetic compartmentalization analyses
The phylogenetic compartmentalization of the Env V1 region, Nef, and Int amino acid sequences between each of the six tissues was analyzed using a modified Slatkin–Maddison test, as described elsewhere.17,26,27 Compartmentalization of sequences between a pair of tissues was considered significant if the ratio of the global mean bootstrap s, where s is the least number of evolutionary steps for a “tissue of origin” character, to the global mean random s was at least two standard errors less than 1.26,28 This procedure was repeated for every possible two-tissue comparison for each of the three viral genes. All sequences for each gene, harvested from each tissue at 2 m.p.i., were aligned with all sequences harvested from the same tissue at 1 w.p.i.17 using the Clustal W method in MegAlign. A modified Slatkin–Maddison test was then performed on each of these alignments as described.17,26,27 Compartmentalization of sequences within a tissue over time was considered significant if the ratio of the mean bootstrap s value, where s is the least number of evolutionary steps for a “timepoint” character, to the mean random s value was two standard errors less than 1.26,28
The phenetic compartmentalization of env V1, nef, and int nucleic acid sequences between each of the six tissues at 2 m.p.i. was compared using a Mantel's test, as described elsewhere.17,26–28 The null hypothesis, of no compartmentalization between tissues, was rejected for p-values of less than, or equal to, 0.05.
Synonymous and nonsynonymous substitution analysis
The selective pressure on the env V1 region, nef, and int in each tissue compartment was analyzed by determining the synonymous (dS) and nonsynonymous (dN) nucleotide substitution rates, as well as the ratio of dS to dN. The mean dS and mean dN values for each sequence of each gene, from each tissue harvested at 2 m.p.i., were determined relative to the SIVsmmFGb stock virus consensus sequence for that gene, as previously described.17 The mean dS and mean dN values for each tissue were used to generate the dS/dN ratio value for each gene in each tissue, across all experimental animals. The Mann–Whitney rank sum test was used to compare the mean dS, mean dN, and dS/dN ratio values between tissues and to compare the mean dS and mean dN values within each tissue. Those Mann–Whitney comparisons with p-values of less than, or equal to, 0.05 were considered statistically significant.
Results
Diversity of Env V1 region, Nef, and Int amino acid sequences, isolated from pigtailed macaque brain regions and lymph node tissues at 2 months postinfection, compared to the SIVsmmFGb stock virus and sequences harvested 1 week postinfection
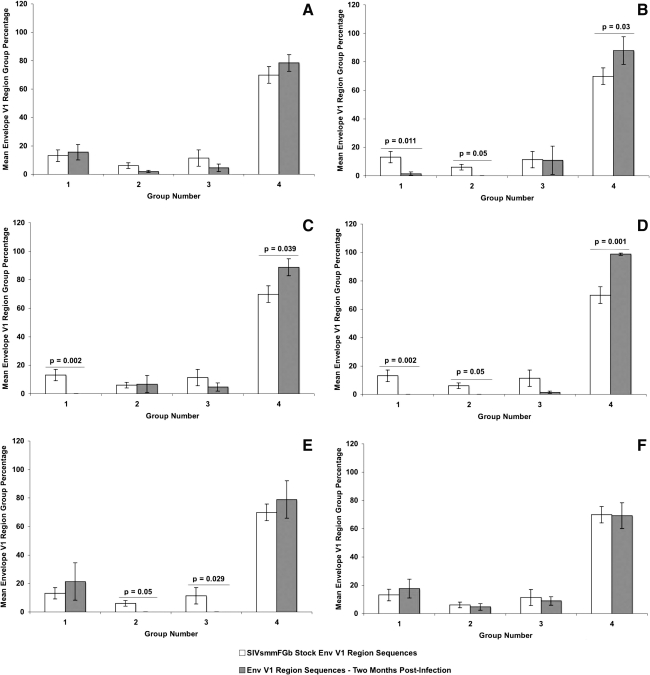
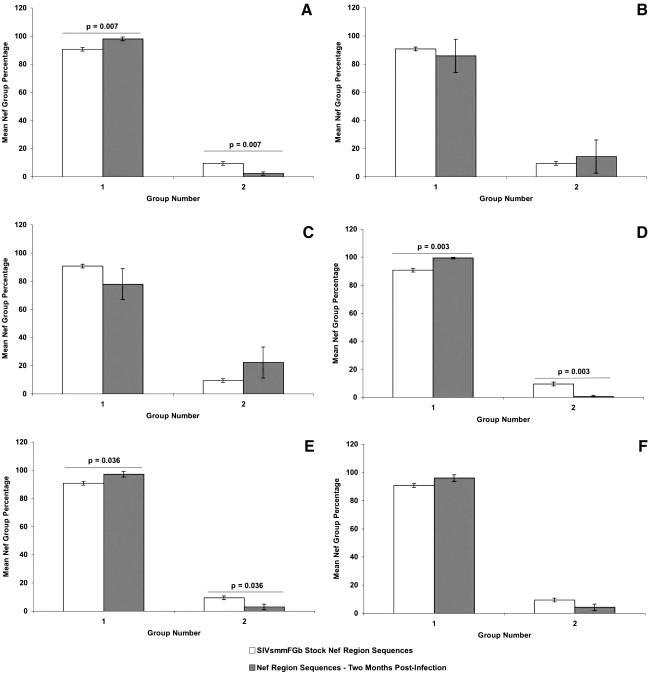
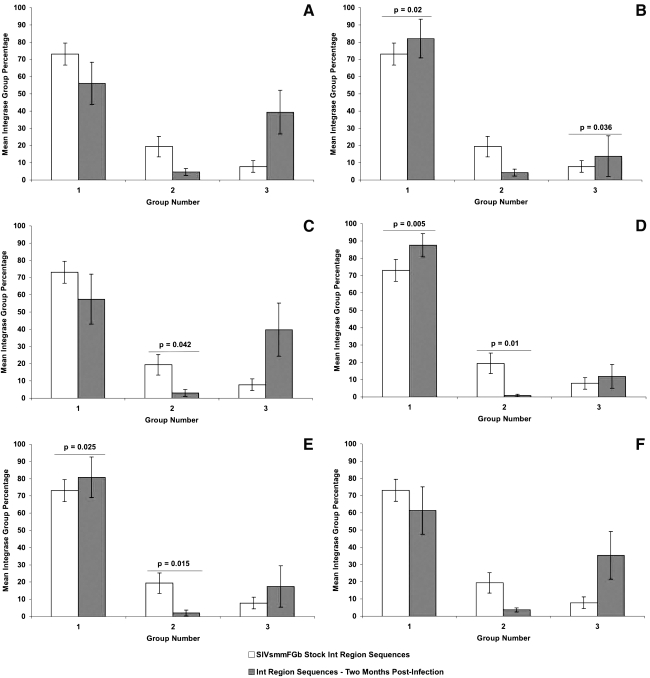
In our previous study, we determined that four groups of Env V1 region, two groups of Nef, and three groups of Int amino acid sequences were present in our SIVsmmFGb stock virus.17 We also analyzed how the prevalence of each group of each gene changed after 1 w.p.i. in the pigtailed macaque host.17 Here, we were interested in determining how the prevalence of those amino acid sequence groups, harvested from infected pigtailed macaques at 2 m.p.i., compared to the prevalence of those groups in the SIVsmmFGb stock virus. For the Env V1 region (Fig. 1), the prevalence of sequence groups 1 and 2 decreased in the basal ganglia, compared to the SIVsmmFGb stock virus, whereas group 4 saw an increase in prevalence in this region of the brain. The hippocampus had a reduced prevalence of Env V1 region sequence groups 2 and 3, compared to the stock virus. An increase in the prevalence of Nef sequence group 1, and a decrease in the prevalence of group 2, was noted in the axillary lymph node, midfrontal cortex, and hippocampus (Fig. 2). The Int sequence group 1 prevalence increased in the basal ganglia, midfrontal cortex, and hippocampus, compared to the stock virus (Fig. 3). The prevalence of Int sequence group 2 decreased in the cerebellum, midfrontal cortex, and hippocampus, whereas the prevalence of group 3 decreased in the basal ganglia.
FIG. 1.
Comparison of Env V1 region group percentages between SIVsmmFGb stock virus and tissues harvested from pigtailed macaques at 2 m.p.i. Env V1 region amino acid sequences obtained from the SIVsmmFGb stock virus were aligned and grouped as previously described.17 Env V1 amino acid sequences from tissues harvested at 2 m.p.i. were grouped as described in Materials and Methods. The percentage of sequences in each group from each tissue was determined for each animal and averaged to yield a mean percentage for each group in each tissue across all animals. The percentage of each group in the SIVsmmFGb stock virus was compared statistically to the mean percentage of each group in the tissues using the Mann–Whitney rank sum test. Statistically significant differences (p < 0.05) are noted. Error bars represent 1 standard error. Results are for Env V1 region sequences from the (A) axillary lymph node, (B) basal ganglia, (C) cerebellum, (D) midfrontal cortex, (E) hippocampus, and (F) mesenteric lymph node.
FIG. 2.
Comparison of Nef group percentages between SIVsmmFGb stock virus and tissues harvested from pigtailed macaques at 2 m.p.i. Nef amino acid sequences obtained from the SIVsmmFGb stock virus were aligned and grouped as previously described.17 Nef amino acid sequences from tissues harvested at 2 m.p.i. were grouped as described in Materials and Methods. The percentage of sequences in each group from each tissue was determined for each animal and averaged to yield a mean percentage for each group in each tissue across all animals. The percentage of each group in the SIVsmmFGb stock virus was compared statistically to the mean percentage of each group in the tissues using the Mann–Whitney rank sum test. Statistically significant differences (p < 0.05) are noted. Error bars represent 1 standard error. Results are for Nef sequences from the (A) axillary lymph node, (B) basal ganglia, (C) cerebellum, (D) midfrontal cortex, (E) hippocampus, and (F) mesenteric lymph node.
FIG. 3.
Comparison of Int group percentages between SIVsmmFGb stock virus and tissues harvested from pigtailed macaques at 2 m.p.i. Int amino acid sequences obtained from the SIVsmmFGb stock virus were aligned and grouped as described.17 Int amino acid sequences from tissues harvested at 2 m.p.i. were grouped as described in Materials and Methods. The percentage of sequences in each group from each tissue was determined for each animal and averaged to yield a mean percentage for each group in each tissue across all animals. The percentage of each group in the SIVsmmFGb stock virus was compared statistically to the mean percentage of each group in the tissues using the Mann–Whitney rank sum test. Statistically significant differences (p < 0.05) are noted. Error bars represent 1 standard error. Results for Int sequences are from the (A) axillary lymph node, (B) basal ganglia, (C) cerebellum, (D) midfrontal cortex, (E) hippocampus, and (F) mesenteric lymph node.
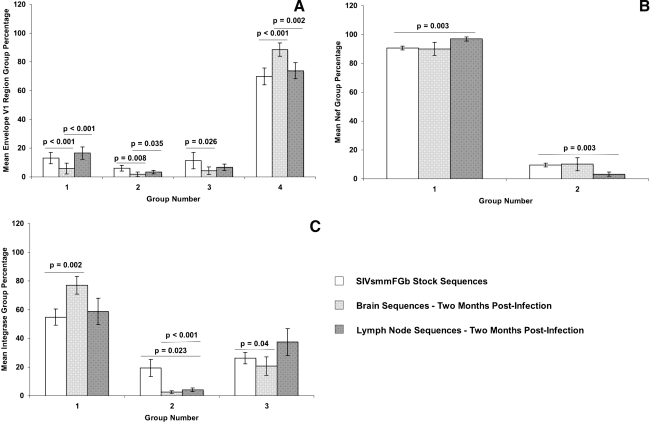
We also compared the average group prevalence for the Env V1 region, Nef, and Int in pooled CNS region sequences to mean group prevalence in the stock virus and the pooled lymph node tissue sequences. Env V1 region sequence group 4 demonstrated an increased prevalence in the pooled CNS sequences, whereas groups 1–3 decreased, compared to the SIVsmmFGb stock virus (Fig. 4A). Env V1 region sequence group 1 and 2 prevalence was lower in the CNS than the lymph nodes, whereas the presence of group 4 was higher in the CNS. For Nef, the presence of sequence group 1 was lower in the pooled CNS sequences, compared to the stock virus, with a corresponding increase in the prevalence of group 2 (Fig. 4B). The prevalence of Nef group 1 increased in the pooled lymph node sequences, compared to the stock virus, whereas the prevalence of group 2 declined. Int sequence groups 2 and 3 had a lower prevalence in the pooled CNS sequences compared to the SIVsmmFGb stock virus, but the prevalence of group 1 was higher in the CNS pool (Fig. 4C). The level of Int group 2 was lower in the lymph node pool relative to the stock virus. There was no significant difference seen in Nef or Int sequence group prevalence between the CNS and lymph node sequence pools.
FIG. 4.
Comparison of sequence group percentages between the SIVsmmFGb stock virus and sequences harvested from the CNS and lymph node at 2 m.p.i. Amino acid sequences for all three genes obtained from the SIVsmmFGb stock virus were aligned and grouped as described.17 Amino acid sequences for the (A) Env V1 region, (B) Nef, and (C) Int harvested from tissues at 2 m.p.i. were grouped as described in Materials and Methods. Sequence group percentages for all CNS tissues from all animals were averaged to yield a mean percentage for each group in the CNS, with a similar procedure conducted for lymph node-derived sequences. The percentage of each group in the SIVsmmFGb stock was compared statistically to the mean group percentage in CNS and lymph nodes using the Mann–Whitney rank sum test. A similar comparison was performed between the means of the CNS and lymph node pools. Statistically significant differences (p < 0.05) are noted and error bars represent 1 standard error.
The prevalence of each sequence group, for each gene in each tissue, was also analyzed in the experimental animals individually. Five of the six animals sacrificed at 2 m.p.i. had group 4 as the predominant Env V1 region group in all tissues analyzed, whereas the most prevalent group in animal PHt1 varied between tissues (Supplemental Fig. 1; see www.liebertonline/aid). For Nef, group 1 sequences were predominant in the lymph nodes, the midfrontal cortex, and the hippocampus of all six animals, as well as in the basal ganglia and cerebellum of most animals (Supplemental Fig. 2; see www.liebertonline.com/aid). Although Int group 1 was generally the most prevalent sequence group, four animals had group 3 dominant, or at similar levels to group 1, in at least one lymphoid tissue or brain region (Supplemental Fig. 3; see www.liebertonline.com/aid).
Phylogenetic compartmentalization analysis of Env V1 region, Nef, and Int amino acid sequences isolated from pigtailed macaque brain regions and lymphoid tissues 2 months postinfection
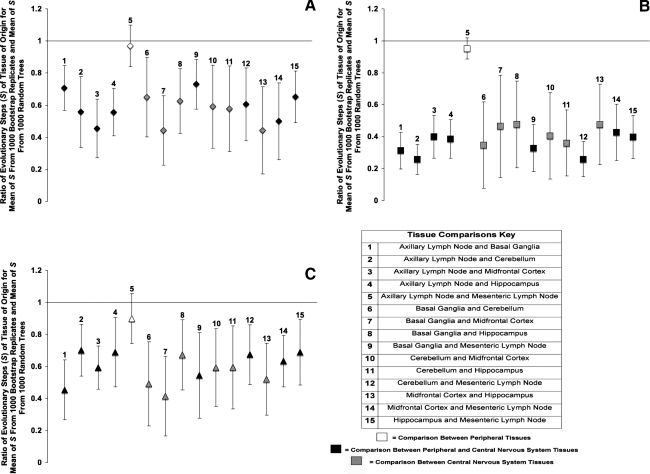
We utilized two methods of analysis to examine the compartmentalization of the Env V1 region, Nef, and Int sequences obtained from the various tissues. The first test, the modified Slatkin–Maddison test, is a phylogenetic method of compartmentalization analysis. Based on the results of this analysis, the Env V1 region amino acid sequences from each region of the brain compartmentalized separately from all other CNS regions and from the lymph nodes, but compartmentalization between the lymph nodes did not appear (Fig. 5A). Similar results were found for the Nef (Fig. 5B) and Int (Fig. 5C) sequences. The Env V1 and Int sequences had a similar degree of compartmentalization, with average s ratio values of 0.61, whereas Nef sequences had the strongest compartmentalization, with an average s ratio of 0.42.
FIG. 5.
Phylogenetic analysis of SIVsmmFGb compartmentalization between tissues harvested from pigtailed macaques at 2 m.p.i. (A) Env V1 region, (B) Nef, and (C) Int compartmentalization was determined using a modified Slatkin–Maddison test as described in Materials and Methods. Error bars represent 2 standard errors of the determined S ratio; significant compartmentalization between tissues is indicated by ratios more than 2 standard errors less than 1.
Phenetic compartmentalization analysis of Env V1 region, Nef, and Int nucleotide sequences isolated from pigtailed macaque brain regions and lymphoid tissues 2 months postinfection
The second test we used to investigate the compartmentalization of Env V1 region, Nef, and Int sequences was the Mantel's test, a phenetic method of analysis. Table 1 summarizes the Mantel's test results on each of 15 two-tissue comparisons of env V1 region, nef, and int nucleotide sequences from all experimental animals. This summary table indicates the number of animals in the cohort that had statistically significant compartmentalization of the env V1 region, nef, or int by Mantel's test in each of the intertissue comparisons. Due to the size of the data sets, a full listing of Pearson's correlation coefficients and p-values for the env V1 region, nef, and int nucleotide sequences can be found in Supplemental Tables 1–3 (see liebertonline.com/aid). The env V1 region sequences obtained from each region of the CNS compartmentalized separately from the other brain regions, and from sequences derived from the lymph nodes, in a majority of animals. However, the env V1 region sequences from the basal ganglia compartmentalized separately only from the axillary lymph node and hippocampus in half of the experimental animals. Compartmentalization of the env V1 region sequences between the lymph nodes appeared in only one test animal. For both nef and int, the sequences compartmentalized separately between CNS regions, as well as between the CNS and the lymph nodes, in a majority of animals. Although none of the experimental animals demonstrated compartmentalization of nef between the lymph nodes, the axillary and mesenteric lymph nodes of one subject did have compartmentalization of int sequences.
Table 1.
Proportion of SIVsmmFGb-Infected Pigtailed Macaques with Compartmentalization Between Compared Tissues as Determined by Mantel's Testa
| Tissue comparison | env V1 | nef | int |
|---|---|---|---|
| A × B | 3/6 | 6/6 | 6/6 |
| A × C | 4/6 | 6/6 | 5/6 |
| A × F | 5/6 | 6/6 | 6/6 |
| A × H | 4/6 | 6/6 | 6/6 |
| A × M | 1/6 | 0/6 | 1/6 |
| B × C | 5/6 | 5/6 | 5/6 |
| B × F | 5/6 | 4/6 | 6/6 |
| B × H | 3/6 | 5/6 | 4/6 |
| B × M | 4/6 | 6/6 | 5/6 |
| C × F | 5/6 | 5/6 | 5/6 |
| C × H | 4/6 | 6/6 | 4/6 |
| C × M | 4/6 | 6/6 | 6/6 |
| F × H | 4/6 | 4/6 | 5/6 |
| F × M | 5/6 | 6/6 | 6/6 |
| H × M | 5/6 | 6/6 | 5/6 |
A, axillary lymph node; B, basal ganglia; C, cerebellum; F, midfrontal cortex; H, hippocampus; M, mesenteric lymph node.
Synonymous and nonsynonymous nucleotide substitution rates of the SIVsmmFGb env V1 region, nef, and int sequences isolated from pigtailed macaque brain regions and lymph node tissues 2 months postinfection
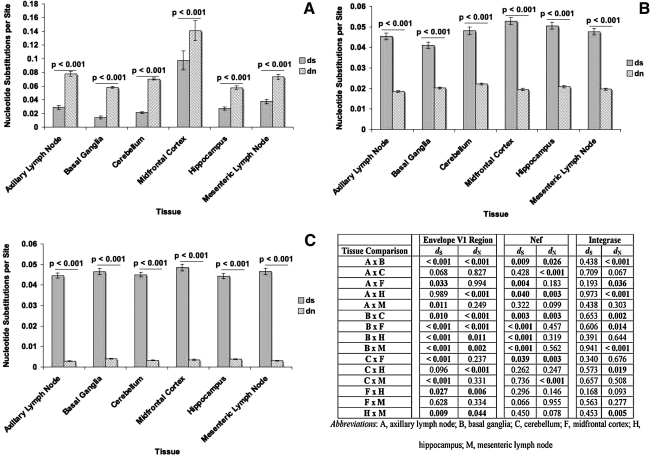
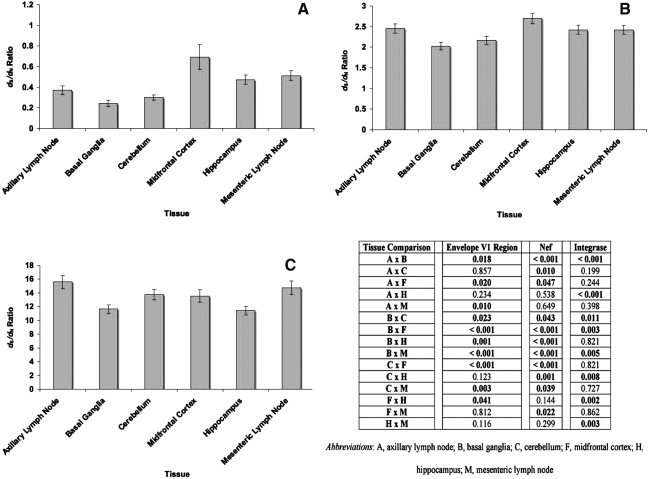
To determine whether the env V1 region, nef, and int genes were undergoing positive or negative selection at 2 m.p.i., we next analyzed dS and dN for all of the sequences, as well as the ratio of dS to dN. The env V1 region sequences harvested from all six of the experimental tissues had significantly higher dN values than dS values (Fig. 6A), as well as dS/dN ratio values of less than 1 (Fig. 7A). The basal ganglia-derived env V1 region sequences had the lowest mean dS value, whereas the sequences obtained from the cerebellum had a lower average dS value than those obtained from the midfrontal cortex and mesenteric lymph node (Fig. 6A and D). The average dS values of the env V1 region sequences from the mesenteric lymph node and midfrontal cortex were significantly higher than the dS values of sequences from the axillary lymph node and the other brain regions. The lowest mean dN value appeared for the env V1 region sequences from the hippocampus. Although the average dN value of the env V1 region sequences from the basal ganglia was higher than those in the hippocampus, it was lower than in both lymph nodes and the remaining two regions of the CNS. The env V1 region sequences from the basal ganglia had the lowest mean dS/dN ratio value, whereas sequences from the cerebellum, axillary lymph node, and hippocampus all had average dS/dN values lower than those sequences from the midfrontal cortex (Fig. 7A and D). The average dS/dN value of the env V1 region sequences from the mesenteric lymph node was higher than the mean dS/dN values of sequences from the basal ganglia, cerebellum, and axillary lymph nodes.
FIG. 6.
dS and dN values for (A) env V1 region, (B) nef, and (C) int sequences isolated from tissues of SIVsmmFGb-infected pigtailed macaques sacrificed at 2 m.p.i. Average dS and dN values for each tissue were determined as described and compared statistically using the Mann–Whitney rank sum test as described in Materials and Methods. Statistically significant differences (p < 0.05) are noted. Error bars represent 1 standard error. dS and dN intertissue comparisons, by the Mann–Whitney rank sum test, for all three genes are provided in the accompanying table; statistically significant differences (p ≤ 0.05) are noted.
FIG. 7.
Average dS/dN values for (A) env V1 region, (B) nef, and (C) int sequences isolated from tissues of SIVsmmFGb-infected pigtailed macaques sacrificed at 2 m.p.i. Mean dS/dN values for each tissue were determined, and statistical comparisons were made, as described in Materials and Methods. Error bars represent 1 standard error. dS dN ratio intertissue comparisons for env V1 region, nef, and int are provided in the accompanying table; statistically significant differences (p ≤ 0.05) by the Mann–Whitney rank sum test are noted.
For the nef sequences, the mean dS values were significantly higher than the mean dN values in all tissues and all tissues had an average nef dS/dN ratio value greater than 1 (Figs. 6B and 7B). The basal ganglia-derived nef sequences had the lowest mean dS value, whereas the mean dS value for nef sequences from the hippocampus was only higher than the mean dS value in the axillary lymph node (Fig. 6B and D). The highest mean dS value was for those nef sequences obtained from the midfrontal cortex. Those nef sequences obtained from the cerebellum had the highest mean dN value, whereas the sequences from the hippocampus and lymph nodes had the lowest mean dN values. The nef sequences from the basal ganglia had the lowest average dS/dN ratio, whereas sequences from the cerebellum had a mean dS/dN ratio lower than the lymph nodes and the remaining regions of the CNS. The highest mean dS/dN value appeared in the nef sequences from the midfrontal cortex (Fig. 7B and D).
The int sequences also had significantly higher mean dS values than mean dN values in all six tissues, and average dS/dN ratios than one (Figs. 6C and 7C), but no differences in mean dS values between tissues were noted (Fig. 6C and D). The midfrontal cortex-derived int sequences had a mean dN value higher than those from the axillary lymph node, but a mean dN value lower than sequences from the basal ganglia. The mean dN value of int sequences from the hippocampus was significantly higher than the cerebellum and the lymph nodes, whereas sequences from the basal ganglia had a mean dN value higher than that of all other tissues except the hippocampus. The average dS/dN values for int sequences obtained from the hippocampus and basal ganglia were significantly lower than the mean dS/dN values of sequences from all other tissues.
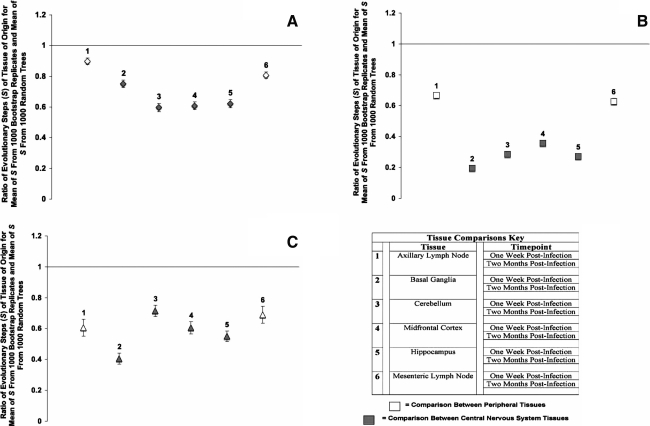
Photogenetic analysis of temporal compartmentalization of envelope VI region, Nef, and integrase amino acid sequence within pigtailed Macaque brain and lymph node tissues between 1 week and 2 months postinfection
We were also interested in determining whether SIVsmmFGb sequences compartmentalized within a particular tissue over time. Thus, we performed a modified Slatkin-Maddison test comparing sequences obtained from a tissue at 1 w.p.i.,17 with sequences obtained from the same tissue at 2 m.p.i. Env V1 region amino acid sequences compartmentalized separately within each tissue, between time points, in all tissue studied (Fig. 8A). While Env V1 region sequences from the basal ganglia had the lowest degree of temporal segregation of all the CNS regions, all brain tissues demonstrated stronger temporal compartmentalization of Env V1 region than either lymph node. Similar results were obtained for Nef, with temporal compartmentalization of this gene in all tissues, but a higher degree of segregation in the CNS over time (Fig. 8B). Nef sequences from the basal ganglia appeared to have the strongest intra-tissue temporal compartmentalization of all the CNS regions, while sequences from the midfrontal cortex had the weakest degree of temporal segregation. All tissues demonstrated temporal compartmentalization of Int (Fig. 8C). The Int sequences from the midfrontal cortex, cerebellum, and lymph nodes had similar degrees of intra-tissue temporal compartmentalization, while basal ganglia-derived Int sequences had the highest degree of temporal segregation over time. The sequences with the lowest degree of temporal segregation were the Env V1 region samples, with an average s ratio value of 0.71, while the Nef sequences had the highest, with a mean s ratio value of 0.40. The Int sequence fell in between, with an average s ratio value of 0.60.
FIG. 8.
Phylogenetic analysis of temporal compartmentalization of SIVsmmFGb sequences within the same tissue harvested from pigtailed macaques at 1 w.p.i. and 2 m.p.i. (A) Env V1 region, (B) Nef, and (C) Int compartmentalization was determined using a modified Slatkin–Maddison test as described in Materials and Methods. Error bars represent 2 standard errors of the determined S ratio; significant compartmentalization between tissues is indicated by ratios more than 2 standard errors less than 1. Some sequences for the Env V1 region, Nef, and Int could not be obtained from animal PQo1 at 1 w.p.i. as described.17
Discussion
Although we have previously studied SIVsmmFGb neuropathogenesis and genetic diversity at 1 w.p.i.,5,6,17 this follow-up study explores the genetic diversity and evolution of SIVsmmFGb at 2 m.p.i. In studies with SIV infection of the CNS, viral replication has been detected in multiple regions of the brain for a period of up to 2 w.p.i.17,20 Subsequently, viral RNA within the CNS becomes undetectable by 3 w.p.i. and active replication, as measured by viral RNA, does not become elevated again until 8–12 w.p.i.16,25,29 The exact mechanism that suppresses SIV RNA expression within the CNS from 3 to 8 w.p.i. has yet to be determined, however, research suggests that beta-interferon induces production of LIP, a dominant negative isoform of the transcription activator LAP, in macrophage and related cells.30–32 Interaction of LIP with the proviral LTR may then block viral transcription, preventing replication and inducing a latent infection in the macrophage.33–38 Based on these SIV models, we would expect SIVsmmFGb replication in the CNS to be completely suppressed before 2 m.p.i. Analyzing SIVsmmFGb genes at 2 m.p.i. allows us to assess the virus population that emerges following completion of the initial burst of replication and evolution during the early weeks of infection. The choice to sample at 2 m.p.i. was balanced between choosing a timepoint late enough for the initial round of viral replication in the CNS to be complete, but early enough that the virus would not have emerged from suppression in most animals.
Prior studies of SIVsmmFGb have shown that rapid progressors do not demonstrate symptoms until at least 3 m.p.i.,5 so we did not specifically monitor the animals in this study for overt clinical symptoms of lentiviral neuropathogenesis. Still, none of the animals in this study were noted to have clinical signs of SIVsmmFGb neuropathogenesis at sacrifice. Although research has not specifically addressed suppression of SIVsmmFGb replication in the CNS, based on the studies of other SIV strains discussed above, we would expect actively replicating SIVsmmFGb in the CNS to be present at low/undetectable levels in most animals at 2 m.p.i. Our rationale for focusing on the SIVsmmFGb env V1 region, nef, and int genes, as well as our choice of tissues to sample, is discussed in our previous report.17 To elaborate on our choice of tissues, prior studies with SIVsmmFGb have shown the development of lesions in the midfrontal cortex, cerebellum, and hippocampus of rapidly progressing animals, indicating these regions of the brain are involved in neuropathogenesis.5
A number of potential issues with our methodology, and the steps we undertook to minimize these biases, are described in our previous report.17 One other potential flaw is that the tissue samples we harvested likely contain a number of different cell types that may support SIV infection. The majority of SIV-infected cells in the CNS are of the monocytic cell lineage (macrophages and microglia), which harbor productive viral infection, when replication is not suppressed.21,39–44 However, astrocytes and oligodendrocytes may also be infected with SIV,45–50 yielding only a transient productive infection, followed by long-term viral latency.39,51,52 Given the limited replication of the virus in non-macrophage-related cells, we would expect only limited selective pressure on SIVsmmFGb viruses infecting these cell types. Thus, infection of oligodendrocytes and astrocytes would not be expected to contribute significantly to the evolution of the SIVsmmFGb genes analyzed in this study. In macrophage-related cells, where virus replicates extensively during the early weeks of infection, viral genes should face increased selective pressure, contributing to the majority of SIVsmmFGb sequence evolution. However, the methods used in our studies do not allow us to determine from which cells our gene sequences were obtained. Given that most infected cells in the CNS are macrophage related, we would expect that most of our sequences originated from these cell types, with astrocyte- or oligodendrocyte-derived sequences representing only a small proportion of our sample. Future studies using techniques such as laser capture microdissection could allow us to collect specific cell types for use in isolating proviral DNA from the CNS.
Other longitudinal studies of HIV and SIV genetic diversity have focused on samples from the peripheral blood lymphocytes, plasma, or CSF,27,53 allowing collection of samples at multiple timepoints from the same animal. However, this type of sampling does not address viral genetic diversity and evolution in tissues such as the brain, which are difficult to sample in a longitudinal manner. Thus, we utilized two pigtailed macaque cohorts: one sacrificed after 1 w.p.i.17 and the other after 2 m.p.i. Variation between host animals may introduce biases not present in longitudinal samples from the same animal, but our procedure is more ethical and feasible. Though longitudinal analyses of SIV env sequence compartmentalization have been conducted in prior research,27 this investigation is unique in studying compartmentalization, evolution, and genetic diversity in multiple CNS and lymphoid tissues.
Selection against Env V1 region sequence group 1 was noted across all experimental tissues at 1 w.p.i., likely due to the lower fitness of sequences in this group relative to the other three groups.17 We expected that the less-fit Env V1 region group 1 sequences would contribute to a smaller proportion of the proviral DNA population over time and the prevalence of these sequences would decline over the course of infection. However, Env V1 region group 1 increased in prevalence of this group in the lymph nodes and hippocampus at 2 m.p.i. One possibility is that group 1 Env V1 region sequences possess mutations that provide a replication advantage with the onset of host adaptive immunity.54–56 Preliminary analyses indicate that group 1 sequences have an increased number of amino acid changes compared to sequences from the other three groups (data not shown), which could allow escape from neutralizing antibody.54–56 However, the increased presence of group 1 sequences may reflect a decline in the proportion of the other Env V1 region groups in the virus population. Given the increased prevalence of Env V1 region group 4 sequences at 1 w.p.i.,17 we expected more virions with these Env V1 regions would be produced as infection continued, leading to increased pressure on these sequences from the adaptive immune response and a decline in their prevalence as infection progressed. However, although group 4 sequences did face a selective disadvantage in the lymph nodes at 2 m.p.i., this group was selected for in the CNS over the course of infection. Preliminary analyses of the Env V1 region group 4 sequences obtained in these studies indicates that the number of sequences in this group with an insertion in the V1 loop, which may affect the capability of the virus to enter target cells or allow viral escape from neutralizing antibody,54–56 increases at 2 m.p.i. (data not shown).
Tissue-specific variation in the fitness of Nef group 1 and 2 sequences was noted in the CNS samples harvested at 1 w.p.i., with group 1 the most prevalent in all tissues.17 In the midfrontal cortex, hippocampus, and lymph nodes, the prevalence of group 1 Nef sequences increased by 2 m.p.i. compared to the SIVsmmFGb stock virus. Nef is capable of selectively downmodulating MHCI molecules,57,58 and if this function was defective in group 2 Nef sequences, this group may face a selective disadvantage in the CNS, where cytotoxic T cells are an important component of the immune response.59 However, Nef sequences from the basal ganglia and cerebellum had an increased prevalence of group 2 and decreased prevalence of group 1 by 2 m.p.i. compared to the stock virus, suggesting selective pressure on Nef varies between regions of the brain.
At 1 w.p.i., selection favored Int group 3.17 We expected this trend could continue through 2 m.p.i., although the increased prevalence of group 3 Int sequences at 1 w.p.i. could result in increased adaptive immune pressure on this group. Consistent with this, Int group 3 prevalence was lower at 2 m.p.i. in all tissues compared to 1 w.p.i.17 Increases in the prevalence of groups 1 and 2 at 2 m.p.i. could result from an advantage of these sequences or merely be a proportional increase due to selection against group 3. In the CNS, the prevalence of Int groups 1 and 3 was higher at 2 m.p.i., whereas group 2 prevalence declined, compared to the SIVsmmFGb stock virus. The decreased prevalence of group 3 Int sequences in the CNS from 1 w.p.i. to 2 m.p.i. suggests selection against this group over time.
The env V1 region sequences formed separate compartments between most tissues at 1 w.p.i.17 We expected that env V1 region sequence compartmentalization would increase at 2 m.p.i. given the continued replication of the virus and selective pressures unique to each microenvironment. The results supported our expectations, showing segregation of env V1 region sequences between all tissues except the axillary and mesenteric lymph nodes at 2 m.p.i. Compartmentalization of env V1 region sequences was weaker in a slight majority of intertissue comparisons at 2 m.p.i. than at 1 w.p.i.,17 suggesting a mixture of convergent and divergent evolution of env V1 region sequences between compartments over time.
At 1 w.p.i., nef sequences formed distinct genetic compartments in all tissues.17 Given our expectation of differences in virus replication and selective pressures between each microenvironment, we expected increases in nef compartmentalization over time. The results confirmed our expectations, showing that nef sequences form separate compartments between different regions of the CNS, and between the CNS regions and the lymph nodes, at 2 m.p.i. Axillary and mesenteric lymph node-derived nef sequences did not compartmentalize separately from each other at 2 m.p.i., perhaps due to the ease of virus transfer between lymph nodes. The degree of nef compartmentalization was largely weaker at 2 m.p.i. than at 1 w.p.i.,17 suggesting convergent evolution of nef sequences between most tissue compartments.
The formation of distinct int sequence compartments between most tissues was noted at 1 w.p.i.17 Given the resistance of int to mutation,7 we expected the intertissue int sequence compartments to remain stable over time. At 2 m.p.i., int sequences formed distinct genetic compartments between all regions of the CNS tissues, and between the CNS regions and the lymph nodes, confirming our expectations. In most two-tissue comparisons, the strength of int compartmentalization increased by 2 m.p.i. compared to 1 w.p.i.,17 suggesting divergent evolution of int sequence populations. However, all intertissue comparisons involving sequences from the hippocampus showed weaker compartmentalization at 2 m.p.i. than at 1 w.p.i.,17 suggesting convergent evolution of int between the hippocampus and the lymph nodes as well as between the hippocampus and the other regions of the CNS.
We expected sequences from each tissue at 2 m.p.i. to compartmentalize separately from those obtained at 1 w.p.i., reflecting additional virus evolution and exposure to selective pressure prior to the suppression of replication. Intratissue temporal compartmentalization was seen for all three genes in all tissues, suggesting that sequences within a tissue diverge over time (Fig. 8). For Nef and Env V1, temporal segregation was strongest between regions of the CNS, suggesting greater divergence of sequences in these regions over time compared to the lymph nodes. Temporal compartmentalization of Int was similar in most of the tissues, suggesting little difference between divergence of Int in the CNS and lymph nodes over time. Genetic divergence of the Env V1 region and Nef in the brain may be necessary to allow efficient viral replication in target cells or immune pressures specific to the CNS. As the highly conserved function of Int does not play a role in virus entry into target cells or avoidance of the immune system, this gene would not be expected to diverge in the CNS relative to the lymph nodes. Preliminary genetic analysis of the SIVsmmFGb Nef amino acid sequences in our studies indicates that amino acid changes in the basic region, N-proximal Y residues, and PAK binding site increase over time in both lymph nodes and all regions of the CNS (data not shown). Nef sequences from the CNS appear to accumulate an increased number of amino acid changes in the AP interaction site over time compared to sequences from the lymph nodes. These changes suggest that a number of Nef functional motifs adapt to enhance the replicative potential of the virus in the CNS and that Nef may play a very important role in driving SIVsmmFGb compartmentalization. Analysis of intratissue temporal compartmentalization by Mantel's test was not feasible given the number of sequences and software limitations.
At 1 w.p.i., negative selection contributed to nef and int compartmentalization, whereas positive selection was a factor in env V1 region segregation.17 We expected that positive selection on the env V1 region sequences would increase over time, with selection for increased coreceptor usage and adaptive immune pressure selecting for escape mutants. Given the conserved functions of nef,60–63 we expected the resistance of this gene to mutation would overcome selective pressures for escape mutants, and that negative selection would continue. Similarly, given the tendency of int to evolve slowly and resist amino acid changes,7 we expected negative selection on that gene to continue. The dS/dN values for sequences harvested at 2 m.p.i. confirmed that positive selection of the env V1 region and negative selection of nef and int continued over the course of infection. The strength of positive selection on env V1 region sequences increased over time in most tissues, based on lower mean dS/dN values at 2 m.p.i. compared to 1 w.p.i.17 Conversely, the strength of the negative selection on nef and int sequences increased over time in most tissues, based on increased mean dS/dN values at 2 m.p.i. compared to 1 w.p.i.17 Although preliminary analysis suggests that nef accumulates mutations in certain functional motifs (data not shown), there may be enough purifying selection on other domains to cause an overall increase in negative selection.
Changes in the mean dS value are affected by the frequency of silent synonymous genetic mutations, whereas dN is affected more by the nonsynonymous substitutions induced by selective pressure.64,65 If both the mean dS value and mean dN value are highest at 2 m.p.i., this suggests the general rate of mutation of a gene increases over the course of infection, likely resulting from both external selective pressure, such as the immune response, and factors intrinsic to the virus, such as generation time and replication fidelity. Increases in both mean dS and mean dN from 1 w.p.i. to 2 m.p.i. occurred for env V1 regions from the midfrontal cortex and lymph nodes, nef sequences from the cerebellum and hippocampus, and int sequences from the basal ganglia and cerebellum.17 If sequences have a higher mean dS value and lower mean dN value at 2 m.p.i. than at 1 w.p.i., this indicates that silent mutations increase over the course of infection, with reduced accumulation of amino acid changes. This suggests that evolution is influenced more by factors intrinsic to the virus than by external selective pressure. This pattern appeared for env V1 region sequences in the hippocampus, nef sequences in the axillary lymph node and midfrontal cortex, and int sequences in the midfrontal cortex, hippocampus, and mesenteric lymph node.17 A higher mean dN value at 2 m.p.i. compared to 1 w.p.i., but lower mean dS, suggests an increased rate of amino acid substitution over time, whereas the rate of silent mutation declines. In this situation, which occurred in basal ganglia-derived env V1 region sequences,17 evolution is influenced more by external selective pressure. Finally, decreased mean dS and dN values at 2 m.p.i., compared to 1 w.p.i., indicate evolution is due to a combination of intrinsic and extrinsic factors, but the general rate of mutation is lower. Decreases in both dS and dN occurred in env V1 region sequences from the cerebellum, int sequences from the axillary lymph node, and nef sequences from the basal ganglia and mesenteric lymph node.17
We also expected an increase in functional differences between env V1 regions harvested from the CNS and the lymph nodes, given the expected variance in selective pressures between the CNS regions and the lymph nodes. At 2 m.p.i., most intertissue comparisons had some variation in mean dS or dN, suggesting functional differences between env V1 regions from different tissues. The number of intertissue comparisons with a significant difference in mean dS, dN, or both increased at 2 m.p.i. compared to 1 w.p.i.,17 suggesting that env V1 regions in the various compartments gained functional differences as infection progressed. At 2 m.p.i., most intertissue comparisons of nef sequences had differences in either dS or dN, suggesting variations in nef function between tissues. Compared with the results at 1 w.p.i.,17 this suggests the differences in nef function between compartments develop over the course of infection. At 2 m.p.i., variations in mean dN were noted in the majority of int intertissue comparisons, suggesting functional differences between certain tissues. An overall net loss of functional differences in int between tissues was noted by 2 m.p.i. compared to 1 w.p.i.17
Although our study did not specifically analyze the immune responses of the infected animals to SIVsmmFGb, the host adaptive immune response likely had some role in the evolution of the viral genes analyzed in this study. In studies with other SIV and HIV strains, CD8+ T cells are known to infiltrate the CNS59 and can respond to Nef and Int proteins66 as well as epitopes within the Env V1 region.67 Relative to protein length, Nef elicits the greatest CTL response of the three genes considered in this study, whereas responses to the Int and the Env V1 region are comparatively moderate and low, respectively.66,67 The high mutation rate and variability of the env gene can alter Env CTL epitopes, making the CD8+ T cell response less effective, over time, than against less variable proteins such as Nef and Int.68 If CD8+ T cells play a similar role in the response to SIVsmmFGb in the CNS, then we might expect that the evolution and compartmentalization of all three genes analyzed in this study may be partly due to differences in CTL activity in different regions of the brain. Unlike Int and Nef, however, the Env protein also induces neutralizing antibodies,68–70 particularly the V1 region.71,72 Although the blood–brain barrier (BBB) normally blocks antibody entry into the CNS, Env can damage the BBB73 and allow extravasation of serum proteins, including IgG.74,75 Although prior studies have demonstrated that plasma levels of anti-SIV p27 antibody have a negative correlation with SIVsmmFGb viral load in the CNS on progression to neurological disease,5 it is currently unknown what role Env-specific antibodies might play in SIVsmmFGb neuropathogenesis. The early appearance of humoral responses to other SIV strains70,76 and the efficacy of these responses against other macrophage-tropic viral strains77 suggest antibodies may be involved in adaptive immunity in the CNS. If SIVsmmFGb elicits similar antibody responses to other SIV strains, we might expect that the humoral response plays a role in env V1 region evolution in the CNS. Future studies will be necessary to analyze the cellular and humoral immune responses to SIVsmmFGb in the CNS and determine how these responses correlate with the evolution and compartmentalization of viral genes. Of particular interest would be to compare immune responses and sequence evolution between SIVsmmFGb-infected animals with different rates of disease progression and the rare animals that do not develop neuropathogenesis. This may allow for the determination of host- or virus-specific factors that influence the development and progression of SIVsmmFGb neuropathogenesis.
Overall, our results reveal the effect of replication and evolution on the diversity and compartmentalization of a neuropathogenic SIV swarm in the lymph nodes and CNS of pigtailed macaques. These results suggest that viruses evolving in different regions of the CNS may vary in their potential to induce neuropathogenesis or escape immune responses. In particular, the diversity and function of Nef appear to differ between regions of the CNS and between the CNS and the periphery, suggesting Nef may determine the presence and severity of neuropathogenesis in a given brain region. We also found convergent evolution of the env V1 region and nef between different compartments and decreased differences in env V1 region and int function and evolution of all three genes within CNS regions over time. These results suggest that selective pressure may drive SIVsmmFGb strains in all CNS regions toward a common phenotype, necessary to induce full neuropathogenesis. We are currently analyzing amino acid sequences from the Env V1 region, Nef, and Int for the presence of mutations in functional domains, which may affect viral fitness. Comparing the prevalence of these mutations over time may allow us to determine sequence changes that may be important in the development of neuropathogenesis. Sequences with potentially interesting mutations, particularly in the Env V1 region and Nef, could then be cloned and functional assays could be performed. We are also interested in determining whether mutations may be present in immune epitopes, particularly in the Env V1 region and Nef protein sequences, which may indicate the effects of immune pressure on viral evolution.
Sequence Data
The nucleotide sequences referenced in this study are available in the GenBank database under the accession numbers FJ396520–FJ396665.
Supplementary Material
Acknowledgments
We are grateful for the assistance of the primate care technicians, vet techs, veterinarians and Research Resources personnel at the Yerkes National Primate Research Center for their assistance in the completion of this project. We also wish to thank Genevieve Niedziela, Ashley St. John, and Chen Chen for their assistance in the early stages of the project. We would also like to thank Anne Piantadosi and Julie Overbaugh for their advice concerning sequencing analysis and appropriate software, as well as Tianwei Yu for advice on statistical analysis. This work was supported by NIH Grants MH067769 (to F.J.N.) and RR00165 to the Yerkes National Primate Research Center.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.UNAIDS. AIDS Epidemic Update. 2007.
- 2.Luo W. Peterlin BM. Activation of the T-cell receptor signalling pathway by Nef from an aggressive strain of simian immunodeficiency virus. J Virol. 1997;71:9531–9537. doi: 10.1128/jvi.71.12.9531-9537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glenn AA. Novembre FJ. A single amino acid change in gp41 is linked to the macrophage-only replication phenotype of a molecular clone of simian immunodeficiency virus derived from the brain of a macaque with neuropathogenic infection. Virology. 2004;325:297–307. doi: 10.1016/j.virol.2004.04.048. [DOI] [PubMed] [Google Scholar]
- 4.Hahn BH. Shaw GM. deCock KM. Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 5.O'Neil SP. Suwyn C. Anderson DC. Niedziela G. Bradley J. Novembre FJ. Herndon JG. McClure HM. Correlation of acute humoral response with brain virus burden and survival time in pigtailed macaques infected with the neurovirulent simian immunodeficiency virus SIVsmmFGb. Am J Pathol. 2004;164:1157–1172. doi: 10.1016/S0002-9440(10)63204-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novembre FJ. De Rosayro J. O'Neil SP. Anderson DC. Klumpp SA. McClure HM. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J Virol. 1998;72:8841–8851. doi: 10.1128/jvi.72.11.8841-8851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skinner LM. Lamers SL. Sanders JC. Eyster ME. Goodenow MM. Klatzman M. Analysis of a large collection of natural HIV-1 integrase sequences, including those from long-term nonprogressors. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:99–110. doi: 10.1097/00042560-199810010-00001. [DOI] [PubMed] [Google Scholar]
- 8.Sharpless N. Gilbert D. Vandercam B. Zhou JM. Verdin E. Ronnett G. Friedman E. Dubois-Dalcq M. The restricted nature of HIV-1 tropism for cultured neural cells. Virology. 1992;191:813–825. doi: 10.1016/0042-6822(92)90257-p. [DOI] [PubMed] [Google Scholar]
- 9.Luciano CA. Pardo CA. McArthur JC. Recent developments in the HIV neuropathies. Curr Opin Neurol. 2003;16:403–409. doi: 10.1097/01.wco.0000073943.19076.98. [DOI] [PubMed] [Google Scholar]
- 10.Price RW. AIDS dementia complex. HIV InSite Knowledge Base Chapter. 1998 [Google Scholar]
- 11.Desrosiers RC. The simian immunodeficiency viruses. Annu Rev Immunol. 1990;8:557–578. doi: 10.1146/annurev.iy.08.040190.003013. [DOI] [PubMed] [Google Scholar]
- 12.McClure HM. Anderson DC. Fultz PN. Ansari AA. Lockwood E. Brodie A. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet Immunol Immunopathol. 1989;21:13–24. doi: 10.1016/0165-2427(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 13.Rausch DM. Murray EA. Eiden LE. The SIV-infected rhesus monkey model for HIV-associated dementia and implication for neurological disease. J Leukocyte Biol. 1999;65:466–474. doi: 10.1002/jlb.65.4.466. [DOI] [PubMed] [Google Scholar]
- 14.Sopper S. Koutsilieri E. Scheller C. Czub S. Riederer P. ter Meulen V. Macaque animal model for HIV-induced neurological disease. J Neural Transmiss. 2002;109:747–766. doi: 10.1007/s007020200062. [DOI] [PubMed] [Google Scholar]
- 15.Whetter LE. Ojukwu IC. Novembre FJ. Dewhurst S. Pathogenesis of simian immunodeficiency virus infection. J Gen Virol. 1999;80:1557–1568. doi: 10.1099/0022-1317-80-7-1557. [DOI] [PubMed] [Google Scholar]
- 16.Clements JE. Babas T. Mankowski JL. Suryanarayana K. Piatak M., Jr Tarwater PM. Lifson JD. Zink MC. The central nervous system as a reservoir for simian immunodeficiency virus (SIV): Steady state levels of SIV DNA in brain from acute through asymptomatic infection. J Infect Dis. 2002;186:905–913. doi: 10.1086/343768. [DOI] [PubMed] [Google Scholar]
- 17.Reeve AB. Patel K. Pearce NC. Augustus KV. Domingues HG. O'Neil SP. Novembre FJ. Reduced genetic diversity in lymphoid and central nervous system tissues and selection-induced tissue-specific compartmentalization of neuropathogenic SIVsmmFGb during acute infection. AIDS Res Hum Retroviruses. 2009;25:583–601. doi: 10.1089/aid.2008.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cichutek K. Merget H. Norley S. Linde R. Kruez W. Gahr M. Kurth R. Development of a quasispecies of human immunodeficiency virus type 1 in vivo. Proc Natl Acad Sci USA. 1992;89:7365–7369. doi: 10.1073/pnas.89.16.7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wain-Hobson S. Human immunodeficiency virus type 1 quasispecies in vivo and ex vivo. Curr Top Microbiol Immunol. 1992;176:181–193. doi: 10.1007/978-3-642-77011-1_12. [DOI] [PubMed] [Google Scholar]
- 20.Williams KC. Corey S. Westmoreland SV. Pauley D. Knight H. deBakker C. Alvarez X. Lackner AA. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: Implications for the neuropathogenesis of AIDS. J Exp Med. 2001;193:905–915. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti L. Hurtrel M. Maire M. Vazeux R. Dormont D. Montagnier L. Hurtrel B. Early viral replication in the brain of SIV-infected rhesus monkeys. Am J Pathol. 1991;139:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper DA. Imrie AA. Penny R. Antibody response to human immunodeficiency virus after primary infection. J Infect Dis. 1987;155:1113–1118. doi: 10.1093/infdis/155.6.1113. [DOI] [PubMed] [Google Scholar]
- 23.Ho DD. Rota TR. Schooley RT. Kaplan JC. Allan JD. Groopman JE. Resnick L. Felsenstein D. Andrews CA. Hirsch MS. Isolation of HTLV-III from cerebrospinal fluid and neural tissues of patients with neurologic syndromes related to the acquired immunodeficiency syndrome. N Engl J Med. 1985;313:1493–1497. doi: 10.1056/NEJM198512123132401. [DOI] [PubMed] [Google Scholar]
- 24.Gaines H. Sonnerberg A. Czajkowski J. Chiodi F. Fenyo EM. von Sydow M. Albert J. Pehrson PO. Moberg L. Asjo B. Forsgren M. Antibody response in primary human immunodeficiency virus infection. Lancet. 1987;I:1249–1253. doi: 10.1016/s0140-6736(87)92696-1. [DOI] [PubMed] [Google Scholar]
- 25.Mankowski JL. Flaherty MT. Spelman JP. Hauer DA. Didier PJ. Amedee AM. Murphey-Corb M. Kirstein LM. Munoz A. Clements JE. Zink MC. Pathogenesis of simian immunodeficiency virus encephalitis: Viral determinants of neurovirulence. J Virol. 1997;71:6055–6060. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen MF. Westmoreland S. Ryzhova EV. Martin-Garcia J. Soldan SS. Lackner A. Gonzalez-Scarano F. Simian immunodeficiency virus envelope compartmentalizes in brain regions independent of neuropathology. J Neurovirol. 2006;12:73–89. doi: 10.1080/13550280600654565. [DOI] [PubMed] [Google Scholar]
- 27.Poss M. Rodrigo AG. Gosink JJ. Learn GH. Panteleeff DDV. Martin HL., Jr Bwayo J. Kreiss JK. Overbaugh J. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins KR. Quinones-Mateu ME. Wu M. Luzze H. Johnson JL. Hirsch C. Toossi Z. Arts EJ. Human immunodeficiency virus type 1 quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J Virol. 2002;76:1697–1706. doi: 10.1128/JVI.76.4.1697-1706.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams K. Alvarez X. Lackner AA. Central nervous system perivascular cells are immunoregulatory cells that connect the CNS with the peripheral immune system. Glia. 2001;36:156–164. doi: 10.1002/glia.1105. [DOI] [PubMed] [Google Scholar]
- 30.Descombers P. Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 31.Honda Y. Rogers L. Nataka K, et al. Type I interferon induces inhibitory 16-kD CCAAT/enhancer binding protein (C/EBP) beta, repressing the HIV-1 long terminal repeat in macrophages: Pulmonary tuberculosis alters C/EBP expression, enhancing HIV-1 replication. J Exp Med. 1998;188:1255–1265. doi: 10.1084/jem.188.7.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiden M. Tanaka N. Qiao Y, et al. Differentiation of monocytes to macrophages switches the Mycobacterium tuberculosis effect on HIV-1 replication from stimulation to inhibition: Modulation of interferon response and CCAAT/enhancer binding protein beta expression. J Immunol. 2000;165:2028–2039. doi: 10.4049/jimmunol.165.4.2028. [DOI] [PubMed] [Google Scholar]
- 33.Henderson AJ. Zou X. Calame KL. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson AJ. Connor RI. Calame KL. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 35.Henderson AJ. Calame KL. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4+ T cells. Proc Natl Acad Sci USA. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barber SA. Herbst DS. Bullock BT. Gama L. Clements JE. Innate immune responses and control of acute simian immunodeficiency virus replication in the central nervous system. J Neurovirol. 2004;10:15–20. doi: 10.1080/753312747. [DOI] [PubMed] [Google Scholar]
- 37.Gessani S. Puddu P. Varano B. Borghi P. Conti L. Fantuzzi L. Gherardi G. Belardelli F. Role of endogenous interferon-beta in the restriction of HIV replication in human monocyte/macrophages. J Leukocyte Biol. 1994;56:358–361. doi: 10.1002/jlb.56.3.358. [DOI] [PubMed] [Google Scholar]
- 38.Hoshino Y. Nakata K. Hoshino S, et al. Maximal HIV-1 replication in alveolar macrophages during tuberculosis requires both lymphocyte contact and cytokines. J Exp Med. 2002;195:495–505. doi: 10.1084/jem.20011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer-Hammerle S. Rothenaigner I. Wolff H. Bell JE. Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111:194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Lackner AA. Smith MO. Munn RJ. Martfeld DJ. Gardner MB. Marx PA. Dandekar S. Localization of simian immunodeficiency virus in the central nervous system of rhesus macaques. Am J Pathol. 1991;139:609–621. [PMC free article] [PubMed] [Google Scholar]
- 41.Lackner AA. Vogel P. Ramos RA. Kluge JD. Marthas M. Early events in tissues during infection with pathogenic (SIVmac239) and nonpathogenic (SIVmac1A11) molecular clones of simian immunodeficiency virus. Am J Pathol. 1994;145:428–439. [PMC free article] [PubMed] [Google Scholar]
- 42.Lane JH. Sasseville VG. Smith MO. Vogel P. Pauley DR. Heyes MP. Lackner AA. Neuroinvasion by simian immunodeficiency virus coincides with increased numbers of perivascular macrophages/microglia and intrathecal immune activation. J Neurovirol. 1996;2:423–432. doi: 10.3109/13550289609146909. [DOI] [PubMed] [Google Scholar]
- 43.Watry D. Lane TE. Streb M. Fox HS. Transfer of neuropathogenic simian immunodeficiency virus with naturally infected microglia. Am J Pathol. 1995;146:914–923. [PMC free article] [PubMed] [Google Scholar]
- 44.Wiley CA. Schrier RD. Nelson JA. Lampert PW. Oldstone MBA. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albright AV. Strizki J. Harouse JM. Lavi E. O'Connor M. Gonzalez-Scarano F. HIV-1 infection of cultured human adult oligodendrocytes. Virology. 1996;217:211–219. doi: 10.1006/viro.1996.0108. [DOI] [PubMed] [Google Scholar]
- 46.Bagasra O. Lavi E. Bobroski L. Khalili K. Pestaner JP. Tawadros R. Pomerantz RJ. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: Identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS. 1996;10:573–585. doi: 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 47.Brack-Werner R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS. 1999;13:1–22. doi: 10.1097/00002030-199901140-00003. [DOI] [PubMed] [Google Scholar]
- 48.Nuovo GJ. Gallery F. MacConnell P. Braun A. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acids and tumor necrosis factor-alpha RNA in the central nervous system. Am J Pathol. 1994;144:659–666. [PMC free article] [PubMed] [Google Scholar]
- 49.Saito Y. Sharer LR. Epstein LG. Michaels J. Mintz M. Louder M. Golding K. Cvetkovich TA. Blumberg BM. Overexpression of Nef as a marker for restricted HIV-1 infection of astrocytes in postmortem pediatric central nervous tissues. Neurology. 1994;44:474–481. doi: 10.1212/wnl.44.3_part_1.474. [DOI] [PubMed] [Google Scholar]
- 50.Trillo-Pazos G. Diamanturos A. Rislove L. Menza T. Chao W. Belem P. Sadiq S. Morgello S. Sharer L. Volsky DJ. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 2003;13:144–154. doi: 10.1111/j.1750-3639.2003.tb00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nath A. Hartloper V. Furer M. Fowke KR. Infection of human fetal astrocytes with HIV-1: Viral tropism and the role of cell to cell contact in viral transmission. J Neuropathol Exp Neurol. 1995;54:320–330. doi: 10.1097/00005072-199505000-00005. [DOI] [PubMed] [Google Scholar]
- 52.Sabri F. Tresoldi E. Di Stefano M. Polo S. Monaco MC. Verani A. Fiore JR. Lusso P. Major E. Chiodi F. Scarlatti G. Nonproductive human immunodeficiency virus type 1 infection of human fetal astrocytes: Independence from CD4 and major chemokine receptors. Virology. 1999;264:370–384. doi: 10.1006/viro.1999.9998. [DOI] [PubMed] [Google Scholar]
- 53.Harrington PR. Connell MJ. Meeker RB. Johnson PR. Swanstrom R. Dynamics of simian immunodeficiency virus populations in blood and cerebrospinal fluid over the full course of infection. J Infect Dis. 2007;196:1058–1067. doi: 10.1086/520819. [DOI] [PubMed] [Google Scholar]
- 54.Gray ES. Moore PL. Choge IA. Decker JM. Bibollet-Ruche F. Li H. Leseka N. Treurnicht F. Mlisana K. Shaw GM. Karim SSA. Williams C. Morris L the CAPRISA 002 Study Team. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones NA. Wei X. Flower DR. Wong ML. Michor F. Saag MS. Hahn BH. Nowak MA. Shaw GM. Borrow P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borrow P. Lewicki H. Wei X. Horwitz MS. Peffer N. Meyers H. Nelson JA. Gairin JE. Hahn BH. Oldstone MBA. Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 57.Swigut T. Iafrate AJ. Muench J. Kirchhoff F. Skowronski J. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J Virol. 2000;74:5691–5701. doi: 10.1128/jvi.74.12.5691-5701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen GB. Gandhi RT. Davis DM. Mandelboim O. Chen BK. Strominger JL. Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 59.Petito CK. Adkins B. CD4+ and CD8+ cells accumulate in the brains of acquired immunodeficiency syndrome patients with human immunodeficiency virus encephalitis. J NeuroVirol. 2003;9:36–44. doi: 10.1080/13550280390173391. [DOI] [PubMed] [Google Scholar]
- 60.Kirchhoff F. Schindler M. Bailer N. Renkema GH. Saksela K. Knoop V. Muller-Trutwin MC. Santiago ML. Bibollet-Ruche F. Dittmar MT. Heeney JL. Hahn BH. Munch J. Nef proteins from simian immunodeficiency virus-infected chimpanzees interact with p21-activated kinase 2 and modulate cell surface expression of various human receptors. J Virol. 2004;78:6864–6874. doi: 10.1128/JVI.78.13.6864-6874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirchhoff F. Munch J. Carl S. Stolte N. Matz-Rensing K. Fuchs D. Haaft PT. Heeney JL. Swigut T. Skowronski J. Stahl-Hennig C. The human immunodeficiency virus type 1 nef gene can to a large extent replace simian immunodeficiency virus Nef in vivo. J Virol. 1999;73:8371–8383. doi: 10.1128/jvi.73.10.8371-8383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindler M. Wurfl S. Benaroch P. Greenough TC. Daniels R. Easterbrook P. Brenner M. Munch J. Kirchhoff F. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J Virol. 2003;77:10548–10556. doi: 10.1128/JVI.77.19.10548-10556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindler M. Munch J. Kutsch O. Li H. Santiago ML. Bibollet-Ruche F. Muller-Trutwin MC. Novembre FJ. Peeters M. Courgnaud V. Bailes E. Roques P. Sodora DL. Silverstri G. Sharp PM. Hahn BH. Kirchhoff F. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 64.Lemey P. Pond SLK. Drummond AJ. Pybus OG. Shapiro B. Barroso H. Taveira N. Rambaut A. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comp Biol. 2007;3:282–292. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gunthard HF. Leigh-Brown AJ. D'Aquila RT. Johnson VA. Kuritzkes DR. Richman DD. Wong JK. Higher selection pressure from antiretroviral drugs in vivo results in increased evolutionary distance in HIV-1 pol. Virology. 1999;259:154–165. doi: 10.1006/viro.1999.9774. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez WR. Addo MM. Rathod A. Fitzpatrick CA. Yu XG. Perkins B. Rosenberg ES. Altfeld M. Walker BD. CD8+ T lymphocyte responses target functionally important regions of protease and integrase in HIV-1 infected subjects. J Translation Med. 2004;2:15–30. doi: 10.1186/1479-5876-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erickson AL. Walker CM. An epitope in the V1 domain of the simian immunodeficiency virus (SIV) gp120 protein is recognized by CD8+ cytotoxic T lymphocytes from an SIV-infected rhesus macaque. J Virol. 1994;68:2756–2759. doi: 10.1128/jvi.68.4.2756-2759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peut V. Kent SJ. Utility of human immunodeficiency virus type 1 envelope as a T-cell immunogen. J Virol. 2007;81:13125–13134. doi: 10.1128/JVI.01408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frost SDW. Wrin T. Smith DM. Pond SLK. Liu Y. Paxinos E. Chappey C. Galovich J. Beauchaine J. Petropoulos CJ. Little SJ. Richman DD. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci USA. 2005;102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richman DD. Wrin T. Little SJ. Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petry H. Pekrun K. Hunsmann G. Jurkiewicz E. Luke W. Naturally occurring V1-Env region variants mediate simian immunodeficiency virus SIVmac escape from a high-titer neutralizing antibodies induced by a protective subunit vaccine. J Virol. 2000;74:11145–11152. doi: 10.1128/jvi.74.23.11145-11152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jurkiewicz E. Hunsmann G. Schaffner J. Nisslein T. Luke W. Petry H. Identification of the V1 region as a linear neutralizing epitope of the simian immunodeficiency virus SIVmac envelope glycoprotein. J Virol. 1997;71:9475–9481. doi: 10.1128/jvi.71.12.9475-9481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanmogne GD. Schall K. Leibhart J. Knipe B. Gendelman HE. Persidsky Y. HIV-1 gp120 compromises blood-brain barrier integrity and enhance monocyte migration across blood-brain barrier: Implication for viral neuropathogenesis. J Cerebral Blood Flow Metab. 2007;27:123–134. doi: 10.1038/sj.jcbfm.9600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petito CK. Cash KS. Blood-brain barrier abnormalities in the acquired immunodeficiency syndrome: Immunohistochemical localization of serum proteins in postmortem brain. Ann Neurol. 1992;32:658–666. doi: 10.1002/ana.410320509. [DOI] [PubMed] [Google Scholar]
- 75.Rhodes RH. Evidence of serum-protein leakage across the blood-brain barrier in the acquired immunodeficiency syndrome. J Neuropathol Exp Neurol. 1991;50:171–183. doi: 10.1097/00005072-199103000-00008. [DOI] [PubMed] [Google Scholar]
- 76.Wei X. Decker JM. Wang S. Hui H. Kappes JC. Wu X. Salazar-Gonzalez JF. Salazar MG. Kilby JM. Saag MS. Komarova NL. Nowak MA. Hahn BH. Kwong PD. Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 77.McEntee MF. Anderson MG. Daniel MD. Adams R. Farzadegan H. Desrosiers RC. Narayan O. Differences in neutralization of simian lentivirus (SIVMAC) in lymphocyte and macrophage cultures. AIDS Res Hum Retroviruses. 1992;8:1193–1198. doi: 10.1089/aid.1992.8.1193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.