Abstract
The prevalence of antiretroviral therapy (ART) resistance mutations present in HIV-1 subtype C pol and env regions of the proviral DNA was analyzed and compared from therapy-naive individuals before (Cohort A) and after (Cohort B) the availability of free ART in Zambia. Mutations present in sequences published in a previous study from Zambian ART-naive individuals infected with subtype C were analyzed using current parameters for the classification of ART drug resistance and compared with Cohorts A and B. No statistically significant differences were observed when comparing mutations present in the pol and env of these cohorts. However, an increase in the number of minor, borderline, or partial resistance mutations as well as the presence of major resistance mutations were observed in Cohort B. These results suggest there is an increasing trend of drug resistance-associated mutations that could be a result of the availability of free ART in Zambia. Moreover, the high prevalence of resistance mutations observed for maraviroc and vicriviroc in both cohorts may suggest a limited efficacy of entry inhibitors on HIV-1 subtype C.
Of the 33 million people infected worldwide with the human immunodeficiency virus type 1 (HIV-1), almost 65% live in sub-Saharan Africa.1 High genetic variability and rapid evolution are the two major factors that contribute to the spread of HIV-1. The main cause of the high genetic heterogeneity, or quasispecies, is the low-fidelity and error-prone reverse transcriptase of the virus. As a result of this continuous divergence, several HIV-1 subtypes or clades have emerged.2 In southern Africa, 98% of HIV-1 infections are subtype C variants; it is the most prevalent subtype, accounting for over 50% of the new infections globally.3
Antiretroviral therapy (ART) has been effective in decreasing morbidity and mortality in developed countries.4 Nevertheless, these regimens have only recently become available in the developing world.5 As ART is rapidly scaled up in Africa and other resource-limited countries, surveillance of the prevalence of ART resistance-associated mutations is necessary to ensure optimal therapy.6 In 2002 the Zambian Ministry of Health initiated an ART program at the country's two largest hospitals, making treatment available to the public sector.7 Access to ART was increased in 2004, when the Zambian Ministry of Health initiated an ART program at primary care sites within Lusaka (capital of Zambia), with financial resources from the U.S. President's Emergency Plan for AIDS Relief (PEPFAR); the Global Fund to Fight AIDS, Tuberculosis and Malaria; and other sources. The first line treatment consists of a three-drug ART, lamivudine (3TC), either nevirapine (NVP) or efavirenz (EFV), and either zidovudine (ZDV) or stavudine (d4T).7 Overall, the rapid scale-up of ART has been associated with good clinical outcomes in primary care settings in Zambia. However, mortality during the first 90 days of therapy is high.7 Currently, the information available on HIV-1 drug-resistant viruses in Zambia is limited, and whether there is transmission of drug-resistant viruses is unclear. Moreover, due to the high prevalence of HIV-1, the rapid scale-up of ART availability still benefits only a portion of the infected individuals in Zambia. By 2009, the National AIDS Council in Zambia (http://www.nac.org.zm) reported that approximately 12% of the population (1.3 million) is infected with HIV-1. Although 350,000 of these HIV-infected individuals require treatment, only 180,000 had access to free ART.
Therapy failure to reverse transcriptase (RT), and protease (PR) inhibitors is mainly caused by mutations in the pol gene. Significant genotypic and phenotypic differences between different HIV-1 subtypes have been observed in drug-resistant variants isolated from both ART-naive and ART-treated patients. Studies have shown a higher prevalence of naturally occurring ART-resistant subtype C variants in the pol gene of ART-naive patients in southern Africa.8,9 There is also a higher prevalence of NVP-resistant subtype C strains than other subtypes.2 Moreover, some of these naturally occurring polymorphisms likely accelerate the emergence of ART resistance.10 In addition to mutations in the pol gene, drug resistance-associated mutations (DRAMs) have also been recently reported in the V3 region of the env against the CCR5 entry inhibitors maraviroc11 and vicriviroc.12 Because drug resistance and resistance-associated mutations may have a profound impact on the clinical management of patients, surveillance of ART resistance in both treated and untreated individuals is essential for the development and implementation of an effective therapy.
The objective of the current study was to determine the effect of the scale-up of ART in Zambia on ART resistance-associated polymorphisms and drug-resistant mutations. To achieve this goal we analyzed and compared the prevalence of ART resistance mutations in the pol and env of proviral DNA from subjects in two cohorts collected before and after ART was freely available in Zambia. All subjects were HIV-1-positive adults infected with subtype C who were ART naive at the time of sample collection. A written informed consent was obtained from all participants and the study was approved by the Institutional Review Boards of the University of Nebraska and University of Zambia. Cohort A, comprising 32 samples collected before the availability of ART, was randomly selected from archived frozen peripheral blood mononuclear cells (PBMCs) from ART-naive HIV-1+ Zambian women obtained between 1998 and 2002 in Lusaka, Zambia. These women were participants of a mother–infant pairs cohort and were diagnosed as HIV+ at the time of delivery. The average age of the women in Cohort A was 26.06 years (range, 18–40 years). In addition to being recently diagnosed as HIV seropositive, these women were ART therapy naive and had no evidence of HIV/AIDS-related opportunistic infection on physical examination. Furthermore, all patients resided within Lusaka and were able to give informed consent. Twenty-six of these samples were collected at the time of delivery (baseline). Due to the low HIV DNA copy number and the difficulty in obtaining a polymerase chain reaction (PCR) product for the baseline time point, a sample from a subsequent time point was analyzed for six individuals included in Cohort A.
Cohort B was composed of 90 samples randomly selected from a surveillance study during 2005 for HIV-1 subtyping and genotyping, and all samples were from HIV-1+ ART-naive adults recruited at the Voluntary Counseling and Testing (VCT) centers. This cohort was composed of 57 females, 29 males, and 4 unknowns with an average age of 33.09 years (range, 20–63). These individuals were able to provide an informed consent and had no reported evidence of HIV/AIDS opportunistic infections at the time the sample was collected. No other clinical information from these individuals was collected. The specimens were collected from various VCT sites located in 15 different towns distributed throughout Zambia. These towns, strategically chosen to represent the Zambian population, included Chirundu, Chipata, Kasama, Kasumbalesa, Kapiri Mposhi, Kitwe, Kawambwa, Kazungula, Lusaka (Chawama and Kanyama area), Livingstone, Ndola, Mansa, Maheba, Mongu, and Solwezi. All 122 samples were analyzed for mutations in the pol PR and RT and the V3 region of the env by direct sequencing.
Due to the difficulties associated with the transport and storage of plasma viral RNA, ART resistance was analyzed from DNA extracted from PBMCs. Although it is known that PBMC samples may include information about archived viral mutations, it has recently been shown that the rates of detection of ART mutations in plasma RNA are similar to those observed in proviral DNA from PBMC samples.13 For both cohorts PBMCs were isolated on site from 2 ml to 5 ml of whole blood treated with ethylenediamine tetraacetic acid (EDTA) followed by a Ficol/Hypaque density centrifugation. The isolated cells were resuspended in 1 ml of phosphate-buffered solution (PBS). DNA extraction from Cohort A PBMC samples was done at the Nebraska Center for Virology using the Gentra Puregene Cell kit (QIAGEN, Valencia, CA) following the manufacturer's indications. DNA extraction from Cohort B PBMCs was performed at the University Teaching Hospital Virology Laboratory in Lusaka, Zambia, using the QIAamp DNA Extraction kit (QIAGEN, Valencia, CA) following the manufacturer's recommended procedure. The extracted DNA was suspended in 100% ethanol and then stored at −80°C for further experimental work.
The C2V4 region of the proviral DNA env was amplified by seminested PCR and sequenced following the conditions previously reported.14 The subtype was determined by neighbor-joining phylogenetic analysis that included group M reference consensus sequences from the Los Alamos HIV Sequence Database and confirmed by reanalyzing the C2V4 sequences using the REGA HIV subtyping tool from HIV Bioinformatics Bioafrica.15 The C2V4 env region was successfully amplified for 28 of the 32 samples in Cohort A. Of those 28 env sequences, 26 (92.8%) were subtype C. One of these non-subtype C sequences clustered with subtype H. However, due to the short length of the latter env sequence this subtype was not assigned as a pure subtype. The analysis also identified the remaining sequence as an AC recombinant with significant clustering (>70%) observed with subtypes A and C, respectively.
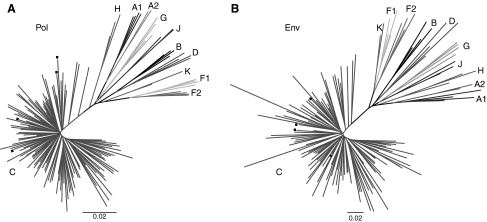
Sequences from Cohort B C2V4 were obtained for 81 of the 90 samples. Seventy-nine (98%) of these sequences were confirmed as subtype C. The REGA HIV subtyping tool classified one of these sequences as subtype A1. No subtype was assigned to the second non-subtype C sequence because no significant clustering was observed with a pure subtype. Results were further corroborated by a neighbor-joining phylogenetic analysis comprising C2V4 subtype C sequences from both cohorts and the most recent (2008) group M HIV-1 reference sequences from the Los Alamos HIV Sequence Database (Fig. 1A). These results confirm that HIV-1 subtype C is the predominant subtype circulating in Zambia. Only subtype C sequences were included in the analysis of entry inhibitor resistance mutations.
FIG. 1.
Phylogenetic analysis of ART-naive individuals infected with HIV subtype C. Nucleotide sequences from the pol (A) and env (B) regions were aligned for all therapy-naive individuals infected with subtype C in conjunction with HIV-1 Group M reference sequences. Each set of reference sequences for a specific subtype is represented by a letter indicating the subtype. Subtype C reference sequences are also identified by solid squares. The scale bar in the unrooted neighbor-joining tree represents 2% evolutionary distance per position in the sequence.
For the analysis of ART mutations in the pol gene, a 1493-bp region of the gag-pol gene including the sequence encoding the Gag protein p7 C terminus, p1, and p6 (Gag codons 406–500), full-length protease, and the first 314 amino acids of RT were amplified by nested PCR using the primers published by Zhang et al.16 The HiFi Hot star high fidelity polymerase (QIAGEN, Valencia, CA) was used to amplify the pol region. For the thermal cycling conditions, an initial 95°C for 5 min denaturing step was followed by 30 cycles of 94°C for 15 s, 50°C for 1 min, and 72°C for 2 min and a final extension of 72°C for 10 min. The size of the amplified PCR products was confirmed by electrophoresis in a 1.5% agarose gel. The positive PCR products were then purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio-Tech, Norcross, GA). The concentration of the final purified PCR products was determined using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, NanoDrop products Wilmington, DE).
The direct sequencing reaction of the pol region was performed from 20–40 ng of template using seven primers (Table 1) and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) following the manufacturer's protocol for PCR and precipitation of sequencing reactions. Sequencing of the pol and env genes was done using the ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA). The seven partially overlapping sequences were assembled and aligned by using the BioEdit 7.0.9 software.17 Resistance-associated mutation analyses based on the Stanford Drug Resistance Database (SDRD), 2009 and the International AIDS Society-USA (IAS-USA) 2007 drug resistance mutation list were generated using the Stanford Drug Resistance Database Calibrated Population Resistance (CPR) tool (http://surveillance.stanford.edu/cpr/index.html). Moreover, the IAS-USA 2008 spring update of drug resistance mutations was also referenced to perform this analysis.18 The subtype from pol sequences was determined using the subtype analyzer (STAR) application19 of the CPR tool and confirmed with the REGA HIV subtyping tool from HIV Bioinformatics Bioafrica.15 As described above for env, most of the pol sequences were classified as subtype C by the REGA HIV subtyping tool, although other subtypes were observed in both cohorts. In Cohort A, 97% of the pol sequences were subtype C with a single sequence identified as an AC recombinant. Likewise, in Cohort B, most of the pol sequences were classified as subtype C (98%) and only two of the 90 sequences were designated as non-subtype C (2%). One of these non-subtype C pol sequences was an AG recombinant, as confirmed by significant bootstrapping with both subtypes. The remaining sequence was identified as a complex mosaic recombinant in which a number of different subtypes were identified, although with low bootstrap confidence. Results for the subtyping analysis were also confirmed by a neighbor-joining phylogenetic analysis comprising pol subtype C sequences from both cohorts and the most recent (2008) group M HIV-1 reference sequences from the Los Alamos HIV Sequence Database (Fig. 1B). Only those sequences classified as subtype C were included in the PR and RT inhibitors genotyping analysis. The prevalence of each mutation was analyzed and also compared between cohorts by a nonparametric Fisher's exact test and a p value ≤0.05 was considered significant. This analysis was performed using the statistical software package SPSS version 17 (SPSS, Chicago, IL).
Table 1.
Sequencing Primers for the Detection of DRAMs in the pol and env Genes of HIV Subtype C
| Position-in HXB2 | Orientation | Sequence |
|---|---|---|
| Protease and reverse transcriptase regions ofpol | ||
| 2066 → 2096 | Sense | 5′-gTACTgAgAgACAggCTAATTTTTTAgggAA-3′a |
| 2219 → 2247 | Antisense | 3′-TTAAgggTTCCCTgTCTTTCggCT-5′ |
| 2486 → 2509 | Sense | 5′-ACCTACACCTgTCAACATAATTgg-3′b |
| 2582 → 2605 | Antisense | 3′-gggCCATCCATTCCTggCTTTAAT-5′ |
| 3001 → 3024 | Sense | 5′-AgggATggAAAggATCACCAgCAA-3′ |
| 3001 → 3024 | Antisense | 3′-TTgCTggTgATCCTTTCCATCCCT-5′ |
| 3345 → 3371 | Antisense | 3′-AATCCCTgCATAAATCTgACTTgCCCA-5′a |
| C2V4Region ofenv | ||
| 6882 → 6904 | Sense | 5′-CCTgCTggTTATgCgATTCTAAA-3′c |
| 7378 → 7350 | Antisense | 3′-CAATAgAAAAATTCTCCTCTACAATTAAA-5′c |
Hypermutated sequences, one from Cohort A and two from Cohort B, were excluded from the analysis to eliminate potential genotypic errors.20 After this elimination, a total of 30 sequences from Cohort A and 86 sequences from Cohort B were analyzed for genotyping of PR and RT. In summary, borderline (SDRD) and minor mutations (IAS-USA) were observed in both cohorts (Table 2) and major mutations were observed only in Cohort B (Table 3). The minor and borderline resistance mutations present in both cohorts are shown in Table 2. For comparison we have also included the ART minor mutations reported by another study conducted during 2000 in Zambia (referred to as the Handema et al. cohort).8 To be consistent in the parameters used for the analysis, PR and RT sequences published in this study (GenBank accession numbers AB081151–AB081175 for protease and AB081176–AB081203 for reverse transcriptase) were analyzed using current drug resistance classification parameters. The prevalence range of the three PR resistance-associated mutations, M36I, H69K, and I93L, was 80–100% in all the three cohorts. These mutations were also present in the subtype C consensus sequence; hence, they were considered to be natural polymorphisms and were excluded from the analysis. After eliminating these three mutations, a total of nine borderline/minor mutations were detected in Cohort A (prevalence range 3–20 %) and 14 in Cohort B (prevalence range 1–21%). These findings contrast with data reported for Zambia by Handema et al., in which only seven minor mutations (prevalence range 4–17%) were observed.8 The lower prevalence of mutations compared to our 1998–2002 cohort could be due to the use of different methods by the authors for the classification of ART resistance-associated mutations when the study was published.8 In the current study we analyzed our data following both the 2008 and 2009 SDRD classification parameters. In fact, when these parameters were used to reanalyze sequences published by Handema et al., nine borderline mutations (prevalence range 4–20%) were detected as seen with our Cohort A. In addition, according to the SDRD 2009 analysis, no PR major mutations were observed in either cohort or the sequences published by Handema et al. However, the PR mutation Q58E, which is considered a borderline mutation by the SDRD 2009, was classified as a major mutation in the SDRD 2008. This mutation was observed only in Cohort B (Table 3). Thus, methods of analysis could affect the interpretation of mutations and a consistent method of analysis needs to be used.
Table 2.
Prevalence of Minor and Borderline ART Mutations Associated with Resistance to PIs and RTIs in Both Cohorts and Compared with Published Data
| |
|
Observed prevalence for each resistance mutation (%) |
|
|
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
|
Handema et al.,8 2000 (N = 25) |
Cohort A, 1998–2002 (N = 30) |
Cohort B, 2005 (N = 66) |
Comparison of prevalence between cohorts, pa |
|||||
| Mutation | Class | SDRM (2009) | IAS-USA (2007)b | SDRM (2009) | IAS-USA (2007)b | SDRM (2009) | IAS-USA (2007)b | Handema to Cohort A | Handema to Cohort B | Cohort A to Cohort B |
| Protease region | ||||||||||
| L10V | PI | —c | 0 | — | 3.33 | — | 0 | 1.000 | N.O. | 0.2586 |
| I13V | PI | — | 4.2 | — | 0 | — | 0 | 0.455 | 0.2252 | N.O. |
| G16E | PI | — | 20 | — | 3.33 | — | 8.14 | 0.082 | 0.1372 | 0.6781 |
| K20R | PI | — | 4 | — | 20 | — | 20.93 | 0.112 | 0.0679 | 1.0000 |
| K20M | PI | — | 0 | — | 0 | — | 1.16 | N.O.d | 1.0000 | 1.0000 |
| L33F | PI | — | 0 | — | 0 | — | 0 | N.O. | N.O. | N.O. |
| M36L | PI | — | 4 | — | 6.67 | — | 2.33 | 1.000 | 1.0000 | 0.2746 |
| M36V | PI | — | 4 | — | 0 | — | 1.16 | 0.455 | 0.4013 | 1.0000 |
| Q58E | PI | — | 0 | — | 0 | — | 1.16 | N.O. | 1.0000 | 1.0000 |
| D60E | PI | — | 4 | — | 10 | — | 12.79 | 0.617 | 0.2922 | 1.0000 |
| I62V | PI | — | 0 | — | 0 | — | 4.65 | N.O. | 0.5728 | 0.5712 |
| L63P | PI | — | 20 | — | 13.33 | — | 20.93 | 0.716 | 1.0000 | 0.4292 |
| I64V | PI | — | 4 | — | 0 | — | 0 | 0.455 | 0.2252 | N.O. |
| I64L | PI | — | 4 | — | 0 | — | 0 | 0.455 | 0.2252 | N.O. |
| A71T | PI | — | 0 | — | 3.33 | — | 1.16 | 1.000 | 1.0000 | 0.4520 |
| V77I | PI | — | 0 | — | 3.33 | — | 5.81 | 1.000 | 0.5858 | 1.0000 |
| V82I | PI | — | 0 | — | 10 | — | 3.49 | 0.242 | 1.0000 | 0.1781 |
| Reverse transcriptase region | ||||||||||
| T69S | NRTI | 0 | — | 0 | — | 1.16 | — | N.O. | 1.000 | 1.0000 |
| V90I | NNRTI | — | 0 | — | 0 | — | 3.49 | N.O. | 1.000 | 0.5674 |
| V179D | NNRTI | 0 | 0 | 0 | 0 | 1.16 | 1.16 | N.O. | 1.000 | 1.0000 |
| D218E | NRTI | 0 | — | 0 | — | 1.16 | — | N.O. | 1.000 | 1.0000 |
p values from the Fisher's exact test were used to compare the prevalence of mutations between cohorts.
IAS-USA (2007) report was updated with mutations that were identified in the IAS-USA 2008 Spring report (L10V, I13V, and L33F).
—, No DRAM was designated for the respective position.
N.O., no observations.
Table 3.
Prevalence of Major ART Mutations Associated with Resistance to PIs and RTIs in Both Cohorts
| |
|
|
Observed prevalence for each resistance mutation (%) |
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
|
Cohort A, 1998–2002 (N = 30) |
Cohort B, 2005 (N = 86) |
Comparison of prevalence between cohorts, pa |
||||
| Gene | Mutation | Class | SDRM (2008) | SDRM (2009) | IAS-USA (2007) | SDRM (2008) | SDRM (2009) | IAS-USA (2007) | Cohort A to Cohort B |
| PR | Q58Eb | PI | 0 | —c | — | 1.16 | — | — | 1.000 |
| RT | K103N | NNRTI | 0 | 0 | 0 | 1.16 | 1.16 | 1.16 | 1.000 |
| RT | M184V | NRTI | 0 | 0 | 0 | 1.16 | 1.16 | 1.16 | 1.000 |
p values from the Fisher's exact test were used to compare the prevalence of mutations between cohorts.
The PR mutation Q58E was listed as a major mutation in the SDRM database in 2008. However, Q58E was not designated as a major mutation by either the SDRM 2009 or the IAS-USA (2007).
—, No DRAM designated for the respective position.
No mutations were observed for the RT region in either Cohort A or the Handema et al. cohort; however, six DRAMs (three NRTIs and three NNRTIs) were present in Cohort B. Two of these were the major mutations (K103N, NNRTI and M184V, NRTI) and were observed in 1% of the samples. The remaining four mutations (T69S, V90I, V179D, and D218E) were minor or borderline/suspicious and were observed in 1–3% of the samples (Table 2). The prevalence of each mutation was also compared between cohorts. For minor and borderline mutations, no statistically significant differences were observed between published results by Handema et al. to Cohort A and to Cohort B or between Cohort A and Cohort B (Table 2). In addition, a trend toward an increase in the number of DRAMs was observed in the 2005 cohort; however, it was not statistically significant. Similar results were observed when comparing the observed prevalence of each primary mutation between Cohort A and Cohort B (Table 3). The similarity of DRAM frequencies may indicate that some of these mutations are natural polymorphisms for HIV-1 subtype C in Zambia. The lack of significant differences between cohorts may also be explained by the low percentage of HIV-infected individuals undergoing ART treatment in Zambia in 2005. Although primary mutations were observed in Cohort B, the low prevalence of these mutations reflects a low percentage of transmission of drug resistance after the availability of free ART in Zambia. However, PIs are not included in the first line treatment offered through Zambia's free ART program, and we suggest that the mutations observed in our cohort are naturally occurring polymorphisms that may have increased in prevalence due to transmission. Although borderline and minor mutations alone do not significantly increase the level of resistance, when combined with other mutations, they can increase the resistance to inhibitors by influencing the enzyme's catalytic efficiency.10 Thus, these mutations should be monitored. This surveillance is also important as a prevalence higher than 15% of ART mutations within a population could have negative implications on treatment efficacy.21
We have also analyzed the env V3 loop sequences isolated from subjects in our cohorts and the Handema et al. study, even though entry inhibitors are not currently included in the ART therapy available in Zambia. It is important to determine whether there are any natural polymorphisms in this region that may compromise the use of this treatment strategy in the future. Mutations associated with resistance to the CCR5 antagonists maraviroc and vicriviroc were included in the analysis because maraviroc is already an FDA-approved drug and vicriviroc is currently in phase three clinical trials,22 mostly with subtype B virus-infected individuals.23,24 Because it is known that about 50% of the patients infected with subtype B virus develop X4 tropic viruses at later stages of infection,25,26 it is not surprising that viruses resistant to these drugs have already been observed by switching their coreceptor usage from R5 to X4.23,24 In contrast to subtype B, subtype C is mainly an R5 tropic virus,22 with very few CXCR4 or dual tropic viruses reported.27 Therefore, the use of these compounds could significantly improve the therapy outcome of individuals infected with subtype C viruses if no DRAMs are present. However, the susceptibility and the development of resistance to these inhibitors by subtype C viruses are not known.12 For maraviroc, the mutations A316T and I323V have been reported to confer partial resistance for subtype B viruses. Complete resistance to this entry inhibitor occurs when both mutations (A316T/I323V) are present.11 Surprisingly, some of these mutations were already present in the subtype C env sequences from both of our cohorts as well as in the cohort analyzed by Handema et al. in 2000. The prevalence of resistance-associated mutations to entry inhibitors is shown in Table 4. The mutation A316T was prevalent in 68% of sequences from the Handema et al. cohort, 80.7% in Cohort A and 64.5% in Cohort B. The high prevalence of this mutation in the three cohorts suggests that A316T is a natural polymorphism in subtype C viruses. However, I323V was observed only in the Handema et al. cohort and Cohort B in a prevalence of 4% and 3.8%, respectively. Because all the Handema et al. cohort and Cohort B samples containing the I323V mutation also had the A316T mutation, these data indicate that these ART-naive individuals also harbor viruses that are resistant to maraviroc. To analyze the presence of resistance against vicriviroc, mutations recently reported from an in vitro study and from a clinical trial were included in the analysis,12,28 with the positions of each of these mutant amino acids based on the HXB2 reference strain.
Table 4.
Prevalence of Resistance-Associated Mutations Associated with Entry Inhibitors in Both Cohortsa
| |
Observed prevalence for each resistance mutation (%) |
Comparison of prevalence between cohorts, pb |
|
|
||||
|---|---|---|---|---|---|---|---|---|
| Handema,c2000 (N = 25) | Cohort A, 1998–2002 (N = 26) | Cohort B, 2005 (N = 79) | Handemacto Cohort A | Handemacto Cohort B | Cohort A to Cohort B | Complete resistance | Partial resistance | |
| Maraviroc11 | ||||||||
| A316T | 68 | 80.7 | 64.5 | 0.3487 | 0.8136 | 0.1487 | − | + |
| I323V | 4 | 0 | 3.8 | 0.4902 | 1.0000 | 0.5727 | − | + |
| A316T/I323Va | 4 | 0 | 3.8 | 0.4902 | 1.0000 | 0.5727 | + | − |
| Vicriviroc12,22 | ||||||||
| K305R12,22 | 4 | 7.7 | 6.33 | 1.0000 | 1.0000 | 1.0000 | − | + |
| S306P22 | 0 | 0 | 0 | N.O.d | N.O. | N.O. | − | + |
| R315Q12 | 100 | 100 | 98.7 | N.O. | 1.0000 | 0.5727 | − | + |
| F318I22 | 0 | 0 | 0 | N.O. | N.O. | N.O. | − | + |
| T320R22 | 4 | 0 | 0 | 0.4902 | 0.2404 | N.O. | − | + |
| G321E22 | 0 | 3.8 | 1.26 | 1.0000 | 1.0000 | 0.4357 | − | + |
| K305R, R315Q | 4 | 7.7 | 5.06 | 1.0000 | 1.0000 | 1.0000 | − | + |
| R315Q, T320R | 4 | 0 | 0 | 0.4902 | 0.2404 | N.O. | − | + |
| K305R, T320R | 4 | 0 | 0 | 0.4902 | 0.2404 | N.O. | − | + |
| K305R, R315Q | 0 | 0 | 0 | N.O. | N.O. | N.O. | − | + |
| T320R | ||||||||
| R315Q, G321E | 0 | 3.8 | 1.26 | 1.0000 | 1.0000 | 0.4357 | − | + |
| Maraviroc + vicriviroc | ||||||||
| A316T + K305R | 4 | 3.8 | 1.26 | 1.0000 | 0.5657 | 1.0000 | − | + |
| R315Q | ||||||||
| A316T + R315Q | 68 | 80.7 | 58.22 | 0.3487 | 0.8116 | 0.1464 | − | + |
| A316T/I323V + | 4 | 0 | 3.8 | 0.4902 | 1.0000 | 0.5727 | +/− | +/− |
| R315Q | ||||||||
| A316T + R315Q | 0 | 3.8 | 1.26 | 1.0000 | 1.0000 | 0.4357 | − | + |
| G321E | ||||||||
Most of these mutations confer partial resistance; only A316T/I323V has been demonstrated to confer complete resistance to maraviroc.
p values from the Fisher's exact test were used to compare the prevalence of mutations between cohorts.
Handema et al.8
N.O., no observations.
Not all these resistance mutations were observed in the sequences from these two studies. For instance, although K305R was reported by both studies, R315Q was observed only by Ogert et al.28 and T320R and G321E were reported only by Tsibris et al.12 When analyzing the prevalence of these mutations in the Handema et al. cohort and Cohorts A and B, the mutation R315Q was observed at a prevalence >95% in the three cohorts and is likely to be a natural polymorphism for subtype C in Zambia. Interestingly, a similar number of mutations associated with vicriviroc resistance was observed in Cohort B (K205R, R315Q, and G321E), Cohort A (K205R, R315Q, and G321E), or the Handema et al. cohort (K305R, R315Q, and T320R), and similar frequencies of these mutations were observed in the three cohorts (Table 4). These mutations were observed in a prevalence range of 0–4% in the Handema et al. cohort, 0–7.7% in Cohort A, and 0–6.3% in Cohort B. Because no signature resistance mutations for vicriviroc have been identified to date,12 the prevalence of combinations of vicriviroc partial mutations K305/R315Q, K305/T320R, and R315Q/G321E was also analyzed. The prevalence range of these combined mutations was 0–4% in the Handema et al. cohort, 0–7.7% in Cohort A, and 0–5.06% in Cohort B.
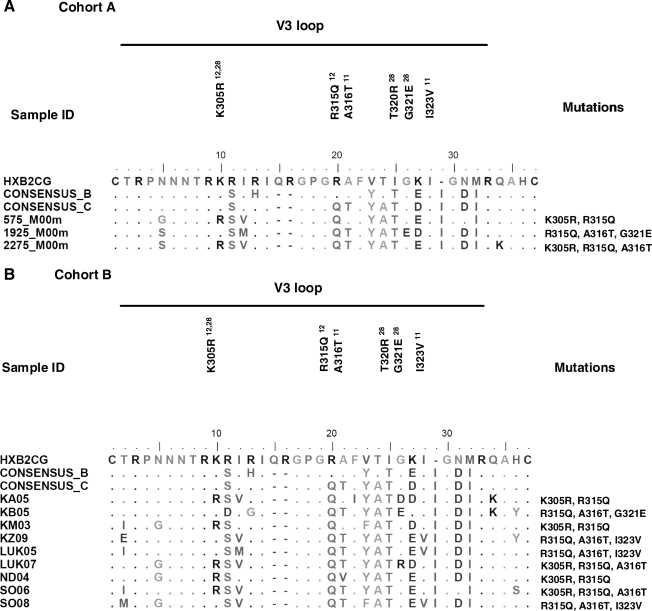
The different combinations of mutations associated with resistance to both maraviroc and viriviroc were also analyzed. The four combinations observed were A316T (maraviroc) + K305/R315Q (vicriviroc), A316T (maraviroc) + R315Q (vicriviroc), A316T/I323V (maraviroc) + R315Q (vicriviroc), and A316T (maraviroc) + R315Q/G321E (vicriviroc). An alignment of the V3 region of sequences in Cohort A and B-containing combination of mutations is shown in Fig. 2. Both the maraviroc mutation A316T and the vicriviroc mutation R315Q were present in each of these combinations and the prevalence of the A316T + R315Q combination was 68% for the Handema et al. cohort, 80.7% for Cohort A, and 58.2% for Cohort B. On the other hand, the other three combinations were observed at a prevalence range of 0–4% in the Handema et al. cohort, 0–3.8% in Cohort A, and 1.3–3.8% in Cohort B. No significant differences were observed when comparing the prevalence of these mutations between cohorts.
FIG. 2.
Entry inhibitor resistance-associated mutations in the V3 loop of ART-naive individuals infected with HIV subtype C. This alignment includes sequences containing a combination of mutations. (A) Three of the 26 env sequences analyzed from Cohort A contained combined mutations. (B) A combination of mutations was also observed in 9 of the 79 env sequences from Cohort B.
Our study did have some limitations. First, due to the low prevalence of major mutations in both the cohorts the sample size may not have been large enough to detect a significant change. Second, since direct sequencing was employed in this study, only those mutations present at levels ≥20% of each sample viral population were detected.29 Third, because it is not possible to accurately determine exactly how long the individuals in our cohorts were actually infected with HIV, even though they were all asymptomatic, we cannot exclude the possibility that mutations may have been gained or lost between the initial infection and the time of sampling.
In summary, most of the mutations observed are natural polymorphisms characteristic of HIV-1 subtype C. No significant differences were observed when comparing drug resistance-associated mutations and polymorphisms present in the pol and env of these two cohorts. However, a trend toward an increase in the number of minor, borderline, or partial resistance mutations as well as the presence of major or complete resistance mutations was observed after the availability of ART in Zambia. The presence of primary mutations conferring resistance to NRTIs and NNRTIs in Cohort B indicates that the transmission of drug resistance was already occurring even though free ART was implemented only recently in Zambia. It is also possible that mutations associated with PI resistance, such as the Q58E, may have occurred in individuals receiving therapy from private clinics before free ART was available. Our observations nevertheless raise concern that subtype C viruses in Zambia have a tendency to increase in DRAMs as well as resistance to ART.
Moreover, we have observed that most of the subtype C Env sequences harbor natural polymorphisms against entry inhibitors. These findings are significant because the high prevalence of these mutations in the population may affect the efficiency of CCR5 entry inhibitors for the treatment of Zambian patients infected with HIV-1 subtype C. Whether this is true for subtype C in other African nations needs to be determined. In addition, because all the vicriviroc mutations have been reported to confer partial resistance, further studies with a larger cohort are needed to determine which individual mutations or combinations confer complete resistance against vicriviroc.
Sequence Data
Sequences are available under GenBank accession numbers GQ427085–GQ427110 (Cohort A env C2V4), GQ427111–GQ427140 (Cohort A pol), GQ433720–GQ433801 (Cohort B env C2V4), and GQ433802–GQ433893 (Cohort B pol).
Acknowledgments
We thank all the study participants and the personnel at the University Teaching Hospital and Voluntary Counseling and Testing centers in Zambia for their collaboration. This study was supported by PHS Grants CA75903 AI048240, T32 AI060547, Fogarty International Grant TW001429, and NCRR COBRE Grant RR15635 to C.W. S.G. is an NIH Ruth L. Kirschstein Fellow; C.G., C.K., and T.M. were Fogarty Fellows.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.WHO. 2008 Report on the global AIDS epidemic Joint United Nations Programme on HIV/AIDS and World Health Organization. 2008.
- 2.Katzenstein D. Diversity, drug resistance, and the epidemic of subtype C HIV-1 in Africa. J Infect Dis. 2006;194(Suppl 1):S45–S50. doi: 10.1086/505353. [DOI] [PubMed] [Google Scholar]
- 3.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 4.Cooper ER. Charurat M. Mofenson L, et al. Combination antiretroviral strategies for the treatment of pregnant HIV-1-infected women and prevention of perinatal HIV-1 transmission. J Acquir Immune Defic Syndr. 2002;29(5):484–494. doi: 10.1097/00126334-200204150-00009. [DOI] [PubMed] [Google Scholar]
- 5.Edgeworth RL. Ugen KE. Immunopathological factors for vertical transmission of HIV-1. Pathobiology. 2000;68(2):53–67. doi: 10.1159/000028115. [DOI] [PubMed] [Google Scholar]
- 6.Buckton AJ. New methods for the surveillance of HIV drug resistance in the resource poor world. Curr Opin Infect Dis. 2008;21(6):653–658. doi: 10.1097/QCO.0b013e3283186d1a. [DOI] [PubMed] [Google Scholar]
- 7.Stringer JS. Zulu I. Levy J, et al. Rapid scale-up of antiretroviral therapy at primary care sites in Zambia: Feasibility and early outcomes. JAMA. 2006;296(7):782–793. doi: 10.1001/jama.296.7.782. [DOI] [PubMed] [Google Scholar]
- 8.Handema R. Terunuma H. Kasolo F, et al. Prevalence of drug-resistance-associated mutations in antiretroviral drug-naive Zambians infected with subtype C HIV-1. AIDS Res Hum Retroviruses. 2003;19(2):151–160. doi: 10.1089/088922203762688667. [DOI] [PubMed] [Google Scholar]
- 9.Bessong PO. Mphahlele J. Choge IA, et al. Resistance mutational analysis of HIV type 1 subtype C among rural South African drug-naive patients prior to large-scale availability of antiretrovirals. AIDS Res Hum Retroviruses. 2006;22(12):1306–1312. doi: 10.1089/aid.2006.22.1306. [DOI] [PubMed] [Google Scholar]
- 10.Coman RM. Robbins AH. Fernandez MA, et al. The contribution of naturally occurring polymorphisms in altering the biochemical and structural characteristics of HIV-1 subtype C protease. Biochemistry. 2008;47(2):731–743. doi: 10.1021/bi7018332. [DOI] [PubMed] [Google Scholar]
- 11.Westby M. Smith-Burchnell C. Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81(5):2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsibris AM. Sagar M. Gulick RM, et al. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J Virol. 2008;82(16):8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soto-Ramirez LE. Rodriguez-Diaz R. Duran AS, et al. Antiretroviral resistance among HIV type 1-infected women first exposed to antiretrovirals during pregnancy: Plasma versus PBMCs. AIDS Res Hum Retroviruses. 2008;24(6):797–804. doi: 10.1089/aid.2007.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H. Orti G. Du Q, et al. Phylogenetic and phenotypic analysis of HIV type 1 env gp120 in cases of subtype C mother-to-child transmission. AIDS Res Hum Retroviruses. 2002;18(18):1415–1423. doi: 10.1089/088922202320935492. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira T. Deforche K. Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21(19):3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H. Zhou Y. Alcock C, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol. 2004;78(4):1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 18.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16(1):62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 19.Myers RE. Gale CV. Harrison A. Takeuchi Y. Kellam P. A statistical model for HIV-1 sequence classification using the subtype analyser (STAR) Bioinformatics. 2005;21(17):3535–3540. doi: 10.1093/bioinformatics/bti569. [DOI] [PubMed] [Google Scholar]
- 20.Gifford RJ. Rhee SY. Eriksson N, et al. Sequence editing by apolipoprotein B RNA-editing catalytic component-B and epidemiological surveillance of transmitted HIV-1 drug resistance. AIDS. 2008;22(6):717–725. doi: 10.1097/QAD.0b013e3282f5e07a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett DE. Myatt M. Bertagnolio S. Sutherland D. Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 22.Dhami H. Fritz CE. Gankin B, et al. The chemokine system and CCR5 antagonists: Potential in HIV treatment and other novel therapies. J Clin Pharm Ther. 2009;34(2):147–160. doi: 10.1111/j.1365-2710.2008.00978.x. [DOI] [PubMed] [Google Scholar]
- 23.Westby M. Lewis M. Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80(10):4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsibris AM. Korber B. Arnaout R, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One. 2009;4(5):e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarlatti G. Tresoldi E. Bjorndal A, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3(11):1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 26.Connor RI. Sheridan KE. Ceradini D. Choe S. Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185(4):621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cilliers T. Nhlapo J. Coetzer M, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol. 2003;77(7):4449–4456. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogert RA. Wojcik L. Buontempo C, et al. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology. 2008;373(2):387–399. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Simen BB. Simons JF. Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199(5):693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 30.Cai F. Chen H. Hicks CB. Bartlett JA. Zhu J. Gao F. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat Methods. 2007;4(2):123–125. doi: 10.1038/nmeth995. [DOI] [PubMed] [Google Scholar]