Abstract
PII, an ancient and widespread signaling protein, transduces nitrogen/carbon/energy abundance signals through interactions with target proteins. We clarify structurally how PII regulates gene expression mediated by the transcription factor NtcA, the global nitrogen regulator of cyanobacteria, shedding light on NtcA structure and function and on how NtcA is activated by 2-oxoglutarate (2OG) and coactivated by the nonenzymatic PII target, protein PipX. We determine for the cyanobacteria Synechococcus elongatus the crystal structures of the PII–PipX and PipX–NtcA complexes and of NtcA in active and inactive conformations (respective resolutions, 3.2, 2.25, 2.3, and 3.05 Å). The structures and the conclusions derived from them are consistent with the results of present and prior site-directed mutagenesis and functional studies. A tudor-like domain (TLD) makes up most of the PipX structure and mediates virtually all the contacts of PipX with PII and NtcA. In the PII–PipX complex, one PII trimer sequesters the TLDs of three PipX molecules between its body and its extended T loops, preventing PipX activation of NtcA. Changes in T loop conformation triggered by 2OG explain PII–PipX dissociation when 2OG is bound. The structure of active dimeric NtcA closely resembles that of the active cAMP receptor protein (CRP). This strongly suggests that with these proteins DNA binding, transcription activation, and allosteric regulation occur by common mechanisms, although the effectors are different. The PipX–NtcA complex consists of one active NtcA dimer and two PipX monomers. PipX coactivates NtcA by stabilizing its active conformation and by possibly helping recruit RNA polymerase but not by providing extra DNA contacts.
Keywords: cyanobacteria, gene expression regulation, nitrogen regulation, signaling, tudor domain
Nitrogen assimilation is crucial for life. In microorganisms and plants a key element controlling it is PII, an ancient, very widespread and conserved signaling protein (1–4). PII integrates nitrogen, carbon, and energy abundance signals by binding as allosteric effectors ATP/ADP and, synergistically with ATP, 2-oxoglutarate (2OG). The latter compound is a key indicator of the carbon/nitrogen balance. Furthermore, PII signaling can involve the covalent modification of residues of its flexible T loop. PII transduces this information by interacting with protein targets, influencing the activity of ammonia channels and nitrogen metabolism-controlling enzymes and modulating the expression of nitrogen assimilation-related genes (1–3).
PII proteins are homotrimers of a 12–13 kDa subunit (2, 3, 5–8). They have a roughly hemispheric body from which the three T loops protrude. In these loops, residues Y51 and S49 are the respective sites of regulatory uridylylation and phosphorylation in enterobacteria and cyanobacteria (3). ADP, MgATP, and 2OG bind in close relation to the T loop (9, 10) and modulate PII binding to its targets (1–3, 11, 12).
Two structures of PII complexes, with the trimeric ammonia channel AmtB (5, 6) and with the hexameric enzyme N-acetylglutamate kinase (NAGK) (7, 8), have recently shown how PII can affect channels and enzymes, revealing 1∶1 stoichiometries of PII subunits:target subunits. The extended T loop blocks the cytoplasmic opening of the AmtB ammonia channel, preventing ammonia passage. A more subtle mechanism operates for NAGK activation by PII. The T loops retract like flexed fingers and bring together both domains of each NAGK subunit, thus favoring a catalytically active enzyme conformation.
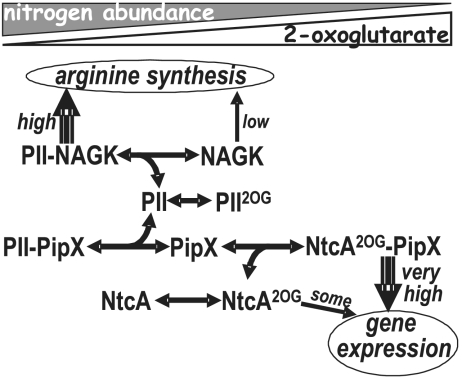
We now show the structural mechanism of gene regulation by PII in an important cyanobacterial system (Fig. 1). In this system PII interacts with PipX (12, 13), an 89-residue protein of previously unknown structure that is a coactivator of 2OG-activated NtcA (12, 14–17). NtcA belongs to the cAMP receptor protein (CRP) transcription factor family (18) and mediates global nitrogen control in cyanobacteria, controlling a very large, nitrogen-responsive regulon encompassing hundreds of genes (18, 19). PII is an abundant protein in cyanobacteria (7) and can sequester the possibly less abundant, and, when PII is absent, toxic PipX (15, 20).
Fig. 1.
PII control of NtcA-mediated gene expression. The interactions of PII with PipX and NAGK, and of PipX with NtcA, depending on nitrogen/2OG abundance, are schematized.
We determine here for the cyanobacteria Synechococcus elongatus crystal structures of the PII–PipX and PipX–NtcA complexes and NtcA alone in active and in apparently inactive conformations. The findings explain why PipX coactivates NtcA, how PII sequesters PipX preventing NtcA activation, also providing structural information on NtcA, and on its activation by 2OG.
A finding of the present studies is the observation that the PipX structure includes as a major component a tudor-like domain (TLD) that mediates the interactions with both PII and NtcA. This exemplifies how this important domain (21) can interact with its protein partners. It is an intriguing domain that consists of a highly bent β sheet forming a β sandwich, and it is already known to recognize methylated arginine and lysine residues in histones (21) and, in bacteria, to be a part of protein complexes involving both RNA polymerase (RNAP) and DNA (22–24).
Results and Discussion
Crystalline Complexes Involving PipX and Their Functional Relevance.
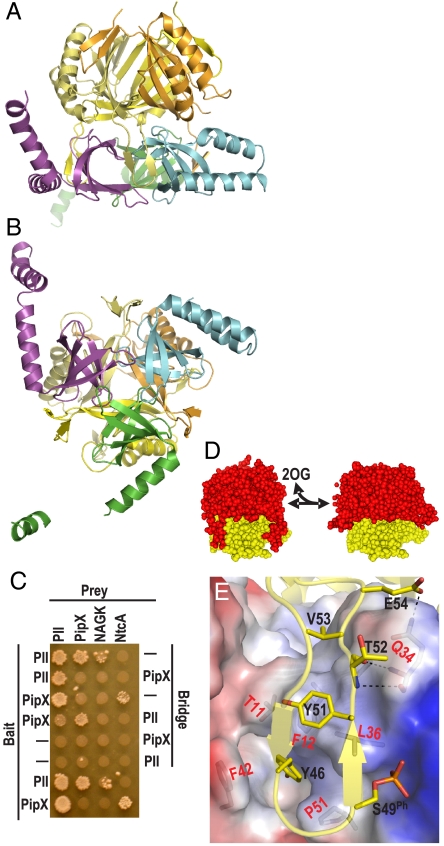
The structures of the PII–PipX and PipX–NtcA complexes, determined at respective resolutions of 3.2 and 2.25 Å (Table S1), reveal that the NtcA-interacting regions of PipX are shielded within the PII–PipX complex, which consists of one PII trimer and three PipX molecules (Fig. 2 A and B), so that a PII–PipX–NtcA ternary complex cannot be formed. Indeed, yeast three-hybrid (Y3H) assays failed to show any PII–NtcA interactions when PipX was used as a potential bridging protein (Fig. 2C). In contrast, these assays showed PipX–PipX interactions when PII was used for bridging (Fig. 2C), as expected from the structure of the crystalline PII–PipX complex, in which three PII–bound PipX molecules interact mutually (Fig. 2 A and B). Pull-down binding assays (see Materials and Methods) agree with the 3∶3 stoichiometry of the PII–PipX complex (Fig. S1A). Support for the relevance of the PII–PipX and PipX–NtcA crystalline complexes is provided by targeted mutagenesis of PII and PipX. In general, the impact of these mutations on the protein–protein interactions, assessed in yeast two-hybrid (Y2H) assays (Figs. 3A and 5E), surface plasmon resonance (SPR) binding assays (Fig. S1B–D), and assays of PipX competition with NAGK for PII (Fig. S1E and see below), generally support the functional relevance of the contacts shown in the crystalline complexes.
Fig. 2.
The PII–PipX complex. Ribbon (A and B) representations of the PII–PipX complex, with PII subunits in different yellow hues and PipX molecules in violet, cyan, and green. The threefold axis is vertical in A and orthogonal to the paper in B. (C) Y3H assays to monitor the effect of a third protein (“Bridge”) on the interactions of the protein fused to the GAL4-Binding-Domain (“Bait”) and the GAL4-Activation-Domain (“Prey”), which is reflected by the growth of yeast diploids. A negative sign indicates a plasmid carrying no gene for either the Bait or the Bridge protein. The last two rows are Y2H assays. (D) Model of how 2OG-triggered T loop alteration should facilitate PipX dissociation. The PipX α-helices have been removed for clarity. Left, PII– (red) PipX (yellow) complex. Right, PII in the conformation corresponding to that observed with MgATP/2OG in this protein from Azospirillum brasilense (10), replacing PII in the PII–PipX complex. (E) T loop interactions with PipX. The T loop is in ribbon representation, showing the side chains of PipX-interacting residues (labeled in black). A phosphate has been modeled over the hydroxyl group of S49 as expected for S49 phosphorylation. PipX is depicted in semitransparent surface representation, with positive/negative surface potential areas in blue/red; side chains of residues that interact with PII are illustrated and they are labeled in red.
Fig. 3.
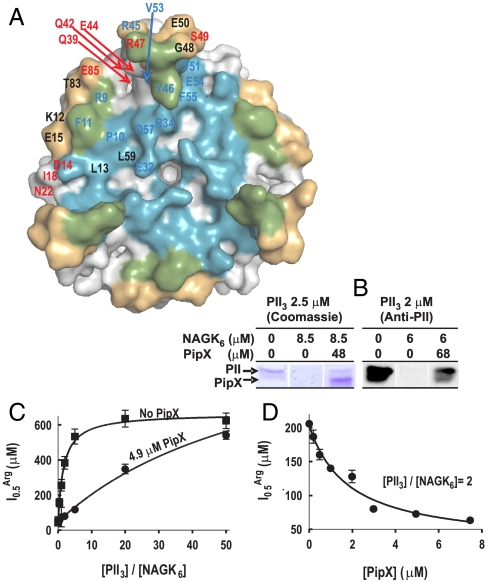
Competition between PipX and NAGK for PII. (A) Surface representation of PII, viewed along the threefold axis from the flat face side of the trimer. The blue, gold, and green areas correspond, respectively, to those regions involved in the interactions with PipX alone, with NAGK alone, or with both PipX and NAGK in the respective complexes. Residue numbers are indicated for one PII subunit in blue or red when their mutation to alanine (except for P10 and V53, which were mutated to L and G, respectively) respectively hampered or did not hamper the interaction with PipX in Y2H assays. The residues in black were not mutated. (B) Ultrafiltration assays of PII binding to NAGK (7) reveal that PipX (added together with NAGK to a PII solution) prevents incorporation of PII into the bulky complex with NAGK, leading to the appearance of PII in the ultrafiltrate [shown by SDS/PAGE and either Coomassie-staining (left), or by western blotting and immunostaining with an anti-PII antibody (33), right]. PII and NAGK concentrations are given in terms of trimers and hexamers, respectively. (C and D) Influence of PipX on the concentration of arginine causing 50% inhibition (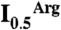 ) in hydroxylamine-based NAGK activity assays (11) also containing PII. The
) in hydroxylamine-based NAGK activity assays (11) also containing PII. The 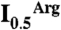 values (with standard error bars) were estimated from plots of activity versus arginine concentration. In C the amount of PII3 (denoting trimers) was varied, whereas in D PipX was varied at a constant PII3/NAGK6 ratio. In both figures NAGK6 (denoting hexamers) concentration was 24 nM.
values (with standard error bars) were estimated from plots of activity versus arginine concentration. In C the amount of PII3 (denoting trimers) was varied, whereas in D PipX was varied at a constant PII3/NAGK6 ratio. In both figures NAGK6 (denoting hexamers) concentration was 24 nM.
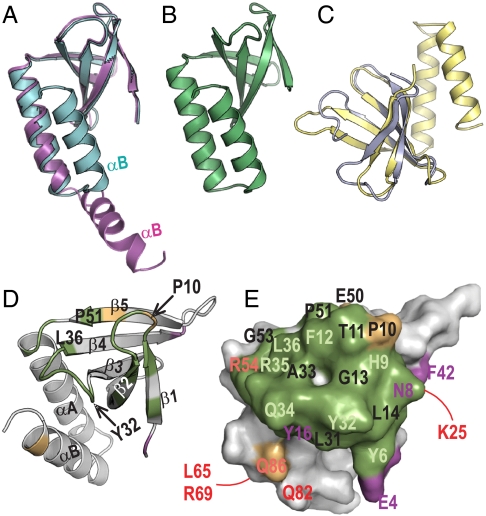
Fig. 5.
Structure of PipX and interacting role of its Tudor-like domain. (A) Superimposition of two PipX molecules from the PII–PipX complex (colored as in this complex), represented in ribbons, to highlight the difference between the extended and flexed positions of the C-terminal helix. (B) One PipX molecule from the PipX–NtcA complex. Its conformation is virtually identical to that of the other PipX molecule in the NtcA complex and to the conformation of the PipX molecule of the PII–PipX complex that has a flexed C-terminal helix. (C) Superimposition of PipX (in yellow) with the N-terminal tudor-like domain of RapA (24). (D) Ribbon and (E) surface representations of PipX in identical orientation to illustrate the regions involved in the interactions with both PII and NtcA (in green), with PII alone (magenta), or with NtcA alone (gold), in the corresponding complexes. In D, secondary structure elements are labeled and a few residues are indicated to facilitate the localization of elements in the surface representation. In E, residue numbers are magenta, dark orange or pale green when their mutation to alanine (Q34, R54, and L65 were mutated to E, C, and Q, respectively) hampered interaction with PII, NtcA, or both, respectively, and in red when the mutation did not hamper interaction with PII or NtcA. The residues numbered in black were not mutated. The lines indicate residues on the back of the molecule.
The Structure of the PII–PipX Complex: Regulatory Implications.
The three PipX molecules of the PII–PipX complex are “caged” in the space between the flat face of the PII body and the vertically extended T loops (Fig. 2 A, D, and E) that have their tip residues projecting inward, gripping the PipX molecules (Fig. 2B). The fact that the T loops are extended in the PII–PipX complex (Fig. 2A and D) and bent in the PII–NAGK complex (7, 8) (Fig. S2) strongly suggest that PipX and NAGK in their respective complexes play an important role in determining the T loop shape, particularly because both complexes are formed in the absence of the PII effector 2OG (11, 12).
When nitrogen is abundant the 2OG level in the cell will be low, and sequestration by PII will decrease PipX availability for complex formation with NtcA (Fig. 1). When nitrogen is less abundant, the increasing 2OG levels will lead to the dissociation of the PII–PipX complex [(12) and see also pull-down experiments of Fig. S3A], and more PipX will be available for NtcA activation. These effects of 2OG may reflect the triggering by 2OG of conformational changes in the T loop (10), resulting in the removal of the steric restrictions imposed by the T loops to the escape of PipX (Fig. 2D Right). ADP promotes an extended T loop conformation (5, 6, 9) that resembles the one in the PII–PipX complex, and, indeed, pull-down (Fig. S3A) and SPR assays (Fig. S3B) suggest that ADP increases the affinity of PII for PipX, whereas ATP had little if any effect on this binding. Given the increased binding of PipX to ADP-bound PII, low-energy conditions in which ADP concentrations are increased might dampen NtcA-mediated gene expression because of enhanced PipX sequestration by PII.
The structure of the PII–PipX complex explains why T loop phosphorylation at residue S49 (1–3) leaves the PII–PipX complex undisturbed (judging from the lack of effect of mutations S49D and S49E, believed to mimic phosphorylation) (12, 13). In the PII–PipX complex structure (Fig. 2E) S49 does not interact with PipX and is exposed amid positive surface potential, with enough room and appropriate charge to accommodate a phosphate (modeled in Fig. 2E). In contrast hydrogen bonds formed by S49 are pivotal for formation of the PII–NAGK complex (7). Consequently, S49 phosphorylation prevents PII–NAGK complex formation (25). PII phosphorylation occurring in S. elongatus and other cyanobacteria under nitrogen-limiting conditions (3), could therefore fine tune PII control of NAGK without affecting PII-modulated gene expression.
Competition of PipX and NAGK for PII.
Formation of a ternary complex of NAGK, PII, and PipX appears to be excluded by the observation that the same face of PII binds either NAGK (7) or PipX (Fig. 2 A and B), with PII residues R9, F11, Y46, R47, and Y51 participating in the interactions with the two proteins (Figs. 2E and 3A). Further, Y3H assays using PII as the bridging protein failed to identify the formation of such ternary complex (Fig. 2C). Rather, the structural observations predict the existence of competition between NAGK and PipX for PII, and this is corroborated here by (i) Y3H assays in which the addition of PipX impaired the interactions between PII and NAGK (Fig. 2C); and (ii) ultrafiltration binding assays (7) (Fig. 3B) in which the addition of PipX to mixtures of PII and NAGK resulted in the detection of PII in the ultrafiltrate, whereas no PII was observed in the ultrafiltrate in the absence of PipX (the PII–PipX complex passes the membrane whereas the bulky PII–NAGK complex is retained). The competition for PII between PipX and NAGK lowers the ability (7, 11) of a given PII concentration to relieve NAGK from arginine inhibition (monitored as the change in the midinhibitory concentration of arginine, Fig. 3 C and D), raising the possibility that PipX, when in large concentration, could influence arginine synthesis.
Structure of the Transcription Factor NtcA in Its 2OG-Activated Form.
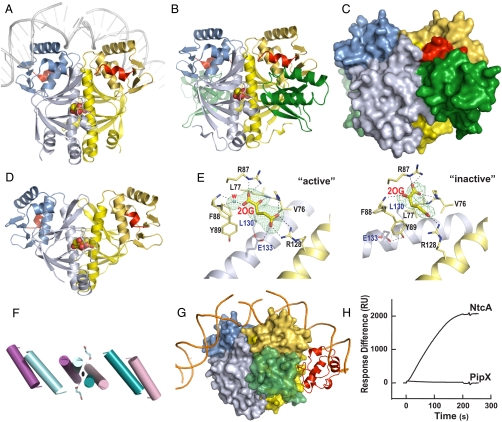
To ascertain the extent to which NtcA resembles the structurally well characterized bacterial transcription factor CRP, with which its sequence can be aligned, and to clarify how NtcA-dependent transcription is activated by 2OG and PipX, we determined the structure of NtcA bound to 2OG alone (Fig. 4A) or in complex with PipX (Fig. 4 B and C). NtcA was found to be a homodimer (Fig. 4A) closely resembling, structurally, the bacterial transcription factor CRP in its active conformation (26) (Fig. S4A). With CRP the N- and C-terminal domains of each subunit of the homodimer have been identified as the regulatory and the DNA-binding domains, respectively (27). NtcA has an extra C-terminal α-helix (shown in red in Fig. 4 A–D) that has no equivalent in CRP: It interacts with PipX and with the regulatory domain of the other NtcA subunit.
Fig. 4.
NtcA structure and activation by 2OG and PipX. Unless otherwise indicated, one NtcA subunit is colored bluish and the other yellow, with the DNA-binding domain shown in darker hues. The C-terminal helix is depicted in red in both NtcA subunits, except in G. (A and D) Structure of the active (A) and inactive (D) NtcA dimer (in ribbon representation) with bound 2OG (in spheres representation). In A the superimposition with CRP–DNA (26) enabled the modeling of bound DNA (in gray). (B and C) The PipX–NtcA complex, with PipX in green in ribbon (B) or in surface (C) representation. (E) Detail of 2OG binding in the active (Active) and apparently inactive (Inactive) NtcA conformations. Electron density maps (Fo-Fc omit maps at 2.5σ) are shown for 2OG (and, in Active, for one water molecule). Broken lines denote hydrogen bonds. (F) Movements of the interfacial helix and the DNA-binding helices upon activation (active, blue; inactive, magenta). 2OG bound to the active form is shown in sticks representation. (G) Model of the PipX–NtcA complex (surface representation) with DNA and the α-CTD of RNAP (in ribbon, red). (H) SPR assay demonstrating the binding of NtcA (6 μg/ml) and lack in binding of PipX (5 μg/ml) to the glnA promoter in the presence of 1 mM 2OG. RU denotes response units of the Biacore instrument.
The effector 2OG may activate NtcA by promoting the same active conformation that is promoted by cAMP and stabilized by DNA in CRP (26–28). In the NtcA dimer we found nonprotein density in the regulatory domains of both subunits near the molecular twofold axis, fitting very well two 2OG molecules (Fig. 4 A, B, and E Active ). These 2OG sites are topographically equivalent to those for cAMP in the CRP dimer (Fig. S4B), each one of them being located in the regulatory domain near to the molecular twofold axis (27). The residues lining this site differ from those in CRP, fitting the chemical and structural characteristics of 2OG. Nevertheless, as for cAMP in active CRP, each 2OG molecule interacts with the long interfacial α-helices of both subunits (Fig. 4E Active). This two-subunit binding of the allosteric effectors appears to play a key role in triggering CRP activation (27, 28) and thus may also trigger the activation of NtcA by 2OG.
CRP and NtcA should bind and bend DNA similarly, given the structural similarity of their active conformations, and the fact that 11 of the 13 residues contacting DNA in CRP (26) are conserved or conservatively replaced in NtcA (Fig. S5A). This strong conservation also parallels the high similarity of the DNA sequences recognized by both transcription factors (26, 29). The change of a G∶C pair by an A∶T pair in the DNA sequence recognized by NtcA relative to that for CRP is largely explained by the occurrence in NtcA of a valine (V187) at the position corresponding to E181 (Fig. S5 B and C). In CRP, E181 makes an acceptor hydrogen bond with the 4-amino group of the cytosine (26), whereas in NtcA the T of the recognized A∶T pair cannot make such bond. Instead, V187 provides in NtcA an appropriate hydrophobic environment for accommodating the 5-methyl group of the T.
Structure of a relaxed, apparently inactive, NtcA form.
In the absence of 2OG, NtcA is transcriptionally inactive (16, 17). We have determined a second NtcA structure (Table S1) that is likely to represent an inactive NtcA form. In a crystal obtained under different crystallization conditions (see NtcA form II in Materials and Methods), one NtcA dimer had a conformation that differed widely from that found in the active NtcA-2OG complex (compare Fig. 4 A and D) or that found in CRP in its active conformation (compare Fig. S4 A and C). The changes in the position of the DNA-binding helix are shown in detail in Fig. S4D. In this apparently inactive conformation (Fig. 4 D and F), the interfacial helices are not parallel, the distance between the DNA-binding helices is increased, and the two subunits are rotated in opposite directions around an axis that crosses perpendicularly the twofold axis. It is interesting that 2OG is still bound to its site in this conformation, but its binding differs from that in activated NtcA, failing to make the contacts with the other subunit interfacial α-helix that appear to be essential for activation (compare Active and Inactive in Fig. 4E). If as with CRP (30) both NtcA subunits interact with RNAP, the change in the relative orientations of the subunits in the NtcA dimer may hamper interaction with RNAP and render NtcA transcriptionally inactive. This NtcA conformation might therefore correspond to, or resemble, that prevailing in the absence of 2OG.
The Structure of the PipX–NtcA Complex Reveals PipX Stabilization of Active NtcA.
The PipX–NtcA complex (Fig. 4 B and C) consists of one NtcA dimer and two PipX molecules. The NtcA dimer binds two 2OG molecules and presents the conformation observed in the “active” NtcA-2OG complex in the absence of PipX (Fig. S6; the root mean square deviation for the superimposition of 390 Cα atoms of active NtcA alone and in the complex with PipX is 0.73 Å). Each PipX molecule lies between the regulatory domain of one NtcA subunit, and the C-terminal domain, the long interfacial α-helix and the regulatory domain of the other NtcA subunit (Fig. 4B, C), gluing together these elements. NtcA stabilization is reflected in the very low B-factors for the complex (Table S1), drastically decreased relative to those for NtcA alone. Given its lock-and-key binding to active NtcA, PipX should be unable to bind to the putative inactive form of the NtcA dimer described here, thus shifting the equilibrium in favor of the active NtcA conformation.
In addition to stabilizing the active NtcA conformation, PipX may also activate NtcA-mediated transcription by providing extra contacts with RNAP. Thus, when the PipX–NtcA and CRP–DNA–αCTD (αCTD, RNAP α subunit C-Terminal Domain) (31) complexes are superimposed by superposing NtcA and CRP, PipX falls next to αCTD without steric clash, rendering mutual contacts possible (Fig. 4G). The possibility of PipX binding to DNA is unlikely, because this superimposition indicates that PipX should be > 25 Å away from the DNA. Further, SPR experiments (see Materials and Methods) that demonstrated NtcA binding to the glnA promoter failed to demonstrate PipX binding to this promoter in the absence of NtcA (Fig. 4H).
The PipX Structure Exemplifies How Tudor Domains Can Act as Protein–Protein Interacting Modules.
The present crystal structures have revealed that PipX consists of a highly bent β-sheet (residues 4–53) that presents the characteristic TLD fold (22–24) and two C-terminal helices (residues 54–89) (Fig. 5 A–D). The TLD mediates all the contacts of PipX with both PII and NtcA (Fig. 5 D and E), thus providing structural foundations for the prior biochemical evidence that bacterial TLDs and tudor domains of eukaryotes are protein–protein interaction modules (21, 23). The finding of a TLD in PipX, a protein that participates in the transcriptional complex involving NtcA, is also consistent with the previous identification of TLDs in bacterial proteins that interact with both RNAP and DNA (22–24). Other than the differences in the position of the C-terminal helix in two PipX molecules of the PII–PipX complex (Figs. 2 A and B and 5A), the five PipX molecules found in the present two complexes exhibit virtually identical structure (Fig. 5 A and B).
As already indicated, the PipX TLD regions that contact PII and NtcA almost completely overlap (Fig. 5E). This is reflected in the competition for PipX between PII and NtcA that is observed in Y3H assays (Fig. 2D). The PipX TLD regions that contact PII or NtcA involve one layer of the β-sandwich (β1–β2 hairpin and β3–β4 connector) and part of the β-sheet convex side (β4 and β5 end) (Fig. 5D). Nevertheless, most (78%) of the PipX surface, which includes the other layer of the sandwich, the outward face of the helix hairpin and both α-helices, remains exposed in the NtcA complex (Fig. 4 B and C) and largely so in the PII–PipX complex (Fig. 2 A and B). Thus, NtcA-bound PipX offers opportunities for interaction with other potential components or regulators of the transcriptional complex, including RNAP, as indicated above. Given the mobile nature of the C-terminal helix (as shown in the PII–PipX complex), this helix might extend and recruit other factors. Thus, in summary, the present analysis reveals how PipX mediates the nitrogen-control relay operated by 2OG and PII in the regulation of NtcA-dependent gene expression. These findings should pave the way toward full understanding of relevant interactions involved in transcriptional regulation of the NtcA-dependent nitrogen regulon.
Final remarks.
The structure of the PipX–PII complex has revealed another novel extended T loop conformation that resembles but is not identical to that of the GlnK T loops in the complex with AmtB (5, 6). The large cavity formed between the T loops and the PII body apparently offers the possibility of “caging” proteins and hiding interacting groups as is the case with PipX, whereas the possibility of changing the conformation of the T loops provides the system with large regulatory potentialities. Furthermore, the question may be raised of whether there are other proteins playing a role resembling that of PipX in organisms where PipX is not present. This possibility appears to be plausible, because PipX sits in its complex with NtcA at a place that corresponds quite accurately with the space left between CRP and CTDRNAP in the DNA-CRP-CTDRNAP complex of Escherichia coli (31), thus highlighting the importance of seeking proteins that could be equated functionally with PipX in other taxonomic groups and of exploring further PipX interactions in cyanobacteria.
Materials and Methods
Full protocols are available in the SI Text.
Production and Crystallization of PII–PipX, PipX–NtcA, and NtcA.
NtcA and PipX with N-terminal His6 tags and tag-free PII from S. elongatus (strain PCC7942) were purified essentially as reported (7, 12). All the solutions used in NtcA purification were supplemented with 4 mM 2OG. To crystallize the PipX–PII complex, a solution containing 0.07 mg/ml of each protein in 20 mM HEPES pH 7.5, 1 mM dithiothreitol, 0.4 M NaCl and 5 mM MgCl2, incubated 5 min at 22 °C, was concentrated to 5.5 mg/ml by ultrafiltration. The complex crystallized as 0.1-mm needles over a six-month period from hanging drops at 21 °C. The drops were prepared by mixing 1 μl of each protein solution and precipitant solution (0.2 M Na formate/15% polyethylene glycol 3.35 K). To crystallize the PipX–NtcA complex, a solution of 0.1 mg/ml PipX and 0.3 mg/ml NtcA in buffer A (50 mM Na citrate, pH 6.5, 0.5 M NaCl, 5 mM MgCl2, 10 mM 2OG, 50 mM L-arginine-HCl and 50 mM Na L-glutamate), incubated 5 min at 22 °C, was concentrated to 3 mg/ml by ultrafiltration. Crystals of ∼0.15 mm maximal dimension grew after six weeks when using 50 mM MES pH 5.5/1 M Na malonate/10 mM MnCl2/2% 2-methyl-2,4-pentanediol (MPD)/5% dimethyl sulfoxide as precipitant. Crystals of NtcA alone were obtained in two different forms from a 4.7 mg/ml NtcA solution in buffer A, using the following crystallization solutions: 1) for form I, 0.1 M Bis-Tris pH 5.5/0.15 M ammonium acetate/11% PEG 10 K; 2) for form II, 0.2 M K citrate pH 8.3/20% PEG 3.35 K. A Hg2+ derivative was prepared from a NtcA crystal grown using as crystallization solution 0.1 M Bis-Tris pH 6.5/36% PEG 400. The crystal was soaked 5 min in crystallization solution containing 10 mM 2OG and 0.5 mM HgCl2, and then for an additional 5-min period in the same solution containing 2 mM HgCl2.
Data Collection and Structure Determination.
See Table S1 and SI Methods for details.
Other Methods.
Pure NAGK was produced as reported (7). We used well established procedures (7) for site-directed mutagenesis of the PII and PipX genes carried in appropriate plasmids (15). All the mutant PII and PipX proteins used in both the SPR studies and in the competition studies with NAGK were purified as the corresponding wild-type protein. Y2H analysis of NAGK and PII mutants was carried out as described (12, 13). The same procedure was followed for Y3H analyses (32), save the cloning of the genes for the indicated bait and bridge proteins (see details in SI Methods) into the respective MCSI and MCSII regions of the pBridge vector (Clontech, Takara Bio Company). The binding of PII to His6-tagged PipX was assayed by SPR as reported (12). SPR was also used to assay the binding of NtcA or of PipX to the glnA DNA promoter sequence (fixed on a Streptavidin sensor chip), and to also assay PipX binding to NtcA bound to chip-attached glnA promoter DNA (see SI Methods for details). The binding of PipX to PII was assayed by SPR using a chip that was first loaded with Strep-tactin II and then with Strep-tagged-PII (12, 25) (see SI Methods). Pull-down assays of PII (5.5–68.2 μM, as PII subunits) binding to His6PipX (30.8 μM) were carried out using His SpinTrap columns (GE Healthcare), to determine the amount of PII that was retained in the column together with PipX.
Supplementary Material
Acknowledgments.
We thank F. Gallego, C. Marco-Marín, L.M. Polo, N. Gougeard (Centro de Investigación Biomédica en Red de Enfermedades Raras), and the staff of the European Synchrotron Research Facility–Grenoble for expert help, I. Fita (Instituto de Biología Molecular de Barcelona), W. Hillen (University Erlangen), G. Montoya (Centro Nacional de Investigaciones Oncológicas), A. Marina (Instituto de Biomedicina de Valencia), and H. Britton for critically reading the manuscript and European Synchrotron Research Facility and European Molecular Biology Laboratory (Grenoble) for supporting our synchrotron use. Supported by grants BFU2008-05021 and BFU2009-07371 (Spanish Ministry of Science and Innovation), Prometeo (Generalitat Valenciana), and Deutsche Forschungsgemeinschaft Fo195/4.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2XG8 (PII–PipX), 2XKO (PipX–NtcA), 2XHK (NtcA native I), 2XKP (NtcA native II), and 2XGX (NtcA Hg2+)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007015107/-/DCSupplemental.
References
- 1.Ninfa AJ, Jiang P. PII signal transduction proteins: Sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–377. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- 3.Forchhammer K. PII signal transducers: Novel functional and structural insights. Trends Microbiol. 2008;16:65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Sant’Anna FH, et al. The PII superfamily revised: A novel group and evolutionary insights. J Mol Evol. 2009;68:322–336. doi: 10.1007/s00239-009-9209-6. [DOI] [PubMed] [Google Scholar]
- 5.Gruswitz F, O’Connell J, III, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc Natl Acad Sci USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conroy MJ, et al. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llácer JL, et al. The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc Natl Acad Sci USA. 2007;104:17644–17649. doi: 10.1073/pnas.0705987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuno Y, Moorhead GB, Ng KK. Structural basis for the regulation of N-acetylglutamate kinase by PII in Arabidopsis thaliana. J Biol Chem. 2007;282:35733–35740. doi: 10.1074/jbc.M707127200. [DOI] [PubMed] [Google Scholar]
- 9.Yildiz O, Kalthoff C, Raunser S, Kuhlbrandt W. Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake. EMBO J. 2007;26:589–599. doi: 10.1038/sj.emboj.7601492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truan D, et al. A new P(II) protein structure identifies the 2-oxoglutarate binding site. J Mol Biol. 2010;400:531–539. doi: 10.1016/j.jmb.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Maheswaran M, Urbanke C, Forchhammer K. Complex formation and catalytic activation by the PII signaling protein of N-acetyl-L-glutamate kinase from Synechococcus elongatus strain PCC 7942. J Biol Chem. 2004;279:55202–55210. doi: 10.1074/jbc.M410971200. [DOI] [PubMed] [Google Scholar]
- 12.Espinosa J, Forchhammer K, Burillo S, Contreras A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol. 2006;61:457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 13.Burillo S, Luque I, Fuentes I, Contreras A. Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J Bacteriol. 2004;186:3346–3354. doi: 10.1128/JB.186.11.3346-3354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa J, Forchhammer K, Contreras A. Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology. 2007;153:711–718. doi: 10.1099/mic.0.2006/003574-0. [DOI] [PubMed] [Google Scholar]
- 15.Espinosa J, Castells MA, Laichoubi KB, Forchhammer K, Contreras A. Effects of spontaneous mutations in PipX functions and regulatory complexes on the cyanobacterium Synechococcus elongatus strain PCC 7942. Microbiology. 2010;156:1517–1526. doi: 10.1099/mic.0.037309-0. [DOI] [PubMed] [Google Scholar]
- 16.Tanigawa R, et al. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp PCC 7942 by 2-oxoglutarate in vitro. Proc Natl Acad Sci USA. 2002;99:4251–4255. doi: 10.1073/pnas.072587199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valladares A, Flores E, Herrero A. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp strain PCC 7120. J Bacteriol. 2008;190:6126–6133. doi: 10.1128/JB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in cyanobacteria. J Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su Z, Olman V, Mao F, Xu Y. Comparative genomics analysis of NtcA regulons in cyanobacteria: Regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic Acids Res. 2005;33:5156–5171. doi: 10.1093/nar/gki817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espinosa J, Castells MA, Laichoubi KB, Contreras A. Mutations at pipX suppress lethality of PII-deficient mutants of Synechococcus elongatus PCC 7942. J Bacteriol. 2009;191:4863–4869. doi: 10.1128/JB.00557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurer-Stroh S, et al. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 22.Steiner T, Kaiser JT, Marinkoviç S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Shaw G, et al. Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription. Structure. 2008;16:1417–1427. doi: 10.1016/j.str.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinrich A, Maheswaran M, Ruppert U, Forchhammer K. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol Microbiol. 2004;52:1303–1314. doi: 10.1111/j.1365-2958.2004.04058.x. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson G, et al. Structure of the CAP-DNA complex at 2.5 angstroms resolution: A complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 27.Weber IT, Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 28.Sharma H, Yu S, Kong J, Wang J, Steitz TA. Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding. Proc Natl Acad Sci USA. 2009;106:16604–16609. doi: 10.1073/pnas.0908380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson CL, et al. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benoff B, et al. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 32.Tirode F, et al. A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J Biol Chem. 1997;272:22995–22999. doi: 10.1074/jbc.272.37.22995. [DOI] [PubMed] [Google Scholar]
- 33.Forchhammer K, Tandeau de Marsac N. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.